SUMMARY
Using a forward genetics ENU mutagenesis screen for recessive mutations that affect circadian rhythmicity in the mouse, we isolated a long period (~26 h) circadian mutant named Overtime (Ovtm). Positional cloning and genetic complementation reveal that Ovtm is encoded by the F-box protein FBXL3 a component of the SKP1-CUL1-F-box-protein (SCF) E3 ubiquitin ligase complex. The Ovtm mutation causes an isoleucine to threonine (I364T) substitution leading to a loss-of-function in FBXL3 which interacts specifically with the CRYPTOCHROME (CRY) proteins. In Ovtm mice, expression of the PERIOD proteins PER1 and PER2 is reduced; however, the CRY proteins CRY1 and CRY2 are unchanged. The loss of FBXL3 function leads to a stabilization of the CRY proteins, which in turn leads to a global transcriptional repression of the Per and Cry genes. Thus, Fbxl3Ovtm defines a molecular link between CRY turnover and CLOCK/BMAL1-dependent circadian transcription to modulate circadian period.
INTRODUCTION
The mechanism of circadian oscillators in mammals is generated by a cell autonomous autoregulatory transcription-translation feedback loop (Lowrey and Takahashi, 2004; Reppert and Weaver, 2002). In the primary negative feedback loop, the bHLH-PAS transcription factors, CLOCK (and its paralog NPAS2) and BMAL1 (ARNTL) dimerize and activate transcription of the Period (Per1, Per2) and Cryptochrome (Cry1, Cry2) genes (Bunger et al., 2000; Gekakis et al., 1998; King et al., 1997; Kume et al., 1999). As the PER proteins accumulate, they form complexes with the CRY proteins, translocate into the nucleus, and interact with the CLOCK/BMAL1 complex to inhibit their own transcription (Lee et al., 2001). This leads to a fall in the inhibitory complex through turnover, and the cycle starts again with a new round of CLOCK/BMAL1- activated transcription. Additional pathways in the circadian gene network such as the second negative feedback loop (involving Rev-erbα) in the positive limb of the oscillator are thought to add robustness to the circadian mechanism (Preitner et al., 2002; Sato et al., 2004). Finally, post-translational modifications play critical roles in regulating the turnover, cellular localization and activity of circadian clock proteins (Eide et al., 2005; Gallego and Virshup, 2007; Lowrey et al., 2000).
Despite this progress, it is clear that a significant number of genes that strongly influence and regulate circadian rhythms in mammals remain to be discovered and identified (Shimomura et al., 2001; Takahashi, 2004). Forward genetic screens have been one of the most effective tools for circadian gene discovery (Takahashi, 2004; Takahashi et al., 1994; Vitaterna et al., 1994), and we have used this approach to screen the mouse genome for circadian rhythm mutants generated in the Neurogenomics Project in the Center for Functional Genomics at Northwestern University (Vitaterna et al., 2006).
Using a forward genetic approach in mice, we report here the isolation and positional cloning of a novel circadian period mutant in mice. This mutation reveals a defect in an orphan member of the F-box protein family, FBXL3, which leads to a stabilization of CRY protein levels and a global repression of Per and Cry gene transcription.
RESULTS
Mutagenesis, screening and identification of the Overtime gene
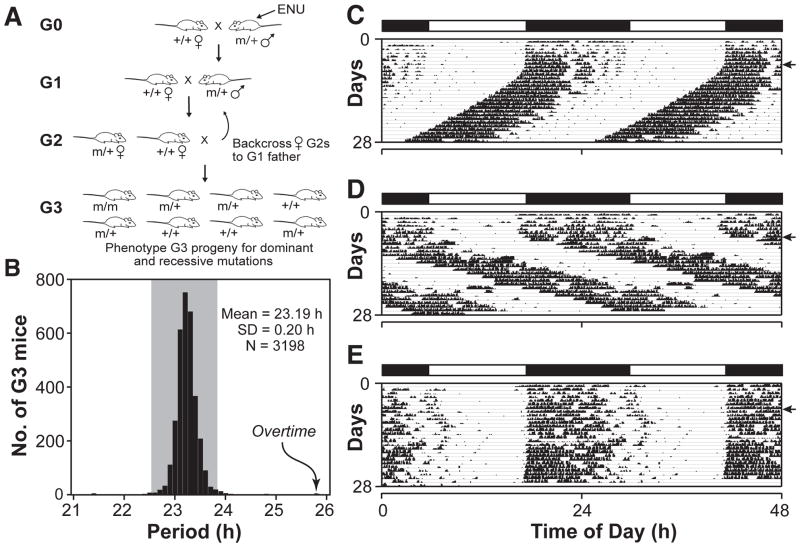
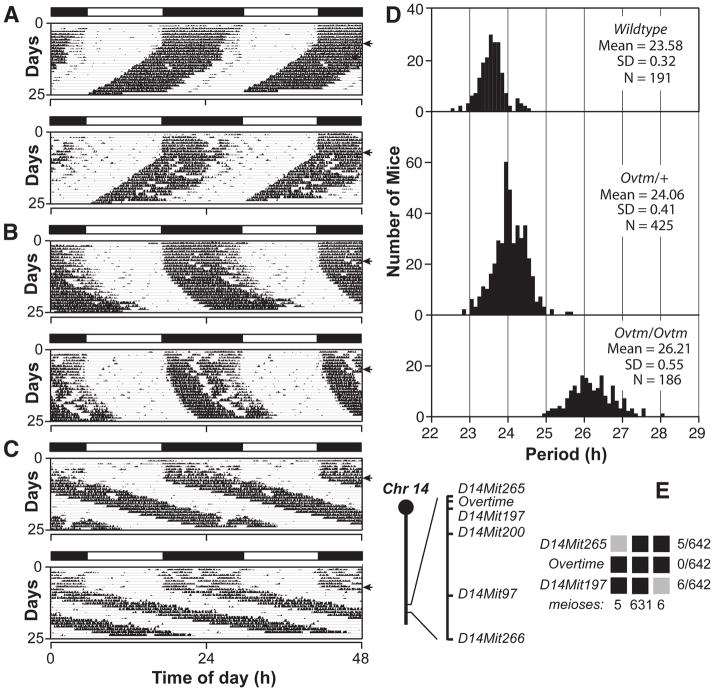
In an N-ethyl-N-nitrosourea (ENU) recessive screen using the BTBR T+ tf/J (BTBR/J) inbred mouse strain (Siepka and Takahashi, 2005a), we identified a long period mutant with a 25.8-h period length, which is more than 10 standard deviations from the mean in this screen (Figure 1). This mutant, named Overtime, is semidominant and autosomal. In crosses with C57BL/6J and C3H/HeJ mouse strains, the homozygous mutant phenotype is distinct with an average circadian period of 26.2 h and a range of about 25–28 h, which is indistinguishable from the original mutant (Figure 2A–D). Ovtm maps to a 1.7 cM interval on chromosome 14 between D14Mit265 (5 recombinants/642 meioses) and D14Mit197 (6 recombinants/642 meioses) (Figure 2E). This region corresponds to a 4 Mb interval and contains 18 open reading frames (Assembly build NCBI m36 December 2005, Gene build June 2006), none of which corresponds to previously known circadian clock genes (Figure 3A) (Lowrey and Takahashi, 2004).
Figure 1. ENU mutagenesis screen.
(A) Mutant mouse production. See Experimental Procedures for description.
(B) Histogram showing circadian period values of 3198 mice in this screen. Gray shading indicates ±3 standard deviations (SD) from the mean. The original Overtime mutant is indicated by the arrow.
(C) Representative actogram of a wildtype BTBR/J mouse. The actogram is double plotted where 48 h of activity is represented on each horizontal line. The mice were kept on a LD12:12 cycle (represented in the bar above) for the first 7 days and then released into constant darkness (DD) for 21 days (indicated by the arrowhead on the right).
(D) Actogram of the original Ovtm G3 mouse in the screen. This animal (an Ovtm homozygote) had a free running period of 25.83 h.
(E) Actogram of an Ovtm G3 littermate. This animal (a heterozygote carrier and founder of the Ovtm strain) had a free running period of 23.98 h.
Figure 2. Semidominant phenotype of Overtime and genetic mapping.
(A) Representative actograms of wildtype, (B) Ovtm/+ and (C) Ovtm/Ovtm [BTBR/J x C57BL/6J] F2 mice. The actograms are plotted as described in Figure 1.
(D) Period distribution of F2 intercross progeny. The three panels from top to bottom represent wildtype, Ovtm/+ and Ovtm/Ovtm mice, respectively. Analysis of variance (ANOVA) of the periods was performed on the three populations of F2 mapping mice (as defined by Ovtm genotype). Significant effects of the Ovtm allele were detected (DF = 2; F= 2112; p < 1 × 10−27).
(E) Ovtm maps between D14Mit265 and D14Mit197. The chromosome 14 schematic lists the distal markers used to map Ovtm. Haplotypes of the 321 Ovtm/Ovtm F2 intercross progeny (642 meioses) are on the right. Black boxes represent BTBR/J alleles, while grey boxes represent heterozygous alleles (BTBR/J and C57BL/6J). The number of recombinants per total meioses is indicated to the right of the haplotype map.
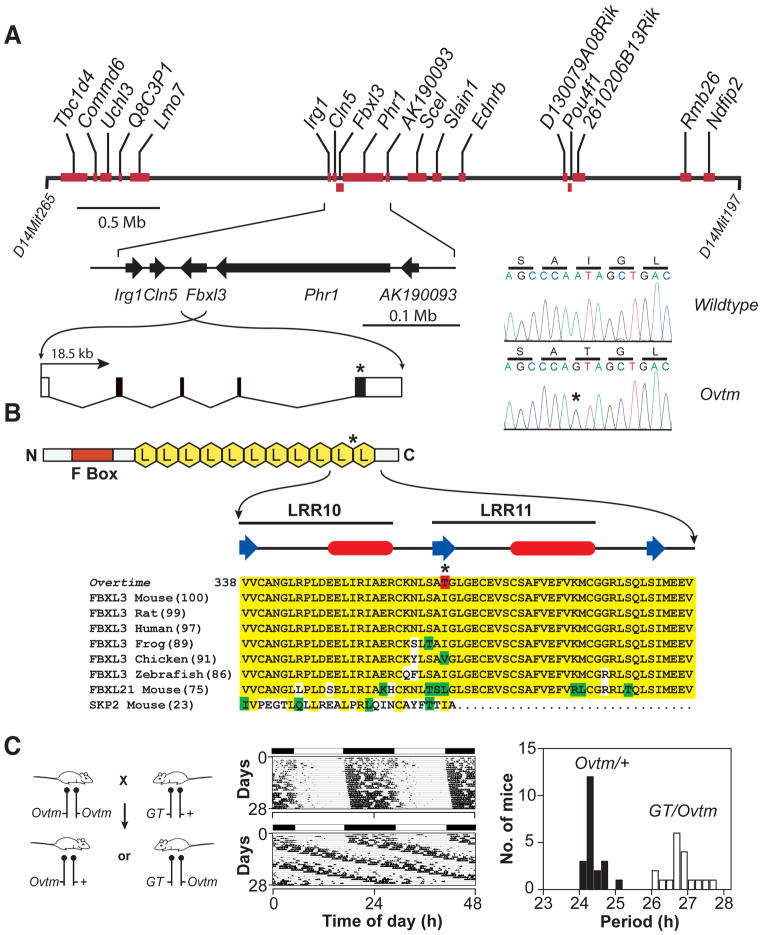
Figure 3. Positional cloning of Overtime and identification of Fbxl3 mutation.
(A) Physical map of the Ovtm interval. Ovtm maps to a 4 Mb region of chromosome 14. Red blocks represent the eighteen candidate genes within the interval. The asterisks indicate the location of the Ovtm mutation.
(B) FBXL3 is an F-box protein. FBXL3 contains one F-box domain (red box) and 11 leucine rich regions (LRR) (yellow hexagons) (Jin et al., 2004). β-strand (blue arrow) and α-helical (red ovals) regions (based on analysis using PROF in the PredictProtein server (Rost et al., 2004)) of LRR10 and LRR11 are indicated above the sequence. In the protein alignment, yellow indicates amino acid identity, green indicates conservative substitutions, white indicates non-conservative substitutions, and red indicates the I364T Ovtm mutation.
(C) Complementation test for Ovtm and Fbxl3. The left panel outlines the mating scheme for the complementation assay. Ovtm mice were crossed to heterozygous Fbxl3 gene trap (GT) mice. Circadian behavior was recorded for 39 progeny. The actograms are representative of Ovtm/+ (top) and GT/Ovtm (bottom) mice. In the period histogram distribution black bars represent Ovtm/+ mice (Mean = 24.20 h; SD = 0.26; N = 21), while white bars represent GT/Ovtm mice (Mean = 26.62 h; SD = 0.41; N = 18). Student’s t-test, unequal variances, shows a significant difference between the two populations (DF= 28; T = −21.47; p = 6.3 × 10−19).
Several genes in the nonrecombinant interval were likely candidates for Ovtm. Protein degradation machinery has as an important role in the clock mechanism (Gallego and Virshup, 2007) and there are several ubiquitin-pathway related genes in this interval (Uch13, Lmo7, Fbxl3, Phr1, 2610206B13Rik and Ndfip2). Because the entire 4 Mb Ovtm interval has a low polymorphism rate and shares the identical haplotype with the mapping strains, the resolution of meiotic mapping could not be increased. This region of the genome is part of the piebald deletion complex which contains several developmentally important genes (Peterson et al., 2002). This may have caused selection of a limited number of ancestral haplotypes in this region of chromosome 14 which would in turn lead to a low polymorphism rate.
We sequenced all annotated exons for the 18 candidate genes in the Ovtm interval and found only a single nonsynonymous point mutation within the coding region of Fbxl3. There is a single base transition from A to G in exon 5 of Fbxl3 in Ovtm mice as compared to wildtype BTBR/J mice. This mutation co-segregated perfectly with the long-period phenotype of Ovtm/Ovtm mice (0 recombinations out of 642 meioses, Figure 2D, E). The point mutation converts amino acid residue 364 from isoleucine to threonine in FBXL3 (Figure 3A, B). This isoleucine residue is highly conserved in FBXL3 from vertebrates and in the mouse paralog FBXL21 (Figure 3B). FBXL3 is a member of the F-box protein family with leucine rich repeats (LRR) which is defined by its founding member, SKP2 (S-phase kinase-associated protein-2) (FBXL1) (Jin et al., 2004). SKP2 is the F-box protein moiety in the SKP1-CUL1-F-box-protein (SCF)SKP2 E3 ubiquitin ligase complex which mediates the recognition and ubiquitination of the CDK2 inhibitor, p27Kip1, to target it for proteasomal degradation (Cardozo and Pagano, 2004). Structural studies show that CUL1 provides a rigid scaffold upon which SKP1 and RBX1 subunits assemble: SKP1 interacts with SKP2 via its N-terminal F-box motif (Zheng et al., 2002). Recent work shows that the LRR region and the C-terminal tail of SKP2 interact with the substrate, p27Kip1, and the accessory protein, CKS1 (Hao et al., 2005). FBXL3 which has 11 LRRs can be aligned with SKP2 which has 10 LLRs based on its protein structure; however the C-termini of SKP2 and FBXL3 are not conserved likely due to the recognition of different substrates (Hao et al., 2005). The Ovtm I364T mutation occurs in the C-terminus of FBXL3 between LRR10 and LRR11 where the alignment with SKP2 becomes divergent (Figure 3B). Because the LRR domains of F-box proteins are involved with substrate recognition with the SCF complex, we hypothesized that the I364T mutation could alter the interaction of FBXL3 with its substrates.
Because we isolated only one mutant allele of Fbxl3 and either a second independent allele, rescue, or functional evidence is required for proof in positional cloning (Takahashi et al., 1994), we used genetic complementation tests to confirm that Ovtm was allelic with Fbxl3. A search of the International Gene Trap Consortium (IGTC) database (http://www.genetrap.org/) revealed three gene traps in Fbxl3, and we obtained one gene trap ES cell line (S13–12G1) from Philippe Soriano (Chen et al., 2004). ES cells were microinjected into blastocysts to produce chimeras and 19 out of 19 mice were chimeric. Because the ES cell contribution in these chimeras was >95%, they were mated directly to Ovtm/Ovtm mice to test for germline transmission and genetic complementation simultaneously. The crosses show that Ovtm and the Fbxl3 gene trap (GT) fail to complement each other, thus providing independent and definitive evidence that Ovtm is an allele of Fbxl3 (Figure 3C). Interestingly, the period length of GT/Ovtm mice is indistinguishable from Ovtm homozygotes suggesting that the Ovtm mutant allele is likely a hypomorphic or loss-of-function allele.
The mRNA expression of Fbxl3 itself in the mouse is ubiquitous, but enriched in the brain (data not shown). Northern blot analysis of Fbxl3 from liver in wildtype (WT) and Ovtm mice did not reveal any significant differences in transcript size or abundance between the genotypes. In addition, there was no obvious circadian rhythm in Fbxl3 mRNA expression in the liver. In situ hybridization of Fbxl3 shows clear RNA expression in the suprachiasmatic nucleus (SCN) and throughout the brain (see Supplemental Material).
Effects of the Overtime mutation on circadian clock gene expression
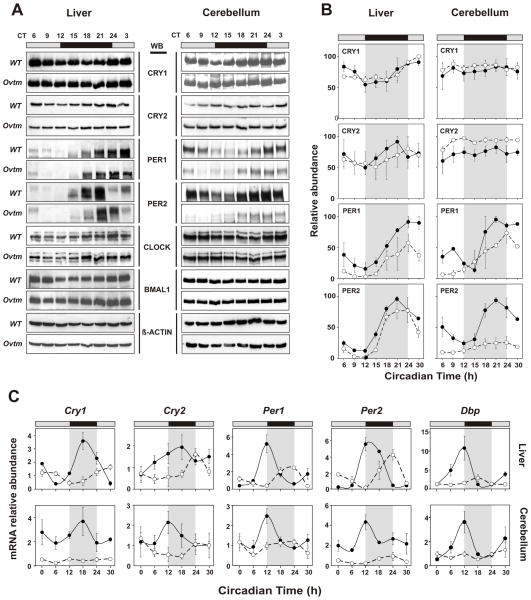
Because FBXL3 is likely a component of an SCF E3 ubiquitin ligase complex, we examined the in vivo expression patterns of circadian clock proteins in mouse tissues to explore whether Ovtm might alter their abundance by affecting degradation. WT and Ovtm mice were housed in running wheel cages to measure activity rhythms and were transferred to constant darkness (DD) for 10–14 days. Animals were killed under infrared illumination every 3 h in DD based on circadian time (CT) to compensate for the differences in circadian period between WT and Ovtm mice. Figure 4A shows the expression patterns of the clock proteins, CRY1, CRY2, PER1, PER2, CLOCK and BMAL1 in liver and cerebellum. In WT mice, there were low amplitude rhythms of CRY1 and CRY2 and high amplitude rhythms of PER1 and PER2 as reported previously (Lee et al., 2001). In Ovtm liver tissue, CRY1 and CRY2 protein patterns were not significantly altered; however, PER1 and PER2 levels were significantly reduced (Figure 4B). In the cerebellum, the effects of Ovtm were more striking. Although CRY1 levels were not different, CRY2 levels were significantly elevated in Ovtm mice consistent with the hypothesis that CRY degradation is impaired. In addition, there were very clear reductions in the levels of PER1 and PER2. The levels of CLOCK and BMAL1 were not detectably changed in either tissue in Ovtm mice (Figure 4A). The reduction of PER1 and PER2 levels in Ovtm mice is significant in both liver and cerebellum, and this finding is unexpected and counterintuitive. We would have expected to see an increase rather than a decrease in protein abundance if the PER proteins were targets of FBXL3 because the Ovtm mutation is a loss-of-function mutation. This suggests that it is unlikely that the PER proteins are targets of FBXL3 and that the reduction in PER levels could occur as a consequence of the negative feedback on CLOCK/BMAL1-dependent transcription.
Figure 4. Altered circadian clock gene expression in Overtime mice.
(A) Protein oscillation profiles of clock genes from liver and cerebellum. Wildtype and Ovtm mutant tissue was collected at indicated circadian times. Western blotting was performed on total protein extracts with indicated antibodies. Each blot shows a representative example from three independent replicates.
(B) Quantification of proteins from liver and cerebellum. Data are taken from 3 independent experiments. Protein levels were quantified using Scion Image (http://www.scioncorp.com). Filled circles with solid line represent normalized values from wildtype, and open circles with dotted line represent normalized values from Ovtm mice. Error bars represent SEM for each time point from three independent replicates. Two-way ANOVA shows significant statistical differences between WT and Ovtm mutant for both PER1 and PER2 expression in cerebellum (p < 0.001, p< 10−5 respectively) and in liver (p< 10−5, p< 0.001 respectively). In contrast to PER, only CRY2 shows a significant difference in expression from cerebellum (p< 10−5) between wildtype and Ovtm mice.
(C) Real time RT-PCR analysis for clock gene expression in wildtype and Ovtm mice. Filled circles with solid line represent wildtype, and open circles with dotted line represent values from Ovtm mice. All cycling genes show a significant reduction of mRNA level from Ovtm mice compared to WT mice in liver and cerebellum except for Per2 in liver (N.S.). Error bars represent SEM for each time point from four independent replicates.
To explore the reasons for the reduction in PER1 and PER2 protein abundance, we profiled the in vivo circadian mRNA expression patterns for Cry1, Cry2, Per1, Per2 and Dbp in the liver and cerebellum of mice maintained in DD. As shown in Figure 4C, the Ovtm mutation caused significant reductions in the mRNA abundance of all of these cycling transcripts with the strongest effects being seen with Cry1 and Per2 in the cerebellum. At the mRNA level, both a delay in the peak time and a reduction in abundance can be seen. Importantly, although CRY1 and CRY2 protein levels were not lower in Ovtm mice, the corresponding mRNA levels for Cry1 and Cry2 are significantly reduced in both tissues. Similar reductions in the mRNA levels for Cry1 and Per2 in Ovtm mice could also be observed in the SCN using in situ hybridization (see Supplemental Material). In addition, mRNA levels for the cycling CLOCK target gene, Dbp (Ripperger and Schibler, 2006), were very strongly reduced in Ovtm mice. Thus, the mRNA profiling experiments point to an interesting and unexpected consequence of the Ovtm mutation: a reduction in steady-state mRNA expression of Cry1, Cry2, Per1, Per2 and Dbp which are all transcriptional targets of the CLOCK/BMAL1 complex (Gekakis et al., 1998; Kume et al., 1999; Ripperger and Schibler, 2006; Yoo et al., 2005).
Comparison of the effects of Ovtm on protein vs. mRNA abundance suggests that there are two different effects on the expression of the CRY and PER proteins. The PER protein levels appear to be reduced as a consequence of reduced transcript levels. By contrast, the CRY protein levels are not reduced even in the face of reduced transcript levels. This suggests that potential reductions in CRY protein levels caused by reduced Cry transcript levels could be compensated by a reduction in protein degradation.
Interaction of OVTM with circadian clock proteins
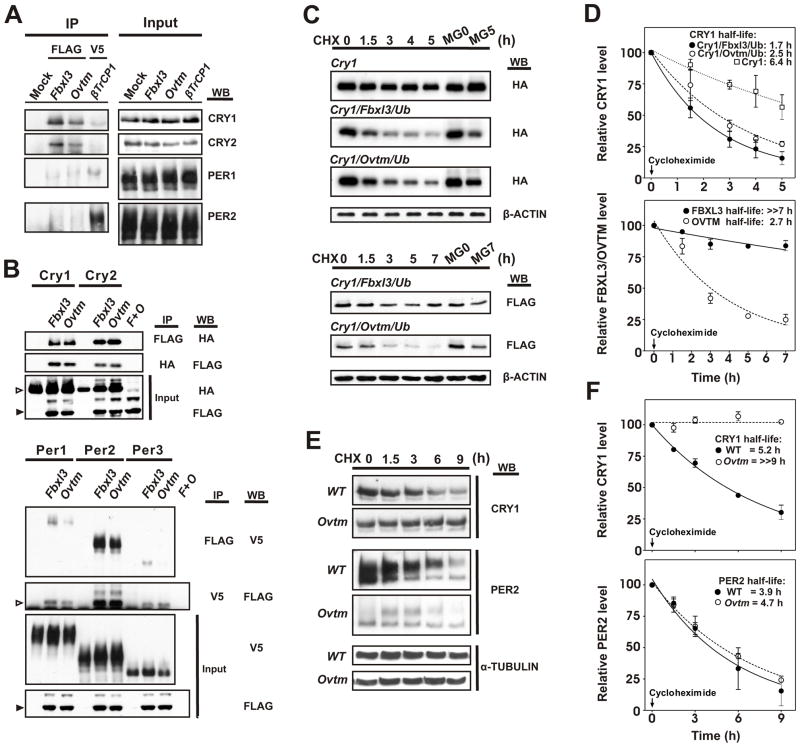
The Pagano laboratory has found that FBXL3 targets CRY proteins for ubiquitination and degradation (personal communication). To confirm these results and determine whether the Ovtm mutation affects interactions with CRY, we examined the interaction of FBXL3 or OVTM with circadian clock proteins by immunoprecipitation assays. First, we used tagged Fbxl3, Ovtm and βTrCP1 expression constructs to transfect NIH 3T3 cells which contain circadian oscillators and express clock proteins (Nagoshi et al., 2004). Cell lysates were immunoprecipitated with anti-FLAG or anti-V5 antibodies, and interactions with CRY1, CRY2, PER1 and PER2 native proteins were probed on Western blots (Figure 5A). Both FBXL3 and OVTM interacted strongly with native CRY1 and CRY2 proteins. Very weak or no interaction of FBXL3 was seen with PER1 and PER2, especially in comparison to that seen between the PERs and βTrCP1, an F-box protein known to interact with the PERs (Eide et al., 2005; Shirogane et al., 2005) (Figure 5A). In all experiments, there was a discernibly stronger interaction of the CRY proteins with FBXL3 relative to OVTM, but the difference was subtle.
Figure 5. Interaction of FBXL3 and OVTM with circadian clock proteins.
(A) NIH 3T3 cells were transfected with FLAG-Fbxl3, FLAG-Ovtm and V5- βTrcp1. 24 h after transfection, cells were incubated with 20μM MG132 for 8 h before collection. Immunoprecipitation was performed with anti-Flag or anti-V5 antibody. Immunoprecipitated proteins were further analyzed by Western blotting with anti-PER and anti-CRY antibody.
(B) Confirmation of interaction between FBXL3 with CRY and PER2. Top panel, 293A cells were transfected with Cry-HA and FLAG-Fbxl3, FLAG-Ovtm. F+O is co-transfection of FLAG-Fbxl3, FLAG-Ovtm. 36 h after transfection, cells were harvested and reciprocal immunoprecipitations were performed with indicated antibodies. Open arrowheads indicate CRY, filled arrow heads indicate FBXL3 or OVTM. Lower panel, Per-V5 were co-transfected with FLAG-Fbxl3, FLAG-Ovtm. Open arrowheads indicate immunoprecipitated FBXL3 or OVTM, filled arrowheads indicate FBXL3 or OVTM from input.
(C) Effects of Fbxl3 and Ovtm on CRY1 and FBXL3 protein stability. Cry1-HA was co-transfected with FLAG-Fbxl3 or FLAG-Ovtm with HA-Ubiquitin (Ub). 24 h after transfection 20 μg/ml cycloheximide (CHX) was added and incubated for the indicated time. Abundance of CRY1, FBXL3 and OVTM was measured by Western blotting. Upper panel: Inhibition of CRY degradation by MG132 treatment plus CHX treatment. Lower panel: Ovtm mutation cause accelerated degradation of FBXL3 through the proteasomal degradation pathway. β-actin was used as a loading control.
(D) Quantitation of the effects of Fbxl3 and Ovtm on CRY1 and FBXL3 protein stability. Data are taken from the three independent experiments. Upper panel: Effects of FBXL3 on CRY1 stability in 293A cells. Open squares: Cry1 only; filled circles: Cry1 co-transfected with Fbxl3; open circles: Cry1 co-transfected with Ovtm mutant. The half life of CRY1 is reduced by either FBXL3 (half life: 1.7 h) or OVTM (half-life: 2.5 h). OVTM co-expression was less effective in CRY1 degradation compared to FBXL3 (the half-life parameter, K, is significantly different, p = 0.16, using model-based nonlinear exponential decay analysis with GraphPad Prism 4). Lower panel: effect of the Ovtm mutation on the half life of FBXL3. Closed circles and open circles represent FBXL3 and OVTM, respectively. In the presence of CRY1, the Ovtm mutation caused a reduction in the protein stability of OVTM (the half-life parameter, K, is significantly different, p = 0.0003). Error bars represent SEM for each time point from 3 independent replicates.
(E) Stability of endogenous CRY1 and PER2 in Ovtm ear fibroblast cells. WT and Ovtm fibroblast cells were plated 48 h before the experiment and synchronized by 1 h treatment with 10 μM forskolin. 20μg/ml CHX was administered during indicated time. Protein extracts from harvested cells were analyzed with Western blotting using anti-CRY1 and anti-PER2 antibody. α–tubulin was used as a loading control.
(F) Half-life measurements of endogenous CRY1 and PER2 proteins in WT and Ovtm fibroblasts. Data are taken from the three independent experiments. Upper panel: Time course of CRY1 levels following addition of CHX. Closed circle are from WT fibroblasts and open circles are from Ovtm cells. CRY1 degradation is much more rapid in WT fibroblasts (half life: 5.2 h) than in Ovtm fibroblasts (≫9 h) (the half-life parameter, K, is significantly different, p < 0.0001). Lower panel: Half-life measurements of PER2 proteins in WT and Ovtm fibroblasts. Closed circles are from WT fibroblasts and open circles are from Ovtm fibroblasts. Error bars represent SEM for each time point from 3 independent replicates.
We also used tagged proteins in co-immunoprecipitation assays in 293A cells which are easily transfected and express relatively low levels of clock proteins. Both FBXL3 and OVTM interacted strongly with CRY1 and CRY2 (Figure 5B). To explore the weak interaction of FBXL3 with PER proteins, tagged Per constructs were also tested. All three PER proteins showed interactions with FBXL3 and OVTM, however, the strongest interactions were seen with PER2. Because PER2 interacts very strongly with CRY1 (Griffin et al., 1999; Kume et al., 1999; Lee et al., 2001), it is likely that the interactions seen here with FBXL3 may be indirect via CRY1.
Effects of OVTM on CRY degradation
To determine whether OVTM is less efficient than FBXL3 in inducing the degradation of CRY1, we compared the effects of FBXL3 and OVTM on the stability of CRY1 following cycloheximide treatment to prevent de novo protein synthesis in transfected cells (Figure 5C). In 293A cells, transfected CRY1 is relatively stable with a half-life of 6.4 h (Figure 5D). Addition of FBXL3 or OVTM increased the turnover of CRY1 significantly with FBXL3 leading to a 1.7 h CRY1 half-life and OVTM leading to a 2.5 h CRY1 half-life (Figure 5D). Thus, FBXL3 expression leads to the degradation of CRY1, and this degradation is blocked by the 26S proteasomal inhibitor MG132 (Figure 5C). OVTM is less effective than FBXL3 in causing CRY1 degradation under these conditions, and the half-life parameter, K, is significantly different (p=0.016) between FBXL3 and OVTM in the model-based nonlinear exponential decay analysis with GraphPad Prism 4. Interestingly, the turnover of FBXL3 is also affected by the OVTM mutation (Figure 5C, bottom). FBXL3 is relatively stable with a half-life of greater than 7 h; whereas, OVTM has a much shorter half-life of 2.7 h (Figure 5D). Therefore, there are two effects of the OVTM mutation: a reduction in proteasome-mediated CRY1 degradation and a decreased stability of the OVTM protein itself, both of which could contribute to a loss-of-function phenotype.
To determine whether these changes in CRY1 stability seen in transfected cells are physiological, we used fibroblasts prepared from either WT or Ovtm mice and determined the half-lives of native CRY1 and PER2 proteins in these cells. Fibroblast cultures were synchronized with 10 μM forskolin and treated with cycloheximide. The half-lives of CRY1 and PER2 were then determined by Western blot against native proteins. As shown in Figure 5E and F, the half-life of CRY1 in Ovtm fibroblasts is extremely long (≫9 h) as compared to WT cells (half-life = 5.2 h). The overall levels of PER2 in Ovtm fibroblasts were very low similar to that seen in the cerebellum. When the half-life of PER2 was determined, however, there was no detectable difference in the half-life of PER2 in WT and Ovtm cells (Figure 5F). Thus, these experiments in fibroblasts from WT and Ovtm mice show that native CRY1, but not PER2, turnover is specifically altered by the Ovtm mutation. These experiments strongly suggest that OVTM is selectively defective in its ability to target CRY1 for degradation by the proteasome.
Effects of Ovtm on Per and Cry transcription
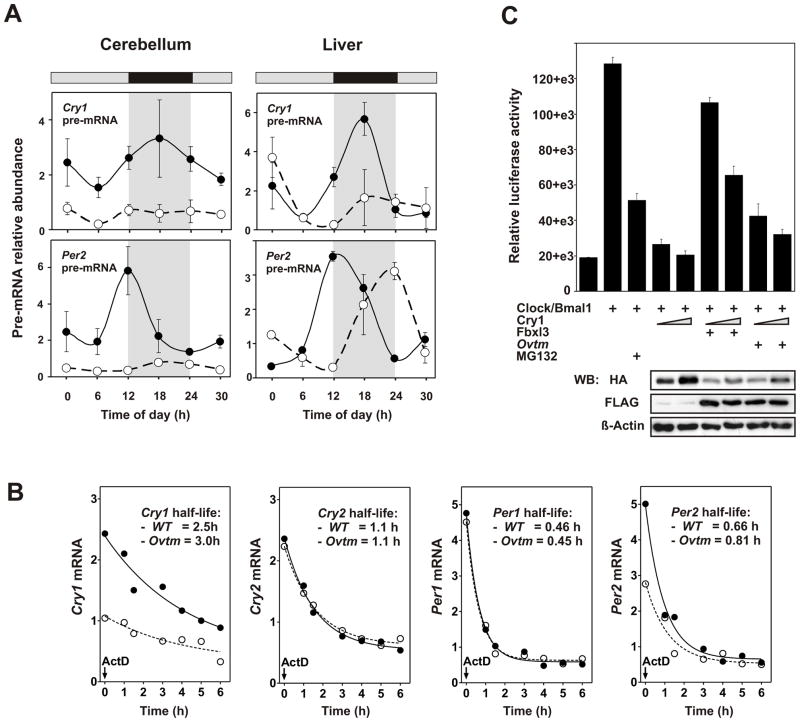
To understand why the steady state transcript levels of Per and Cry are reduced in Ovtm mice, we estimated transcription rates by measurement of pre-mRNA levels and mRNA stability by half-life experiments. In both liver and cerebellum, Cry1 and Per2 pre-mRNA levels are strongly correlated with steady-state mRNA abundance levels with lower pre-mRNA levels observed in Ovtm tissue (Figure 6A). As seen for steady state mRNA levels, the reduction in Cry1 and Per2 pre-mRNA levels was most clear in the cerebellum. Thus Ovtm strongly reduces the transcription of Cry1 and Per2 in vivo consistent with the hypothesis that transcription rates account for the reduction in steady state mRNA abundance. To reinforce this hypothesis, we also measured the half-lives of mRNAs for Cry1, Cry2, Per1 and Per2. WT and Ovtm fibroblasts were treated with Actinomycin D to inhibit de novo transcription, and mRNA half-life was determined by quantitative PCR. As shown in Figure 6B, although there are differences in the initial mRNA levels of Cry1 and Per2 as seen in the cerebellum, the half-lives of all four transcripts are not significantly different in WT and Ovtm cells using model-based nonlinear exponential decay analysis with GraphPad Prism 4. Thus, taken together these experiments argue that the reduced mRNA levels of the Cry and Per genes in Ovtm mouse tissues are due to a reduction in transcription of these genes.
Figure 6. Effects of Ovtm on Cry and Per transcription, mRNA stability and CLOCK/BMAL1-mediated transcriptional activation.
(A) Real time RT-PCR analysis of Per2 and Cry1 pre-mRNA in cerebellum and liver from WT and Ovtm mice. Error bars represent ±SEM for each time point.
(B) mRNA half-life measurements of Cry1, Cry2, Per1 and Per2. WT and Ovtm fibroblast cells were plated 48 h before, then treated with 5 μg/ml Actinomycin D for indicated amount of time. Total RNA was isolated from the harvested cells and quantitative PCR was performed as described previously. There were no significant differences in mRNA half-lives between WT and Ovtm cells.
(C) Effects of FBXL3 on CLOCK/BMAL1-mediated transcriptional activation. pGL3Basic-E2 was transfected into 293A cells with the indicated DNA constructs (plus sign). Each value represents the mean ± SEM of three replicates from a single experiment. The results shown are representative of at least three independent experiments. The lower panel shows Western blots from the same extracts used in the luciferase assays. Anti-HA and anti-FLAG antibodies were used to detect CRY1-HA and FBXL3, OVTM-FLAG, respectively.
To address the functional consequences of the mutation, we assessed whether Fbxl3 or Ovtm could affect Clock/Bmal1-mediated transcriptional activation. We asked whether Fbxl3 or Ovtm could modulate the inhibitory effects of Cry1 on Clock/Bmal1-induced transcription using a Per2 E2 enhancer driving luciferase (Yoo et al., 2005). In Figure 6C, Clock/Bmal1 transfection activates luciferase activity ~6-fold. As recently reported (Kwon et al., 2006), treatment with the proteasomal inhibitor MG132 significantly reduces the magnitude of Clock/Bmal1 activation. As expected, Cry1 co-transfection strongly inhibited Clock/Bmal1 activation (Griffin et al., 1999; Kume et al., 1999). The addition of Fbxl3 strongly diminished the Cry1-dependent inhibitory effects on Clock/Bmal1 activation; while addition of Ovtm was much less effective than that seen with Fbxl3. Western blots confirmed that the expression levels of FBXL3 and OVTM were comparable. However, Fbxl3 and Ovtm transfection reduced the levels of CRY1 consistent with the CRY1 degradation experiments in Figure 5C. Thus, FBXL3 and OVTM both reduced CRY1 expression levels and the reduction in CRY1 is correlated with a reduction in CRY1-dependent inhibition of CLOCK/BMAL1 activation (linear regression analysis of CRY1 level vs. PER::LUC activity: slope is significant, p = 0.0181, Pearson’s correlation coefficient R = −0.67). Thus, one possible explanation for the difference in CLOCK/BMAL1-dependent transcription in WT and Ovtm tissues could be that FBXL3 normally provides a steady state turnover of CRY via proteasomal degradation that is reduced by the Ovtm mutation. This could then lead to an accumulation of CRY to suppress CLOCK/BMAL1-dependent transcription.
DISCUSSION
We have shown that the ENU-induced Overtime mutant is caused by an I364T mutation in the mouse FBXL3 protein, a member of the F-box protein with leucine rich repeats family (Jin et al., 2004). The OVTM protein is less efficient than FBXL3 in degrading CRY1 thus providing genetic evidence that FBXL3 appears likely to be a primary F-box protein within an SCF E3 ubiquitin ligase complex (Cardozo and Pagano, 2004) that targets the CRY proteins for degradation in the proteasome. The I364T OVTM mutation lengthens circadian periodicity ~2.5 h in mice. We propose that the phenotypic effects of the Ovtm mutation occur primarily through two mechanisms: (1) loss of FBXL3 function leading to stability of CRY1 protein; and (2) repression of CLOCK/BMAL1-dependent transcriptional activation. These two processes lead to a striking reduction in the expression of the PER proteins which is caused by a reduction in transcription of the Per genes. By contrast, the levels of CRY are not reduced by the Ovtm mutation despite lower rates of Cry transcription. Indeed in the cerebellum, the level of CRY2 protein is significantly higher as expected (perhaps due to the slightly higher relative rate of transcription of Cry2 compared to Cry1 in the cerebellum in Ovtm vs. WT mice). However, the levels of CRY1 protein were not elevated in Ovtm mice. Because the majority of CRY protein is cytoplasmic (Lee et al., 2001), we also examined levels of CRY1 in the nucleus but did not detect an increase in CRY levels in nuclear fractions from the liver of Ovtm mice in comparison to WT (data not shown). In retrospect, because the transcription of the Cry1 gene is strongly attenuated by the Ovtm mutation, it is surprising that CRY1 protein levels are not lower. The low rate of CRY1 protein degradation in Ovtm tissues must offset the lower synthesis of CRY1 so that the steady state abundance of CRY1 protein is similar in Ovtm and WT mice. Importantly however, because the turnover rate of CRY1 is reduced, the clearance of CRY1 will be prolonged even if the initial steady-state abundance levels are comparable. This would then lead to a prolongation of the CRY-dependent repression phase of the circadian cycle. If such a prolongation extended CRY repression for 2–3 hours, the period lengthening phenotype seen in Ovtm mice would follow as a consequence. It will be of great interest to test existing biochemical models of the circadian oscillator (Forger and Peskin, 2003; Leloup and Goldbeter, 2004) for these phenotypic effects. In particular, it will be important to determine whether global changes in the set point for transcription rates of the clock genes occur as a result of decreases in the degradation of the CRY proteins and whether the changes in CRY turnover rates can account for the period lengthening phenotype of Ovtm mice. In addition, it will be interesting to explore the reasons underlying the tissue-specific differences observed in Ovtm mice.
These results highlight the significance of an SCFFBXL3 E3 ubiquitin ligase complex in regulating the stability and kinetics of CRY degradation. The specificity of the FBXL3 interaction with the CRY proteins is striking and suggests that additional F-box proteins may regulate other circadian clock components. The first example of an F-box protein playing a role in circadian rhythmicity was the Arabidopsis gene ZEITLUPE (ZTL) which encodes an F-box protein with an N-terminal LOV domain and C-terminal kelch repeats (Somers et al., 2000). ZTL targets the Arabidopsis clock protein, TOC1, for degradation by the proteasome and is thought to regulate circadian period by controlling TOC1 stability (Mas et al., 2003). In addition, the F-box protein, FKF1, mediates the cyclic degradation of CDF1, a repressor of the photoperiodic gene CONSTANS (Imaizumi et al., 2005), and the F-box protein, AFR, is a positive regulator of phytochrome A-mediated light signaling in Arabidopsis (Harmon and Kay, 2003). FBXL3 is the second example of a mammalian F-box protein regulating the circadian clock proteins. The first example is βTrCP which has been shown to interact directly with the PER proteins (Eide et al., 2005; Shirogane et al., 2005). Evidence for βTrCP in the circadian pathway first emerged from Drosophila in which it was shown that Slimb, the ortholog of βTrCP, regulated circadian expression of PER and TIM (Grima et al., 2002; Ko et al., 2002). Interestingly, in Neurospora, the ortholog of βTrCP, FWD1, regulates the degradation of the clock protein, FREQUENCY (He et al., 2003). More recently, JETLAG, the Drosophila ortholog of Fbxl15, has been shown to play a critical role in light-induced TIM degradation by the proteasome (Koh et al., 2006). In Drosophila two different SCF complexes appear to control TIM levels: a circadian pathway involving Slimb and a light-dependent pathway involving JET (Koh et al., 2006). It will be interesting see whether similar types of mechanisms are conserved in mammals. Because of the differences in the roles of the PER and CRY proteins in Drosophila and in mammals (Allada et al., 2001; Young and Kay, 2001), where CRY is primarily a circadian repressor (not a photoreceptor), FBXL3 appears to function in a circadian SCF complex-mediated pathway. Unlike Drosophila PER, the PER1 and PER2 proteins in mammals are transcriptionally induced by light in the SCN. It will be interesting to see whether βTrCP functions in a circadian or in a light-dependent SCF pathway for PER degradation (analogous to the TIM protein in Drosophila).
Using a forward genetics approach in mice, we have identified, FBXL3, as a new molecular component of the negative feedback loop that generates circadian rhythmicity. Our results both confirm biochemical studies from the laboratory of Pagano (personal communication) showing that the SCFFBXL3 E3 ubiquitin ligase complex targets CRY1 for ubiquitination and provide in vivo evidence that links CRY stability to CLOCK/BMAL1-mediated transcriptional activation. In Ovtm mice the fine equilibrium between activation and repression of clock gene transcription is disturbed due to the mutation. Future studies will further clarify how this equilibrium between activators and repressors leads to the generation of circadian periodicity.
EXPERIMENTAL PROCEDURES
Mouse Strains
BTBR T+ tf/J (Stock #002282), C57BL/6J (Stock # 000664), C3H/HeJ (Stock # 000659) and 129S1/SvImJ (Stock #002448) mice were purchased from The Jackson Laboratory (Bar Harbor, ME). CD-1 (Strain code 022) female mice were purchased from Charles River Laboratories (Wilmington, MA). All mice were housed under LD12:12 unless otherwise noted. All animal care and experimental treatments were in accordance with Northwestern University guidelines for animal care and use.
Mutagenesis and breeding scheme
N-Ethyl-N-nitrosourea (ENU) (Sigma cat #N3385) was prepared as described previously (Siepka and Takahashi, 2005a). Six-week-old male BTBR/J mice were injected with 250 mg/kg body weight of ENU. After a 6-week recovery period, the ENU treated mice were mated with wildtype BTBR/J females to generate generation 1 (G1) males. G1 males were mated with wildtype BTBR/J females to produce G2 females. Four G2 females were backcrossed to their G1 fathers to produce G3 mice for phenotyping. Five G3 mice from every G2 backcross (20 mice per G1 pedigree) were phenotyped to ensure an 85% probability of detecting a recessive mutation. 3198 G3 mice from 216 G1 pedigrees were successfully screened from a total of 3609 G3 mice produced.
Measurement of free-running period
Mice (6–10 weeks of age) were placed in individual running wheel cages and activity recorded using the ClockLab data collection system (Actimetrics, Wilmette, IL). After one week on LD12:12, the mice were released into constant darkness (DD) for an additional 3 weeks. Free running period was measured by linear regression analysis of activity onsets from data collected during the DD portion of the assay using ClockLab software (Siepka and Takahashi, 2005b).
Genetic mapping
Ovtm/Ovtm male mice were mated to C3H/HeJ female mice to create [Ovtm/+ x C3H/HeJ] F1 mice. The F1 females were backcrossed to produce 156 [(Ovtm/+ x C3H/HeJ) x Ovtm/Ovtm] N2 mice for mapping. Alternately, Ovtm/Ovtm female mice were mated to C57BL/6J male mice to create [Ovtm/+ x C57BL/6J] F1 mice. These F1 siblings were intercrossed to produce 1480 [Ovtm x C57BL/6J] F2 mice for mutation mapping. Wheel-running behavior of all mapping mice was collected and analyzed as described above. Only presumptive Ovtm homozygous mice (321 genotyped for a total of 642 meioses) were used for genetic mapping as described in the Supplemental Materials.
Sequencing of candidate genes
Sequencing was performed either in our laboratory or at the Harvard Partners Genome Center, Cambridge MA. Genomic DNA was extracted from tail tips from 2 wildtype BTBR/J mice and two Ovtm/Ovtm mice. All annotated exons from MGSC 36 within the 4 MB interval between D14Mit265 and D14Mit197, which contains Ovtm, were amplified by PCR using genomic DNA templates. Most exons were small enough to be amplified and sequenced in a single amplicon (~600 bp). Some larger exons however, were sequenced by multiple overlapping 600 bp amplicons. The following genes and the number of exons and amplicons sequenced were as follows: Tcb1d4 (19 exons, 20 amplicons), Commd6 (4 exons, 3 amplicons), Uch13 (9 exons, 8 amplicons), Q8C3P1 (1 exon, 2 amplicons), Lmo7 (27 exons, 31 amplicons), Irg1 (5 exons, 8 amplicons), Cln5 (4 exons, 5 amplicons), Fbxl3 (5 exons, 7 amplicons), Phr1 (86 exons, 88 amplicons), AK190093 (3 exons, 0 amplicons—due to the highly repetitive nature of the sequence), Scel (31 exons, 30 amplicons), Slain1 (7 exons, 9 amplicons), Ednrb (8 exons, 8 amplicons), D130079A08Rik (2 exons, 3 amplicons), Pou4f1 (3 exons, 4 amplicons), 2610206B13Rik (6 exons, 9 amplicons), 1700009P03Rik (22 exons, 21 amplicons), Ndfip2 (8 exons, 7 amplicons). A table of primers sequences used in this project is available on request. Note that all primers also contained M13 tags (M13 Forward 5′-TGTAAAACGACGGCCAGT-3′ and M13 reverse 5′-AACAGCTATGACCATG-3′) at their 5′ ends. M13 universal primers were used for all sequencing reactions. Sequencing data were analyzed using Sequencher 4.5 software (Gene Codes Corporation, Ann Arbor, MI).
Overtime genotyping
Mice were genotyped for the Ovtm mutation using real-time PCR to detect single nucleotide polymorphisms (SNPs). PCR reactions were carried out in 6 μl volumes using 10–25 ng template genomic DNA with 1x Master Mix for SYBR® Green (Part # 4309155, Applied Biosystems Inc, Foster City, CA) 5 pmoles forward (wildtype forward primer = 5′-GTTGCAAAAATTTGTCAGCaAt-3′ or Ovtm forward primer = 5′-GTTGCAAAAATTTGTCAGCaAc-3′) and 5 pmoles reverse primer (5′-CCCCACACATCTTCACAAACT-3′) in MicroAmp 96- or 384-well Optical Reaction Plates (Applied Biosystems, Inc.). Thermo-cycling reaction conditions were as follows: 40 cycles of 95°C for 15 seconds and 60°C for 30 seconds. Data was collected and analyzed with either an ABI Prism 7700 Sequence Detector or an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Inc.).
Gene trap mouse production
Mouse embryonic stem cell gene trap line S13–12G1 was obtained from the Soriano Lab Gene Trap Resource at the Fred Hutchinson Cancer Research Center, Seattle WA (http://www.fhcrc.org/science/labs/soriano/trap.html). The cell line contains a ROSAFARY gene trap vector (Chen et al., 2004) that disrupts expression of Fbxl3 to create a null allele of Fbxl3. The maintenance of ES cells, production of chimeric mice and genotyping of the Fbxl3 gene trap are described in the Supplemental Materials.
Immunoblotting and immunoprecipitation
Immunoblotting was performed as described previously (Yoo et al., 2004). Antibodies against PER1, PER2, CRY1, CRY2, CLOCK, BMAL1 were made as described previously (Lee et al., 2001). Immunoprecipitation was performed 32 h after transfection including an 8 h treatment of 20 μM MG132 (EMD Biosciences). Harvested cells were homogenized in EB (20mM HEPES pH7.5, 100mM NaCl, 0.05% TritonX-100, 1mM EDTA, 20mM NaF, 1mM Na3OV4, Complete mini, Roche) and centrifuged at maximum speed for 10 min at 4 °C. The supernatants were transferred to fresh tubes and incubated with 2 μg of anti-FLAG or anti-V5 antibodies for 2 h at 4°C. 10 μl 50% protein-A slurry (GE) was added, and the incubation continued for an additional 1.5 h.
Supernatants were discarded after centrifugation at 3000 rpm at 4 °C for 1 min and protein-A beads were washed 3 times with 1 ml EB. Pellets were resuspended in 20 μl of 2 SDS sample buffer and boiled for 3 min. Protein samples were separated by 10% or 6% SDS-polyacrylamide gel then transferred to a nitrocellulose membrane (NEN). 5% nonfat dry milk in Tris-buffered saline containing 0.05% Tween20 was used for blocking buffer. Anti-HA (Roche), anti-FLAG (Sigma), anti-V5 (Invitrogen) were used to detect CRY1, FBXL3 (or OVTM) and βTRCP1, respectively.
RNA isolation and real time PCR
Total RNA was isolated from frozen tissues with Trizol reagent (Invitrogen) then treated with DNase (Ambion). 0.5 μg of DNase-treated total RNA was reverse-transcribed using Taqman reverse transcription reagent (Roche). The cDNA equivalent to 10 ng of total RNA was PCR-amplified in either an ABI Prism 7700 Sequence Detector or an ABI 7900HT Fast Real-Time PCR System (Applied Biosystems, Inc.). RNA expression was quantified by using SYBR green real-time PCR analysis. Analysis was performed as described previously (Wilsbacher et al., 2002). Primer sequences are described in the Supplemental Material.
Cell culture and transfection
Primary fibroblasts were isolated from the ear tissue of wildtype and Ovtm mutant mice. Briefly, minced ear tissue was incubated with collagenase III (1 mg/ml) and trypsin (0.05%). After 10 min incubation at 37°C minced tissue was allowed to adhere onto a 100 mm culture dish. Approximately 10 days after culture in DMEM (Mediatech) supplemented with 10% fetal bovine serum, ear fragments were removed and outgrowing fibroblasts were replated into T75 flasks by trypsinization. NIH 3T3 and 293A (ATCC) cells were cultured in DMEM (Mediatech) supplemented with 10% fetal bovine serum. For immunoprecipitations, 1 × 106 cells were plated into the 100 mm dishes one day before transfection, Effectene reagent (Qiagen) was used to transfect DNA according to the manufacturer’s protocol. For the luminescence assay, 293A cells were plated the day before transfection at 2 × 105 cells per well in 6-well plates. Cells were transfected with indicated vectors by using Effectene reagent. 48 h after transfection, cells were lysed and luminescence was measured from 20 μl of lysate in the Luciferase Assay System (Promega) using a luminometer (AutoLumet Plus; Berthold Technologies).
Supplementary Material
Acknowledgments
We thank Michele Pagano and Pat Nolan for communicating unpublished results, Steven Reppert for clock gene expression plasmids, Lawrence H. Pinto for help with mutagenesis; Renee McGurk, Min Cheng, Dawn Olson and Jennifer Wakowiak for mouse production, phenotyping, genotyping and DNA sequencing; Ethan Buhr, Kazuhiro Shimomura and Ming-Lee Chow for assistance with experiments; Andrew Schook for in situ hybridization experiments; Kate Montgomery and Alex Tamburino at Harvard Partners Genome Center for DNA sequencing and analysis of candidate genes. Research supported by NIH grants U01 MH61915, P50 MH074924 and R01 MH078024 to J.S.T. and R01 NS053616 to C.L. J.S.T. is an Investigator in the Howard Hughes Medical Institute.
References
- Allada R, Emery P, Takahashi JS, Rosbash M. Stopping time: the genetics of fly and mouse circadian clocks. Annu Rev Neurosci. 2001;24:1091–1119. doi: 10.1146/annurev.neuro.24.1.1091. [DOI] [PubMed] [Google Scholar]
- Bunger MK, Wilsbacher LD, Moran SM, Clendenin C, Radcliffe LA, Hogenesch JB, Simon MC, Takahashi JS, Bradfield CA. Mop3 is an essential component of the master circadian pacemaker in mammals. Cell. 2000;103:1009–1017. doi: 10.1016/s0092-8674(00)00205-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardozo T, Pagano M. The SCF ubiquitin ligase: insights into a molecular machine. Nat Rev Mol Cell Biol. 2004;5:739–751. doi: 10.1038/nrm1471. [DOI] [PubMed] [Google Scholar]
- Chen WV, Delrow J, Corrin PD, Frazier JP, Soriano P. Identification and validation of PDGF transcriptional targets by microarray-coupled gene-trap mutagenesis. Nat Genet. 2004;36:304–312. doi: 10.1038/ng1306. [DOI] [PubMed] [Google Scholar]
- Eide EJ, Woolf MF, Kang H, Woolf P, Hurst W, Camacho F, Vielhaber EL, Giovanni A, Virshup DM. Control of mammalian circadian rhythm by CKIepsilon-regulated proteasome-mediated PER2 degradation. Mol Cell Biol. 2005;25:2795–2807. doi: 10.1128/MCB.25.7.2795-2807.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forger DB, Peskin CS. A detailed predictive model of the mammalian circadian clock. Proc Natl Acad Sci U S A. 2003;100:14806–14811. doi: 10.1073/pnas.2036281100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego M, Virshup DM. Post-translational modifications regulate the ticking of the circadian clock. Nat Rev Mol Cell Biol. 2007;8:139–148. doi: 10.1038/nrm2106. [DOI] [PubMed] [Google Scholar]
- Gekakis N, Staknis D, Nguyen HB, Davis FC, Wilsbacher LD, King DP, Takahashi JS, Weitz CJ. Role of the CLOCK protein in the mammalian circadian mechanism. Science. 1998;280:1564–1569. doi: 10.1126/science.280.5369.1564. [DOI] [PubMed] [Google Scholar]
- Griffin EA, Jr, Staknis D, Weitz CJ. Light-independent role of CRY1 and CRY2 in the mammalian circadian clock. Science. 1999;286:768–771. doi: 10.1126/science.286.5440.768. [DOI] [PubMed] [Google Scholar]
- Grima B, Lamouroux A, Chelot E, Papin C, Limbourg-Bouchon B, Rouyer F. The F-box protein slimb controls the levels of clock proteins period and timeless. Nature. 2002;420:178–182. doi: 10.1038/nature01122. [DOI] [PubMed] [Google Scholar]
- Hao B, Zheng N, Schulman BA, Wu G, Miller JJ, Pagano M, Pavletich NP. Structural basis of the Cks1-dependent recognition of p27(Kip1) by the SCF(Skp2) ubiquitin ligase. Mol Cell. 2005;20:9–19. doi: 10.1016/j.molcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Harmon FG, Kay SA. The F box protein AFR is a positive regulator of phytochrome A-mediated light signaling. Curr Biol. 2003;13:2091–2096. doi: 10.1016/j.cub.2003.11.019. [DOI] [PubMed] [Google Scholar]
- He Q, Cheng P, Yang Y, Yu H, Liu Y. FWD1-mediated degradation of FREQUENCY in Neurospora establishes a conserved mechanism for circadian clock regulation. Embo J. 2003;22:4421–4430. doi: 10.1093/emboj/cdg425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imaizumi T, Schultz TF, Harmon FG, Ho LA, Kay SA. FKF1 F-box protein mediates cyclic degradation of a repressor of CONSTANS in Arabidopsis. Science. 2005;309:293–297. doi: 10.1126/science.1110586. [DOI] [PubMed] [Google Scholar]
- Jin J, Cardozo T, Lovering RC, Elledge SJ, Pagano M, Harper JW. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004;18:2573–2580. doi: 10.1101/gad.1255304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TD, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. Positional cloning of the mouse circadian Clock gene. Cell. 1997;89:641–653. doi: 10.1016/s0092-8674(00)80245-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature. 2002;420:673–678. doi: 10.1038/nature01272. [DOI] [PubMed] [Google Scholar]
- Koh K, Zheng X, Sehgal A. JETLAG resets the Drosophila circadian clock by promoting light-induced degradation of TIMELESS. Science. 2006;312:1809–1812. doi: 10.1126/science.1124951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume K, Zylka MJ, Sriram S, Shearman LP, Weaver DR, Jin X, Maywood ES, Hastings MH, Reppert SM. mCRY1 and mCRY2 are essential components of the negative limb of the circadian clock feedback loop. Cell. 1999;98:193–205. doi: 10.1016/s0092-8674(00)81014-4. [DOI] [PubMed] [Google Scholar]
- Kwon I, Lee J, Chang SH, Jung NC, Lee BJ, Son GH, Kim K, Lee KH. BMAL1 shuttling controls transactivation and degradation of the CLOCK/BMAL1 heterodimer. Mol Cell Biol. 2006;26:7318–7330. doi: 10.1128/MCB.00337-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C, Etchegaray JP, Cagampang FR, Loudon AS, Reppert SM. Posttranslational mechanisms regulate the mammalian circadian clock. Cell. 2001;107:855–867. doi: 10.1016/s0092-8674(01)00610-9. [DOI] [PubMed] [Google Scholar]
- Leloup JC, Goldbeter A. Modeling the mammalian circadian clock: sensitivity analysis and multiplicity of oscillatory mechanisms. J Theor Biol. 2004;230:541–562. doi: 10.1016/j.jtbi.2004.04.040. [DOI] [PubMed] [Google Scholar]
- Lowrey PL, Shimomura K, Antoch MP, Yamazaki S, Zemenides PD, Ralph MR, Menaker M, Takahashi JS. Positional syntenic cloning and functional characterization of the mammalian circadian mutation tau. Science. 2000;288:483–492. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowrey PL, Takahashi JS. Mammalian circadian biology: elucidating genome-wide levels of temporal organization. Annu Rev Genomics Hum Genet. 2004;5:407–441. doi: 10.1146/annurev.genom.5.061903.175925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mas P, Kim WY, Somers DE, Kay SA. Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature. 2003;426:567–570. doi: 10.1038/nature02163. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Saini C, Bauer C, Laroche T, Naef F, Schibler U. Circadian gene expression in individual fibroblasts: cell-autonomous and self-sustained oscillators pass time to daughter cells. Cell. 2004;119:693–705. doi: 10.1016/j.cell.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Peterson KA, King BL, Hagge-Greenberg A, Roix JJ, Bult CJ, O’Brien TP. Functional and comparative genomic analysis of the piebald deletion region of mouse chromosome 14. Genomics. 2002;80:172–184. doi: 10.1006/geno.2002.6818. [DOI] [PubMed] [Google Scholar]
- Preitner N, Damiola F, Lopez-Molina L, Zakany J, Duboule D, Albrecht U, Schibler U. The orphan nuclear receptor REV-ERBalpha controls circadian transcription within the positive limb of the mammalian circadian oscillator. Cell. 2002;110:251–260. doi: 10.1016/s0092-8674(02)00825-5. [DOI] [PubMed] [Google Scholar]
- Reppert SM, Weaver DR. Coordination of circadian timing in mammals. Nature. 2002;418:935–941. doi: 10.1038/nature00965. [DOI] [PubMed] [Google Scholar]
- Ripperger JA, Schibler U. Rhythmic CLOCK-BMAL1 binding to multiple E-box motifs drives circadian Dbp transcription and chromatin transitions. Nat Genet. 2006;38:369–374. doi: 10.1038/ng1738. [DOI] [PubMed] [Google Scholar]
- Rost B, Yachdav G, Liu J. The PredictProtein server. Nucleic Acids Res. 2004;32:W321–326. doi: 10.1093/nar/gkh377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato TK, Panda S, Miraglia LJ, Reyes TM, Rudic RD, McNamara P, Naik KA, FitzGerald GA, Kay SA, Hogenesch JB. A functional genomics strategy reveals Rora as a component of the mammalian circadian clock. Neuron. 2004;43:527–537. doi: 10.1016/j.neuron.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Shimomura K, Low-Zeddies SS, King DP, Steeves TD, Whiteley A, Kushla J, Zemenides PD, Lin A, Vitaterna MH, Churchill GA, et al. Genome-wide epistatic interaction analysis reveals complex genetic determinants of circadian behavior in mice. Genome Res. 2001;11:959–980. doi: 10.1101/gr.171601. [DOI] [PubMed] [Google Scholar]
- Shirogane T, Jin J, Ang XL, Harper JW. SCFbeta-TRCP controls clock-dependent transcription via casein kinase 1-dependent degradation of the mammalian period-1 (Per1) protein. J Biol Chem. 2005;280:26863–26872. doi: 10.1074/jbc.M502862200. [DOI] [PubMed] [Google Scholar]
- Siepka SM, Takahashi JS. Forward genetic screens to identify circadian rhythm mutants in mice. Methods Enzymol. 2005a;393:219–229. doi: 10.1016/S0076-6879(05)93007-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siepka SM, Takahashi JS. Methods to record circadian rhythm wheel running activity in mice. Methods Enzymol. 2005b;393:230–239. doi: 10.1016/S0076-6879(05)93008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers DE, Schultz TF, Milnamow M, Kay SA. ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell. 2000;101:319–329. doi: 10.1016/s0092-8674(00)80841-7. [DOI] [PubMed] [Google Scholar]
- Takahashi JS. Finding new clock components: past and future. J Biol Rhythms. 2004;19:339–347. doi: 10.1177/0748730404269151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS, Pinto LH, Vitaterna MH. Forward and reverse genetic approaches to behavior in the mouse. Science. 1994;264:1724–1733. doi: 10.1126/science.8209253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, King DP, Chang AM, Kornhauser JM, Lowrey PL, McDonald JD, Dove WF, Pinto LH, Turek FW, Takahashi JS. Mutagenesis and mapping of a mouse gene, Clock, essential for circadian behavior. Science. 1994;264:719–725. doi: 10.1126/science.8171325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaterna MH, Pinto LH, Takahashi JS. Large-scale mutagenesis and phenotypic screens for the nervous system and behavior in mice. Trends Neurosci. 2006;29:233–240. doi: 10.1016/j.tins.2006.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsbacher LD, Yamazaki S, Herzog ED, Song EJ, Radcliffe LA, Abe M, Block G, Spitznagel E, Menaker M, Takahashi JS. Photic and circadian expression of luciferase in mPeriod1-luc transgenic mice in vivo. Proc Natl Acad Sci U S A. 2002;99:489–494. doi: 10.1073/pnas.012248599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Ko CH, Lowrey PL, Buhr ED, Song EJ, Chang S, Yoo OJ, Yamazaki S, Lee C, Takahashi JS. A noncanonical E-box enhancer drives mouse Period2 circadian oscillations in vivo. Proc Natl Acad Sci U S A. 2005;102:2608–2613. doi: 10.1073/pnas.0409763102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SH, Yamazaki S, Lowrey PL, Shimomura K, Ko CH, Buhr ED, Siepka SM, Hong HK, Oh WJ, Yoo OJ, et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc Natl Acad Sci U S A. 2004;101:5339–5346. doi: 10.1073/pnas.0308709101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young MW, Kay SA. Time zones: a comparative genetics of circadian clocks. Nat Rev Genet. 2001;2:702–715. doi: 10.1038/35088576. [DOI] [PubMed] [Google Scholar]
- Zheng N, Schulman BA, Song L, Miller JJ, Jeffrey PD, Wang P, Chu C, Koepp DM, Elledge SJ, Pagano M, et al. Structure of the Cul1-Rbx1-Skp1-F boxSkp2 SCF ubiquitin ligase complex. Nature. 2002;416:703–709. doi: 10.1038/416703a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.