Abstract
A screening for dye-decolorizing alkali-thermophilic microorganisms resulted in a Bacillus sp. strain isolated out of the wastewater drain of a textile finishing company. An NADH-dependent azoreductase of this strain, Bacillus sp. strain SF, was found to be responsible for the decolorization of azo dyes. This enzyme was purified by a combination of ammonium sulfate precipitation and anion-exchange and affinity chromatography and had a molecular mass of 61.6 kDa and an isoelectric point at pH 5.3. The pH optimum of the azoreductase depended on the substrate and was within the range of pHs 8 to 9, while the temperature maximum was reached at 80°C. Decolorization only took place in the absence of oxygen and was enhanced by FAD, which was not consumed during the reaction. A 26% similarity of this azoreductase to chaperonin Cpn60 from a Bacillus sp. was found by peptide mass mapping experiments. Substrate specificities of the azoreductase were studied by using synthesized model substrates based on di-sodium-(R)-benzyl-azo-2,7-dihydroxy-3,6-disulfonyl-naphthaline. Those dyes with NO2 substituents, especially in the ortho position, were degraded fastest, while analogues with a methyl substitution showed the lowest degradation rates.
In the last few years, environmental legislation, e.g., about the appearance of color in discharges, combined with the increasing cost of water for the industrial sector, has made the treatment and reuse of dyeing effluents increasingly attractive to the industry. The common method for the treatment of wastewater in the textile finishing industry is physicochemical flocculation in combination with biological treatment (17). The conventional treatment of colored effluents produces a lot of sludge, but does not remove all dyes, thus preventing recycling of the treated wastewater. In activated sludge treatments, dyeing effluents, e.g., reactive azo dyes and naphthaline-sulfonic acids as well as aromatic amino derivatives, represent an extensive nonbiodegradable class of compounds (18) and can even inhibit activated sludge organisms. Thus, the development of more effective biological systems for the treatment of these types of wastewater has resulted in considerable interest by the industry.
Wood-rotting fungi (e.g., Phanerochaete chrysosporium, Trametes sp., and Aspergillus sp.) have been found to effectively degrade a variety of azo dyes under aerobic conditions (16). Dye-degrading fungi have been used frequently in bioreactors for the decolorization and degradation of azo dyes (37). While fungal treatment of effluents is usually very time-consuming (3), immobilized enzymes could have potential for dye decolorization and recycling of effluents (1) without the need for the addition of growth substrates. The enzymes involved in the degradation of azo dyes are mainly peroxidases (12) and laccases (1). However, a significant cost reduction for these enzymes would be required in order to make this process economically more attractive.
Aerobic bacteria have been described to oxidatively decolorize many dyes from several classes, among which azo dyes always turned out to be the most recalcitrant compounds (16). In contrast, under anaerobic conditions, the decolorization of many azo dyes via reduction of the azo bond has been shown for anaerobic (e.g., Bacteroides sp., Eubacterium sp., and Clostridium sp.) as well as facultative anaerobic (e.g., Proteus vulgaris and Streptococcus faecalis) bacteria (5, 11, 30, 36). The main interest in this field has been focused on bacteria from the human intestine that are involved in the metabolism of azo dyes ingested as food additives (9). The fecal enzyme activity of azoreductase is commonly considered a marker for procarcinogenic activity (14). The nonspecificity of the azoreductase reaction is also demonstrated by many reports on the decolorization of azo dyes by sewage sludge under anaerobic conditions (6, 28). It seems that almost all azo compounds tested are biologically reduced under anaerobic conditions, although there are some indications that metal-ion-containing dyes sometimes have reduced decolorization rates (8).
Earlier studies with facultative anaerobic bacteria repeatedly suggested that reduced flavins generated by cytosolic flavine-dependent reductases (flavine reductases) were responsible for the unspecific reduction of azo dyes (31, 35). The ability of cytosolic flavine reductases to act in vitro as azoreductases was recently demonstrated by experiments using a recombinant flavine reductase in different genetic backgrounds (32). Cell extracts generally showed much higher rates of anaerobic reduction of azo dyes than did preparations of resting cells (23, 36). This has generally been explained by the low permeability of the cell membranes for the highly polar sulfonated azo compounds (32).
A different model for the nonspecific reduction of azo dyes by bacteria which does not require transport of the azo dyes or reduced flavins through the cell membrane was recently suggested for Sphingomonas xenophaga BN6. It was proposed that in this system quinines act as redox mediators which are reduced by a quinone reductase located in the cell membrane of Sphingomonas xenophaga BN6 and that the hydroquinones formed reduce the azo dyes in the culture supernatants in a purely chemical redox reaction (19). Additionally, some publications exist which describe the extracellular reduction of azo dyes by anaerobic bacteria (30), although it remains unclear how an extracellular azoreductase would have access to NADH, which is necessary for the reduction of azo dyes.
The reduction of azo compounds has also been shown to occur under aerobic conditions. Degradation pathways of 4-carboxy-4′-sulfoazobenzene, with reductive cleavage of the azo double bond as the initial degradation step, have been established (4). The aerobic azoreductases from the carboxy-orange-degrading Pseudomonas strains K22 and KF46 are monomeric flavine-free enzymes that use NADPH and NADH as cofactors and reductively cleave several sulfonated azo dyes (38, 39). The enzymes (21 and 30 kDa) show different substrate specificities, while both require the presence of hydroxyl groups in the aromatic ring of the substrate.
Although several microorganisms seem to have potential for azo dye degradation, very few strains can withstand the conditions of dying effluents in terms of pH and temperature. In this study, we describe an azoreductase from a newly isolated alkali-thermophilic Bacillus sp. growing at pH 9.5 and 65°C.
MATERIALS AND METHODS
Screening of dye-decolorizing microorganisms.
Three Bacillus sp. isolates (Bacillus sp. strain SF, Bacillus sp. strain LF, and Bacillus pallidus), previously isolated from a wastewater drain of a textile finishing company (27), were incubated on petri dishes for 1 week at 50°C with the following medium (sterile): 250 ml of tap water, 9.25 g of standard I nutrient agar (Merck KGaA), 1.05 g of NaHCO3, and 10 mg of dye. The pH was controlled to 7.0 and 9.5, and growth and decolorization were monitored visually. Further experiments were carried out in 300-ml baffled Erlenmeyer flasks on a rotary shaker at 65°C and 160 rpm. The cultivation medium consisted of the following (in grams per liter): yeast extract (Merck), 3.0; peptone from casein (Merck), 3.0 NaHCO3, 4.2; KH2PO4, 1.0; and 1 ml of trace element solution SL-6 (29) per liter. The pH was adjusted to 9.0 and the dye concentration was 0.1 g l−1. Flasks containing 90 ml of medium were inoculated with 10 ml of microorganism suspension from precultures, and dyes were added from stock solutions. The dyes Mordant Black 9 and Mordant Black 96 were from Dyestar (Frankfurt, Germany), Acid Blue 225 and Disperse Red 86 were from Ciba (Basel, Switzerland), and all other dyes were from Sigma. All dye names are given according to the color index (http://www.colour-index.org) when structures are available. Growth was monitored under a microscope, and dye degradation was measured by spectrophotometric determination of the absorbance at the wavelength maximum for each dye. Blanks were run by adding 100 mg of thiomersal [sodium ethyl-(2-mercaptobenzoato-(2)-O,S)-mercurate] per liter to prevent growth, taking into consideration the possible adsorption of the dyestuff on biomass.
Cultivation of Bacillus sp. strain SF.
Bacillus strain SF was grown in 300-ml baffled Erlenmeyer flasks on a rotary shaker at 60°C and 160 rpm in a medium consisting of the following components (in grams per liter): KH2PO4, 3.5; Na2HPO4 · 7H2O, 7.5; yeast extract (Merck), 10.0; peptone from casein (Merck), 20.0; NH4SO4, 2.5; MgSO4 · 7H2O, 4.5; MnSO4 · H2O, 0.2; iron citrate · 5H2O, 0.7; and 2.5% (vol/vol) SL-6 trace element solution, as described previously (29). SL-6 trace element solution contained the following (in milligrams per liter): ZnSO4 · 7H2O, 100.0; MnCl2 · 4H2O, 30.0; H3BO3, 300.0; CuCl2 · 2H2O, 10.0; NiCl2 · 6.0H2O, 20.0; Na2MoO4 · 2H2O, 30.0; and CoCl2 · 6H2O, 200.0. To avoid precipitation of salts during sterilization in an autoclave, MnSO4 · H2O, MgSO4 · 7H2O, and iron citrate · 5H2O were autoclaved separately, and the two solutions were combined sterilely after they were cooled in a laminar flow hood.
Ten-liter fermentations were carried out in a Chemap-CF 2000 fermentor, using the cultivation medium described above. The fermentor was equipped with pH and oxygen controls. Precultures were grown in baffled Erlenmeyer flasks as described above.
Downstream processing.
Cells were harvested at the end of the exponential phase of growth and centrifuged for 15 min at 3,000 × g, and the pellet was suspended in an equal volume of 50 mM NaH2PO4 buffer (pH 7.0). Cell disruption was carried out by using a sonification unit (Sonoplus HD 70; Bandelin, Berlin, Germany), with monitoring of progress under the microscope. The cell debris was removed by centrifugation for 20 min at 23,700 relative centrifugal force, and the remaining cell lysate was stored at 4°C. During fermentation of Bacillus strain SF, samples of 50 ml were taken every 30 min and were treated similarly.
Assay for azoreductase activity.
Assays were carried out in cuvettes (path length = 1 cm) with a total volume of 1 ml. All substances were dissolved in 50 mM phosphate buffer (pH 7.0) at 50°C and degassed with N2 for 5 min. Four hundred microliters of phosphate buffer was mixed with 200 μl of sample and 200 μl of Reactive Black 5 (100 mg liter−1), resulting in a final concentration of 16.3 μM dye. The reaction was started by the addition of 200 μl of NADH (7.09 mg ml−1; final concentration, 2 mM) and was monitored photometrically at 597 nm. The slope of the initial linear decrease of absorption (ΔA min−1) was used to calculate the azoreductase activity based on the molar absorption coefficient of Reactive Black 5 (ɛ = 35.5 mmol−1 cm−1). One nanokatal was defined as the reduction of 1 nmol of dye per s.
The same assay was used to determine the effects of inhibitors which were added to the phosphate buffer to give the concentrations indicated below. Dye decolorization in the presence of FAD was measured similarly by the addition of a 1.0 mM FAD solution to 50 mM phosphate buffer (pH 7.0) to give a final concentration of 0.2 mM FAD in the assay.
The degradation of Mordant Black 9 and model dyes for the determination of degradation products and substrate specificities, respectively, was carried out as described above, with 20 μM dye and 1 mM NADH.
pH and temperature optima and stability of the azoreductase.
For determination of the pH dependence of azoreductase, the buffer used for the assay was set to pH values of 5.0 to 8.0. For pH values 8.5 and 9.0, a 50 mM Na2CO3-NaHCO3 buffer system was used. NADH solution and dyestuff solution were prepared with the corresponding buffers. For the determination of the temperature optimum, all solutions were brought to the corresponding temperature before mixing and the spectrophotometer was temperature controlled during measurement.
Enzyme stability was tested at three different pH values and three different temperatures. For this test, 1.5 ml of enzyme solution was mixed with 4.5 ml of buffer and incubated in a thermo-controlled water bath. Samples were taken periodically, immediately frozen at −20°C, and subsequently analyzed. Buffers used for incubation were as follows: at pH 7.0, Na2HPO4-KH2PO4; at pH 8.5, NaBO3-NaOH-HCl; at pH 10.0, Na2CO3-NaHCO3.
Oxygen measurement.
For determination of the oxygen dependence of the azoreductase, a thermo-constant mini oxygen gauge (Rank Brothers Ltd.) was used. The volume of the reaction vessel was 1.5 ml and the volumes for the azoreductase assay were adjusted accordingly. The device was calibrated with a saturated Na2SO3 solution (0% O2 saturation) and by 10 min of aeration (100% O2 saturation). All reactions were carried out in 50 mM phosphate buffer at pH 7.0 at 50°C, with 2.0 mM NADH, 0.2 mM Reactive Black 5, and 0.2 mM FAD.
Protein purification.
The cell lysate of Bacillus strain SF was subjected to fractionated ammonium sulfate precipitation at 40% saturation to remove impurities, followed by 70% saturation in a second step to precipitate the azoreductase. This was carried out by the addition of 72.6 g of (NH4)2SO4 to 300 ml of cell lysate. The precipitated proteins after the second precipitation step were collected by centrifugation, and the pellet was dissolved in 300 ml of phosphate buffer (50 mM, pH 7.0). The solution was desalted by dialysis against phosphate buffer (50 mM, pH 7.0) overnight, until a conductivity of <10 mS cm−1 was measured (WTW microprocessor conductivity meter LF 96). Thereafter, the solution was concentrated by ultrafiltration in an Amicon stirred cell (model 8400; pressurized with N2 at 2 × 105 N/m2) equipped with a cellulose-polysulfonate membrane with a 10-kDa cutoff.
For anion-exchange fractionation, 9.5-ml aliquots of the resulting solution were applied to a DEAE-cellulose DE 52 column installed in an ΔKTA fast-performance liquid chromatography system (Amersham Pharmacia Biotech). Proteins were eluted at 6 ml min−1 with the sample buffer and a linear gradient from 0 to 1 M NaCl. Azoreductase active fractions were pooled (approximately 50 ml) and desalted for affinity chromatography.
Two-milliliter aliquots of the sample were fractionated by affinity chromatography through a HiTrap Blue affinity column from Amersham Pharmacia Biotech. The column contained Cibacron Blue F3G-A immobilized on 6% highly cross-linked spherical agarose (Sepharose High Performance) by the triazine method, binding specifically, e.g., enzymes requiring adenylyl-containing cofactors (including NADH and NADPH), albumin, coagulation factors, and interferon. Proteins were eluted at 1 ml min−1 with the sample buffer and a linear gradient from 0 to 1 M NaCl. Fractions with azoreductase activity were pooled and concentrated with Centricon 10 concentrators equipped with cellulose-polysulfonate membranes with a 10-kDa cutoff.
The protein content of samples was measured by the Bradford method. Samples (160 μl) were pipetted into separate microtiter plate wells, and 40 μl of the reagent (obtained from Bio-Rad) per sample was added and mixed thoroughly. After 5 to 10 min of incubation at room temperature, the absorbances were measured. Bovine serum albumin was used for calibration.
Gel electrophoresis.
Polyacrylamide gel electrophoresis (PAGE) was carried out as described previously (20), with some modifications for native PAGE, i.e., both the detergent component sodium dodecyl sulfate (SDS) and 2-mercaptoethanol (β-mercaptoethanol) were omitted. Twelve percent polyacrylamide gels were cast, and broad-range molecular weight markers from Pharmacia were used as standards. For activity staining, carboxymethylcellulose (CMC) was mixed into the separating gel at a concentration of 0.2% (wt/vol) to enable the dye Reactive Black 5 to bind to the gel. For both SDS-PAGE and native PAGE, the Coomassie blue R-250 staining method was used. If visualization was not satisfactory, the silver staining procedure was subsequently applied.
Staining of CMC native PAGE gels was done by the following procedure. CMC native gels were removed from the electrophoresis chamber and glass plates. The gel was immediately immersed in approximately 50 ml of 50 μM Reactive Black 5 solution for 25 min. After removal of the staining solution, the gel was put into a desiccator and a vacuum was applied. Subsequently, the desiccator was scavenged with N2, and a degassed NADH solution (2 mM NADH in 50 mM phosphate buffer, pH 7.0) was added under the N2 atmosphere. To make sure that no oxygen remained in the test vessel, vacuum generation and N2 scavenging were repeated alternately several times. Bands of the active enzyme appeared after approximately 10 min as white spots within a blue background.
Isoelectric focusing was carried out as described elsewhere (14).
Synthesis of model dyes.
Anilines with substitutions of trifluoromethyl-, methyl-, chloro-, bromo-, and fluoro- groups at the o-, m-, and p- positions and 2,7-dihydroxynaphtaline-3,6-disulfonic acid disodium salt were supplied by Sigma Aldrich. Urea, sodium nitrite, and sodium hydroxide were supplied by Merck, and 32% hydrochloric acid was purchased from Riedel de Häen.
Model azo dyes were synthesized from the corresponding aromatic amines in two steps as described elsewhere (2). Precipitation of the dyes was facilitated by acidification with hydrochloric acid. For purification, the dyes were recrystallized from hot water and freeze-dried.
Determination of dye content.
For confirmation of the purity of dyes, a method for the determination of the dye content was developed based on the reduction of azo bonds by excesses of sodium dithionite. Five milliliters of an oxygen-free solution of sodium dithionite (approximately 30 mM, prepared freshly every day) was mixed with 15 ml of a 100 mM formaldehyde solution to yield a more stable hydroxymethanesulfinate (26) after complete conversion. This solution was titrated with I2-KI titrimetric solution (50 mM), with soluble starch as an indicator, resulting in the consumption of about 6 ml of the solution.
For determination of the normality of the I2-KI solution, 25 g of pure KI was dissolved in about 40 ml of deionized H2O to which was added 12.7 g of I2. The flask was closed and shaken until the iodine was completely dissolved. Only then was the volumetric flask filled up to exactly 1 liter, giving an approximate concentration of 0.5 M. The exact normality was determined by titration with a solution of As2O3 buffered with 2 g of sodium hydrogen-carbonate liter−1 in which 1 ml of the iodine solution was equivalent to 4.946 mg of As2O3. The blue complex of iodine with starch was used to indicate the end point of the titration. During titration, the complete conversion of Na2S2O4 was only found in the presence of an excess of the dye. However, extrapolation of the straight line of absorbance versus concentration of Na2S2O4 to an absorbance of 0.0 (complete reduction of the dyes) gave the exact amount of dyestuff in the sample.
The molar absorption coefficients (ɛ) of the dyes were determined by measuring the absorbance at the wavelength of maximum absorbance (λmax) of each dye. The slope of the referring trend line gives the molar absorption coefficient.
HPLC and MS analysis.
FAD measurements were done based on the method of Wahlund (34) in a Kontron 422S high-performance liquid chromatography (HPLC) apparatus equipped with a Hypersil RP 18 column with methanol-H2O (30:70 [vol/vol]) and 5 mmol of tetrabutylammonium hydrogen sulfate (TBAHS) as the mobile phase at a flow rate of 1 ml min−1 at 40°C. FAD was detected at 260 and 450 nm (λmax of FAD) with a Kontron UV 430 detector.
Dye degradation products were separated in a Dionex HPLC system (Dionex Corporation, Sunnyvale, Calif.) consisting of a P580 pump, an ASI-100 automated sample injector, and a PDA-100 photodiode array detector. Chromatographic separation was performed through a Hypersil ODS 5-μm (250 by 4 mm [inside diameter]) column. The following operating conditions were used for all analyses: injection volume, 20 μl; column temperature, 40°C; mobile phase A, 100% H2O; mobile phase B, 50% methanol-50% H2O; gradient at 0 to 20 min, 100% (A) to 50% (B); gradient at 20 to 25 min, 50% (B); gradient at 25 to 26 min, 50 to 0% (B); gradient at 26 to 35 min, 100% (A); flow rate, 0.9 ml/min; detection wavelength, 530 nm. Mass spectral (MS) analysis was performed in an SL ion trap mass spectrometer (1100 Series LC/MSD Trap; Agilent, Waldbronn, Germany) equipped with an electrospray ionization source and connected to the HPLC system described above. The spectra were recorded in negative ionization mode. Full-scan spectra were collected from m/z 50 to 1,200, with a capillary temperature of 350°C. The nebulizer was set to 70.00 lb/in2, the dry gas was set to 12.00 liter min−1, and the HV capillary was set to 3,000 V. Extracted ion chromatograms were obtained by selecting ions (M-Na) at e.g., m/z 359 for Mordant Black 9.
RESULTS
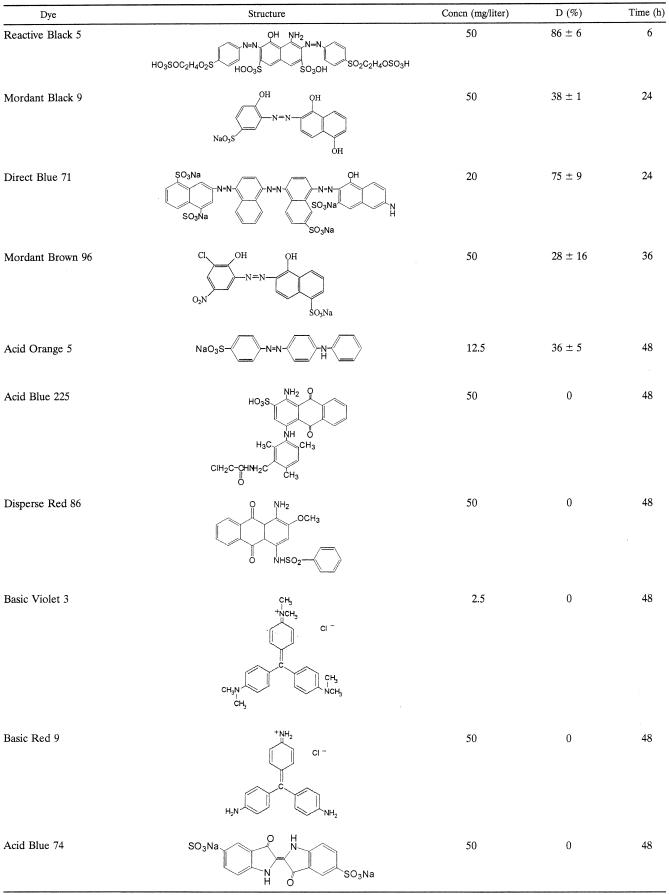
Three Bacillus species (Bacillus sp. strain SF, Bacillus sp. strain LF, and B. pallidus) isolated out of textile wastewater drains were grown on agar plates containing dyes. Of these organisms, only Bacillus strain SF showed clearing zones. Next, the strains were cultivated in liquid cultures at pH 9.5 and 65°C based on a yeast extract-peptone medium with dyes as the only additional carbon source. Bacillus strain SF decolorized a wide range of azo dyes at different rates (Table 1), while the other strains did not attack these dyes. The triarylmethane dyes Basic Violet 3 and Basic Red 9, Acid Blue 74 (indigo carmine), and the anthraquinone dyes Acid Blue 225 and Disperse Red 86 were not decolorized at all by any of the Bacillus sp. The same substrate specificity was observed when a cell lysate of Bacillus strain SF was incubated with dyes in the presence of NADH, while the culture filtrate and a previously purified catalase-peroxidase (13) did not show any decolorization activity (data not shown). Furthermore, decolorization rates with the cell lysate of Bacillus strain SF depended on the concentration of NADH, indicating the presence of azoreductase (EC 1.7.1.6 [azobenzene reductase]) activity.
TABLE 1.
Decolorization (D) of various textile dyes by Bacillus strain SF
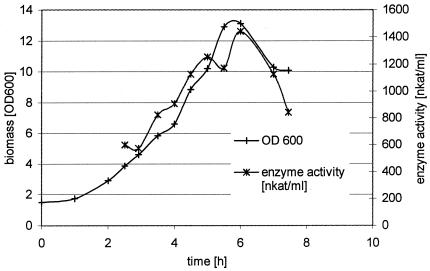
Bacillus strain SF was cultivated in a 10-liter bioreactor with a yeast extract-peptone medium, and a specific growth rate (μ) of 0.41 h−1 was determined. The production of azoreductase was coupled to growth, and both parameters reached a maximum at around 6 h (Fig. 1). For subsequent fermentations, cells were harvested after this phase and downstream processing yielded about 300 ml of cell lysate.
FIG. 1.
Fermentation of Bacillus strain SF and production of azoreductase.
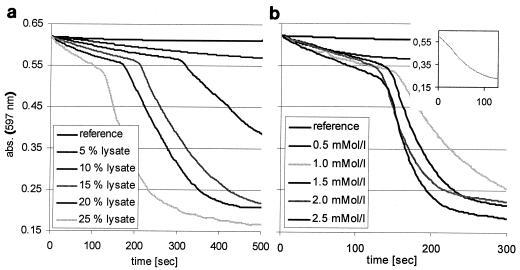
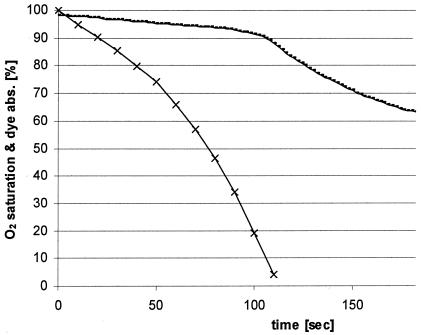
For all tested dyes, decolorization by the cell lysate of Bacillus strain SF in the presence of NADH started with a slow linear fading of color followed by rapid decolorization after about 150 s (Fig. 2a). There was a linear dependence of the azoreductase activity and NADH concentration up to about 1.5 mM (Fig. 2b). Above this concentration, further addition of NADH did not increase the reaction velocity. At a constant concentration of NADH, the duration of the delay of decolorization depended on the amount of cell lysate used (Fig. 2a). These findings led to the assumption that a certain amount of NADH was consumed by another reaction and only thereafter was NADH available for dye decolorization. This hypothesis was confirmed by the fact that rapid decolorization started immediately when the cell lysate was preincubated with NADH for 5 min before the addition of the dye (Fig. 2, inset). Obviously, the second reaction consuming NADH was dependent on the presence of oxygen, since the delay was not observed when all solutions were degassed with N2. This fact was confirmed by monitoring of the concentration of oxygen during decolorization of Reactive Black 5 (Fig. 3).
FIG. 2.
Decolorization of Reactive Black 5 with Bacillus strain SF cell lysate. (a) Different amounts of cell lysate and 2 mM NADH. (b) Different concentrations of NADH at an azoreductase activity of 6.8 × 102 nkat ml−1 (16.3 μM Reactive Black 5, 50 mM phosphate buffer, pH 7.0; 50°C). Inset, preincubation with 2 mM NADH.
FIG. 3.
Oxygen dependence (×) of dye decolorization (-) by the azoreductase of Bacillus strain SF (2.0 mM NADH, 0.2 mM FAD, azoreductase activity of 6.1 × 102 nkat ml−1, 0.2 mM Reactive Black 5, 50 mM phosphate buffer, pH 7.0).
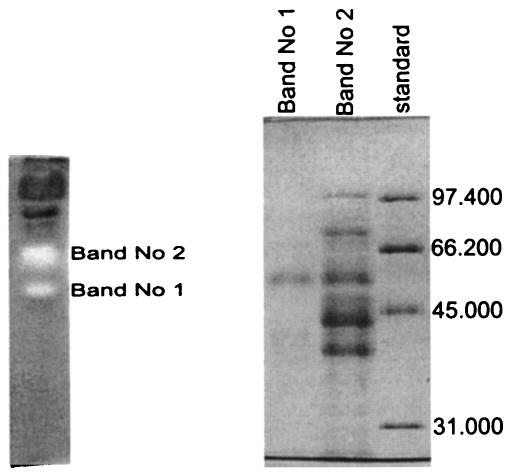
Based on these results, an assay was developed for the monitoring of azoreductase activity during enzyme purification. Samples were diluted to achieve a decolorization rate between 0.1 and 0.3 ΔA min−1, a range for which a linear dependence between cell lysate concentrations was found. In addition, a new method for the detection of azoreductase in polyacrylamide gels was developed. This assay was based on the decolorization of the dye Reactive Black 5, which was incorporated into the gels bound to CMC as a carrier. The most important step in the development of this method was to ensure an oxygen-free atmosphere. With this method, two white bands were detected on the gel for the cell lysate, indicating the presence of azoreductase activity. Bands 1 and 2 were cut out and the eluted samples were reapplied to an SDS gel (Fig. 4), resulting in a single band for band 1, of about 62 kDa, and a pattern of at least six bands for band 2, including a band of 62 kDa. This 62-kDa protein with azoreductase activity was chosen for purification.
FIG. 4.
Native gel with newly developed staining procedure for azoreductase of Bacillus strain SF (left). The azoreductase from the native gel was resolved by SDS-12.5% PAGE (right); numbers indicate molecular weights.
In the first step, the azoreductase was subjected to fractionated ammonium sulfate precipitation at 40 and 60% saturation. The sample resulting from precipitation was desalted by repeated ulfrafiltration and was fractionated by anion-exchange chromatography with a DEAE column. A purification factor of 20.2 was achieved with this purification step (Table 2), and the azoreductase was the most prominent band on SDS gels. Since size exclusion chromatography did not give satisfactory results for further purification of the enzyme, affinity chromatography was used. A column with Cibacron Blue F3G-A immobilized on spherical agarose with an affinity to, e.g., enzymes requiring adenylyl-containing cofactors (including NADH and NADPH), was used for the final purification step of the azoreductase. In contrast to many reports for which NADH was used for elution of the bound proteins, our best results were achieved with 1 M NaCl used for elution of the azoreductase.
TABLE 2.
Purification of azoreductase from Bacillus strain SF
| Purification step | Amt of protein (mg) | Activity (nkat) | Sp act (nkat/mg) | Purification factor (fold) | Yield (%) |
|---|---|---|---|---|---|
| Extract | 493.1 | 9,842 | 20 | 1.0 | 100.0 |
| (NH4)2SO4 precipitation | 193.2 | 8,104 | 42 | 2.1 | 82.3 |
| Anion-exchange chroma- tography | 2.1 | 1,767 | 850 | 42.6 | 18.0 |
| Affinity chromatography | 0.04 | 198 | 4,972 | 249.1 | 2.0 |
Characterization of the azoreductase of Bacillus strain SF.
The resulting pure protein had a molecular mass of 61.6 ± 1.4 kDa, as calculated from SDS gels (Fig. 4). The isoelectric point of the azoreductase was measured to be at pH 5.3. The pH optimum of the azoreductase was determined with four different azo dyes by measuring the activity within the range of pH values 5.0 to 9.0. An optimum at pH 7.0 was found for Mordant Black 9, Mordant Brown 96, and Reactive Black 5, while for Direct Blue 71 the maximum activity was measured at pH 8.0. The azoreductase activity increased almost linearly in the temperature range from 20 to 45°C. Thereafter, the activity slightly increased, reaching its optimum at 80°C. The enzyme was quite stable at pH 7 and 50°C, with a half-life of 1 day, while the stability decreased significantly at 60°C to a half-life of 3 h. However, at this temperature and pH 10, the half-life was still 1 h, while it was 7.6 h at this pH and 30°C.
Fifty percent inhibition of the azoreductase was detected with 0.08% SDS and 2.2 μM KCN. Inhibitors of the electron transport chain in cells, including rotenone, antimycin A, and NaN3, did not show any effect up to concentrations of 500 mg liter−1. Preincubation of the reaction mixture with CO did not have any effect on the decolorization rate either.
Substrate specificity of the azoreductase from Bacillus strain SF.
A linear correlation between azoreductase activity and the concentration of Reactive Black 5 up to 13.5 μM was observed when NADH was provided in excess (k = 2.56 × 10−2 s−1). Under aerobic conditions, azoreductase activity increased >10-fold, from 5.8 × 102 to 6.7 × 103 nkat ml−1, upon addition of 0.25 mM FAD, while only a small increase, to 8.4 × 103 nkat ml−1, was measured when the concentration of FAD was raised to 0.5 mM. By using HPLC, we determined the concentration of FAD in the assay to be 0.29 ± 0.02 mM and 0.31 ± 0.01 mM before and after the reaction, respectively, indicating that FAD was not consumed during the reaction with NADH, but acted as a mediator.
While samples were in the presence of oxygen, no decolorization was seen for samples containing NADH, FAD, and various dyes, but the same samples were decolorized in the absence of oxygen at similar rates by high concentrations of FAD or by the azoreductase, except for Direct Blue 71, which was degraded faster by FAD (Table 3).
TABLE 3.
Comparison of decolorization by azoreductase from Bacillus strain SF and nonenzymatic decolorization by FAD in the absence of oxygen
| Dye | Decolorization (ΔA/min)
|
|
|---|---|---|
| FAD | Azoreductase | |
| Mordant Black 9 | 0.117 | 0.116 |
| Direct Blue 71 | 0.125 | 0.025 |
| Reactive Black 5 | 0.123 | 0.116 |
| Mordant Brown 96 | 0.013 | 0.011 |
The degradation of the mono-azo dye Mordant Black 9 by the azoreductase from Bacillus strain SF was monitored both spectrophotometrically and by HPLC-MS. After complete decolorization of the dye, the corresponding amines obviously formed degradation products. After separation of the two products and the cofactor by HPLC, two peaks were identified by coupled MS as 3-amino-4-hydroxybenzenesulfonicacid ([M-2H]−, 187.6 m/z) and NADH ([M-2Na-H]−, 662.0 m/z), while 1-amino-3,7-dihydroxynaphthalene was not detectable by this MS method.
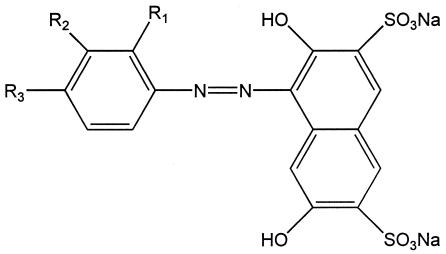
Various model dyes with different substituents (R1-, R2-, and R3-) in the ortho, meta, and para positions of the benzyl ring were synthesized (Fig. 5). The o-fluorinated isomer was missing from the series since it was not stable in an aqueous solution. After dissolving of the isomer, the reddish-brown solution turned black within a few minutes and a precipitate was formed. After preparation of the dyes, the molar extinction coefficients were determined (Table 4). Dyes with NO2 substituents were clearly degraded fastest. As for many other compounds, dyes with a substituent in the ortho position were degraded faster than those with a substituent in the para position (Table 4). In comparison to a dye without any substitutions, the methylated dyes showed the same or, in the case of the m-methyl dye, lower degradation rates.
FIG. 5.
Basic structure of synthesized model dye di-sodium-(R)-benzyl-azo-2,7-dihydroxy-3,6-disulfonyl-naphthaline.
TABLE 4.
Molecular weights, absorbance maximums, molar extinction coefficients, and reaction rates (expressed as activities) with azoreductase of synthesized model dyes
| Substituent | Molecular weight (g mol−1) | λmax (nm) | ɛ (cm−1 mmol−1) | Activity (nkat ml−1) |
|---|---|---|---|---|
| H- | 436.4 | 482 | 35.1 | 17.0 |
| o-methyl | 450.4 | 490 | 30.2 | 21.4 |
| m-methyl | 450.4 | 485 | 33.0 | 5.8 |
| p-methyl | 450.4 | 490 | 28.4 | 13.0 |
| o-bromine | 515.3 | 483 | 27.0 | 106.9 |
| m-bromine | 515.3 | 480 | 30.4 | 47.3 |
| p-bromine | 515.3 | 488 | 54.2 | 15.0 |
| m-fluorine | 454.4 | 479 | 21.2 | 61.6 |
| p-fluorine | 454.4 | 482 | 29.9 | 100.4 |
| o-CF3 | 504.4 | 473 | 23.4 | 150.2 |
| m-CF3 | 504.4 | 474 | 25.5 | 73.7 |
| p-CF3 | 504.4 | 474 | 28.6 | 68.6 |
| o-nitro | 481.4 | 486 | 30.8 | 1,134 |
| p-nitro | 481.4 | 485 | 40.3 | 543 |
| o-chlorine | 470.8 | 482 | 30.1 | 127.5 |
DISCUSSION
The newly isolated alkali-thermophilic Bacillus sp. strain SF decolorizes a number of azo dyes. This activity, however, cannot be attributed to the catalase-peroxidase previously isolated from this organism, which did not attack any of the tested dyes (13). An intracellular azoreductase (EC 1.7.1.6 [azobenzene reductase]) is responsible for the decolorization activity, as previously shown for other azo-dye-degrading microorganisms (11, 31, 36). Two bands showing azoreductase activity were found after separation of the cell lysate from Bacillus strain SF in gels combined with activity staining. A new method for activity staining was developed since a previously described procedure by Zimmermann et al. (39) had failed for this organism. CMC was included in the gel as a carrier for dyes, which were decolorized by active protein bands under an oxygen-free atmosphere.
In general, azoreductase activity was only measured in the absence of oxygen. Interestingly, prior to dye decolorization oxygen was consumed in solutions containing NADH, azoreductase, and a dye. Similarly, other authors (10) reported that azoreductase activity was only observed in oxygen-free solutions (7). Chang et al. mentioned a direct influence of oxygen on the degradation rates of azo dyes by Pseudomonas luteola, although they did not measure any influence with the crude cell extract.
The azoreductase from Bacillus strain SF was purified by a combination of ammonium sulfate precipitation, anion-exchange chromatography, and affinity chromatography to an overall purification level of 250-fold. Zimmermann et al. (38) reported an 80-fold purification with a 17% yield for the azoreductase from Pseudomonas sp., indicating a higher enzyme concentration in the cell lysate but a lower specific activity than the Bacillus strain SF azoreductase (50 nkat mg−1 compared to 5 × 103 nkat mg−1). However, a comparison of activity values from the literature is difficult since most studies are based on the decolorization of different dyes.
The application of affinity chromatography for the purification of azoreductase has previously been described (24, 25, 33, 39). Compared to previous reports (25), for which a 55-fold purification was achieved with this technique, only a 5-fold purification was calculated for the azoreductase from Bacillus strain SF, probably due to denaturation and/or irreversible adsorption of the enzyme in the Cibacron Blue F3G-A column. A clear separation of azoreductase was reported for a Red Sepharose CL-6B column (33).
The molecular mass of the azoreductase from Bacillus strain SF was determined to be 61.6 ± 1.4 kDa, and a pI of 5.3 was measured. Previously, molecular masses of 21 and 30 kDa were reported for azoreductases from Pseudomonas sp. (38, 39) and a mass of 28 kDa was reported for Enterobacter agglomerans (24). Temperature optima for azoreductases reported in the literature range from 40 to 45°C (15, 22). In this work, for the first time, an azoreductase from an alkali-thermophilic bacterium was investigated, and this azoreductase from Bacillus strain SF had a temperature optimum at 80°C. Like its catalase-peroxidase (27), the azoreductase of Bacillus strain SF had a pH optimum at pH 7, although the organism had a growth optimum above pH 9. Interestingly, the half-lives of the azoreductase from Bacillus strain SF under different conditions were shorter than those determined for the catalase-peroxidase previously isolated from this organism. The half-life of the catalase-peroxidase at pH 7.0 was >200 h, while the azoreductase showed a half-life of only 24 h.
The azoreductase from Bacillus strain SF was inhibited by CN− and by SDS, as was previously shown for an azoreductase from Bacillus cereus (22). CO did not affect azoreductase activity, which contradicts with previously obtained results (10). Similarly, other typical inhibitors of respiration chain enzymes, such as rotenone, antimycin A, and N3−, did not have any effect on the azoreductase, indicating that this enzyme is not part of the respiration chain.
Several authors reporting on azoreductases claimed that FAD enhances azo bond reduction (10, 21, 22). With the present work, we found that under anaerobic conditions, some reduction of azo dyes takes place in the presence of FAD and NADH without enzyme. This reaction started when dissolved oxygen was completely removed from the solution. Several authors (10, 21, 22) reported that the addition of FAD in catalytic amounts increased azoreductase activity due to its electron-mediating function. Furthermore, it was suggested that the enzyme itself contained a flavine prosthetic group (22), and it was assumed that FAD as a mediator was not degraded and/or metabolized during the reaction (25). By using HPLC analysis, we confirmed that FAD was not consumed during decolorization of dyes by the azoreductase from Bacillus strain SF.
Surprisingly, in the presence of NADH, Direct Blue 71 was decolorized much faster by FAD than by the enzyme alone. Compared to all the other dyes used for this study, this dye was probably too big to enter the active site of the enzyme. This observation is in contrast to previous observations (10) which did not measure any dye decolorization by NADH and FAD. Oxygen probably had not been completely removed from the solution in these previous investigations, which we found to be essential for any reaction to take place.
We observed a linear dependency between the concentration of Reactive Black 5 up to 13.5 μM and the decolorization rate of azoreductase from Bacillus strain SF, while other groups (21, 38) reported substrate saturation kinetics for this concentration range and other azoreductases. The substrate specificity was tested with 15 newly synthesized azo dyes, and dyes with NO2 substituents showed the highest decolorization rates. As for several other substituents, a stronger influence was seen with the substituent in the ortho position than in the para position. In comparison to a dye without substitutions, the methylated dyes showed the same or, in the case of the m-methyl dye, lower decolorization rates. Substitution with electron-withdrawing groups, especially in the ortho position, clearly enhanced decolorization rates. A comparison of the degradabilities of dyes with p-methyl, p-bromine, p-fluorine, and p-CF3 substituents revealed increased rates with stronger electronegative substituents. Electron-withdrawing groups might lead to a decreased electron density at the N=N double bond (especially with ortho and para substituents). Hence, electron transfer to these groups is relieved. In addition, the transfer of electrons to the dye molecule leads to a negatively charged intermediate, which might be stabilized by more electronegative substituents. This hypothesis would also explain the very high reactivities of the dye molecules with nitro substituents. In addition to the −I effect, the NO2 group also causes a very strong +M effect, stabilizing the intermediately occurring negative charge by mesomeric stabilization.
In summary, we have shown that an azoreductase is responsible for azo dye reduction by Bacillus strain SF. Future investigations should focus on the application of this highly alkali-thermostable organism for the treatment of textile dyeing effluents.
Acknowledgments
This study was supported by European Union projects BIOEFFTEX and COST847.
We thank Doris Steifer for her brilliant technical assistance.
REFERENCES
- 1.Abadulla, E., K. H. Robra, G. Gubitz, L. Silva, and A. Cavaco-Paulo. 2000. Enzymatic decolorization of textile dyeing effluents. Textile Res. J. 70:409-414. [Google Scholar]
- 2.Abrahart, E. N. 1977. Dyes and their intermediates, p. 73-87. Edward Arnold Ltd., London, United Kingdom.
- 3.Banat, I. M., P. Nigam, D. Singh, and R. Marchant. 1996. Microbial decolorization of textile dye containing effluents: a review. Biores. Technol. 58:217-227. [Google Scholar]
- 4.Blümel, S., M. Contzen, M. Lutz, A. Stolz, and H. J. Knackmuss. 1998. Isolation of a bacterial strain with the ability to utilize the sulfonated azo compound 4-carboxy-4′-sulfoazobenzene as the sole source of carbon and energy. Appl. Environ. Microbiol. 64:2315-2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bragger, J. L., A. W. Lloyd, S. H. Soozandehfar, S. F. Bloomfield, C. Marriot, and G. P. Martin. 1997. Investigations into the azo reducing activity of a common colonic microorganism. Int. J. Pharm. 157:61-71. [Google Scholar]
- 6.Carliell, C. M., S. J. Barclay, N. Naidoo, C. A. Buckley, D. A. Mulholland, and E. Senior. 1994. Anaerobic decolorisation of reactive dyes in conventional sewage treatment processes. Water San. 20:341-344. [Google Scholar]
- 7.Chang, J. S., C. Chou, Y. C. Lin, P. J. Lin, J. Y. Ho, and T. Lee Hu. 2001. Kinetic characteristics of bacterial azo-dye decolorization by Pseudomonas luteola. Water Res. 35:2841-2850. [DOI] [PubMed] [Google Scholar]
- 8.Chung, K. T., and S. E. Stevens. 1993. Degradation of azo dyes by environmental microorganism and helminths. Environ. Toxicol. Chem. 12:2121-2132. [Google Scholar]
- 9.Chung, K. T., S. E. Stevens, and C. E. Cerniglia. 1992. The reduction of azo dyes by the intestinal microflora. Crit. Rev. Microbiol. 18:175-190. [DOI] [PubMed] [Google Scholar]
- 10.Fujita, S., and J. Peisach. 1982. The stimulation of microsomal azoreduction by flavins. Biochim. Biophys. Acta 719:178-189. [DOI] [PubMed] [Google Scholar]
- 11.Gingell, R., and R. Walker. 1971. Mechanisms of azo reduction by Streptococcus faecalis. II. The role of soluble flavins. Xenobiotica 1:231-239. [DOI] [PubMed] [Google Scholar]
- 12.Goszczynski, S., A. Paszczynski, M. B. Pastigrigsby, R. L. Crawford, and D. L. Crawford. 1994. New pathway for degradation of sulfonated azo dyes by microbial peroxidases of Phanerochaete chrysosporium and Streptomyces chromofuscus. J. Bacteriol. 176:1339-1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gudelj, M., G. Fruhwirth, A. Paar, F. Lottspeich, K. H. Robra, A. Cavaco-Paulo, and G. M. Gübitz. 2001. A catalase-peroxidase from a newly isolated thermoalkalophilic Bacillus sp. with potential for the treatment of textile bleaching effluents. Extremophiles 5:423-429. [DOI] [PubMed] [Google Scholar]
- 14.Haberer, P., M. du Toit, L. M. T. Dicks, F. Ahrens, and W. H. Holzapfel. 2003. Effect of potentially probiotic lactobacilli on faecal enzyme activity in minipigs on a high-fat, high-cholesterol diet-a preliminary in vivo trial. Int. J. Food Microbiol. 87:287-291. [DOI] [PubMed] [Google Scholar]
- 15.Hu, T. L. 2001. Kinetics of azoreductase and assessment of toxicity of metabolic products from azo dyes by Pseudomonas luteola. Water Sci. Technol. 43:261-269. [PubMed] [Google Scholar]
- 16.Kandelbauer, A., and G. M. Gübitz. Bioremediation for the decolorization of textile dyes, a review. In E. Lichtfouse, S. Dudd, and D. Robert (ed.), Environmental chemistry, in press. Springer-Verlag, Heidelberg, Germany.
- 17.Krull, R., M. Hemmi, P. Otto, and D. C. Hempel. 1998. Combined biological and chemical treatment of highly concentrated residual dyehouse liquors. Water Sci. Technol. 38:339-346. [Google Scholar]
- 18.Krull, R., and D. C. Hempel. 1994. Biodegradation of naphthalenesulphonic acid-containing sewages in a two-stage treatment plant. Bioproc. Eng. 10:229-234. [Google Scholar]
- 19.Kudlich, M., A. Keck, J. Klein, and A. Stolz. 1997. Localization of the enzyme system involved in the anaerobic degradation of azo dyes by Sphingomonas sp. BN6 and effect of artificial redox mediators on the rate of azo reduction. Appl. Environ. Microbiol. 63:3691-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 21.Mallett, K., J. King, and R. Walker. 1982. A continuous spectrophotometric determination of hepatic microsomal azo reductase activity and its dependence on cytochrome P-450. Biochem. J. 201:589-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsudomi, N., K. Kobayashi, and S. Akuta. 1977. Purification and some properties of new coccine (NC)-reductase from Bacillus cereus T-105 strain. Agric. Biol. Chem. 41:2323-2329. [Google Scholar]
- 23.Mechsner, K., and K. Wuhrmann. 1982. Cell permeability as a rate factor in the microbial reduction of sulfonated azo dyes. Eur. J. Appl. Microbiol. Biotechnol. 15:123-126. [Google Scholar]
- 24.Moutaouakkil, A., Y. Zeroual, F. Zohra Dzayri, M. Talbi, K. Lee, and M. Blaghen. 2003. Purification and partial characterization of azoreductase from Enterobacter agglomerans. Arch. Biochem. Biophys. 413:139-146. [DOI] [PubMed] [Google Scholar]
- 25.Nakanishi, M., C. Yatome, N. Ishida, and Y. Kitade. 2001. Putative ACP phosphodiesterase gene (acpD) encodes an azoreductase. J. Biol. Chem. 276:46394-46399. [DOI] [PubMed] [Google Scholar]
- 26.Nickless, G. 1968. Inorganic sulfur chemistry, p. 220-227. Elsevier, Amsterdam, The Netherlands.
- 27.Paar, A., S. Costa, T. Tzanov, M. Gudelj, K.-H. Robra, A. Cavaco-Paulo, and G. M. Gübitz. 2001. Thermoalkalistable catalases from newly isolated Bacillus sp. for the treatment and recycling of textile bleaching effluents. J. Biotechnol. 89:147-154. [DOI] [PubMed] [Google Scholar]
- 28.Pagga, U., and D. Brown. 1986. The degradation of dyestuffs. II. Behaviour of dyestuffs in aerobic biodegradation tests. Chemosphere 15:479-491. [Google Scholar]
- 29.Pfennig, N. 1974. Rhodopseudomonas globiformis sp. n., a new species of Rhodospirillaceae. Arch. Microbiol. 100:197-206. [Google Scholar]
- 30.Rafii, F., W. Franklin, and C. E. Cerniglia. 1990. Azoreductase activity of anaerobic bacteria isolated from human intestinal microflora. Appl. Environ. Microbiol. 56:2146-2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roxon, J. J., A. J. Ryan, and S. E. Wright. 1967. Enzymatic reduction of tartrazine by Proteus vulgaris from rats. Food Chem. Toxic. 5:645-656. [DOI] [PubMed] [Google Scholar]
- 32.Russ, R., J. Rau, and A. Stolz. 2000. The function of cytoplasmic flavin reductases in the bacterial reduction of azo dyes. Appl. Environ. Microbiol. 66:1429-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Suzuki, Y., T. Yoda, A. Ruhul, and W. Sugiura. 2001. Molecular cloning and characterization of the gene coding for azoreductase from Bacillus sp. OY1-2 isolated from soil. J. Biol. Chem. 276:9059-9065. [DOI] [PubMed] [Google Scholar]
- 34.Wahlund, K. G. 1975. Reversed-phase ion-pair partition chromatography of carboxylates and sulphonates. J. Chromatogr. 115:411-422. [DOI] [PubMed] [Google Scholar]
- 35.Walker, R. 1970. The metabolism of azo compounds: a review of the literature. Food Cosmet. Toxicol. 8:659-676. [DOI] [PubMed] [Google Scholar]
- 36.Wuhrmann, K., K. Mechsner, and T. Kappeler. 1980. Investigation on rate-determining factors in the microbial reduction of azo dyes. Eur. J. Appl. Microbiol. Biotechnol. 9:325-338. [Google Scholar]
- 37.Zhang, F., J. S. Knapp, and K. N. Tapley. 1999. Development of bioreactor system for decolorization of orange II using white rot fungus. Enzyme Microb. Technol. 24:49-53. [Google Scholar]
- 38.Zimmermann, T., F. Gasser, H. G. Kulla, and T. Leisinger. 1984. Comparison of two bacterial azoreductases acquired during adaptation to growth on azo dyes. Arch. Microbiol. 138:37-43. [DOI] [PubMed] [Google Scholar]
- 39.Zimmermann, T., H. G. Kulla, and T. Leisinger. 1982. Properties of purified orange II azoreductase, the enzyme initiating azo dye degradation by Pseudomonas KF46. Eur. J. Biochem. 129:197-203. [DOI] [PubMed] [Google Scholar]