Abstract
Mammals synchronize their circadian activity primarily to the cycles of light and darkness in the environment. This is achieved by ocular photoreception relaying signals to the suprachiasmatic nucleus (SCN) in the hypothalamus. Signals from the SCN cause the synchronization of independent circadian clocks throughout the body to appropriate phases. Signals that can entrain these peripheral clocks include humoral signals, metabolic factors, and body temperature. At the level of individual tissues, thousands of genes are brought to unique phases through the actions of a local transcription/translation-based feedback oscillator and systemic cues. In this molecular clock, the proteins CLOCK and BMAL1 cause the transcription of genes which ultimately feedback and inhibit CLOCK and BMAL1 transcriptional activity. Finally, there are also other molecular circadian oscillators which can act independently of the transcription-based clock in all species which have been tested.
Introduction
As the sun sets, nocturnal rodents begin to forage, nocturnal birds of prey begin their hunt while diurnal birds of prey sleep, filamentous fungi begin their daily production of spores, and cyanobacteria begin nitrogen fixation in an environment of low O2 after the day’s photosynthesis. As the sun rises the next morning many plants have positioned their leaves to catch the first rays of light and many humans sit motionless in cars on a nearby gridlocked highway. It is now understood that the obedience to temporal niches in these and all organisms is governed by a molecular circadian clock. These clocks are not driven by sunlight, but are rather synchronized by the 24 hour patterns of light and temperature produced by the earth’s rotation. The term circadian is derived from “circa” which means “approximately” and “dies” which means “day”. A fundamental feature of all circadian rhythms is their persistence in the absence of any environmental cues. This ability of clocks to “free-run” in constant conditions at periods slightly different than 24 hours, but yet synchronize, or “entrain”, to certain cyclic environmental factors allows organisms to anticipate cyclic changes in the environment. Another fundamental feature of circadian clocks is the ability to be buffered against inappropriate signals and to be persistent under stable ambient conditions. This robust nature of biological clocks is well illustrated in the temperature compensation observed in all molecular and behavioral circadian rhythms. Here temperature compensation refers to the rate of the clock being nearly constant at any stable temperature which is physiologically permissive. The significance of temperature compensation is especially evident in poikilotherimic animals that contain clocks that need to maintain 24 hour rhythmicity in a wide range of temperatures. Combined, the robust oscillations of the molecular clocks (running at slightly different rates in different organisms) and their unique susceptibility to specific environmental oscillations contribute to and fine-tune the wide diversity of temporal niches observed in nature.
However, the circadian clock governs rhythmicity within an organism far beyond the sleep: activity cycle. In humans and most mammals there are ~24 hour rhythms in body temperature, blood pressure, circulating hormones, metabolism, retinal electroretinogram (ERG) responses, as well as a host of other physiological parameters (1–4). Importantly, these rhythms persist in the absence of light:dark cycles and in many cases in the absence of sleep:wake cycles. On the other side of the coin, a number of human diseases display a circadian component, and in some cases, human disorders and diseases have been shown to occur as a consequence faulty circadian clocks. This is evident in sleep disorders such as Delayed Sleep Phase Syndrome (DSPS) and Advanced Sleep Phase Syndrome (ASPS) in which insomnia or hypersomnia result from a misalignment of one’s internal time and desired sleep schedule (5). In familial ASPS (FASPS), the disorder cosegregates both with a mutation in the core circadian clock gene PER2 and independently with a mutation in the PER2-phosphorylating kinase, CK1 δ (6,7). Intriguingly, transgenic mice engineered to carry the same single amino acid change in PER2 observed in FASPS patients recapitulate the human symptoms of a shortened period (8). Although, these mutations are likely not the end of the story for these disorders, they give insight into the way molecular clocks affect human well-being. Jet lag and shift work sleep disorder are other examples of health issues where the internal circadian clock is desynchronized from the environmental rhythms. In addition to sleep-related disorders, circadian clocks are also directly linked with feeding and cellular metabolism, and a number of metabolic complications may result from miscommunication with the circadian clock and metabolic pathways (2). For example, loss-of-function of the clock gene, Bmal1, in pancreatic beta cells can lead to hypoinsulinaemia and diabetes (9). Finally, some health conditions show evidence of influence of the circadian clock or a circadian clock-controlled process. For example, myocardial infarction and asthma episodes show strong nocturnal or early morning incidence (10,11). Also, susceptibility to UV light-induced skin cancer and chemotherapy treatments varies greatly across the circadian cycle in mice (12,13).
In mammals the suprachiasmatic nucleus (SCN) of the hypothalamus is the master circadian clock for the entire body (14,15). However, the SCN is more accurately described as a “master synchronizer” than a strict pacemaker. Most tissues and cell types have been found to display circadian patterns of gene expression when isolated from the SCN (16–19). Therefore, the SCN serves to synchronize the individual cells of the body to a uniform internal time more like the conductor of an orchestra rather than the generator of the tempo themselves. The mammalian SCN is entrained to light cycles in the environment by photoreceptors found exclusively in the eyes (20). The SCN then relays phase information to the rest of the brain and body via a combination of neural, humoral, and systemic signals which will be discussed in more detail later. Light information influencing the SCN’s phase, the molecular clock within the SCN, and the SCN’s ability to set the phase of behavior and physiology throughout the body constitute the three necessary components for a circadian system to be beneficial to an organism: 1) environmental input, 2) a self-sustained oscillator, and 3) an output mechanism.
Mechanism of the molecular circadian clock
Transcriptional feedback circuits
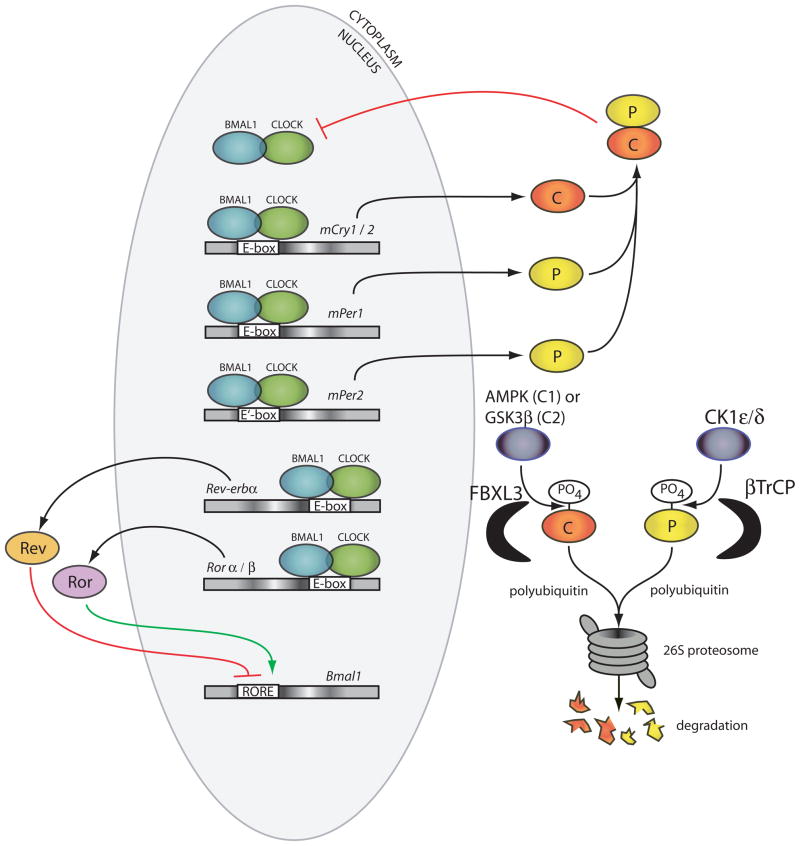
The molecular clock mechanism in mammals is currently understood as a transcriptional feedback loop involving at least ten genes (Figure 1). The genes Clock and Bmal1 (or Mop3) encode bHLH-PAS (basic helix-loop-helix; Per-Arnt-Single-minded, named after proteins in which the domains were first characterized) proteins that form the positive limb of the feedback circuit (reviewed in (21)). The CLOCK:BMAL1 heterodimer initiates the transcription by binding to specific DNA elements, E-boxes (5′-CACGTG-3′) and E′-boxes (5′-CACGTT-3′) in the promoters of target genes (22–24). This set of activated genes includes members of the negative limb of the feedback loop including the Per (Per1 and, Per2) and Cry (Cry1 and Cry2) genes (22,25,26). The resulting PER and CRY proteins dimerize and inhibit further CLOCK:BMAL1 transcriptional activity allowing the cycle to repeat from a level of low transcriptional activity (27–30). The chromatin remodeling necessary for this cyclic transcriptional activity is achieved by a combination of clock-specific and ubiquitous histone-modifying proteins, and can be observed in the rhythmic acetylation/deacetylation of histones (H3 and H4) at multiple clock target genes (31,32). The CLOCK protein itself possesses a histone acetyl transferase (HAT) domain which is necessary for the rescue of rhythms in Clock mutant fibroblasts (33). The CLOCK:BMAL1 complex also recruits the methyltransferase MLL1 to cyclically methylated histone H3 and HDAC inhibitor JARID1a to further facilitate transcriptional activation (34,35). Deacetylation takes place, in part, due to recruitment by PER1 of the SIN3-HDAC (SIN3-histone deacetylase) complex to CLOCK:BMAL1-bound DNA, and more members of the circadian deacetylation process are sure to be elucidated (36). Intriguingly, the rhythmic deacetylation of histone H3 at the promoters of circadian genes is regulated by the deacetylase SIRT1, which is sensitive to NAD+ levels (37,38). This is interesting considering that the NAD+:NADH ratio has been shown to regulate CLOCK:BMAL1’s ability to bind DNA in vitro (39). Thus, cellular metabolism may prove to play an important role in regulating the transcriptional state, and therefore the phase, of the clock.
Figure 1.
Schematic of the molecular clock of mammals. CLOCK:BMAL1 heterodimers (green and blue ovals) bind DNA of clock target genes at E-boxes or E′-boxes and initiate the transcription of their RNA. The resulting PER and CRY proteins (red and yellow ovals) dimerize in the cytoplasm and translocate to the nucleus where they inhibit CLOCK:BMAL1 proteins from initiating further transcription.
Degradation of the negative limb proteins PER and CRY is required to terminate the repression phase and restart a new cycle of transcription. The stability/degradation rate of the PER and CRY proteins is key to setting the period of the clock. The first mammal identified as a circadian mutant was the tau mutant hamster which displays a free-running period of 20 hours, compared to a wild-type free-running period of 24 hours (40). This shortened period results from a mutation in the enzyme casein kinase 1ε (CK1ε), a kinase which phosphorylates the PER proteins (41). Another casein kinase, CK1δ, was later found to phosphorylate the PER proteins and that this CK1ε/δ-mediated phosphorylation targets the PER proteins for ubiquitination by βTrCP and degradation by the 26S proteasome (42–45). Similar to PER, mutant animals with unusual free-running periods (although longer than wild-type in these cases) led to elucidation of the degradation pathway of CRY proteins. In two independent examples, a chemically induced mutation responsible for long period phenotypes in mice was found in the F-box gene Fbxl3 (46,47). FBXL3 polyubiquitinates CRY proteins, thereby targeting them for proteosomal degradation (48). Interestingly, CRY1 and CRY2 are targeted for ubiquitination by unique phosphorylation events and kinases. CRY1 is phosphorylated by AMPK1 and CRY2 by a sequential DYRK1A/GSK-3β cascade (49–51).
The paralogs of the Per genes (Per1 and Per2) and the Cry genes (Cry1 and Cry2) have non-redundant roles. Three independent null alleles of Per1 yielded mice with free-running periods 0.5 – 1 hr shorter than wild-types, but a loss of Per2 produced mice with a 1.5 hr period reduction (52–55). However, the behavior of the Per2 null mice only remained rhythmic for less than a week before becoming arrhythmic (54,55). Knockout alleles of the Cry paralogs produced opposite effects. Cry1−/− mice ran 1 hr shorter than wild-type mice while Cry2−/− mice ran 1 hr longer (56–58). At the molecular level, further unique properties of the individual paralogs appear, specifically paralog compensation. Paralog compensation means that when one gene of a family is lost or reduced, the expression of a paralog of that gene is increased to partially compensate. A reduction in Per1 or Cry1 produced an increase in Per2 or Cry2, respectively (59). However, reductions or loss of Per2 or Cry2 did not produce compensatory expression of their respective paralogs (59). Perhaps network features such as these give insight into the differences seen at the behavioral level of the individual null alleles. Importantly, at both the behavioral and molecular level, at least one member of each family is critical for circadian rhythmicity, as Per1−/−;Per2−/− mice and Cry1−/−;Cry2−/− mice display no signs of intrinsic circadian rhythmicity (54–58).
Our laboratory has recently interrogated on a genome-wide level the cis-acting regulatory elements (cistrome) of the entire CLOCK:BMAL1 transcriptional feedback loop in the mouse liver (60). This has revealed a global circadian regulation of transcription factor occupancy, RNA polymerase II recruitment and initiation, nascent transcription and chromatin remodeling.. We find that the circadian transcriptional cycle of the clock consists of three distinct phases — a poised state, a coordinated de novo transcriptional activation state, and a repressed state. Interestingly only 22% of mRNA cycling genes are driven by de novo transcription, suggesting that both transcriptional and post-transcriptional mechanisms underlie the mammalian circadian clock. We also find that circadian modulation of RNAPII recruitment and chromatin remodeling occurs on a genome-wide scale far greater than that seen previously by gene expression profiling. (60). This reveals both the extensive reach of the circadian clock and potential functions of the clock proteins outside of the clock mechanism.
The members of the negative limb, in particular the PERs, act as the state variable in the mechanism (61). Briefly, this means that the levels of these proteins determine the phase of the clock. In the night, when levels of the PER proteins are low, acute administration of light causes an induction in Per1 and Per2 transcription (62–64). With light exposure in the early night, behavioral phase delays are observed, and this corresponds to light-induced increases of both PER1 and PER2 proteins observed in the SCN (65). In the second half of the night only PER1 levels rise with light exposure, and this corresponds to phases of the night when light-induced phase advances occur (65). These delays in behavior when light is present in the early night and advances in the late night/early morning are sufficient to support entrainment of an animal to a light:dark cycle. If a master clock is running shorter than 24 hr, the sensitive delay region of the state variables will receive light and will slightly delay daily, thus tracking dusk. If the clock is running at a period longer than 24 hr, the advance region will be affected and cause a daily advance in rhythms, and the animal’s behavior will track dawn. The light activation of the Per genes is achieved through CREB/MAPK signaling acting on cAMP-response elements (CRE) in the Per promoters (66).
The CLOCK:BMAL1 dimers also initiate the transcription of a second feedback loop which acts in coordination with the loop described above. This involves the E-box mediated transcription of the orphan nuclear-receptor genes Rev-Erbα/β and RORα/β (67–69). The REV-ERB and ROR proteins then compete for Retinoic acid-related Orphan receptor Response Element (RORE) binding sites within the promoter of Bmal1 where ROR proteins initiate Bmal1 transcription and REV-ERB proteins inhibit it (67,69). This loop was originally acknowledged as an accessory loop due to the subtle phenotypes observed in mice with individual null alleles of any one of these genes. While a traditional double-knockout is lethal during development, inducible double knockout strategies have allowed the deletion of Rev-Erbα and β in an adult animal. This has revealed that the Rev-erbs are necessary for normal period regulation of circadian behavioral rhythmicity (70). A separate set of PAR bZIP genes which contain D-box elements in their promoters make up another potential transcriptional loop. These include genes in the HLF family (71), DBP (72), TEF (73), and Nfil3 (74). If one considers just the rate of transcription/translation and the E-box transcription loop described for the Per/Cry genes alone, it would be easy to imagine the whole cycle taking significantly less than a day or even less than several hours. It has been proposed that the three known binding elements together provide the necessary delay to cycle at near 24 hr: E-box in the morning, D-box in the day, and RORE elements in the evening (75). Although no genes, or even gene families, in these D-box accessory loops are required for clock function, they may serve to make the core oscillations more robust and add precision to the period (67,76).
Non-transcriptional rhythms
In some specific examples, the minimum elements required for molecular 24 hr rhythms do not include transcription or translation. In the cyanobacterium Synechococcus, 24 hr rhythms of phosphorylation of the KaiC protein are observed when the proteins KaiA, KaiB, and KaiC are isolated in a test tube in the presence of ATP (77). The auto-phosphorylation and –dephosphorylation of KaiC is mediated by the phosphorylation promoting KaiA and the dephosphorylation promoting KaiB (78–80). Later, circadian rhythms which are independent of transcription were discovered in organisms as diverse as algae and humans. In Ostreococcus tauri algae transcription stops in the absence of light; however, the 24 hr oxidation cycles of the antioxidant proteins peroxiredoxins continue in constant darkness (81). Similarly, in human red blood cells, which lack nuclei, peroxiredoxins are oxidized with a circadian rhythm (82). These transcription-lacking oscillators are also temperature compensated and entrainable to temperature cycles fulfilling other necessary attributes of true circadian clocks (77,82,83). It should be noted, however, that in nucleated cells the transcriptional clock influences the cytoplasmic peroxiredoxin clock (82). The peroxiredoxin oscillators are remarkably conserved among all phyla that have been examined (84). It is likely that there are more molecular circadian rhythms that can persist without the transcriptional oscillator left to be discovered and that the communication between these and transcriptional molecular clocks will reveal a whole new level of regulation of circadian functions within a single cell.
Peripheral clocks
The transcriptional feedback loop described above can be observed not only in the SCN, but in nearly every mammalian tissue (85). If viewed at the single cell level, the molecular clockwork of transcription and translation can be observed as autonomous single cell oscillators (86,87). In addition to the core clock genes, hundreds or even thousands of genes are expressed with a circadian rhythm in various tissues, but this is not to say there are hundreds of clock genes. Imagine that the core circadian genes act like the gears of a mechanical clock that has hundreds of hands pointing to all different phases but moving at the same rate. Various cellular pathways and gene families pay attention to the hand of the clock in the proper phase for their individual function. It is the same set of core clock components (gears) that drive the phase messengers (hands of the clock) which vary greatly depending on the cell type.
The extent to which the global transcription in a cell was controlled by the circadian clock was not appreciated until the implement of genome-wide tools (88). Between 2 and 10% of the total genome is transcribed in a circadian manner in various mouse tissues (89–94). In a study comparing gene expression profiles of ~10,000 genes and expressed sequence tags (EST) in the SCN and liver, 337 genes were found to be cyclic in the SCN and 335 in the liver with an overlap of only 28 genes cycling in both (91). Another study found a similar overlap of only 37 rhythmic genes between the liver and heart while each tissue expressed more than 450 genes (out of 12,488 analyzed) with a circadian rhythm (93). The differences in the exact number of genes found to be cycling in a given tissue between studies is almost certainly the result of experimental and analytical variation. Indeed more recent genome-wide transcriptome analyses have revealed many thousands of cycling transcripts in the liver (88). Circadian gene expression in each tissue is tissue-specific and optimized to best accommodate that tissue’s respective function throughout a circadian cycle.
The clock-controlled genes in various tissues are involved in diverse gene pathways depending on the tissue. In the retina for example, nearly 300 genes show rhythmic expression in darkness, and this includes genes involved in photoreception, synaptic transmission, and cellular metabolism (92). The number of oscillating genes jumps to an astonishing ~2600 genes in the presence of a light:dark cycle, and these are phased around the cycle suggesting they are not merely driven by the light. Importantly, these robust transcriptional oscillations are lost in the absence of the core clock gene Bmal1 (92). In the liver, between 330 and 450 genes are expressed with a circadian rhythm (91,93). In a creative use of conditional transgene expression, Ueli Schibler and colleagues knocked down the expression of the CLOCK:BMAL1 transcriptional oscillator exclusively in the liver. Remarkably, 31 genes in the clock-less liver continued to oscillate presumably using systemic signals from the rest of the animal (95).
These systemic signals originating from the phase of the SCN that can drive and entrain rhythms of gene expression, and thus physiology, of peripheral oscillators are still being uncovered. They include signals from feeding, circulating humoral factors, and fluctuations in body temperature. The phase of the circadian rhythms of gene expression the liver can be uncoupled from the rest of the body by providing food only when the animal would typically be asleep (96,97). This food induced resetting of peripheral oscillators is achieved, at least in part, by the ability of glucocorticoids in the circulatory system to control the phase of peripheral clocks (97,98). The Clara cells of the lung which are involved in detoxification of inhalants and production of various pulmonary secretions can also be entrained by glucocorticoids (99).
It is likely that just as various peripheral oscillators have fine-tuned their circadian transcriptomes, they also use unique combinations of physiologic phase cues for synchronization to the SCN’s phase. The different rates of reentrainment among peripheral tissues to a new light:dark cycle suggests these distinctive properties (18). However, there may be signals which are sufficient to control the phase of most tissues. For example, physiologic fluctuations in temperature can entrain all peripheral oscillators which have been examined (100–102). The body temperature of mammals exhibits a circadian oscillation driven by the SCN regardless of sleep: activity state (103–106). Thus light synchronizes the SCN to the external environment and the SCN controls circadian fluctuations of body temperature. This SCN output serves as an input to the circadian clocks of peripheral tissues whose outputs are the various physiological and transcriptional rhythms seen within the local cells throughout the body. Fittingly, the SCN seems to be resistant to physiologic changes in body temperature (100,101,107). This would be an important feature of the system so that the phase of the SCN would not be influenced by the very parameter it was controlling. However, it is possible that the SCN may be sensitive to many cycles of cyclic temperature changes and that the SCN of some species may be more temperature sensitive than others (108,109). The intercellular coupling in the SCN responsible for these differences and possible mechanisms for temperature entrainment of peripheral tissues will be discussed in the following sections.
Further differences exist between the central pacemaker (SCN) and peripheral tissues at the level of the core molecular clock itself. The Clock gene was discovered as a hypomorphic mutation which caused the behavior of the animal and the molecular rhythms of the SCN to free-run at extremely long periods and become arrhythmic without daily entrainment cues (110,111). However, if Clock is removed from the system as a null allele, the SCN itself and the behavior of the animal remain perfectly rhythmic (112). This is because the gene Npas2 acts as a surrogate for the loss of Clock and compensates as the transcriptional partner of Bmal1 (113). This compensatory role of Npas2 only functions in the SCN, as the loss of Clock abolishes the circadian rhythmicity of the molecular oscillations in peripheral clocks (114). The SCN remains robustly rhythmic in the case of a loss of any single member of the negative limb of the transcriptional feedback cycle (115). The rhythms of peripheral clocks and dissociated cells remain rhythmic with the loss of Cry2; however, circadian rhythmicity is lost in peripheral tissues when Cry1, Per1, or Per2 are removed (115). This importance of the Per1 gene in these cellular rhythms is interesting in light of the subtle effect of the Per1 null allele on behavior (53,55). Adding further complexity, the combined removal of Per1 and Cry1 (two necessary negative limb components in peripheral tissues and single cells) reveals mice with normal free-running periods (116). Clearly differences exist between peripheral and the central oscillator both at the level of transcriptional circuitry and intercellular communication.
The SCN is the master synchronizer in mammals
The discovery of self-sustained circadian clocks in the cells of tissues throughout the body does not mean that the SCN should no longer be considered the “master” circadian clock. Although it does not drive the molecular rhythms in these cells, the SCN is necessary for the synchronization of phases among tissues to distinct phases (117). The SCN does drive circadian rhythms of behavior such as activity:rest cycles and physiological parameters such as body temperature rhythms, as the 24 hr component to these rhythms is lost when the SCN is lesioned (14,103). The behavioral rhythms of an SCN-lesioned animal can be restored by transplantation of donor SCN into the third ventricle (118). The definitive proof that the SCN is the master clock for an animal’s behavior came when Michael Menaker and colleagues transplanted SCN from tau mutant hamsters into SCN-lesioned wild-type hosts. The behavior of the host invariably ran with the free-running period of the donor SCN graft (119).
The suprachiasmatic nuclei are paired structures of the ventral hypothalamus, with each half containing about 10,000 neurons in mice and about 50,000 neurons in humans (120,121). The most dorsal neurons of the SCN and their dorsal reaching efferents straddle the ventral floor of the third ventricle, and the most ventral neurons border the optic chiasm. Light information reaches the SCN from melanopsin-containing retinal ganglion cells (also called “intrinsically photosensitive retinal ganglion cells” or “ipRGCs”) via the retinohypothalamic tract (RHT)(122–124). The SCN receives retinal signals from rods, cones, and/or melanopsin; however, all light information which sets the SCN’s phase is transmitted through the ipRGCs (125–127). Within the SCN there are two main subdivisions known as the dorsomedial “shell” and the ventrolateral “core” (128). These designations were originally defined due to distinct neuropeptide expression. The dorsomedial region is marked by high arginine-vasopressin (AVP) expression, and the ventrolateral region has high expression of vasoactive intestinal peptide (VIP) (129–131). This peptide expression is in addition to a mosaic of other peptides for which the expression and anatomical distinction varies among various species. For example, the mouse SCN also expresses gastrin-releasing peptide, enkephalin, neurotensin, angiotensin II, and calbindin, but the exact functions of each of these are unknown (132).
Another hallmark feature of the SCN is its circadian pattern of spontaneous action potentials (reviewed in (133)). The phase of neuronal firing is entrained by the light:dark cycle, but it also persists in constant darkness and as an ex vivo slice culture (134–136). Similar to the induction of the Per genes by nocturnal light exposure, light pulses during the dark also cause an immediate induction of firing in the SCN (137). Just as the transcriptional clock can be observed in single cells, dissociated SCN neurons continue to fire action potentials with a circadian rhythm for weeks in vitro, although their phases scatter from one another (138).
Synchrony of neurons within the SCN to each other is of paramount importance for the generation of a coherent output signal. At the onset of each circadian cycle, expression of the clock genes Per1 and Per2 starts in the most dorsomedial cells (AVP expressing) and the expression then spreads across each SCN towards the central and ventrolateral (VIP expressing) regions (139–141). This medial-to-lateral, mirrored expression pattern is evident when gene expression in the SCN is viewed through in situ hybridization of fixed tissue or with visualization of gene reporters from a single organotypic culture (141,142). VIP signaling in particular seems key to maintaining synchrony among SCN neurons. Mice lacking VIP or its receptor VPAC2 display erratic free-running behavior and the rhythms of individual neurons within a single SCN are no longer held in uniform phase (143–145). Rhythmic application of a VPAC2 receptor agonist to VIP−/− SCN neurons restores rhythmicity to arrhythmic cells and entrains the cells to a common phase (144). Application of purified VIP peptide into the SCN of animals in vivo causes phase shifts in free-running behavioral rhythms (146). This VIP action on VPAC2 receptors is mediated through cAMP signaling (147,148) which itself has been demonstrated as a determinant of phase and period in multiple tissues (149). The period of the whole SCN, and thus behavior, is determined by an averaging or an intermediate value of the periods of the individual neurons. In chimeric mice in which the SCN were comprised of various proportions of ClockΔ19 (long free-running periods) and wild-type neurons, the free-running period of the mouse’s behavior was determined by the proportion of wild-type to mutant cells (150).
Interestingly, the synaptic communication between cells in the SCN is necessary for the robust molecular oscillations of the core clock genes within individual cells. When intercellular communication via action potentials is lost by blocking voltage-gated Na+ channels with tetrodotoxin (TTX), the circadian oscillations of Per1 and Per2 are greatly reduced and the synchrony of cells within the tissue lose phase coherence (142). When TTX is then removed, robust molecular oscillations resume and the cells resynchronize with the same intercellular phase profile as before the treatment (142). The amplitude of the molecular clock in an intact SCN allows the cells to overcome genetic and physiologic perturbations to which peripheral clocks are susceptible. For example, dissociated SCN neurons from Cry1−/− or Per1−/− mice lack circadian rhythm of clock gene expression; however, the intact SCN harboring these same mutations is as rhythmic as wild-type SCN with only period phenotypes (115). Even in the case of a severe clock gene mutation such as Bmal1−/− which causes a loss of circadian rhythmicity at the behavioral and single cell level, the synaptic communication in an intact Bmal1−/− SCN allows for coordinated, but stochastic, expression of PER2 among SCN neurons (151).
The robustness of the intact SCN is also important for its ability to remain in appropriate phase in the presence of rhythmic physiologic perturbations. This is especially relevant in cases when an animal is exposed to situations that might uncouple aspects of behavior from a natural light:dark cycle. For example when food availability is restricted to a time of the day when an animal is typically asleep and certain peripheral clocks shift their phase accordingly (as discussed in the previous section), the phase of the SCN remains tightly entrained to the light:dark cycle (96,97). While body temperature fluctuations can entrain the rhythms of peripheral circadian clocks, the SCN can maintain its phase in the presence of physiologic temperature fluctuations (100,101,107). This is especially evident in cultured SCN where the tissue becomes sensitive to physiologic temperature changes when communication between cells is lost. Cells which hold their phase in the presence of temperature cycles as large as 2.5°C in an intact SCN show exquisite sensitivity to temperature cycles as small as 1.5°C when decoupled (101,132). It should be noted that the above temperature data was collected in mice and in other species, such as rats, the temperature sensitivity of the SCN may be much greater (108,109).
Most neurons in the SCN produce the neurotransmitter γ-amminobutyric acid (GABA) (152). Daily administration of GABA to cultured dissociated SCN neurons can synchronize rhythms of spontaneous firing and a single administration can shift their phase (153). GABA has also been implicated in conveying phase information between the dorsal and ventral portions exhibiting opposite acute effects on cells from these regions (154). However, other reports suggest that GABA signaling is not necessary for intra-SCN synchrony, and even that GABA receptor antagonism increases firing rhythm amplitude (155). In fact, rhythmic application of a VPAC2 agonist in a Vip−/− SCN was able to synchronize neuronal rhythms in the presence of chronic GABA signaling blockade (155).
Along with internal synchrony, peptides and diffusible factors from the SCN are also important in the signaling from the SCN to the rest of the brain. The arrhythmic behavior of an SCN lesioned animal could be rescued (at least partially) by the transplantation of a donor SCN encapsulated in a semi-permeable membrane which allowed for passage of diffusible factors, but not neural outgrowth (156). The identity of this factor or factors is still being discovered. The SCN-secreted peptides transforming growth factor α (TGF-α), prokineticin 2 (PK2), and cardiotrophin like cytokine (CLC) induce acute activity suppression and are rhythmically produced by the SCN (157–159). Perhaps more behavioral activity inhibiting and maybe some activity inducing factors will be identified in the future. It is likely that just as there is a mosaic of peptides produced locally in the SCN, the output signal involves a cocktail of secreted peptides along with direct neuronal efferents.
Temperature and circadian clocks
The influence of temperature on circadian clocks is important to discuss here both because of the ubiquity of temperature regulatory mechanisms in circadian clocks but also as potential targets for chronotherapeutics. First, as mentioned in the introduction to this chapter, all circadian rhythms are temperature compensated. This fundamental property allows the clock to maintain a stable period of oscillation regardless of the ambient temperature. A circadian clock would not be reliable if its period changed every time the sun went down or ran at a different period in the winter than in the summer. Temperature compensation is expressed as the coefficient Q10 which represents the ratio of the rate of a reaction at temperatures 10°C apart. The Q10 of periods of various circadian rhythms of many species of broad phyla are between 0.8 and 1.2. Most chemical reactions within cells are affected by temperature; for example, most enzymatic reactions increase in rate as temperature is increased. In fact, the kinases CK1ε and δ increase their rate of phosphorylation of some protein targets at higher temperatures as would be expected; however, their rates of phosphorylation of clock proteins are stable at those same temperatures (160). This temperature compensation is yet another example of the robustness of the molecular clock to retain precision in varying conditions. Even with broad reduction in global transcription, the clocks in mammalian cells remain rhythmic with only slightly shorter periods (161).
The mechanisms of temperature compensation are still not understood, but great strides have been taken using the Neurospora crassa fungus. These organisms are routinely exposed to wide variations in temperature in their natural environment. The levels of the clock protein FRQ (which plays the negative limb role in fungus as PER and CRY do in mammals) are elevated at warmer temperatures and a long-form splice variant is observed at warm temperatures (162–164). Mutants of the kinase CK-2, which phosphorylates FRQ, display either better temperature compensation than wild-type or opposite “overcompensation” (165). In our own work we observed an impairment in temperature compensation of PER2 rhythms in the SCN and pituitary of mice when the Heat Shock Factors (HSF) were pharmacologically blocked (101). These results fit with a model in which positive and negative effects of temperature on rates of cellular activity balance out to a net null effect. However, other findings suggest that this balancing model may be more complicated than necessary. Other extremely simple circadian rhythms, such as the in vitro phosphorylation of KaiC in Synechococcus, demonstrate beautiful temperature compensation with the presence of just the three proteins and ATP (77). Also, the transcription/translation-free rhythms of oxidation in peroxiredoxins in human red blood cells are temperature compensated (82). These results suggest that very simple oscillators may be temperature compensated purely by the robustness inherent in the individual processes rather than requiring balancing agents.
Although, circadian clocks run at the same period at various temperatures, this does not mean that circadian clocks ignore temperature. Most species, particularly poikilotherimic organisms, are exposed to wide daily temperature oscillations, and they use the change in temperature as an entraining cue. In fact, in Neurospora if a temperature cycle and light:dark cycle are out of phase, the fungus will entrain to the temperature cycle more strongly than to the light (163). In the fruit fly Drosophila melanogaster, the entrainment of global transcription rhythms appears to use a coordinated combination of light:dark cycles and temperature cycles so that the phase of light entrainment slightly leads the phase set by temperature of the same genes (166). The importance of temperature changes is most strikingly observed at the behavioral level. In standard laboratory conditions with a light:dark cycle at a stable temperature, the flies show strong crepuscular activity with a large inactive period during the middle of the day. When more natural lighting is paired with a temperature cycle, the flies show a strong afternoon bout of activity and behaviorally act like a different species (167).
Environmental temperature cycles act as extremely weak behavioral entrainment cues in warm-blooded animals, or “homeothermic” animals, which maintain their body temperature regardless of ambient temperature (168). However, the internal body temperature of homeothermic animals undergoes circadian fluctuations with amplitudes of approximately 1°C and 5°C depending on the species (169). As mentioned earlier, the surgical ablation of the SCN abolishes the circadian component to body temperature fluctuation along with behavioral and sleep rhythms in mice, rats, and ground squirrels (103,105,106). Although it is hard to isolate effects that activity, sleep, and the SCN have on body temperature oscillations, both human and rodent examples exist. In humans, the circadian oscillation of rectal temperature persists if a person is restricted to 24 hour bed rest and is deprived of sleep (1). In hibernatory animals, such as the ground squirrel, a low amplitude SCN-driven body temperature rhythm is observed during bouts of hibernation in which there is an absence of activity for days at a time (106,170).
As discussed in the Peripheral Clocks section, these rhythms of body temperature fluctuation are sufficient to entrain the peripheral oscillators of homeothermic animals in all cases that have been reported (100–102,171). The most recent evidence suggests that this effect on the molecular clock mammals by temperature cycles is regulated by the heat shock pathway. Briefly, after heat exposure the Heat Shock Factors (HSF1, HSF2, and HSF4) initiate the transcription of genes with Heat Shock Elements (HSE) in their promoters (172). The genes of Heat Shock Proteins (HSP) contain HSEs and once translated these proteins chaperone, or sequester the HSFs from further transcription. This feedback loop maintains a transient response to temperature changes. Although commonly associated with heat tolerance to extreme temperatures, the dynamic range heat shock pathway can include temperature changes within the physiologic range (173). Blocking HSF transcription transiently with the pharmacological agent KNK437 mimicked the phase shifts caused by a cool temperature pulse and blocked the phase shifting effects of warm pulses (101). Also, a brief exposure to warm temperatures caused an acute reduction of Per2 levels followed by an induction when returned to a cooler temperature in the liver (95). Along with being a temperature sensor for phase setting, it is also evident that the HSF family and the circadian clock are more intimately related. Although the levels of HSF proteins have not been found to have a circadian oscillation, their binding to target motifs certainly does even in the absence of temperature cycles (174). Additionally, the promoter of the Per2 gene contains HSEs that are conserved among multiple species, and a number of hsp genes oscillate with a phase similar to Per2 (95). Finally, deletion of the Hsf1 gene lengthens the free-running behavioral period of mice by about 30 min, and pharmacologic blockade of HSF-mediated transcription ex vivo causes the molecular clock to run >30 hr in SCN and peripheral tissues (101,174). Clearly the heat shock response pathway exerts both phase and period influence on the circadian clock. It will be exciting to see how this relationship is further elucidated in the future.
Conclusions and summary
The circadian system of all organisms contain a core oscillator, a way by which this clock can be set by the environment, and output behaviors or processes whose phases are determined by the core clock. This can be observed as an animal in its environment synchronizes its behavior to the sun or as a cell in the liver synchronizes its metabolic state to the phase of the SCN. The precision of the system allows for perfectly timed oscillations throughout the body of a well-functioning organism, or sets the stage for mistimed events and disease in a malfunctioning system. Much has been learned about the molecular function of the clock itself and the ways by which clocks within a single organism communicate, but more insights are uncovered monthly. The field is now at the level where serious therapeutic strategies can be developed and implemented for the treatment of sleep and metabolic disorders, optimizing timing of drug delivery, and the cooption of circadian elements to control various cellular pathways and vice versa.
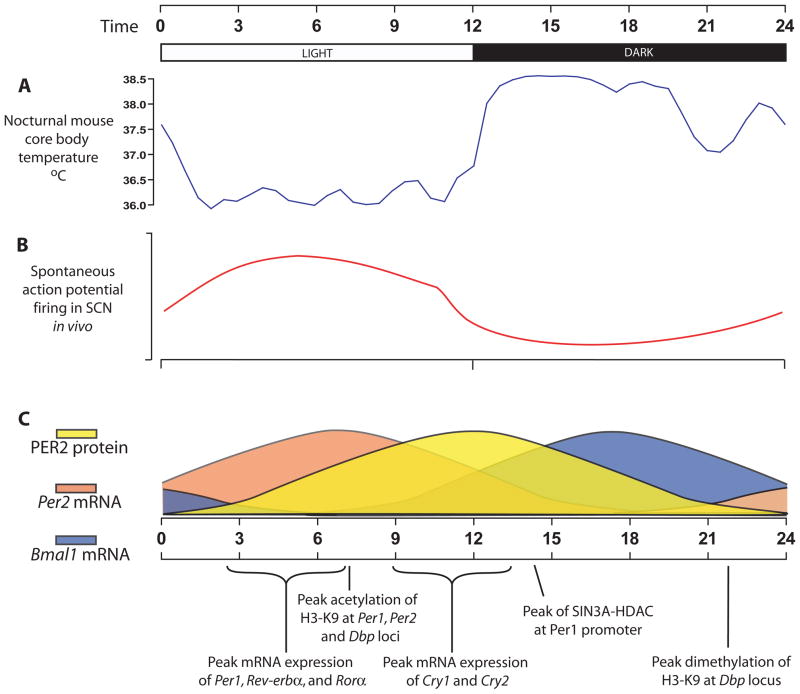
Figure 2.
Timing of circadian events in nocturnal rodents. A) Mouse core body temperature as measured by radio telemetry. B) Spontaneous firing rhythms from a cultured rat SCN as adapted from (175). C) Molecular clock events are plotted schematically without axes for clarity. Yellow sine wave represents the phase of PER2 protein abundance in the mouse SCN. Orange wave represents the phase of mPer2 mRNA abundance in the mouse SCN and the blue wave represents the phase of Bmal1 mRNA abundance. Chromatin information relates to the promoter regions of the Per genes and Dbp as reported by (31,32). Sin3A-HDAC phase from (36).
References
- 1.Aschoff J. Circadian control of body temperature. Journal of Thermal Biology. 1983;8:143–147. [Google Scholar]
- 2.Green CB, Takahashi JS, Bass J. The meter of metabolism. Cell. 2008;134:728–742. doi: 10.1016/j.cell.2008.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cameron MA, Barnard AR, Lucas RJ. The electroretinogram as a method for studying circadian rhythms in the mammalian retina. J Genet. 2008;87:459–466. doi: 10.1007/s12041-008-0068-5. [DOI] [PubMed] [Google Scholar]
- 4.Eckel-Mahan KL, Storm DR. Circadian rhythms and memory: not so simple as cogs and gears. EMBO Rep. 2009;10:584–591. doi: 10.1038/embor.2009.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reid KJ, Zee PC. Circadian rhythm disorders. Semin Neurol. 2009;29:393–405. doi: 10.1055/s-0029-1237120. [DOI] [PubMed] [Google Scholar]
- 6.Toh KL, Jones CR, He Y, Eide EJ, Hinz WA, Virshup DM, Ptácek LJ, Fu YH. An hPer2 phosphorylation site mutation in familial advanced sleep phase syndrome. Science. 2001;291:1040–1043. doi: 10.1126/science.1057499. [DOI] [PubMed] [Google Scholar]
- 7.Xu Y, Padiath QS, Shapiro RE, Jones CR, Wu SC, Saigoh N, Saigoh K, Ptácek LJ, Fu YH. Functional consequences of a CKIdelta mutation causing familial advanced sleep phase syndrome. Nature. 2005;434:640–644. doi: 10.1038/nature03453. [DOI] [PubMed] [Google Scholar]
- 8.Xu Y, Toh KL, Jones CR, Shin JY, Fu YH, Ptacek LJ. Modeling of a human circadian mutation yields insights into clock regulation by PER2. Cell. 2007;128:59–70. doi: 10.1016/j.cell.2006.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marcheva B, Ramsey KM, Buhr ED, Kobayashi Y, Su H, Ko CH, Ivanova G, Omura C, Mo S, Vitaterna MH, Lopez JP, Philipson LH, Bradfield CA, Crosby SD, JeBailey L, Wang X, Takahashi JS, Bass J. Disruption of the clock components CLOCK and BMAL1 leads to hypoinsulinaemia and diabetes. Nature. 2010;466:627–631. doi: 10.1038/nature09253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muller JE, Stone PH, Turi ZG, Rutherford JD, Czeisler CA, Parker C, Poole WK, Passamani E, Roberts R, Robertson T. Circadian variation in the frequency of onset of acute myocardial infarction. N Engl J Med. 1985;313:1315–1322. doi: 10.1056/NEJM198511213132103. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson R. Circadian rhythms and sleep-related breathing disorders. Sleep Med. 2007;8:681–687. doi: 10.1016/j.sleep.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 12.Gaddameedhi S, Selby CP, Kaufmann WK, Smart RC, Sancar A. Control of skin cancer by the circadian rhythm. Proc Natl Acad Sci U S A. 2011;108:18790–18795. doi: 10.1073/pnas.1115249108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorbacheva VY, Kondratov RV, Zhang R, Cherukuri S, Gudkov AV, Takahashi JS, Antoch MP. Circadian sensitivity to the chemotherapeutic agent cyclophosphamide depends on the functional status of the CLOCK/BMAL1 transactivation complex. Proc Natl Acad Sci U S A. 2005;102:3407–3412. doi: 10.1073/pnas.0409897102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A. 1972;69:1583–1586. doi: 10.1073/pnas.69.6.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moore RY, Eichler VB. Loss of a circadian adrenal corticosterone rhythm following suprachiasmatic lesions in the rat. Brain Res. 1972;42:201–206. doi: 10.1016/0006-8993(72)90054-6. [DOI] [PubMed] [Google Scholar]
- 16.Balsalobre A, Damiola F, Schibler U. A serum shock induces circadian gene expression in mammalian tissue culture cells. Cell. 1998;93:929–937. doi: 10.1016/s0092-8674(00)81199-x. [DOI] [PubMed] [Google Scholar]
- 17.Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 18.Yamazaki S, Numano R, Abe M, Hida A, Takahashi R, Ueda M, Block GD, Sakaki Y, Menaker M, Tei H. Resetting central and peripheral circadian oscillators in transgenic rats. Science. 2000;288:682–685. doi: 10.1126/science.288.5466.682. [DOI] [PubMed] [Google Scholar]
- 19.Abe M, Herzog ED, Yamazaki S, Straume M, Tei H, Sakaki Y, Menaker M, Block GD. Circadian rhythms in isolated brain regions. J Neurosci. 2002;22:350–356. doi: 10.1523/JNEUROSCI.22-01-00350.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nelson R, Zucker I. Absence of extraocular photoreception in diurnal and nocturnal rodents exposed to direct sunlight. Comp Biochem Physiol A. 1981;69:145–148. [Google Scholar]
- 21.Lowrey PL, Takahashi JS. Genetics of circadian rhythms in Mammalian model organisms. Adv Genet. 2011;74:175–230. doi: 10.1016/B978-0-12-387690-4.00006-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- 23.Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohno T, Onishi Y, Ishida N. A novel E4BP4 element drives circadian expression of mPeriod2. Nucleic Acids Res. 2007;35:648–655. doi: 10.1093/nar/gkl868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hogenesch JB, Gu YZ, Jain S, Bradfield CA. The basic-helix-loop-helix-PAS orphan MOP3 forms transcriptionally active complexes with circadian and hypoxia factors. Proc Natl Acad Sci U S A. 1998;95:5474–5479. doi: 10.1073/pnas.95.10.5474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- 27.Griffin EA, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- 28.Sangoram AM, Saez L, Antoch MP, Gekakis N, Staknis D, Whiteley A, Fruechte EM, Vitaterna MH, Shimomura K, King DP, Young MW, Weitz CJ, Takahashi JS. Mammalian circadian autoregulatory loop: a timeless ortholog and mPer1 interact and negatively regulate CLOCK-BMAL1-induced transcription. Neuron. 1998;21:1101–1113. doi: 10.1016/s0896-6273(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 29.Field MD, Maywood ES, O’Brien JA, Weaver DR, Reppert SM, Hastings MH. Analysis of clock proteins in mouse SCN demonstrates phylogenetic divergence of the circadian clockwork and resetting mechanisms. Neuron. 2000;25:437–447. doi: 10.1016/s0896-6273(00)80906-x. [DOI] [PubMed] [Google Scholar]
- 30.Sato TK, Yamada RG, Ukai H, Baggs JE, Miraglia LJ, Kobayashi TJ, Welsh DK, Kay SA, Ueda HR, Hogenesch JB. Feedback repression is required for mammalian circadian clock function. Nat Genet. 2006;38:312–319. doi: 10.1038/ng1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Etchegaray JP, Lee C, Wade PA, Reppert SM. Rhythmic histone acetylation underlies transcription in the mammalian circadian clock. Nature. 2003;421:177–182. doi: 10.1038/nature01314. [DOI] [PubMed] [Google Scholar]
- 32.Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- 33.Doi M, Hirayama J, Sassone-Corsi P. Circadian regulator CLOCK is a histone acetyltransferase. Cell. 2006;125:497–508. doi: 10.1016/j.cell.2006.03.033. [DOI] [PubMed] [Google Scholar]
- 34.Katada S, Sassone-Corsi P. The histone methyltransferase MLL1 permits the oscillation of circadian gene expression. Nat Struct Mol Biol. 2010;17:1414–1421. doi: 10.1038/nsmb.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.DiTacchio L, Le HD, Vollmers C, Hatori M, Witcher M, Secombe J, Panda S. Histone lysine demethylase JARID1a activates CLOCK-BMAL1 and influences the circadian clock. Science. 2011;333:1881–1885. doi: 10.1126/science.1206022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Duong HA, Robles MS, Knutti D, Weitz CJ. A molecular mechanism for circadian clock negative feedback. Science. 2011;332:1436–1439. doi: 10.1126/science.1196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134:329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U. SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell. 2008;134:317–328. doi: 10.1016/j.cell.2008.06.050. [DOI] [PubMed] [Google Scholar]
- 39.Rutter J, Reick M, Wu LC, McKnight SL. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science. 2001;293:510–514. doi: 10.1126/science.1060698. [DOI] [PubMed] [Google Scholar]
- 40.Ralph MR, Menaker M. A mutation of the circadian system in golden hamsters. Science. 1988;241:1225–1227. doi: 10.1126/science.3413487. [DOI] [PubMed] [Google Scholar]
- 41.Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Camacho F, Cilio M, Guo Y, Virshup DM, Patel K, Khorkova O, Styren S, Morse B, Yao Z, Keesler GA. Human casein kinase Idelta phosphorylation of human circadian clock proteins period 1 and 2. FEBS Lett. 2001;489:159–165. doi: 10.1016/s0014-5793(00)02434-0. [DOI] [PubMed] [Google Scholar]
- 43.Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- 45.Vanselow K, Vanselow JT, Westermark PO, Reischl S, Maier B, Korte T, Herrmann A, Herzel H, Schlosser A, Kramer A. Differential effects of PER2 phosphorylation: molecular basis for the human familial advanced sleep phase syndrome (FASPS) Genes Dev. 2006;20:2660–2672. doi: 10.1101/gad.397006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Siepka SM, Yoo SH, Park J, Song W, Kumar V, Hu Y, Lee C, Takahashi JS. Circadian mutant Overtime reveals F-box protein FBXL3 regulation of cryptochrome and period gene expression. Cell. 2007;129:1011–1023. doi: 10.1016/j.cell.2007.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Godinho SI, Maywood ES, Shaw L, Tucci V, Barnard AR, Busino L, Pagano M, Kendall R, Quwailid MM, Romero MR, O’neill J, Chesham JE, Brooker D, Lalanne Z, Hastings MH, Nolan PM. The after-hours mutant reveals a role for Fbxl3 in determining mammalian circadian period. Science. 2007;316:897–900. doi: 10.1126/science.1141138. [DOI] [PubMed] [Google Scholar]
- 48.Busino L, Bassermann F, Maiolica A, Lee C, Nolan PM, Godinho SI, Draetta GF, Pagano M. SCFFbxl3 controls the oscillation of the circadian clock by directing the degradation of cryptochrome proteins. Science. 2007;316:900–904. doi: 10.1126/science.1141194. [DOI] [PubMed] [Google Scholar]
- 49.Lamia KA, Sachdeva UM, DiTacchio L, Williams EC, Alvarez JG, Egan DF, Vasquez DS, Juguilon H, Panda S, Shaw RJ, Thompson CB, Evans RM. AMPK regulates the circadian clock by cryptochrome phosphorylation and degradation. Science. 2009;326:437–440. doi: 10.1126/science.1172156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Harada Y, Sakai M, Kurabayashi N, Hirota T, Fukada Y. Ser-557-phosphorylated mCRY2 is degraded upon synergistic phosphorylation by glycogen synthase kinase-3 beta. J Biol Chem. 2005;280:31714–31721. doi: 10.1074/jbc.M506225200. [DOI] [PubMed] [Google Scholar]
- 51.Kurabayashi N, Hirota T, Sakai M, Sanada K, Fukada Y. DYRK1A and glycogen synthase kinase 3beta, a dual-kinase mechanism directing proteasomal degradation of CRY2 for circadian timekeeping. Mol Cell Biol. 2010;30:1757–1768. doi: 10.1128/MCB.01047-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zheng B, Albrecht U, Kaasik K, Sage M, Lu W, Vaishnav S, Li Q, Sun ZS, Eichele G, Bradley A, Lee CC. Nonredundant roles of the mPer1 and mPer2 genes in the mammalian circadian clock. Cell. 2001;105:683–694. doi: 10.1016/s0092-8674(01)00380-4. [DOI] [PubMed] [Google Scholar]
- 53.Cermakian N, Monaco L, Pando MP, Dierich A, Sassone-Corsi P. Altered behavioral rhythms and clock gene expression in mice with a targeted mutation in the Period1 gene. EMBO J. 2001;20:3967–3974. doi: 10.1093/emboj/20.15.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bae K, Jin X, Maywood ES, Hastings MH, Reppert SM, Weaver DR. Differential functions of mPer1, mPer2, and mPer3 in the SCN circadian clock. Neuron. 2001;30:525–536. doi: 10.1016/s0896-6273(01)00302-6. [DOI] [PubMed] [Google Scholar]
- 55.Zheng B, Larkin DW, Albrecht U, Sun ZS, Sage M, Eichele G, Lee CC, Bradley A. The mPer2 gene encodes a functional component of the mammalian circadian clock. Nature. 1999;400:169–173. doi: 10.1038/22118. [DOI] [PubMed] [Google Scholar]
- 56.Thresher RJ, Vitaterna MH, Miyamoto Y, Kazantsev A, Hsu DS, Petit C, Selby CP, Dawut L, Smithies O, Takahashi JS, Sancar A. Role of mouse cryptochrome blue-light photoreceptor in circadian photoresponses. Science. 1998;282:1490–1494. doi: 10.1126/science.282.5393.1490. [DOI] [PubMed] [Google Scholar]
- 57.Vitaterna MH, Selby CP, Todo T, Niwa H, Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T, Miyazaki J, Takahashi JS, Sancar A. Differential regulation of mammalian period genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl Acad Sci U S A. 1999;96:12114–12119. doi: 10.1073/pnas.96.21.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van der Horst GT, Muijtjens M, Kobayashi K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van Leenen D, Buijs R, Bootsma D, Hoeijmakers JH, Yasui A. Mammalian Cry1 and Cry2 are essential for maintenance of circadian rhythms. Nature. 1999;398:627–630. doi: 10.1038/19323. [DOI] [PubMed] [Google Scholar]
- 59.Baggs JE, Price TS, DiTacchio L, Panda S, Fitzgerald GA, Hogenesch JB. Network features of the mammalian circadian clock. PLoS Biol. 2009;7:e52. doi: 10.1371/journal.pbio.1000052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS. Transcriptional Architecture and Chromatin Landscape of the Core Circadian Clock in Mammals. Science. 2012;338:349–354. doi: 10.1126/science.1226339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Edery I, Rutila JE, Rosbash M. Phase shifting of the circadian clock by induction of the Drosophila period protein. Science. 1994;263:237–240. doi: 10.1126/science.8284676. [DOI] [PubMed] [Google Scholar]
- 62.Albrecht U, Sun ZS, Eichele G, Lee CC. A differential response of two putative mammalian circadian regulators, mper1 and mper2, to light. Cell. 1997;91:1055–1064. doi: 10.1016/s0092-8674(00)80495-x. [DOI] [PubMed] [Google Scholar]
- 63.Shearman LP, Zylka MJ, Weaver DR, Kolakowski LF, Jr, Reppert SM. Two period homologs: circadian expression and photic regulation in the suprachiasmatic nuclei. Neuron. 1997;19:1261–1269. doi: 10.1016/s0896-6273(00)80417-1. [DOI] [PubMed] [Google Scholar]
- 64.Shigeyoshi Y, Taguchi K, Yamamoto S, Takekida S, Yan L, Tei H, Moriya T, Shibata S, Loros JJ, Dunlap JC, Okamura H. Light-induced resetting of a mammalian circadian clock is associated with rapid induction of the mPer1 transcript. Cell. 1997;91:1043–1053. doi: 10.1016/s0092-8674(00)80494-8. [DOI] [PubMed] [Google Scholar]
- 65.Yan L, Silver R. Resetting the brain clock: time course and localization of mPER1 and mPER2 protein expression in suprachiasmatic nuclei during phase shifts. Eur J Neurosci. 2004;19:1105–1109. doi: 10.1111/j.1460-9568.2004.03189.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Travnickova-Bendova Z, Cermakian N, Reppert SM, Sassone-Corsi P. Bimodal regulation of mPeriod promoters by CREB-dependent signaling and CLOCK/BMAL1 activity. Proc Natl Acad Sci U S A. 2002;99:7728–7733. doi: 10.1073/pnas.102075599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- 68.Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 69.Guillaumond F, Dardente H, Giguère V, Cermakian N. Differential control of Bmal1 circadian transcription by REV-ERB and ROR nuclear receptors. J Biol Rhythms. 2005;20:391–403. doi: 10.1177/0748730405277232. [DOI] [PubMed] [Google Scholar]
- 70.Cho H, Zhao X, Hatori M, Yu RT, Barish GD, Lam MT, Chong LW, DiTacchio L, Atkins AR, Glass CK, Liddle C, Auwerx J, Downes M, Panda S, Evans RM. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature. 2012;485:123–127. doi: 10.1038/nature11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Falvey E, Fleury-Olela F, Schibler U. The rat hepatic leukemia factor (HLF) gene encodes two transcriptional activators with distinct circadian rhythms, tissue distributions and target preferences. EMBO J. 1995;14:4307–4317. doi: 10.1002/j.1460-2075.1995.tb00105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lopez-Molina L, Conquet F, Dubois-Dauphin M, Schibler U. The DBP gene is expressed according to a circadian rhythm in the suprachiasmatic nucleus and influences circadian behavior. EMBO J. 1997;16:6762–6771. doi: 10.1093/emboj/16.22.6762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fonjallaz P, Ossipow V, Wanner G, Schibler U. The two PAR leucine zipper proteins, TEF and DBP, display similar circadian and tissue-specific expression, but have different target promoter preferences. EMBO J. 1996;15:351–362. [PMC free article] [PubMed] [Google Scholar]
- 74.Mitsui S, Yamaguchi S, Matsuo T, Ishida Y, Okamura H. Antagonistic role of E4BP4 and PAR proteins in the circadian oscillatory mechanism. Genes Dev. 2001;15:995–1006. doi: 10.1101/gad.873501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ukai-Tadenuma M, Yamada RG, Xu H, Ripperger JA, Liu AC, Ueda HR. Delay in feedback repression by cryptochrome 1 is required for circadian clock function. Cell. 2011;144:268–281. doi: 10.1016/j.cell.2010.12.019. [DOI] [PubMed] [Google Scholar]
- 76.Liu AC, Tran HG, Zhang EE, Priest AA, Welsh DK, Kay SA. Redundant function of REV-ERBalpha and beta and non-essential role for Bmal1 cycling in transcriptional regulation of intracellular circadian rhythms. PLoS Genet. 2008;4:e1000023. doi: 10.1371/journal.pgen.1000023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakajima M, Imai K, Ito H, Nishiwaki T, Murayama Y, Iwasaki H, Oyama T, Kondo T. Reconstitution of circadian oscillation of cyanobacterial KaiC phosphorylation in vitro. Science. 2005;308:414–415. doi: 10.1126/science.1108451. [DOI] [PubMed] [Google Scholar]
- 78.Iwasaki H, Nishiwaki T, Kitayama Y, Nakajima M, Kondo T. KaiA-stimulated KaiC phosphorylation in circadian timing loops in cyanobacteria. Proc Natl Acad Sci U S A. 2002;99:15788–15793. doi: 10.1073/pnas.222467299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kitayama Y, Iwasaki H, Nishiwaki T, Kondo T. KaiB functions as an attenuator of KaiC phosphorylation in the cyanobacterial circadian clock system. EMBO J. 2003;22:2127–2134. doi: 10.1093/emboj/cdg212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nishiwaki T, Iwasaki H, Ishiura M, Kondo T. Nucleotide binding and autophosphorylation of the clock protein KaiC as a circadian timing process of cyanobacteria. Proc Natl Acad Sci U S A. 2000;97:495–499. doi: 10.1073/pnas.97.1.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.O’Neill JS, van Ooijen G, Dixon LE, Troein C, Corellou F, Bouget FY, Reddy AB, Millar AJ. Circadian rhythms persist without transcription in a eukaryote. Nature. 2011;469:554–558. doi: 10.1038/nature09654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.O’Neill JS, Reddy AB. Circadian clocks in human red blood cells. Nature. 2011;469:498–503. doi: 10.1038/nature09702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tomita J, Nakajima M, Kondo T, Iwasaki H. No transcription-translation feedback in circadian rhythm of KaiC phosphorylation. Science. 2005;307:251–254. doi: 10.1126/science.1102540. [DOI] [PubMed] [Google Scholar]
- 84.Edgar RS, Green EW, Zhao Y, van Ooijen G, Olmedo M, Qin X, Xu Y, Pan M, Valekunja UK, Feeney KA, Maywood ES, Hastings MH, Baliga NS, Merrow M, Millar AJ, Johnson CH, Kyriacou CP, O’Neill JS, Reddy AB. Peroxiredoxins are conserved markers of circadian rhythms. Nature. 2012;485:459–464. doi: 10.1038/nature11088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Stratmann M, Schibler U. Properties, entrainment, and physiological functions of mammalian peripheral oscillators. J Biol Rhythms. 2006;21:494–506. doi: 10.1177/0748730406293889. [DOI] [PubMed] [Google Scholar]
- 86.Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 87.Welsh DK, Yoo SH, Liu AC, Takahashi JS, Kay SA. Bioluminescence imaging of individual fibroblasts reveals persistent, independently phased circadian rhythms of clock gene expression. Curr Biol. 2004;14:2289–2295. doi: 10.1016/j.cub.2004.11.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hogenesch JB, Ueda HR. Understanding systems-level properties: timely stories from the study of clocks. Nat Rev Genet. 2011;12:407–416. doi: 10.1038/nrg2972. [DOI] [PubMed] [Google Scholar]
- 89.Kornmann B, Preitner N, Rifat D, Fleury-Olela F, Schibler U. Analysis of circadian liver gene expression by ADDER, a highly sensitive method for the display of differentially expressed mRNAs. Nucleic Acids Res. 2001;29:E51–51. doi: 10.1093/nar/29.11.e51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Akhtar RA, Reddy AB, Maywood ES, Clayton JD, King VM, Smith AG, Gant TW, Hastings MH, Kyriacou CP. Circadian cycling of the mouse liver transcriptome, as revealed by cDNA microarray, is driven by the suprachiasmatic nucleus. Curr Biol. 2002;12:540–550. doi: 10.1016/s0960-9822(02)00759-5. [DOI] [PubMed] [Google Scholar]
- 91.Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- 92.Storch KF, Paz C, Signorovitch J, Raviola E, Pawlyk B, Li T, Weitz CJ. Intrinsic circadian clock of the mammalian retina: importance for retinal processing of visual information. Cell. 2007;130:730–741. doi: 10.1016/j.cell.2007.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Storch KF, Lipan O, Leykin I, Viswanathan N, Davis FC, Wong WH, Weitz CJ. Extensive and divergent circadian gene expression in liver and heart. Nature. 2002;417:78–83. doi: 10.1038/nature744. [DOI] [PubMed] [Google Scholar]
- 94.Miller BH, McDearmon EL, Panda S, Hayes KR, Zhang J, Andrews JL, Antoch MP, Walker JR, Esser KA, Hogenesch JB, Takahashi JS. Circadian and CLOCK-controlled regulation of the mouse transcriptome and cell proliferation. Proc Natl Acad Sci U S A. 2007;104:3342–3347. doi: 10.1073/pnas.0611724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kornmann B, Schaad O, Bujard H, Takahashi JS, Schibler U. System-driven and oscillator-dependent circadian transcription in mice with a conditionally active liver clock. PLoS Biol. 2007;5:e34. doi: 10.1371/journal.pbio.0050034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 97.Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Balsalobre A, Brown SA, Marcacci L, Tronche F, Kellendonk C, Reichardt HM, Schütz G, Schibler U. Resetting of circadian time in peripheral tissues by glucocorticoid signaling. Science. 2000;289:2344–2347. doi: 10.1126/science.289.5488.2344. [DOI] [PubMed] [Google Scholar]
- 99.Gibbs JE, Beesley S, Plumb J, Singh D, Farrow S, Ray DW, Loudon AS. Circadian timing in the lung; a specific role for bronchiolar epithelial cells. Endocrinology. 2009;150:268–276. doi: 10.1210/en.2008-0638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brown SA, Zumbrunn G, Fleury-Olela F, Preitner N, Schibler U. Rhythms of mammalian body temperature can sustain peripheral circadian clocks. Curr Biol. 2002;12:1574–1583. doi: 10.1016/s0960-9822(02)01145-4. [DOI] [PubMed] [Google Scholar]
- 101.Buhr ED, Yoo SH, Takahashi JS. Temperature as a universal resetting cue for mammalian circadian oscillators. Science. 2010;330:379–385. doi: 10.1126/science.1195262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Granados-Fuentes D, Saxena MT, Prolo LM, Aton SJ, Herzog ED. Olfactory bulb neurons express functional, entrainable circadian rhythms. Eur J Neurosci. 2004;19:898–906. doi: 10.1111/j.0953-816x.2004.03117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Eastman CI, Mistlberger RE, Rechtschaffen A. Suprachiasmatic nuclei lesions eliminate circadian temperature and sleep rhythms in the rat. Physiol Behav. 1984;32:357–368. doi: 10.1016/0031-9384(84)90248-8. [DOI] [PubMed] [Google Scholar]
- 104.Scheer FA, Pirovano C, Van Someren EJ, Buijs RM. Environmental light and suprachiasmatic nucleus interact in the regulation of body temperature. Neuroscience. 2005;132:465–477. doi: 10.1016/j.neuroscience.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 105.Filipski E, King VM, Li X, Granda TG, Mormont MC, Liu X, Claustrat B, Hastings MH, Lévi F. Host circadian clock as a control point in tumor progression. J Natl Cancer Inst. 2002;94:690–697. doi: 10.1093/jnci/94.9.690. [DOI] [PubMed] [Google Scholar]
- 106.Ruby NF, Dark J, Burns DE, Heller HC, Zucker I. The suprachiasmatic nucleus is essential for circadian body temperature rhythms in hibernating ground squirrels. J Neurosci. 2002;22:357–364. doi: 10.1523/JNEUROSCI.22-01-00357.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Abraham U, Granada AE, Westermark PO, Heine M, Kramer A, Herzel H. Coupling governs entrainment range of circadian clocks. Mol Syst Biol. 2010;6:438. doi: 10.1038/msb.2010.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ruby NF, Burns DE, Heller HC. Circadian rhythms in the suprachiasmatic nucleus are temperature-compensated and phase-shifted by heat pulses in vitro. J Neurosci. 1999;19:8630–8636. doi: 10.1523/JNEUROSCI.19-19-08630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Herzog ED, Huckfeldt RM. Circadian entrainment to temperature, but not light, in the isolated suprachiasmatic nucleus. J Neurophysiol. 2003;90:763–770. doi: 10.1152/jn.00129.2003. [DOI] [PubMed] [Google Scholar]
- 110.Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Vitaterna MH, Ko CH, Chang AM, Buhr ED, Fruechte EM, Schook A, Antoch MP, Turek FW, Takahashi JS. The mouse Clock mutation reduces circadian pacemaker amplitude and enhances efficacy of resetting stimuli and phase-response curve amplitude. Proc Natl Acad Sci U S A. 2006;103:9327–9332. doi: 10.1073/pnas.0603601103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Debruyne JP, Noton E, Lambert CM, Maywood ES, Weaver DR, Reppert SM. A clock shock: mouse CLOCK is not required for circadian oscillator function. Neuron. 2006;50:465–477. doi: 10.1016/j.neuron.2006.03.041. [DOI] [PubMed] [Google Scholar]
- 113.DeBruyne JP, Weaver DR, Reppert SM. CLOCK and NPAS2 have overlapping roles in the suprachiasmatic circadian clock. Nat Neurosci. 2007;10:543–545. doi: 10.1038/nn1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.DeBruyne JP, Weaver DR, Reppert SM. Peripheral circadian oscillators require CLOCK. Curr Biol. 2007;17:R538–539. doi: 10.1016/j.cub.2007.05.067. [DOI] [PubMed] [Google Scholar]
- 115.Liu AC, Welsh DK, Ko CH, Tran HG, Zhang EE, Priest AA, Buhr ED, Singer O, Meeker K, Verma IM, Doyle FJ, 3rd, Takahashi JS, Kay SA. Intercellular coupling confers robustness against mutations in the SCN circadian clock network. Cell. 2007;129:605–616. doi: 10.1016/j.cell.2007.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Oster H, Baeriswyl S, Van Der Horst GT, Albrecht U. Loss of circadian rhythmicity in aging mPer1−/−mCry2−/− mutant mice. Genes Dev. 2003;17:1366–1379. doi: 10.1101/gad.256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, Menaker M, Takahashi JS. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Drucker-Colín R, Aguilar-Roblero R, García-Hernández F, Fernández-Cancino F, Bermudez Rattoni F. Fetal suprachiasmatic nucleus transplants: diurnal rhythm recovery of lesioned rats. Brain Res. 1984;311:353–357. doi: 10.1016/0006-8993(84)90099-4. [DOI] [PubMed] [Google Scholar]
- 119.Ralph MR, Foster RG, Davis FC, Menaker M. Transplanted suprachiasmatic nucleus determines circadian period. Science. 1990;247:975–978. doi: 10.1126/science.2305266. [DOI] [PubMed] [Google Scholar]
- 120.Cassone VM, Speh JC, Card JP, Moore RY. Comparative anatomy of the mammalian hypothalamic suprachiasmatic nucleus. J Biol Rhythms. 1988;3:71–91. doi: 10.1177/074873048800300106. [DOI] [PubMed] [Google Scholar]
- 121.Swaab DF, Fliers E, Partiman TS. The suprachiasmatic nucleus of the human brain in relation to sex, age and senile dementia. Brain Res. 1985;342:37–44. doi: 10.1016/0006-8993(85)91350-2. [DOI] [PubMed] [Google Scholar]
- 122.Moore RY, Lenn NJ. A retinohypothalamic projection in the rat. J Comp Neurol. 1972;146:1–14. doi: 10.1002/cne.901460102. [DOI] [PubMed] [Google Scholar]
- 123.Berson DM, Dunn FA, Takao M. Phototransduction by retinal ganglion cells that set the circadian clock. Science. 2002;295:1070–1073. doi: 10.1126/science.1067262. [DOI] [PubMed] [Google Scholar]
- 124.Hattar S, Liao HW, Takao M, Berson DM, Yau KW. Melanopsin-containing retinal ganglion cells: architecture, projections, and intrinsic photosensitivity. Science. 2002;295:1065–1070. doi: 10.1126/science.1069609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Freedman MS, Lucas RJ, Soni B, von Schantz M, Muñoz M, David-Gray Z, Foster R. Regulation of mammalian circadian behavior by non-rod, non-cone, ocular photoreceptors. Science. 1999;284:502–504. doi: 10.1126/science.284.5413.502. [DOI] [PubMed] [Google Scholar]
- 126.Panda S, Sato TK, Castrucci AM, Rollag MD, DeGrip WJ, Hogenesch JB, Provencio I, Kay SA. Melanopsin (Opn4) requirement for normal light-induced circadian phase shifting. Science. 2002;298:2213–2216. doi: 10.1126/science.1076848. [DOI] [PubMed] [Google Scholar]
- 127.Guler AD, Ecker JL, Lall GS, Haq S, Altimus CM, Liao HW, Barnard AR, Cahill H, Badea TC, Zhao H, Hankins MW, Berson DM, Lucas RJ, Yau KW, Hattar S. Melanopsin cells are the principal conduits for rod-cone input to non-image-forming vision. Nature. 2008;453:102–105. doi: 10.1038/nature06829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Morin LP. SCN organization reconsidered. J Biol Rhythms. 2007;22:3–13. doi: 10.1177/0748730406296749. [DOI] [PubMed] [Google Scholar]
- 129.Samson WK, Said SI, McCann SM. Radioimmunologic localization of vasoactive intestinal polypeptide in hypothalamic and extrahypothalamic sites in the rat brain. Neurosci Lett. 1979;12:265–269. doi: 10.1016/0304-3940(79)96073-7. [DOI] [PubMed] [Google Scholar]
- 130.Vandesande F, Dierickx K. Identification of the vasopressin producing and of the oxytocin producing neurons in the hypothalamic magnocellular neurosecretroy system of the rat. Cell Tissue Res. 1975;164:153–162. doi: 10.1007/BF00218970. [DOI] [PubMed] [Google Scholar]
- 131.Dierickx K, Vandesande F. Immunocytochemical localization of the vasopressinergic and the oxytocinergic neurons in the human hypothalamus. Cell Tissue Res. 1977;184:15–27. doi: 10.1007/BF00220524. [DOI] [PubMed] [Google Scholar]
- 132.Abrahamson EE, Moore RY. Suprachiasmatic nucleus in the mouse: retinal innervation, intrinsic organization and efferent projections. Brain Res. 2001;916:172–191. doi: 10.1016/s0006-8993(01)02890-6. [DOI] [PubMed] [Google Scholar]
- 133.Herzog ED. Neurons and networks in daily rhythms. Nat Rev Neurosci. 2007;8:790–802. doi: 10.1038/nrn2215. [DOI] [PubMed] [Google Scholar]
- 134.Yamazaki S, Kerbeshian MC, Hocker CG, Block GD, Menaker M. Rhythmic properties of the hamster suprachiasmatic nucleus in vivo. J Neurosci. 1998;18:10709–10723. doi: 10.1523/JNEUROSCI.18-24-10709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Groos G, Hendriks J. Circadian rhythms in electrical discharge of rat suprachiasmatic neurones recorded in vitro. Neurosci Lett. 1982;34:283–288. doi: 10.1016/0304-3940(82)90189-6. [DOI] [PubMed] [Google Scholar]
- 136.Green DJ, Gillette R. Circadian rhythm of firing rate recorded from single cells in the rat suprachiasmatic brain slice. Brain Res. 1982;245:198–200. doi: 10.1016/0006-8993(82)90361-4. [DOI] [PubMed] [Google Scholar]
- 137.Nakamura W, Yamazaki S, Nakamura TJ, Shirakawa T, Block GD, Takumi T. In vivo monitoring of circadian timing in freely moving mice. Curr Biol. 2008;18:381–385. doi: 10.1016/j.cub.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 138.Welsh DK, Logothetis DE, Meister M, Reppert SM. Individual neurons dissociated from rat suprachiasmatic nucleus express independently phased circadian firing rhythms. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 139.Yan L, Okamura H. Gradients in the circadian expression of Per1 and Per2 genes in the rat suprachiasmatic nucleus. Eur J Neurosci. 2002;15:1153–1162. doi: 10.1046/j.1460-9568.2002.01955.x. [DOI] [PubMed] [Google Scholar]
- 140.Hamada T, Antle MC, Silver R. Temporal and spatial expression patterns of canonical clock genes and clock-controlled genes in the suprachiasmatic nucleus. Eur J Neurosci. 2004;19:1741–1748. doi: 10.1111/j.1460-9568.2004.03275.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Asai M, Yamaguchi S, Isejima H, Jonouchi M, Moriya T, Shibata S, Kobayashi M, Okamura H. Visualization of mPer1 transcription in vitro: NMDA induces a rapid phase shift of mPer1 gene in cultured SCN. Curr Biol. 2001;11:1524–1527. doi: 10.1016/s0960-9822(01)00445-6. [DOI] [PubMed] [Google Scholar]
- 142.Yamaguchi S, Isejima H, Matsuo T, Okura R, Yagita K, Kobayashi M, Okamura H. Synchronization of cellular clocks in the suprachiasmatic nucleus. Science. 2003;302:1408–1412. doi: 10.1126/science.1089287. [DOI] [PubMed] [Google Scholar]
- 143.Harmar AJ, Marston HM, Shen S, Spratt C, West KM, Sheward WJ, Morrison CF, Dorin JR, Piggins HD, Reubi JC, Kelly JS, Maywood ES, Hastings MH. The VPAC(2) receptor is essential for circadian function in the mouse suprachiasmatic nuclei. Cell. 2002;109:497–508. doi: 10.1016/s0092-8674(02)00736-5. [DOI] [PubMed] [Google Scholar]
- 144.Aton SJ, Colwell CS, Harmar AJ, Waschek J, Herzog ED. Vasoactive intestinal polypeptide mediates circadian rhythmicity and synchrony in mammalian clock neurons. Nat Neurosci. 2005;8:476–483. doi: 10.1038/nn1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Colwell CS, Michel S, Itri J, Rodriguez W, Tam J, Lelievre V, Hu Z, Liu X, Waschek JA. Disrupted circadian rhythms in VIP- and PHI-deficient mice. Am J Physiol Regul Integr Comp Physiol. 2003;285:R939–949. doi: 10.1152/ajpregu.00200.2003. [DOI] [PubMed] [Google Scholar]
- 146.Piggins HD, Antle MC, Rusak B. Neuropeptides phase shift the mammalian circadian pacemaker. J Neurosci. 1995;15:5612–5622. doi: 10.1523/JNEUROSCI.15-08-05612.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.An S, Irwin RP, Allen CN, Tsai C, Herzog ED. Vasoactive intestinal polypeptide requires parallel changes in adenylate cyclase and phospholipase C to entrain circadian rhythms to a predictable phase. J Neurophysiol. 2011;105:2289–2296. doi: 10.1152/jn.00966.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Atkinson SE, Maywood ES, Chesham JE, Wozny C, Colwell CS, Hastings MH, Williams SR. Cyclic AMP signaling control of action potential firing rate and molecular circadian pacemaking in the suprachiasmatic nucleus. J Biol Rhythms. 2011;26:210–220. doi: 10.1177/0748730411402810. [DOI] [PubMed] [Google Scholar]
- 149.O’Neill JS, Maywood ES, Chesham JE, Takahashi JS, Hastings MH. cAMP-dependent signaling as a core component of the mammalian circadian pacemaker. Science. 2008;320:949–953. doi: 10.1126/science.1152506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Low-Zeddies SS, Takahashi JS. Chimera analysis of the Clock mutation in mice shows that complex cellular integration determines circadian behavior. Cell. 2001;105:25–42. doi: 10.1016/s0092-8674(01)00294-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ko CH, Yamada YR, Welsh DK, Buhr ED, Liu AC, Zhang EE, Ralph MR, Kay SA, Forger DB, Takahashi JS. Emergence of noise-induced oscillations in the central circadian pacemaker. PLoS Biol. 2010;8:e1000513. doi: 10.1371/journal.pbio.1000513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Okamura H, Bérod A, Julien JF, Geffard M, Kitahama K, Mallet J, Bobillier P. Demonstration of GABAergic cell bodies in the suprachiasmatic nucleus: in situ hybridization of glutamic acid decarboxylase (GAD) mRNA and immunocytochemistry of GAD and GABA. Neurosci Lett. 1989;102:131–136. doi: 10.1016/0304-3940(89)90067-0. [DOI] [PubMed] [Google Scholar]
- 153.Liu C, Reppert SM. GABA synchronizes clock cells within the suprachiasmatic circadian clock. Neuron. 2000;25:123–128. doi: 10.1016/s0896-6273(00)80876-4. [DOI] [PubMed] [Google Scholar]
- 154.Albus H, Vansteensel MJ, Michel S, Block GD, Meijer JH. A GABAergic mechanism is necessary for coupling dissociable ventral and dorsal regional oscillators within the circadian clock. Curr Biol. 2005;15:886–893. doi: 10.1016/j.cub.2005.03.051. [DOI] [PubMed] [Google Scholar]
- 155.Aton SJ, Huettner JE, Straume M, Herzog ED. GABA and Gi/o differentially control circadian rhythms and synchrony in clock neurons. Proc Natl Acad Sci U S A. 2006;103:19188–19193. doi: 10.1073/pnas.0607466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Silver R, LeSauter J, Tresco PA, Lehman MN. A diffusible coupling signal from the transplanted suprachiasmatic nucleus controlling circadian locomotor rhythms. Nature. 1996;382:810–813. doi: 10.1038/382810a0. [DOI] [PubMed] [Google Scholar]
- 157.Kramer A, Yang FC, Snodgrass P, Li X, Scammell TE, Davis FC, Weitz CJ. Regulation of daily locomotor activity and sleep by hypothalamic EGF receptor signaling. Science. 2001;294:2511–2515. doi: 10.1126/science.1067716. [DOI] [PubMed] [Google Scholar]
- 158.Kraves S, Weitz CJ. A role for cardiotrophin-like cytokine in the circadian control of mammalian locomotor activity. Nat Neurosci. 2006;9:212–219. doi: 10.1038/nn1633. [DOI] [PubMed] [Google Scholar]
- 159.Cheng MY, Bullock CM, Li C, Lee AG, Bermak JC, Belluzzi J, Weaver DR, Leslie FM, Zhou QY. Prokineticin 2 transmits the behavioural circadian rhythm of the suprachiasmatic nucleus. Nature. 2002;417:405–410. doi: 10.1038/417405a. [DOI] [PubMed] [Google Scholar]
- 160.Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, Kito R, Nakao K, Kishimoto W, Yoo SH, Shimomura K, Takao T, Takano A, Kojima T, Nagai K, Sakaki Y, Takahashi JS, Ueda HR. CKIepsilon/delta-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci U S A. 2009;106:15744–15749. doi: 10.1073/pnas.0908733106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Dibner C, Sage D, Unser M, Bauer C, d’Eysmond T, Naef F, Schibler U. Circadian gene expression is resilient to large fluctuations in overall transcription rates. EMBO J. 2009;28:123–134. doi: 10.1038/emboj.2008.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Liu Y, Garceau NY, Loros JJ, Dunlap JC. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell. 1997;89:477–486. doi: 10.1016/s0092-8674(00)80228-7. [DOI] [PubMed] [Google Scholar]
- 163.Liu Y, Merrow M, Loros JJ, Dunlap JC. How temperature changes reset a circadian oscillator. Science. 1998;281:825–829. doi: 10.1126/science.281.5378.825. [DOI] [PubMed] [Google Scholar]
- 164.Diernfellner AC, Schafmeier T, Merrow MW, Brunner M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev. 2005;19:1968–1973. doi: 10.1101/gad.345905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Mehra A, Shi M, Baker C, Colot H, Loros J, Dunlap J. A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell. 2009;137:749–760. doi: 10.1016/j.cell.2009.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Boothroyd CE, Wijnen H, Naef F, Saez L, Young MW. Integration of light and temperature in the regulation of circadian gene expression in Drosophila. PLoS Genet. 2007;3:e54. doi: 10.1371/journal.pgen.0030054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Vanin S, Bhutani S, Montelli S, Menegazzi P, Green EW, Pegoraro M, Sandrelli F, Costa R, Kyriacou CP. Unexpected features of Drosophila circadian behavioural rhythms under natural conditions. Nature. 2012;484:371–375. doi: 10.1038/nature10991. [DOI] [PubMed] [Google Scholar]
- 168.Rensing L, Ruoff P. Temperature effect on entrainment, phase shifting, and amplitude of circadian clocks and its molecular bases. Chronobiol Int. 2002;19:807–864. doi: 10.1081/cbi-120014569. [DOI] [PubMed] [Google Scholar]
- 169.Refinetti R, Menaker M. The circadian rhythm of body temperature. Physiol Behav. 1992;51:613–637. doi: 10.1016/0031-9384(92)90188-8. [DOI] [PubMed] [Google Scholar]
- 170.Grahn DA, Miller JD, Houng VS, Heller HC. Persistence of circadian rhythmicity in hibernating ground squirrels. Am J Physiol. 1994;266:R1251–1258. doi: 10.1152/ajpregu.1994.266.4.R1251. [DOI] [PubMed] [Google Scholar]
- 171.Barrett R, Takahashi J. Temperature compensation and temperature entrainment of the chick pineal cell circadian clock. J Neurosci. 1995;15:5681–5692. doi: 10.1523/JNEUROSCI.15-08-05681.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Morimoto R. Regulation of the heat shock transcriptional response: cross talk between a family of heat shock factors, molecular chaperones, and negative regulators. Genes Dev. 1998;12:3788–3796. doi: 10.1101/gad.12.24.3788. [DOI] [PubMed] [Google Scholar]
- 173.Sarge KD, Murphy SP, Morimoto RI. Activation of heat shock gene transcription by heat shock factor 1 involves oligomerization, acquisition of DNA-binding activity, and nuclear localization and can occur in the absence of stress. Mol Cell Biol. 1993;13:1392–1407. doi: 10.1128/mcb.13.3.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Reinke H, Saini C, Fleury-Olela F, Dibner C, Benjamin IJ, Schibler U. Differential display of DNA-binding proteins reveals heat-shock factor 1 as a circadian transcription factor. Genes Dev. 2008;22:331–345. doi: 10.1101/gad.453808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Ding JM, Chen D, Weber ET, Faiman LE, Rea MA, Gillette MU. Resetting the biological clock: mediation of nocturnal circadian shifts by glutamate and NO. Science. 1994;266:1713–1717. doi: 10.1126/science.7527589. [DOI] [PubMed] [Google Scholar]