Abstract
Mounting evidence indicates that miRNAs play important roles in the control of glial cell development in the central nervous system. Suppression of miRNA formation disrupts the initial generation of oligodendrocyte progenitor cells from the ventricular neuroprogenitor cells in embryonic spinal cord. miRNAs also regulate the later events of oligodendrocyte development including cell proliferation, maturation and myelin formation. In addition, miRNAs are essential for the development of astrocytes, and inhibition of miRNA genesis completely blocks astrogliogenesis in the spinal cord.
Keywords: Oligodendrocytes, astrocytes, spinal cord, miRNA, regulation
During early development, neurons and glia are sequentially produced from neural epithelial or progenitor cells in the ventricular zone throughout the entire central nervous system (CNS), with neurogenesis preceding gliogenesis (Miller 2002; Richardson and others 2006; Rowitch and others 2004). It has been well documented that different domains of progenitor cells generate distinct types of neurons and glia (oligodendrocytes or astrocytes) (Lu and others 2002; Takebayashi and others 2002; Zhou and Anderson 2002). In the developing spinal cord, neural epithelial cells in the motor neuron progenitor domain (pMN) produce motor neurons from E9.0 to E12.5 followed by oligodendrocyte progenitor cells (OPCs or OLPs) from E12.5 to E15.5 (Figure 1, Rowitch and others 2004). Other progenitor domains give rise to various types of interneurons followed by astroglia. During gliogenesis, early OPCs delaminate from the ventricular zone and subsequently undergo non-directional migration to populate all regions of the CNS before they differentiate into mature myelinating oligodendrocytes (Lu and others 2000; Zhou and Anderson 2000). In contrast, astroglia appear to reach their final destinations through radial migration and differentiate locally (Houchstim and others 2008).
Figure 1.
Progenitor domains and their progeny cells in the ventral spinal cord. Different progenitor domains express distinct combinations of transcription factors, and give rise to distinct neuronal and glial cell types during development. The motor neuron precursor (pMN) domain expresses Nkx6.1, Pax6 and Olig2, and produce motor neurons (MN) followed by oligodendrocytes (OL). The p1, p2, p3 and possibly other domains generate interneurons (V0-V3) followed by astrocytes (VA1, VA2, VA3).
miRNAs are essential for the initiation of oligodendrogenesis in the spinal cord
Although much has been known about the spatiotemporal events of neurogenesis and gliogenesis in the developing CNS, one remaining outstanding question in developmental neurobiology is how the binary switch from neurogenesis to gliogenesis is temporally controlled at the molecular level. The molecular pathways that instruct neural progenitor cells to turn off neurogenesis and at the same time to turn on gliogenesis remain to be identified. Our recent studies have demonstrated that microRNA (miRNA) molecules play a crucial role in the initiation of gliogenic process.
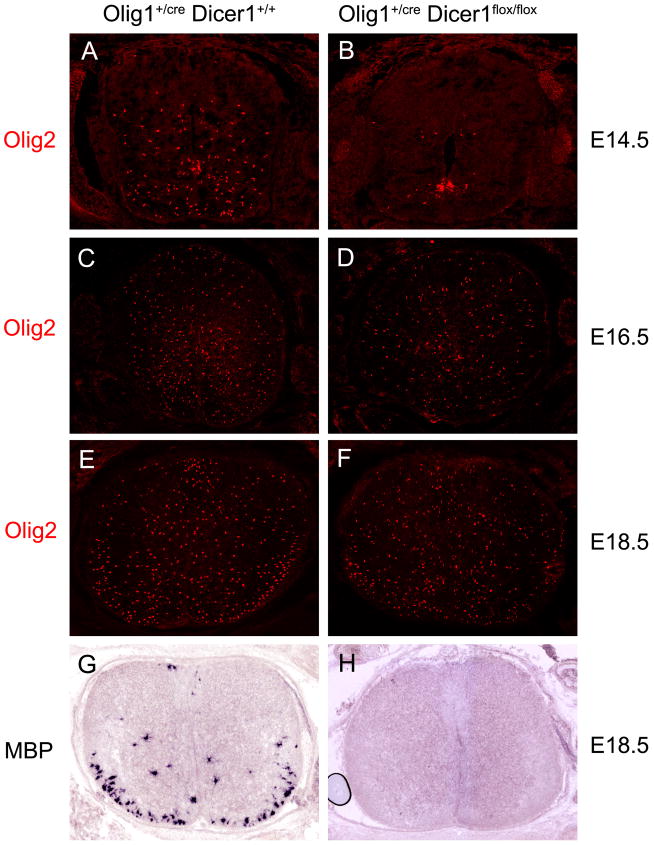
miRNAs are small non-coding RNA molecules that are processed from longer RNA precursors through cleavage by the ubiquitously expressed Dicer RNase (Carthew and Sontheimer 2009). They function as the negative regulators of post-transcriptional gene expression (Ouellet and others 2006) by binding to the 3′untranslational region of specific mRNA targets, directing their degradation and/or repressing their translation (He and Hannon 2004). Recent studies have demonstrated that miRNAs control lineage-specific development and functioning of various cell types in both animals and plants (Banerjee and Slack 2002; De Pietri Tonelli and others 2008). In the developing central nervous system, miRNAs display abundant and selective expression in undifferentiated neural progenitor cells in ventricular zone and their progeny cells (neurons and glia) (Cao and others 2007; Farrell and others 2010), raising the possibility that miRNA-regulated gene expression participates in the early fate specification and differentiation of neurons and glia. This possibility has been recently examined in Olig1creDicerflox/flox conditional mutants in which the Dicer gene (aka, miRNAs) was selectively ablated in the Olig1-expressing ventral spinal neuroepithelium including the pMN and p3 domains (Fig 1). In these conditional mutants, expression of several neural progenitor genes including Olig2 in this region is normal and motor neuron production is not affected, indicating that miRNAs are not critical for the early neural patterning and neurogeneis in the developing spinal cord (Zheng and others 2010). However, generation of early OPC cells from the pMN domain during early gliogenic stages (E12.5 to E14.5) was nearly completely blocked (Fig 2A–B) (Zheng and others 2010). More recent follow-up studies demonstrate that at later stages, a small number of OPCs are produced from the dorsal region of the mutant spinal cord (Fig 2C–D), and these dorsally derived OPC cells continue to proliferate for an expansion of progenitor cells (Cai and others 2005; Valstedt and others 2005). By the time of birth, the number of OPCs in the mutants is comparable to that in the normal embryos (Fig 2E–F). However, expression of mature oligodendrocyte markers MBP and PLP is not detected in the Dicer mutants (Fig 2G–H). The lack of MBP/PLP expression is likely to be secondary to the defective OPC generation, as the differentiation defect is also observed in other genetic mutants (e.g. Nkx6−/−, Gli2−/−) with retarded OPC generation from the pMN domain (Qi and others 2001; Liu and others 2003; Cai and others 2005). Together, these results indicate that miRNA formation is essential for the initiation of OPC generation in the ventral spinal cord. Intriguingly, the inhibition of oligodendrogenesis is not associated with the extension of motor neuron generation (no supernumerary motor neurons is detected in the Dicer mutants), suggesting that neurogenesis and gliogenesis can be uncoupled and may be controlled by distinct molecular mechanisms.
Figure 2.
Inhibition of oligodendrogenesis in Olig1cre/Dicer flox/flox mutant spinal cord. A–F. Transverse spinal cord sections from E14.5 (A–B), E16.5 (C–D) and E18.5 (E–F) normal and Olig1Cre/+/Dicer flox/flox embryos were stained with Olig2 antibody. Generation of ventral OPCs, but not dorsal OPCs, was inhibited in the mutant spinal cords. G–H. Spinal cord tissues from E18.5 wild-type and mutant spinal cords were subjected to in situ hybridization with MBP riboprobe. MBP expression was absent in the mutant tissue.
miRNAs regulate oligodendrocyte proliferation, differentiation and myelin maintenance in the CNS
As OPCs migrate away from the ventricular zone into the surrounding regions, they continue to proliferate in response to the endogenous mitogen PDGF-A (Calver and others 1998; Fruttiger and others 1999). The proliferation of OPCs appears to be modulated by miRNAs as well. MiR-219 negatively regulates the expression of PDGFRα in OPCs, and inhibition of miRNA formation in Dicer mutants leads to increased OPC numbers (Dugas and others 2010; Zhao and others 2010). In contrast, miR-17-92 cluster enhances OPC proliferation, and ablation of this miRNA cluster in OPCs results in a modest decrease in the number of OPCs (Budde and others 2010). The modulation of OPC division by miR17-92 cluster is mediated by activation of the PTEN/Akt pathway. As the cluster contains four miRNAs, it is not known yet which specific miRNA is directly involved in the regulation of OPC proliferation.
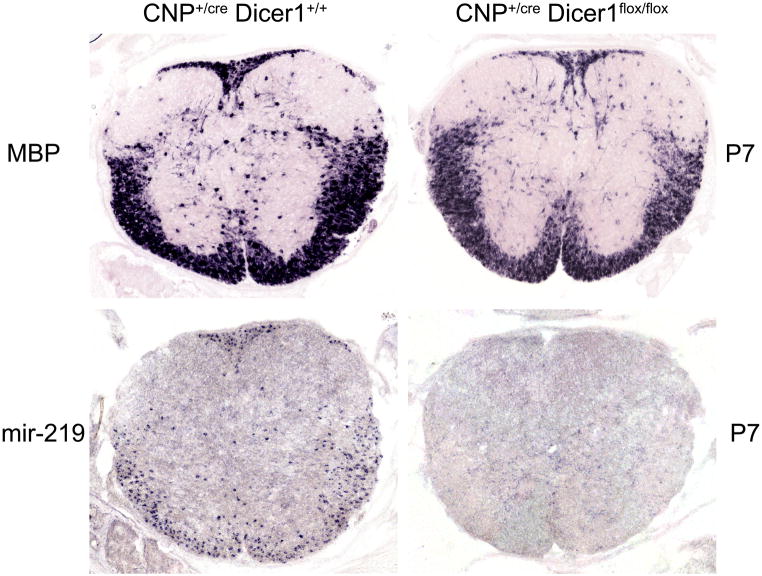
After OPCs migrate to the white matter region and adhere to axons, they start to undergo a series of morphological and molecular changes to become mature myelinating OLs. Two recent studies demonstrated that expression of several miRNAs, most notably miR-219 and miR-308, is selectively up-regulated in differentiating white matter OPCs (Dugas and others 2010; Zhao and others 2010). Over-expression of these miRNAs in OPC cells promotes their differentiation in culture. Conversely, inhibition of the function of these miRNAs suppressed OPC differentiation and maturation in culture. These miRNAs seem to regulate oligodendrocyte maturation by repressing the expression of inhibitory factors such as Hes5, Id2/4, Sox6 and PDGFRα. Consistently, ablation of Dicer in oligodendrocytes with Olig1Cre or Olig2Cre results in a dramatic reduction of oligodendrocyte differentiation and myelin formation (Dugas and others 2010; Zhao and others 2010). However, in view of the requirement of miRNAs for the initial generation of OPC cells (Fig 2), it is possible that the dramatic phenotypes observed in Olig1CreDicerflox/flox and Olig2CreDicerflox/flox might be attributed to the delayed and/or reduced OPC generation as well. In support of this notion, conditional disruption of Dicer in OPC cells the CNPCreDicerflox/flox mutants only have mild effect on oligodendrocyte differentiation (Dugas and others 2010; Fig 3), even though the expression of miR-219 is not detected in differentiating OPCs (Fig 3). Therefore, it remains plausible that miRNAs may fine-tune the timing of oligodendrocyte differentiation during normal development, in keeping with the previous observations that miR-219 target genes Hes5 and Sox6 regulate the timing of oligodendrocyte maturation (Kondo and others 2000) and inactivation of these genes leads to premature oligodendrocyte maturation (Liu and others 2006; Stolt and others 2006). Genetic mutations of these maturation-specific miRNAs will further define their roles in the control of oligodendrocyte differentiation and myelination.
Figure 3.
Dicer ablation in OPCs slightly inhibited oligodendrocyte differentiation in the spinal cord. Cross sections of spinal cord tissues from control and CNPcre/Dicer flox/flox mutant pups were subjected to in situ hybridization with miR-219 (A–B) and MBP (C–D) riboprobes. Expression of miR-219 was completely inhibited but MBP expression was only slightly reduced.
In addition, miRNA expression in mature oligodendrocytes is also required for the proper maintenance of myelin composition and structure. Conditional deletion of Dicer in mature myelinating oligodendrocytes using PLP-CreERT results in a dysregulation of the expression of miR-219 target gene Elov7 that is involved in lipid homeostasis, leading to increased demyelination and inflammatory neuronal damage (Shin and others 2009).
miRNAs are required for astrogliogenesis in the spinal cord
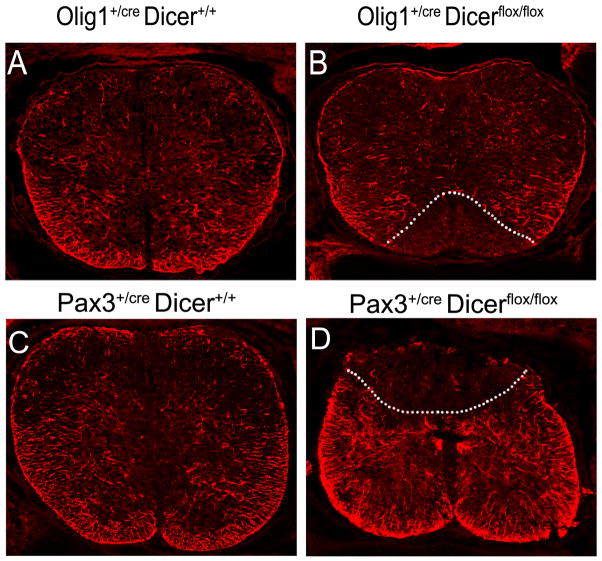
In the developing spinal cord, astrocytes are produced from p1, p2, p3 and possibly other domains as well (Fig 1; Houchstim and others 2008). In Olig1Cre/+ Dicerflox/floxanimals, expression of three general astrocyte markers (GFAP, ID3 and S100-β) was completely absent in a triangular region immediately flanking the floor plate (Fig 4A–B; Zheng and others 2010), suggesting the development of astrocytes derived from the ventral-most p3 domain was completely inhibited. Consistently, expression of VA3 astrocyte (a subset of astrocytes derived from p3 domain) marker Slit1 (Houchstim and others 2008) was also suppressed in the mutant spinal cord (Zheng and others 2010). Similarly, conditional ablation of Dicer from the dorsal neuroepithelium in Pax3cre Dicerflox/flox mutants selectively eliminates the dorsal expression of astrocyte marker GFAP (Fig 4C–D). Together, these results clearly indicate that miRNAs play an essential role in astrogliogenesis in the spinal cord, and provide further evidence for the radial migration and distribution of astroglia in the spinal cord. However, due to the lack of early markers for astrocyte lineage, it is difficult to determine whether disruption of miRNA formation inhibits the initial generation or the terminal differentiation of astroglial cells. Also, it would be interesting and important to identify the specific miRNAs and their target genes responsible for the development of astrocytes in the spinal cords.
Figure 4.
miRNA formation is required for astrogliogenesis in the spinal cord. Transverse spinal cord sections at the thoracic level from E18.5 wild-type, Olig1cre/Dicer flox/flox (A–B) and Pax3cre/Dicer flox/flox (C–D) mutant embryos were immunostained with anti-GFAP. The regions devoid of GFAP signal in the mutants are outlined by dashed lines.
Conclusion
miRNAs have been implicated in governing various aspects of cell development in plants and animals. Thus, it is not surprising to find that miRNAs control the development of glial cells at multiple stages including fate specification, cell proliferation, terminal differentiation, myelin homeostasis and maintenance. As the roles of miRNAs in glial genesis are gradually being unraveled, there are still many important issues that remain to be addressed: (1). Although miRNAs are known to be essential for the initiation of gliogenesis, it is not clear what specific miRNA species are involved in the commencement of early gliogenic process; (2) miRNAs are indispensable for the onset of both oligodendrogenesis and astrogliogenesis in the spinal cord, but it remains to be investigated whether miRNAs are similarly required for gliogenesis in the forebrain and other CNS regions; (3) What signals or factors control the selective and dynamic expression of miRNA molecules in cells of glial lineage during development? Answering these questions will provide important insights into the molecular control of glial lineage progression and possibly the strategic design for promoting cell replacement and repair in CNS injuries and diseases.
Acknowledgments
This work is supported by CNSF (31071879, 31000945), NIH (NS37717), NMSS (RG 3275) and Kentucky Spinal Cord and Head Injury Trust.
References
- Banerjee D, Slack F. Control of developmental timing by small temporal RNAs: a paradigm for RNA-mediated regulation of gene expression. Bioessays. 2002;24:119–29. doi: 10.1002/bies.10046. [DOI] [PubMed] [Google Scholar]
- Briscoe J, Pierani A, Jessell T, Ericson J. A homeodomain protein code specifies progenitor cell identity and neuronal fate in the ventral neural tube. Cell. 2000;101:435–45. doi: 10.1016/s0092-8674(00)80853-3. [DOI] [PubMed] [Google Scholar]
- Budde H, Schmitt S, Fitzner D, Opitz L, Salinas-Riester G, Simons M. Control of oligodendroglial cell number by the miR-17-92 cluster. Development. 2010;137(13):2127–32. doi: 10.1242/dev.050633. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45(1):41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Calver AR, Hall AC, Yu WP, Walsh FS, Heath JK, Betsholtz C, Richardson WD. Oligodendrocyte population dynamics and the role of PDGF in vivo. Neuron. 1998;20(5):869–82. doi: 10.1016/s0896-6273(00)80469-9. [DOI] [PubMed] [Google Scholar]
- Cao XW, Pfaff SL, Gage FH. A functional study of miR-124 in the developing neural tube. Genes Dev. 2007;21:531–6. doi: 10.1101/gad.1519207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carthew R, Sontheimer E. Origins and Mechanisms of miRNA and siRNAs. Cell. 2009;136:642–55. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Pietri Tonelli D, Pulvers J, Haffner C, Murchison E, Hannon G, Huttner W. miRNAs are essential for survival and differentiation of newborn neurons but not for expansion of neural progenitors during early neurogenesis in the mouse embryonic neocortex. J Neurosci. 2008;135:3911–21. doi: 10.1242/dev.025080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugas JC, Cuellar TL, Scholze A, Ason B, Ibrahim A, Emery B, Zamanian JL, Foo LC, McManus MT, Barres BA. Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron. 2010;65:597–611. doi: 10.1016/j.neuron.2010.01.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell BC, Power EM, Dermott KW. Developmentally regulated expression of Sox9 and microRNAs 124, 128 and 23 in neuroepithelial stem cells in the developing spinal cord. Int J Dev Neurosci. 2010;29(1):31–6. doi: 10.1016/j.ijdevneu.2010.10.001. [DOI] [PubMed] [Google Scholar]
- Fruttiger M, Karlsson L, Hall AC, Abramsson A, Calver AR, Boström H, Willetts K, Bertold CH, Heath JK, Betsholtz C, Richardson WD. Defective oligodendrocyte development and severe hypomyelination in PDGF-A knockout mice. Development. 1999;126(3):457–67. doi: 10.1242/dev.126.3.457. [DOI] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5:522–31. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Houchstim C, Deneen B, Lukaszewicz A, Zhou Q, Anderson D. Identification of positionally distinct astrocyte subtypes whose identities are specified by a homeodomain code. Cell. 2008;133:510–22. doi: 10.1016/j.cell.2008.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J. 2000;19(9):1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–9. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25(20):4833–42. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R, Cai J, Hu X, Tan M, Qi Y, German M, Rubenstein J, Sander M, Qiu M. Region-specific and stage-dependent regulation of Olig gene expression and oligodendrogenesis by Nkx6.1 homeodomain transcription factor. Development. 2003;130(25):6221–31. doi: 10.1242/dev.00868. [DOI] [PubMed] [Google Scholar]
- Lu Q, Yuk D, Alberta J, Zhu Z, Pawlitsky I, Chan J, McMahon A, Stiles C, Rowitch D. Sonic Hedgehog-regulated oligodendrocyte lineage genes encoding bHLH proteins in the mammalian central nervous system. Neuron. 2000;25:317–29. doi: 10.1016/s0896-6273(00)80897-1. [DOI] [PubMed] [Google Scholar]
- Lu Q, Sun T, Zhu Z, Ma N, Garcia M, Stiles C, Rowitch D. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog Neurobiol. 2002;67:451–67. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Ouellet DL, Perron MP, Gobeil LA, Plante P, Provost P. MicroRNAs in gene regulation: when the smallest governs it all. J Biomed Biotechnol. 2006;2006(69616):1–20. doi: 10.1155/JBB/2006/69616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Cai J, Wu Y, Wu R, Lee J, Fu H, Rao M, Sussel L, Rubenstein J, Qiu M. Control of oligodendrocyte differentiation by the Nkx2.2 homeodomain transcription factor. Development. 2001;128 :2723–33. doi: 10.1242/dev.128.14.2723. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–8. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowitch D. Glial specification in the vertebrate neural tube. Nat Rev Neurosci. 2004;5:409–19. doi: 10.1038/nrn1389. [DOI] [PubMed] [Google Scholar]
- Shin D, Shin JY, McManus MT, Ptácek LJ, Fu YH. Dicer ablation in oligodendrocytes provokes neuronal impairment in mice. Ann Neurol. 2009;66(6):843–57. doi: 10.1002/ana.21927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolt CC, Schlierf A, Lommes P, Hillgärtner S, Werner T, Kosian T, Sock E, Kessaris N, Richardson WD, Lefebvre V, Wegner M. SoxD proteins influence multiple stages of oligodendrocyte development and modulate SoxE protein function. Dev Cell. 2006;11:697–709. doi: 10.1016/j.devcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–63. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45(1):55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Zhao X, He X, Han X, Yu Y, Ye F, Chen Y, Hoang T, Xu X, Mi QS, Xin M, Wang F, Appel B, Lu QR. MicroRNA-mediated control of oligodendrocyte differentiation. Neuron. 2010;65:612–26. doi: 10.1016/j.neuron.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng K, Li H, Zhu Y, Zhu Q, Qiu M. MicroRNAs are essential for the developmental switch from neurogenesis to gliogenesis in the developing spinal cord. J Neurosci. 2010;30(24):8245–50. doi: 10.1523/JNEUROSCI.1169-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Wang S, Anderson D. Identification of a novel family of oligodendrocyte lineage-specific basic helix-loop-helix transcription factors. Neuron. 2000;25:331–43. doi: 10.1016/s0896-6273(00)80898-3. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Anderson D. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]