Abstract
Hypoxia, or low oxygen tension, is a unique environmental stress that induces global changes in a complex regulatory network of transcription factors and signaling proteins in order to coordinate cellular adaptations in metabolism, proliferation, DNA repair, and apoptosis. Several lines of evidence now establish microRNAs (miRNAs), which are short non-coding RNAs that regulate gene expression through post-transcriptional mechanisms, as key elements in this response to hypoxia. Oxygen deprivation induces a distinct shift in a specific group of miRNAs, termed hypoxamirs, and emerging evidence indicates that hypoxia regulates several facets of hypoxamir transcription, maturation, and function. Transcription factors such as hypoxia-inducible factor (HIF) are upregulated under conditions of low oxygen availability and directly activate the transcription of a subset of hypoxamirs. Conversely, hypoxia selectively represses other hypoxamirs through less well characterized mechanisms. In addition, oxygen deprivation has been directly implicated in epigenetic modifications such as DNA demethylation that control specific miRNA transcription. Finally, hypoxia also modulates the activity of key proteins that control posttranscriptional events in the maturation and activity of miRNAs. Collectively, these findings establish hypoxia as an important proximal regulator of miRNA biogenesis and function. It will be important for future studies to address the relative contributions of transcriptional and posttranscriptional events in the regulation of specific hypoxamirs and how such miRNAs are coordinated order to integrate into the complex hierarchical regulatory network induced by hypoxia.
Keywords: Hypoxia, MicroRNA, Hypoxamir, HIF
Introduction
The processes that govern cellular adaptation to hypoxia, or low oxygen availability, are complex and incompletely defined. Recently, several lines of evidence directly implicate microRNAs (miRNAs), which are short non-coding RNAs that regulate gene expression through posttranscriptional mechanisms, in these molecular events [1–3]. Approximately 1–2% of the transcriptome in eukaryotic organisms consists of miRNAs [4, 5]. They coordinate complex posttranscriptional regulatory events relevant to a variety of fundamental cellular processes, including proliferation, apoptosis, and differentiation. Mature miRNAs are approximately 18–23 nucleotides (nt) in length and control gene expression through degradation and repression of translation of primary messenger RNA (mRNA) [6].
Hypoxic stress regulates the expression of an expanding but specific subset of miRNAs, termed hypoxamirs [3, 7]. Although the specific miRNA hypoxic signature can vary based on the cellular or physiological scenario, a core group of hypoxamirs appears to be modulated consistently by hypoxia in diverse contexts [2] (Table 1). Among these, multiple hypoxamirs directly target important gene transcripts that coordinate metabolic reprogramming, DNA repair, apoptosis, and angiogenesis, among many other cellular adaptations to low oxygen availability [8–11].
Table 1.
Verified Hypoxamirs in Mammalian Cells
| Upregulated by Hypoxia | Downregulated by Hypoxia |
|---|---|
|
| |
| Let-7b,e,I [33] | Let-7a,c,d,f [33] |
| miR-7 [105] | miR-15b [105] |
| miR-21 [2] | miR-16 [33] |
| miR-23a,b [2] | miR-19a [33] |
| miR-24 [2] | miR-20a,b [33] |
| miR-26a,b [33] | miR-29b [105] |
| miR-27a [105] | miR-30e [105] |
| miR-30b [33] | miR-92 [106] |
| miR-93 [2] | miR-101 [105] |
| miR-98 [105] | miR-122a [105] |
| miR-103 [2] | miR-135a [110] |
| miR-106a [2] | miR-141 [105] |
| miR-125b [2] | miR-186 [2] |
| miR-130 [106] | miR-197 [105] |
| miR-146a,b [8] | miR-199a [111] |
| miR-148a,b [105] | miR-200b [112] |
| miR-151 [33] | miR-224 [33] |
| miR-181a,b,c [2, 33] | miR-320 [105] |
| miR-188 [33] | miR-374 [33] |
| miR-191 [105] | miR-422b [105] |
| miR-192 [2] | miR-449a,b [113] |
| miR-195 [105] | miR- 565 [105] |
| miR-199a-5p [107] | |
| miR-204 [108] | |
| miR-205 [109] | |
| miR-210 [2, 33, 87] | |
| miR-213 [2] | |
| miR-335 [109] | |
| miR-373 [105] | |
| miR-424 [11, 104] | |
| miR-429 [105] | |
| miR-451 [95] | |
| miR-491 [109] | |
| miR-498 [105] | |
| miR-563 [105] | |
| miR-572 [105] | |
| miR-628 [105] | |
| miR-637 [105] | |
Hypoxic insults can be acute, transient, and/or localized, and miRNAs are uniquely suited to participate in the rapid, adaptive responses to oxygen deprivation. The post-transcriptional regulatory mechanisms employed by miRNAs facilitate an expedient, precise, and quickly reversible fine-tuning of the cellular response to environmental stress. In addition, the network of hypoxamirs and their direct targets offer the potential for a dynamic and robust biological response to hypoxia through the coordinated regulation of host genes and miRNA clusters, with each miRNA potentially controlling multiple and often functionally related mRNA transcripts. Thus, in order to maintain such exquisite coordination of these adaptive programs, important molecular crosstalk has evolved between hypoxic signaling cascades and miRNA biogenesis and function. In this review, we will summarize our current understanding of such hypoxia-mediated regulation of miRNA expression via alterations in miRNA biogenesis, maturation, processing, and potentially degradation.
Cellular Adaptations to Hypoxia
Hypoxia evokes stereotyped and highly coordinated cellular responses in the acute setting in order to preserve cell viability [12]. These adaptive responses are orchestrated by a variety of global molecular regulators, including the hypoxia inducible factors (HIF), a “master” regulator of the hypoxic response [13]. HIF is a heterodimeric transcription factor (TF) consisting of HIFα and HIFβ subunits. Unlike HIFβ, which is stable regardless of cellular oxygen tension, HIFα is rapidly degraded under normoxic conditions. The HIFα isoforms--HIF1α, HIF2α, and HIF3α splice variants--undergo efficient prolyl hydroxylation of an oxygen-dependent degradation domain (ODD) by prolyl hydroxylation domain (PHD) proteins. This posttranslational modification (PTM) leads to HIF1α binding to the von Hippel-Lindau tumor suppressor protein (VHL) and its subsequent ubiquitination and degradation by the 26S proteasome. In contrast, under hypoxic conditions, hydroxylation of proline residues in the ODD of HIFα is significantly diminished. As a result, HIFα accumulates in the cytosol and translocates to the nucleus where it forms a functional HIF heterodimer with HIFβ. The HIF heterodimeric complex then binds to consensus hypoxia responsive elements (HREs) in the promoters of a number of target genes to activate wide-scale gene programs that coordinate a switch to glycolysis, angiogenesis, erythropoeisis, and apoptosis. Therefore, HIFα fulfills roles as a critical oxygen sensor and regulator of the hypoxic adaptive response.
Recent evidence indicates HIF alone is insufficient to implement the full program of adaptive changes required for cell survival under hypoxic stress. Rather, HIF-independent pathways involving p53 [14–17], mTOR [18–20], endoplasmic reticulum (ER) stress, and the unfolded protein response (UPR) [21, 22] play important complementary roles that promote cell survival in conditions of low oxygen availability. These HIF-independent pathways facilitate important energy conservation and cell survival measures in the setting of low oxygen levels. Accumulating evidence indicates that miRNAs interface with both HIF-driven and HIF-independent pathways to form a highly interconnected regulatory network during hypoxic stress.
The Diverse Roles of miRNAs in Cellular Adaptation to Hypoxia
Although individual miRNAs mediate relatively modest inhibitory effects on protein translation via 30–50% reductions in target protein levels [23, 24], they collectively constitute an important component of the cellular response to hypoxia through combinatorial and coordinated regulation of key targets. An exhaustive discussion of the constantly expanding portfolio of hypoxamir functions is beyond the scope of this report. Yet, it is worth reviewing the broad themes that characterize their general actions in overall hypoxic reprogramming (Table 2), including the reinforcement of HIF adaptive responses, and the regulation of the levels of proteins involved in metabolism, cell cycle progression, DNA repair, apoptosis, and angiogenesis.
Table 2.
Summary of Prototypical Functions of Hypoxamirs in Hypoxic Adaptation
| Hypoxia-Regulated miRNA | Target mRNA | Regulatory Effect | References |
|---|---|---|---|
| Induced Hypoxamirs: | |||
| miR-21 | PDCD4 | Anti-apoptosis | [114] |
| SPRY2 | Cell proliferation, migration | [115] | |
| PPARα | Cell Proliferation, migration | [115] | |
| miR-23a | Fas Ligand | Anti-apoptosis | [116] |
| miR-26a | GSK-3β | Pro-apoptosis | [117] |
| miR-27a,b | SEMA6A | Angiogenesis | [118] |
| miR-130 | DDX6 | Enhanced translation of HIF-1α | [113] |
| miR-146a,b | TRAP6 IRAF1 |
Anti-inflammatory effects | [31] |
| miR-155 | HIF-1α | Resolution of HIF effects | [29] |
| miR-181c | TNFα | Anti-inflammatory/ anti-apoptotic effects | [32] |
| miR-204 | LC3-II | Inhibition of autophagy | |
| miR-210 | ISCU | [8, 9] | |
| NDUFA4 | Impairment of mitochondrial function | [120] | |
| SDHD | Impairment of mitochondrial function | [120] | |
| COX10 | Impairment of mitochondrial function | [121] | |
| EFNA3 | Impairment of mitochondrial function | [62] | |
| RAD52 | Inhibition of cell migration | [1] | |
| E2F3 | Inhibition of DNA repair | [30] | |
| GPD1L | Cell cycle arrest HIF-1α stabilization |
[26] | |
| miR-373 | RAD23B | Inhibition of DNA repair | [1] |
| miR-424 | CUL2 | HIF-1α stabilization | [11] |
| Repressed Hypoxamirs: | |||
| miR-15/16 | BCL2 | Anti-apoptosis | [122] |
| miR-34a | Notch | Epithelial-to-mesenchymal transition | [123] |
| miR-122 | PSMD10 | Promotes UPR | [124] |
| miR-135a | FLAP | Promotes leukotriene formation | [107] |
| miR-199a | HIF-1α | Derepression of HIF-1α | [111] |
| miR-200b | Ets-1 | Angiogenesis | [125] |
| miR-449a | SERPINE1 | Fibrosis | [110] |
A number of hypoxamirs promote HIF expression and/or activity [7] through positive feedback circuits that support HIF-dependent hypoxic adaptation [13, 25]. Some of these miRNAs are direct transcriptional targets of HIF itself during hypoxia. For example, the master hypoxamir miR-210 is potently induced by hypoxia in a HIF-dependent manner and stabilizes HIF-1α by targeting glycerol-3-dehydrogenase like 1 [26]. Hypoxia–dependent miRNAs also modulate HIFα at the transcriptional level. In hypoxic endothelial cells, induction of miR-424 expression stabilizes HIFα isoforms through targeting of cullin2 (CUL2) [11]. In addition, miRNAs that target the HIFα transcript, such as miR-20b and miR-199a, are dynamically repressed under conditions of hypoxia, thereby increasing HIFα expression and promoting HIF-mediated transcriptional activation [27, 28]. Recent work indicates that hypoxia-regulated miRNAs may also participate in negative feedback regulatory loops: for example, the HIF-dependent hypoxamir miR-155 is potently induced by hypoxia and may contribute to the resolution of HIF-effects in hypoxic intestinal epithelial cells by targeting the HIFα mRNA transcript [29].
Emerging data indicate that miRNAs also coordinate important adaptive responses to hypoxia downstream of HIF in a manner that is independent of, but synergistic with, its direct transcriptional effects. For example, HIF directly promotes the expression of metabolic enzymes such as pyruvate dehydrogenase 1 and lactate dehydrogenase A that support a shift toward glycolysis [25]. The HIF-induced hypoxamir miR-210 complements these effects by suppressing mitochondrial respiration by alternative mechanisms, such as targeting iron-sulfur cluster assembly and COX proteins [8, 9], among other targets that influence mitochondrial function (Table 2). MiR-210 also lowers cellular energy requirements by inducing growth arrest of hypoxic cells by targeting E2F transcription factor 3 (E2F3), a cell-cycle regulator that is not directly transcriptionally regulated by HIF [30]. Cells exposed to low levels of oxygen also limit DNA repair to conserve energy resources. Both miR-210 and miR-373, which is also a direct transcriptional target of HIF, restrict DNA repair responses by decreasing levels of DNA repair proteins, such as RAD52 and RAD23B [1].
Several miRNAs have also been implicated in modulating other important pro-survival cellular responses during hypoxia involving apoptosis and inflammation. For example, miR-146a,b are potently induced during hypoxia and have been shown to repress nuclear factor-kappa B (NF-κB)-mediated inflammatory responses by targeting TNF receptor-associated factor 6 (TRAF6) and IL1 receptor-associated kinase 1 (IRAK1) [31]. Similarly, miR-181c limits activation of inflammatory pathways and apoptosis in hypoxic neuronal cells by targeting the TNFα mRNA transcript [32]. In addition to controlling cell death, miR-181c may also contribute metabolic reprogramming.
During hypoxia, activation of angiogenic pathways seeks to restore a homeostatic balance between oxygen demand and supply. HIF directly targets vascular endothelial growth factor (VEGF), a master regulator of angiogenesis. In addition, miRNA networks downstream of HIF contribute to the regulation of VEGF and angiogenesis under hypoxic conditions. In a study examining miRNA-directed regulation of VEGF in nasopharyngeal carcinoma cells [33], a computational approach identified 96 miRNA that were predicted to target VEGF. A significantly smaller number of these miRNAs was selectively repressed under hypoxic conditions. When these miRNAs were experimentally studied, a complex pattern of regulation emerged. Co-transfection of miRNAs that do not share 3′UTR binding sites (miR-20a and 361) resulted in additive repressive effects on VEGF expression, while co-transfection of miRNAs with common binding sites (miR-20a and 106b) yielded the same or a slightly lower effect compared to individual transfections. These findings suggest that miRNAs likely function in a combinatorial and hierarchical fashion during hypoxic stress. As such, miRNAs represent important nodes in the regulatory networks that coordinate hypoxic reprogramming of cellular metabolism, growth, as well as DNA repair responses and angiogenesis.
Biogenesis of miRNAs
The generation of mature miRNAs is a complex, multistep process (Figure 1) that is regulated by hypoxia at virtually every level. We will, therefore, briefly review the transcription and processing of miRNAs to provide a conceptual framework for the emerging evidence on hypoxic regulation of miRNA expression and activity. The biogenesis of miRNA commences in the nucleus with the transcription of the primary microRNA (pri-miRNA) by RNA polymerase II or III [34, 35]. Pri-miRNAs are cleaved by the microprocessor complex, which consists of Drosha and the Di George Syndrome critical region 8 gene (DGCR8) protein [36] to form the precursor miRNA (pre-miRNA) [37–39]. Exportin 5 then translocates the pre-miRNA to the cytosol [40], where Dicer, cleaves pre-miRNAs to generate a double-stranded 22 nt intermediate miRNA moiety [41]. In the final step, the guide strand of the miRNA duplex, which contains the seed sequence, associates with Argonaute 2 (Ago2), Dicer, TRBP, and PACT proteins to form the miRNA-induced silencing complex (miRISC) [41, 42]. The miRISC then binds to the 3′ untranslated region (3′UTR) of the target mRNA and mediates sequence-specific gene silencing through mRNA destabilization and translational repression [43].
Figure 1. Overview of miRNA biogenesis and hypoxia-mediated regulation of miRNA generation and activity.
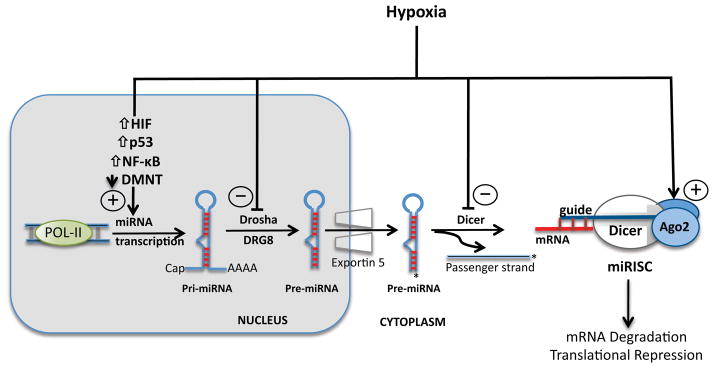
The generation of mature miRNAs is a multi-step process that begins with transcription of the primary miRNA transcripts (pri-miRNA) by Pol II in the nucleus. Following cleavage by the Drosha- DGCR8 complex, the precursor miRNA (pre-miRNA) is transported to the cytosol where it is cleaved by Dicer in preparation for loading of the guide strand onto the miRISC complex. Hypoxia regulates miRNA on multiple levels including transcription of the pri-miRNA, processing of precursor miRNA intermediates, and post-translational modifications of the miRISC complex.
Although the mechanisms that underlie degradation of miRNA species remain poorly understood, emerging data indicate that decay of miRNA may represent an additional level of regulation of miRNA activity [44]. The unprotected 5′and 3′ ends of miRNAs may render them susceptible to cleavage and degradation by exoribonucleases [44]. Several studies indicate that cells also actively secrete miRNAs in association with membrane vesicles (exososmes), apoptotic bodies, and protein complexes [45–47]. These extracellular miRNAs resist degradation and are present in the circulation as well as a number of body fluids, suggesting that they may play important functions in distant tissue sites [46]. Although a notion that has not been fully established, it is plausible that the active and selective secretion of miRNAs may also serve as an additional regulatory mechanism to control miRNA compartmentalization and function [46].
Regulation of miRNAs by Hypoxia
Experimental studies have identified several important mechanisms by which hypoxia regulates miRNA expression and activity (Figure 1). An early study examining protein levels of key enzymes that modulate miRNA maturation did not demonstrate any significant change in Ago2, Dicer, or Drosha in human trophoblasts under hypoxic conditions [48], interpreted by many to suggest that hypoxia regulates miRNA primarily at the transcriptional level. However, the use of primary human trophoblast cells and placental tissue in this study limited the relevance of these findings to a specific developmental context. In addition, this investigation examined a single time point of 48 hours and only focused on dynamic regulation of Ago2 and its interacting protein DP103. As such, although early studies on hypoxia-mediated regulation of miRNA largely focused on TFs that consistently and robustly control hypoxamir transcription, such as HIF [2], subsequent work quickly recognized that transcriptional control of miRNA expression comprises only part of the narrative of hypoxamir regulation. Hypoxia prompts specific changes in miRNA expression in a time frame that is too rapid to be explained solely by TF action. Additionally, the complement of hypoxamirs varies depending on the cellular context as well as degree and duration of the hypoxic insult [2, 49, 50]--observations that do not correlate entirely with hypoxia-dependent TF alterations. Emerging evidence now indicates that hypoxia modulates several other phases of miRNA biogenesis, maturation, and function. Here, we will discuss hypoxia-mediated regulation of miRNAs under broad classifications of transcriptional and non-transcriptional mechanisms (Table 3).
Table 3.
Summary of Transcriptional and Non-transcriptional Hypoxic Regulation of miRNA Biogenesis and Activity
| Hypoxia-Related Regulatory Factor | Hypoxamir | Regulatory Effect |
|---|---|---|
|
| ||
| Transcriptional Regulation: | ||
| A. Transcription Factors: | ||
| 1. HIF | ↑miR-210,-373,-181 [1] ↑miR-155 [29] |
Mitochondrial function, DNA repair, cell cycle |
| 2. NF-κB | ↑miR-210 [44] | Mitochondrial function, DNA repair, cell cycle |
| 3. p53 | ↑miR-210 [74] | Mitochondrial function, DNA repair, cell cycle |
| 4. PU.1 | ↑miR-474 [11] | HIF stabilization |
| 5. Akt2 | ↑miR-21[116] ↑miR-210 [74] |
Mitochondrial function, DNA repair, cell cycle |
| B. Epigenetic Modifications: | ||
| 1. DNA demethylation | ↑miR-210 [81] | Mitochondrial function, DNA repair, cell cycle |
| miRNA Processing/Maturation: | ||
| 1. Dicer/Drosha Inhibition | ↓ miR-21,-22,-30c, let7f [82, 83] | ? |
| 2. Ago2 | ↑miR-451 [84] | ? |
Transcriptional regulation of miRNAs by hypoxic stress
Our overall understanding of the transcriptional regulation of miRNA biogenesis is still evolving. Similar to mRNA, miRNA transcription is driven primarily by Pol-II, although recent evidence indicates that Pol-III maybe involved in the expression of a select subset of miRNAs [35]. The expression of miRNAs that reside within the exons or introns of coding genes usually depends on the promoter of the host gene; however, up to 1/3 of intronic miRNAs may have independent promoters [51]. The functional significance of these observations is borne out in studies that demonstrate a discordance between host gene mRNA and resident miRNA(s) expression patterns [49].
Intergenic miRNAs possess independent promoters that lie up to several kilobases (kb) upstream of the transcription start site [52]. Although relatively few miRNA promoters have been characterized experimentally, multiple studies examining global miRNA promoter architecture identify important similarities to promoters of protein-coding genes [52–54]. For instance, miRNA promoters identified by combining mapping of nucleosome occupancy and chromatin signatures with experimental validation demonstrated that intronic and intergenic miRNA promoters have significant sequence similarity to traditional protein coding gene promoters [53]. Not surprisingly, miRNA promoters are also replete with TF consensus binding sites, and bioinformatic approaches demonstrate a complex and diverse TF-miRNA network [55, 56].
Furthermore, TFs bind these consensus sequences and exert fundamental regulatory actions on miRNA transcription specifically in the context of cellular hypoxia. Given these observations, as well as the established role of TFs in adaptation to hypoxia, the transcriptional regulation of miRNAs by hypoxia-induced TFs is an active area of investigation. HIFs play a predominant role in the coordination of transcriptional changes during hypoxic stress [13], but several other TFs such as NF-κB, PU.1, and p53, also execute important regulatory roles in the context of oxygen deprivation [57]. Therefore, the transcriptional regulation of miRNA expression under hypoxic conditions can be broadly categorized as HIF-dependent and HIF-independent (Figure 2).
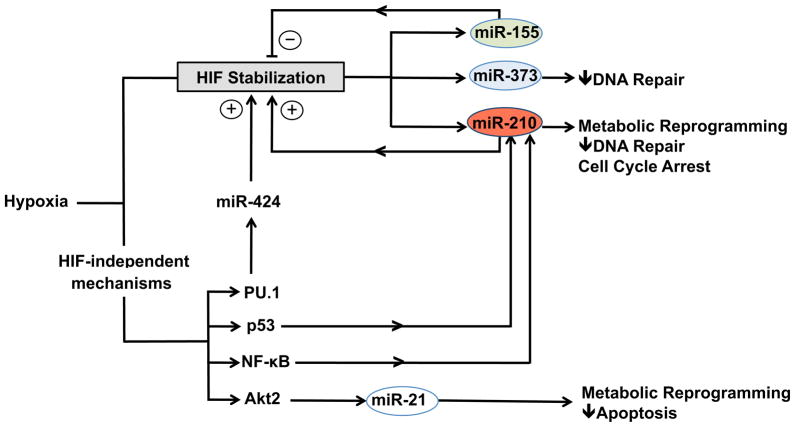
Figure 2. Summary of crosstalk between HIF and HIF-independent regulatory pathways and miRNAs in hypoxic cellular adaptation.
A complex hierachical regulatory network exists between transcription factors and miRNAs in cells exposed to hypoxia. HIF stabilization results in the induction of key miRNA targets such as miR-210 (a master hypoxamir), miR-373, and miR-155 which coordinate metabolic reprogramming, DNA repair, and apoptosis responses. HIF-independent transcriptional pathways involving p53 and NF-κB reinforce upregulation of miR-210 and other hypoxamirs such as miR-21.
Direct HIF regulation of hypoxamir transcription
Recent evidence indicates that HIF induces the transcription of multiple hypoxamirs through direct binding to HREs in their respective promoters [2, 49, 58]. In a study investigating the miRNA signature of hypoxia in multiple breast and colon cancer cell lines, a subset of 23 hypoxamirs was consistently induced greater than two-fold. In silico analysis of the putative promoter sequence of these hypoxamirs demonstrated a significant enrichment for the presence of HREs compared to the promoters of 23 randomly selected, unrelated miRNAs [2, 58]. Experimental validation was performed on a group of these hypoxamirs, including, miR-24–1, miR-26, miR-103, miR-181, miR-210, and miR-213, using genetic approaches, promoter-reporter assays, and chromatin immunoprecipitation (ChIP). Overexpression of stable HIF isoforms robustly induced expression of miR-103, miR-210, and miR-213. In addition, HIF-1α was noted to bind directly and transactivate HREs in the promoters of miR-24–1, miR-26, and miR-181. Finally, HIF is dynamically recruited to the promoters of miR-26 and miR-210. As such, by HRE recognition alone, HIF may exert multitiered effects on a highly interconnected network of genes and miRNAs in the context of hypoxia. However, the strongest data for direct involvement of HIF in the transcriptional regulation of miRNA under hypoxia only exists for miR-26 and miR-210 in this study. For the remainder of the validated miRNAs, data on HIF recruitment to and/or transactivation of respective miRNA promoters was not presented to support the direct involvement of HIF in their transcriptional regulation.
MiR-210 was one of the first hypoxamirs discovered as a direct transcriptional target of HIF [2], and its strikingly robust induction by hypoxia (up to 30–100-fold) across multiple mammalian cell types is unique among all HIF-dependent miRNAs [8]. In humans, the miR-210 gene is located within the intronic sequence of a hypoxia-related transcript AK123483 [59] and is also located in close proximity to 2 other genes, HRAS and RASSF7, which are induced during hypoxia [60]. Although not yet experimentally verified, it is possible that miR-210 participates in autoregulatory loops that involve its host gene based on prior observations of the behavior of other intronic miRNAs. Under normoxic conditions, miR-210 is expressed at low levels, especially in certain cell types such as endothelial cells. This transcription of miR-210 appears to be driven by the transcription factor AP-1 under basal conditions; however, given its relative low expression in this context, it is unclear whether miR-210 adequately engages its main targets under normoxia [61]. Upon exposure to hypoxia, miR-210 transcription is dynamically induced in all known mammalian cell types, primarily through HIF-1α interaction with the HRE located within its promoter [44, 58–60]. Similar studies of HIF-2a have demonstrated conflicting results with regard to its activity in upregulating miR-210 [44]. In turn, miR-210 exerts broad pleiotropic effects (Figure 2, Table 2) by targeting genes involved in RNA/DNA processing and repair, mitochondrial function, apoptosis, cell cycle progression, and cell survival [44, 61–63]. As previously discussed, miR-210 may also stabilize HIF1α by targeting glycerol-3-dehydrogenase like 1, an enzyme that hydroxylates HIFα and contributes to its degradation [26].
Beyond miR-210, a number of other HIF-regulated hypoxamirs have been studied in detail. During hypoxia, miR-373 is potently induced in a HIF-1α-dependent manner and targets the RAD23B transcript [1]. The RAD23B gene has been implicated in nucleotide excision repair and homology-dependent repair. Given that miR-210 also targets the transcript of a related gene RAD52 that is also involved in these pathways, miR-373 may participate in a hypoxamir regulatory network modulating DNA repair during hypoxic stress [1]. Interestingly, miR-21, which is induced by hypoxia, contains an HRE in its promoter [2], but is regulated independent of HIF under conditions of low oxygen tension [64]. Rather, induction of miR-21 during hypoxia occurs downstream of Akt2 activation (Figure 2) [65] and promotes resistance to hypoxia in primary immortalized lung fibroblasts and a number of mammary and ovarian cancer cell lines [66]. The hypoxamir miR-155 also possesses a functional HRE in its promoter and is potently induced by hypoxia in epithelial cells [29]. Unlike miR-210, which serves as a downstream effector of HIF, miR-155 directly targets HIF mRNA and contributes to resolution of HIF effects under conditions of chronic hypoxia.
Recent work has also elucidated an important convergence of HIF-miRNA pathways relevant to tumor biology and hypoxia. MiR-10b, a microRNA that has emerged as an important molecular determinant of tumor cell migration and invasion, has been recently linked to HIF-mediated transcriptional regulation through the corepressor protein CCN5. CCN5 modulates breast tumor cell migration and invasion in vitro through a HIF-1α-dependent mechanism involving the TWIST1 transcription factor [67]. MiR10b has also been implicated in tumor invasion and prognosis in the context of pancreatic adenocarcinoma [68].
HIF-independent transcriptional regulation of hypoxamirs
Emerging evidence also implicates a number of other TFs in hypoxic adaptive responses (Figure 2, Table 3) [57]. For example, hypoxia can potently induce expression of TP53, which encodes p53. The complex mechanisms underlying the increased levels of p53 in response to hypoxia remain incompletely understood; however, there appear to be HIF-dependent and HIF-independent effects [14]. Similar to HIF, p53 regulates the expression of both protein-coding genes as well as non-coding RNA transcripts. For instance, p53 directly induces several miRNAs implicated in tumor suppression and cell cycle arrest, including miR-34 [69], miR-15a [70], and miR-16-1 [70]. In addition, p53 controls the transcription of miR-200 subfamilies that modulate the epithelial-to-mesenchymal transition, a phenomena that is central to tumor metastasis [71, 72].
The p53-miRNA network has been more extensively examined in the context of oncogenesis and tumor biology [73]; however, recent work indicates that p53 may also regulate specific miRNA transcriptional events during hypoxia independent of HIF signaling (Figure 2, Table 3). Under hypoxic conditions, p53 accumulates in HIF-β knockout mouse embryonic fibroblasts (MEFs) that lack intact HIF signaling and directly induces miR-210. Furthermore, p53-mediated expression of miR-210 protects cardiomyocytes exposed to hypoxic stress [74]; however, robust induction of miR-210 by p53 in this study required deep hypoxia. At very low levels of oxygen tension (0.5% O2), miR-210 expression was induced approximately 25-fold compared to normoxic conditions. More modest levels of hypoxia (5% O2) induced miR-210 to a significantly lesser extent, approximately 8-fold lower than the changes observed in exposure to 0.5% O2 [74]. In addition to highlighting the apparent need for very low levels of oxygen tension for induction of hypoxamirs by p53, this study also emphasizes the complex interplay that exists between HIF and p53 during cellular adaptations to hypoxia. Although hypoxia potently increased miR-210 in HIF-β knockout MEFs, the induction of miR-210 by p53 appeared to be HIF-dependent. Notably, p53 induces other miRNAs, such as miR-107 and miR-192, during tumorigenesis that have also been implicated as hypoxamirs [73]. However, direct experimental evidence demonstrating a functional link between p53 and the induction of these miRNAs in the context of hypoxia is currently lacking.
Hypoxic stress can also activate NF-κB [57], a well-studied TF connected to inflammation that regulates an expanding network of miRNAs [75]. A systematic screening approach in human monocytes identified several NF-κB-regulated miRNAs, including miR-146a and miR-155, that have subsequently been shown to exert divergent effects on inflammation [31]. While miR-146a was implicated in a negative feedback loop that limits inflammation by targeting TNF receptor-associated factor 6 (TRAF6) [76], miR-155 facilitates downstream pro-inflammatory signaling [77]. In addition, other NF-κB-induced miRNAs such as miR-21 also possess promoter NF-κB binding sites and function to modulate inflammatory responses [78]. Recent work demonstrates that NF-κB may also exert important regulatory effects on the transcription of hypoxamirs. The miR-210 promoter contains a conserved NF-κB binding site approximately 200 bp upstream of the primary stem-loop structure in the pri-miRNA [44]. ChIP, as well as promoter-reporter assays and gene knockdown studies, indicate that NF-κB directly interacts with and transactivates the miR-210 promoter under hypoxic conditions [44]. The functional importance of HIF-independent transcriptional regulation of miRNAs under conditions of low oxygen availability is also underscored by the emergence of miR-424 as an important component of the response to hypoxic stress in endothelial cells [11]. Hypoxic endothelial cells activate transcription of miR-424 through HIF-independent mechanisms that involve the PU.1, runt-related transcription factor 1 (RUNX-1) and CCAAT/enhancer binding protein α (C/EBPα). The downstream effects of miR-424 center on stabilizing HIFα by targeting the CUL2 scaffolding protein of the ubiquitin ligase complex [11].
Given the high degree of structural similarity between promoters and enhancers of miRNA “genes” and protein-coding regions, epigenetic modifications are also emerging as an important transcriptional regulatory mechanism in miRNA biogenesis. Silencing of miRNAs through DNA methylation of CpG islands in promoter regions is already known to hold important pathobiological and prognostic implications in a wide spectrum of malignancies [79, 80]. Although data on epigenetic regulation of genomic regions that encode miRNAs in hypoxia are limited, a recent study demonstrated that hypoxic stress decreased DNA methyltransferase (DMNT) activity in neural progenitor cells [81]. This decline in DMNT activity correlated with demethylation of a CpG island associated with the miR-210 coding region and a corresponding increase in miR-210 expression (Figure 2). The molecular pathways driving alterations in DMNT activity and DNA methylation status have not been fully elucidated, and further work is needed to determine if hypoxia specifically regulates higher order chromatin structure through other mechanisms such as histone modifications.
Regulation of miRNA processing by hypoxia
Beyond transcriptional control, regulation of hypoxamir expression and function appears to exist at virtually every level of miRNA biogenesis and processing, including posttranscriptional modifications mediated by the microprocessor complex and miRISC stability and activity. First, hypoxia regulates processing complexes containing Drosha and Dicer to mediate dynamic changes in hypoxamir maturation and function (Figure 1). For example, a recent study examining regulation of miRNAs in an experimental animal model of hypoxia-induced pulmonary hypertension demonstrated that hypoxia modulates levels of key miRNA biogenesis proteins [82]. Exposure of rats to hypoxia for as little as 48 hours repressed the expression of Drosha and Dicer mRNA and protein levels in lung tissue samples. When primary pulmonary fibroblasts were subjected to corresponding hypoxic conditions in vitro, a similar decrease in Drosha mRNA was noted. The downstream consequences of these changes were reflected in distinct and dynamic shifts in miRNA expression under conditions of hypoxia. A small group of miRNAs, including miR-21, −22, −30c, and let7f, were downregulated in vitro and in vivo in this model. Interestingly, miR-21 has been shown to be induced during hypoxia by our group and others, likely through HIF-independent mechanisms that involve Akt2 activation [66]. However, without direct evidence of dynamic shifts in the levels of precursor forms of these miRNAs in response to low oxygen tension, it is difficult to link their reduced expression directly to the changes in Drosha and Dicer protein levels observed during hypoxia. In addition, consistent with prior studies, hypoxic conditions also induce a complement of miRNAs, including miR-22, miR-322, and miR-451 [82]. Another study partially corroborated these findings and established the importance of hypoxia in the regulation of Dicer activity and consequent miRNA function in endothelial cells [83]. Consistent with prior data, hypoxia produced a distinct shift in miRNA expression in human umbilical vein endothelial cells (HUVECs). While 365 miRNAs were induced, a smaller subset of 36 miRNAs was significantly downregulated in hypoxic HUVECs. Interestingly, Dicer mRNA and protein levels were decreased 2–3 fold in a VHL-dependent manner. These changes were accompanied by a corresponding decrease in Dicer functional activity and an accumulation of precursors (pri-miRNAs and pre-miRNAs) of repressed miRNAs. Collectively, these data indicate that oxygen deprivation decreases expression and activity of key miRNA processing proteins. However, it remains unclear how these changes lead to a selective repression of a subset of mature miRNA while a substantial number of hypoxamirs are potently induced under the same conditions.
Hypoxia also mediates important PTMs that modulate the activity of proteins involved in the biogenesis and functionality of miRNAs (Figure 1). A recent study demonstrated that hypoxia increases the expression of type I collagen prolyl-4-hydroxylase [C-P4H(I)], an enzyme that promotes prolyl-hydroxylation and accumulation of Ago2, a crucial component of the miRISC. This PTM is required for the association of Ago2 with heat shock protein 90 (Hsp90) and the subsequent efficient loading of guide strand miRNAs into miRISC. Together, these discrete hypoxia-mediated events enhance the endonuclease activity of miRNAs in RISC during hypoxia [84]. These findings provide a plausible mechanism for the dynamic induction of multiple hypoxamirs under conditions of low oxygen tension. Notably, it is difficult to reconcile the discrepancy between the accentuating effects of hypoxia on miRISC activity, which promotes miRNA bioactivity, and its inhibition of Drosha/Dicer-mediated miRNA maturation. Future studies will be necessary to explain how these divergent effects may integrate to produce selective but uniform shifts in hypoxamir expression and function.
Summary and Future Direction
The cellular response to hypoxic stress requires the precise coordination of a complex regulatory network. The emergence of miRNAs as important contributors to cellular adaptation to low oxygen tension has prompted interest in understanding how hypoxia directly regulates miRNA transcription, biogenesis, and maturation. Experimental evidence now indicates that hypoxia exerts important modulatory effects on miRNA at multiple levels of biogenesis. As our understanding of hypoxamirs advances, it will be equally important to ascertain the functional relevance and regulation of repressed miRNAs during conditions of low oxygen availability. Studies demonstrating that hypoxic conditions decrease levels and activity of key miRNA-processing proteins provide some indication of potential mechanisms by which hypoxia downregulates miRNAs; however, the molecular details of how of hypoxia exerts divergent effects on the expression of select subsets of miRNAs while globally modulating levels and activity of miRNA processing proteins remain largely undefined.
Future studies that interrogate the complexity of adaptive mechanisms in miRNA biology should offer further insight. A growing body of evidence indicates that HIF may function as a transcriptional repressor under conditions of low oxygen availability, as in the case of its repression of the equilibrative nucleoside transporter during hypoxia [85] and its inhibition of adenosine kinase leading to hypoxia-mediated vascular leakage [86]. HIF also blocks expression of the Na-K-2Cl ion cotransporter in hypoxic intestinal epithelial cells [87]. Thus, it is plausible that HIF may also transcriptionally repress a subset of miRNAs while otherwise inducing the expression of others (Figure 3). In addition, recent work demonstrates that hypoxia is associated with rare alternative spicing mechanisms such as intron retention that may have a direct impact on miRNA expression [88]. Although other environmental stresses, such as heat treatment, induce specific alternative splicing events that modulate levels of miR-400 in lower organisms such as plants [89], it is not clear as to whether hypoxic conditions activate similar alternative splicing pathways that directly regulate miRNA expression and whether these mechanisms are operative in higher order eukaryotes. Recent studies have also revealed that hypoxia induces adenosine-to-inosine (A-to-I) RNA sequence editing [90]. Given its emerging biological importance in human diseases [91, 92], such RNA editing of hypoxamir seed sequences or mRNA target sequences may represent a novel regulatory mechanism of miRNA function in hypoxia. Finally, multiple lines of evidence indicate that miRNA bioactivity and stability are influenced by interactions with RNA binding proteins [93]. This additional level of posttranscriptional regulation may also contribute to the unique but specific shifts in hypoxamir expression patterns noted during exposure to low oxygen levels.
Figure 3. Potential novel mechanisms of hypoxia-mediated regulation of miRNA transcription and maturation.
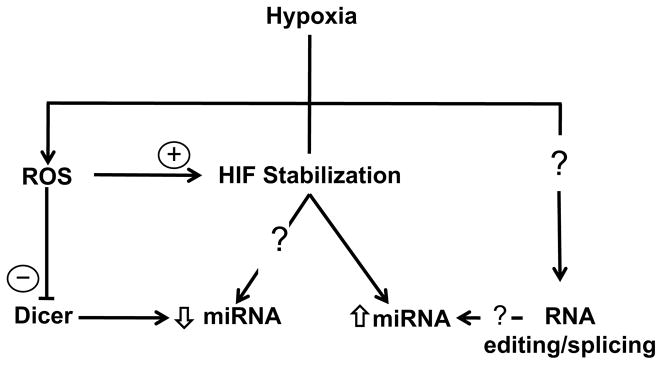
Recent evidence directly implicate oxidant stress and reactive oxygen species (ROS) in HIF stabilization and modulation of Dicer activity. In addition, RNA processing mechanisms operative in other cellular stress responses, such as RNA alternative splicing and editing, may control hypoxamir expression.
The emerging concept of the TF-hypoxamir regulatory network may also offer important perspectives on the complexity of miRNA function and regulation during hypoxia. Multiple groups have undertaken bioinformatic approaches to construct integrated TF-miRNA-protein target maps [55, 56] that emphasize several important points potentially applicable to the hypoxic adaptive response. First, TF and hypoxamirs likely form functionally-related hubs and clusters that permit a synergistic amplification in effect size and pleotropic actions [94]. A TF and hypoxamir may combinatorially regulate a specific protein target. Simultaneously, this TF-miRNA pair may be highly connected to other network nodes that target functionally related proteins in the same or related biological pathway. As such, higher order feedforward and feedback regulatory loops may account for the diverse and intricate patterns of miRNA regulation and action during hypoxic adaptation. Interestingly, over 70% of mature miRNAs also have conserved TF-binding sites of uncertain functional significance [95]. It is currently unclear if TFs interact with mature miRNAs independent of promoter events, and if such interactions have bidirectional implications on TF and miRNA function and stability.
Recent evidence also identifies potential links between reduction/oxidation (redox) perturbations during hypoxic stress and regulation of miRNA biogenesis (Figure 3). The redox hypothesis maintains that non-energetic ROS, such as superoxide and hydrogen peroxide (H2O2), serve as important signaling intermediates in cellular adaptation to acute and chronic hypoxia [96]. Although a detailed discussion of the redox perturbations in hypoxia is beyond the scope of this review, accumulating data indicate dynamic shifts in ROS levels occur during periods of low oxygen tension [96]. The duration and severity of hypoxia, as well as the cellular context, appear to influence whether ROS levels increase or decrease, creating some debate about the exact role and relative importance of redox status in hypoxic cellular responses. Despite these controversies, multiple studies suggest that bidirectional regulatory crosstalk likely exists between ROS and miRNA pathways. For instance, miR-21 and miR-128 directly modulate ROS levels in human bronchial epithelial and medulloblastoma cells, respectively [97, 98]. Simultaneously, a growing body of evidence suggests that shifts in ROS levels may result in broad changes in miRNA expression profiles. In murine hippocampal neuronal cells, H2O2 induced changes in the expression of greater than 100 miRNAs, including miR-135b and miR-708 [99]. Similarly, oxidative stress produced global changes in expression of miRNAs in RAW 264.7 macrophages [100].
The molecular mechanisms underlying these effects remain poorly understood; however, ROS may exert these actions through effects on transcription factors and enzymes that control miRNA transcription and maturation (Figure 3). The generation of mitochondrial ROS during hypoxia appears to be required for the stabilization of HIF [101, 102], which controls the expression of multiple hypoxamirs (Figure 1, Table 3). Lung epithelial cells pre-treated with mitochondrial complex I and III inhibitors that block mitochondrial ROS generation fail to accumulate HIF during exposure to hypoxia (O2 1.5%) [102]. In addition, H2O2 activates the HIF-1α promoter in pulmonary arterial smooth muscle cells through a functional NF-κB site [103]. ROS may also modulate Dicer activity and levels. Hydrogen peroxide downregulates Dicer protein levels and activity in JAR trophoblasts and a range of other cell lines [104]. Although these reports raise the intriguing possibility that ROS mediate important aspects of miRNA biogenesis, further studies are needed to define a direct mechanistic link between oxidant stress and coordinated changes in miRNA expression and maturation.
Additionally, further work is needed to decipher the hierarchical regulatory relationships that must certainly exist among hypoxia-regulated miRNAs. Do hypoxamirs participate in auto-regulatory loops or directly repress other miRNAs in an hierarchical system? Does hypoxia modulate target affinity for a single miRNA or even a network of hypoxamirs? The presence of genomic miRNA clusters, polycistronic miRNA genes, and shared/overlapping targets imply that combinatorial regulation is a central feature of miRNA biology. However, at present better computational tools are needed to help predict integrated miRNA effects and regulation during hypoxic cellular adaptation. Overall, addressing these unresolved issues in hypoxamir biology will directly inform our understanding of their role in an expanding complement of adaptive responses and hypoxic and ischemic human diseases.
Supplementary Material
Acknowledgments
We thank S. Tribuna for expert administrative assistance.
Funding Sources
This work was supported by the NIH Training Grant 5T32HL007208-34 (S.N.); NIH grant HL096843, the Lerner, Harris, and Watkins Funds, Gilead Research Scholars Fund, and the Pulmonary Hypertension Association (S.Y.C.); and NIH grants HL061795, HL48743, HL107192, HL70819, and HL108630 (J.L.). The funding sources for SN, SYC, and JL did not participate in the preparation of this report or the decision to submit it for publication.
Abbreviations
- 3′UTR
3′ untranslated region
- Ago2
Argonaute 2
- C/EBPα
CCAAT/enhancer binding protein α
- CCN5
cysteine-rich 61/connective tissue growth factor/nephroblastoma overexpressed 5
- (ChIP)
chromatin immunoprecipitation
- CUL2
cullin2
- DGCR8
Di George Syndrome critical region 8
- DMNT
DNA methyltransferase
- E2F3
E2F transcription factor 3
- ER
endoplasmic reticulum
- Hsp90
heat shock protein 90
- HIF
hypoxia-inducible factor
- HUVECs
human umbilical vein endothelial cells
- HRE
hypoxia responsive element
- kb
kilobases
- IRAK1
IL1 receptor-associated kinase 1
- mTOR
mammalian target of rapamycin
- miRNA
microRNA
- miRISC
miRNA-induced silencing complex
- mRNA
messenger RNA
- NF-κB
nuclear factor-kappa B
- nt
nucleotides
- ODD
oxygen dependent domain
- PERK
PKR-like ER kinase
- PTM
post-translational modification
- premiRNA
precursor miRNA
- pri-miRNA
primary microRNA
- PHD
prolyl hydroxylation domain
- ROS
reactive oxygen species
- RUNX-1
runt-related transcription factor 1
- TF
transcription factor
- C-P4H(I)
type I collagen prolyl-4-hydroxylase
- TRAF6
TNF receptor-associated factor 6
- TWIST1
twist related protein 1
- UPR
unfolded protein response
- VHL
von Hippel-Lindau tumor suppressor protein
Footnotes
Disclosures
None
References
- 1.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kulshreshtha R, Ferracin M, Wojcik SE, Garzon R, Alder H, Agosto-Perez FJ, Davuluri R, Liu CG, Croce CM, Negrini M, Calin GA, Ivan M. A microRNA signature of hypoxia. Mol Cell Biol. 2007;27:1859–1867. doi: 10.1128/MCB.01395-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pocock R. Invited review: Decoding the microRNA response to hypoxia. Pflugers Arch. 2011;461:307–315. doi: 10.1007/s00424-010-0910-5. [DOI] [PubMed] [Google Scholar]
- 4.Bentwich I, Avniel A, Karov Y, Aharonov R, Gilad S, Barad O, Barzilai A, Einat P, Einav U, Meiri E, Sharon E, Spector Y, Bentwich Z. Identification of hundreds of conserved and nonconserved human microRNAs. Nat Genet. 2005;37:766–770. doi: 10.1038/ng1590. [DOI] [PubMed] [Google Scholar]
- 5.Berezikov E, Cuppen E, Plasterk RH. Approaches to microRNA discovery. Nat Genet. 2006;38(Suppl):S2–7. doi: 10.1038/ng1794. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 7.Loscalzo J. The cellular response to hypoxia: Tuning the system with microRNAs. J Clin Invest. 2010;120:3815–3817. doi: 10.1172/JCI45105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan SY, Zhang YY, Hemann C, Mahoney CE, Zweier JL, Loscalzo J. MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 2009;10:273–284. doi: 10.1016/j.cmet.2009.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Favaro E, Ramachandran A, McCormick R, Gee H, Blancher C, Crosby M, Devlin C, Blick C, Buffa F, Li JL, Vojnovic B, Pires das Neves R, Glazer P, Iborra F, Ivan M, Ragoussis J, Harris AL. MicroRNA-210 regulates mitochondrial free radical response to hypoxia and krebs cycle in cancer cells by targeting iron sulfur cluster protein ISCU. PLoS One. 2010;5:e10345. doi: 10.1371/journal.pone.0010345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosby ME, Kulshreshtha R, Ivan M, Glazer PM. MicroRNA regulation of DNA repair gene expression in hypoxic stress. Cancer Res. 2009;69:1221–1229. doi: 10.1158/0008-5472.CAN-08-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghosh G, Subramanian IV, Adhikari N, Zhang X, Joshi HP, Basi D, Chandrashekhar YS, Hall JL, Roy S, Zeng Y, Ramakrishnan S. Hypoxia-induced microRNA-424 expression in human endothelial cells regulates HIF-alpha isoforms and promotes angiogenesis. J Clin Invest. 2010;120:4141–4154. doi: 10.1172/JCI42980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Majmundar AJ, Wong WJ, Simon MC. Hypoxia-inducible factors and the response to hypoxic stress. Mol Cell. 2010;40:294–309. doi: 10.1016/j.molcel.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Semenza GL. Hypoxia-inducible factors in physiology and medicine. Cell. 2012;148:399–408. doi: 10.1016/j.cell.2012.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sermeus A, Michiels C. Reciprocal influence of the p53 and the hypoxic pathways. Cell Death Dis. 2011;2:e164. doi: 10.1038/cddis.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.An WG, Kanekal M, Simon MC, Maltepe E, Blagosklonny MV, Neckers LM. Stabilization of wild-type p53 by hypoxia-inducible factor 1alpha. Nature. 1998;392:405–408. doi: 10.1038/32925. [DOI] [PubMed] [Google Scholar]
- 16.Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, Koch CJ, Ratcliffe P, Moons L, Jain RK, Collen D, Keshert E. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- 17.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sarbassov DD, Ali SM, Sabatini DM. Growing roles for the mTOR pathway. Curr Opin Cell Biol. 2005;17:596–603. doi: 10.1016/j.ceb.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Sofer A, Lei K, Johannessen CM, Ellisen LW. Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol. 2005;25:5834–5845. doi: 10.1128/MCB.25.14.5834-5845.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Koritzinsky M, Magagnin MG, van den Beucken T, Seigneuric R, Savelkouls K, Dostie J, Pyronnet S, Kaufman RJ, Weppler SA, Voncken JW, Lambin P, Koumenis C, Sonenberg N, Wouters BG. Gene expression during acute and prolonged hypoxia is regulated by distinct mechanisms of translational control. EMBO J. 2006;25:1114–1125. doi: 10.1038/sj.emboj.7600998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao L, Ackerman SL. Endoplasmic reticulum stress in health and disease. Curr Opin Cell Biol. 2006;18:444–452. doi: 10.1016/j.ceb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 22.Xu C, Bailly-Maitre B, Reed JC. Endoplasmic reticulum stress: Cell life and death decisions. J Clin Invest. 2005;115:2656–2664. doi: 10.1172/JCI26373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 25.Greer SN, Metcalf JL, Wang Y, Ohh M. The updated biology of hypoxia-inducible factor. EMBO J. 2012;31:2448–2460. doi: 10.1038/emboj.2012.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelly TJ, Souza AL, Clish CB, Puigserver P. A hypoxia-induced positive feedback loop promotes hypoxia-inducible factor 1alpha stability through miR-210 suppression of glycerol-3-phosphate dehydrogenase 1-like. Mol Cell Biol. 2011;31:2696–2706. doi: 10.1128/MCB.01242-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cascio S, D’Andrea A, Ferla R, Surmacz E, Gulotta E, Amodeo V, Bazan V, Gebbia N, Russo A. miR-20b modulates VEGF expression by targeting HIF-1 alpha and STAT3 in MCF-7 breast cancer cells. J Cell Physiol. 2010;224:242–249. doi: 10.1002/jcp.22126. [DOI] [PubMed] [Google Scholar]
- 28.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bruning U, Cerone L, Neufeld Z, Fitzpatrick SF, Cheong A, Scholz CC, Simpson DA, Leonard MO, Tambuwala MM, Cummins EP, Taylor CT. MicroRNA-155 promotes resolution of hypoxia-inducible factor 1alpha activity during prolonged hypoxia. Mol Cell Biol. 2011;31:4087–4096. doi: 10.1128/MCB.01276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giannakakis A, Sandaltzopoulos R, Greshock J, Liang S, Huang J, Hasegawa K, Li C, O’Brien-Jenkins A, Katsaros D, Weber BL, Simon C, Coukos G, Zhang L. miR-210 links hypoxia with cell cycle regulation and is deleted in human epithelial ovarian cancer. Cancer Biol Ther. 2008;7:255–264. doi: 10.4161/cbt.7.2.5297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang L, Dong LY, Li YJ, Hong Z, Wei WS. The microRNA miR-181c controls microglia-mediated neuronal apoptosis by suppressing tumor necrosis factor. J Neuroinflammation. 2012;9:211. doi: 10.1186/1742-2094-9-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hua Z, Lv Q, Ye W, Wong CK, Cai G, Gu D, Ji Y, Zhao C, Wang J, Yang BB, Zhang Y. MiRNA-directed regulation of VEGF and other angiogenic factors under hypoxia. PLoS One. 2006;1:e116. doi: 10.1371/journal.pone.0000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Borchert GM, Lanier W, Davidson BL. RNA polymerase III transcribes human microRNAs. Nat Struct Mol Biol. 2006;13:1097–1101. doi: 10.1038/nsmb1167. [DOI] [PubMed] [Google Scholar]
- 36.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 38.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 39.Denli AM, Tops BB, Plasterk RH, Ketting RF, Hannon GJ. Processing of primary microRNAs by the microprocessor complex. Nature. 2004;432:231–235. doi: 10.1038/nature03049. [DOI] [PubMed] [Google Scholar]
- 40.Bohnsack MT, Czaplinski K, Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.O’Toole AS, Miller S, Haines N, Zink MC, Serra MJ. Comprehensive thermodynamic analysis of 3′ double-nucleotide overhangs neighboring watson-crick terminal base pairs. Nucleic Acids Res. 2006;34:3338–3344. doi: 10.1093/nar/gkl428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y, Fei M, Xue G, Zhou Q, Jia Y, Li L, Xin H, Sun S. Elevated levels of hypoxia-inducible microRNA-210 in pre-eclampsia: New insights into molecular mechanisms for the disease. J Cell Mol Med. 2012;16:249–259. doi: 10.1111/j.1582-4934.2011.01291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dinger ME, Mercer TR, Mattick JS. RNAs as extracellular signaling molecules. J Mol Endocrinol. 2008;40:151–159. doi: 10.1677/JME-07-0160. [DOI] [PubMed] [Google Scholar]
- 46.Zhu H, Fan GC. Extracellular/circulating microRNAs and their potential role in cardiovascular disease. Am J Cardiovasc Dis. 2011;1:138–149. [PMC free article] [PubMed] [Google Scholar]
- 47.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J Biol Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Donker RB, Mouillet JF, Nelson DM, Sadovsky Y. The expression of Argonaute2 and related microRNA biogenesis proteins in normal and hypoxic trophoblasts. Mol Hum Reprod. 2007;13:273–279. doi: 10.1093/molehr/gam006. [DOI] [PubMed] [Google Scholar]
- 49.Guimbellot JS, Erickson SW, Mehta T, Wen H, Page GP, Sorscher EJ, Hong JS. Correlation of microRNA levels during hypoxia with predicted target mRNAs through genome-wide microarray analysis. BMC Med Genomics. 2009;2:15. doi: 10.1186/1755-8794-2-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voellenkle C, Rooij J, Guffanti A, Brini E, Fasanaro P, Isaia E, Croft L, David M, Capogrossi MC, Moles A, Felsani A, Martelli F. Deep-sequencing of endothelial cells exposed to hypoxia reveals the complexity of known and novel microRNAs. RNA. 2012;18:472–484. doi: 10.1261/rna.027615.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corcoran DL, Pandit KV, Gordon B, Bhattacharjee A, Kaminski N, Benos PV. Features of mammalian microRNA promoters emerge from polymerase II chromatin immunoprecipitation data. PLoS One. 2009;4:e5279. doi: 10.1371/journal.pone.0005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ozsolak F, Poling LL, Wang Z, Liu H, Liu XS, Roeder RG, Zhang X, Song JS, Fisher DE. Chromatin structure analyses identify miRNA promoters. Genes Dev. 2008;22:3172–3183. doi: 10.1101/gad.1706508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Monteys AM, Spengler RM, Wan J, Tecedor L, Lennox KA, Xing Y, Davidson BL. Structure and activity of putative intronic miRNA promoters. RNA. 2010;16:495–505. doi: 10.1261/rna.1731910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang J, Lu M, Qiu C, Cui Q. TransmiR: A transcription factor-microRNA regulation database. Nucleic Acids Res. 2010;38:D119–22. doi: 10.1093/nar/gkp803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Croft L, Szklarczyk D, Jensen LJ, Gorodkin J. Multiple independent analyses reveal only transcription factors as an enriched functional class associated with microRNAs. BMC Syst Biol. 2012;6:90. doi: 10.1186/1752-0509-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cummins EP, Taylor CT. Hypoxia-responsive transcription factors. Pflugers Arch. 2005;450:363–371. doi: 10.1007/s00424-005-1413-7. [DOI] [PubMed] [Google Scholar]
- 58.Kulshreshtha R, Ferracin M, Negrini M, Calin GA, Davuluri RV, Ivan M. Regulation of microRNA expression: The hypoxic component. Cell Cycle. 2007;6:1426–1431. [PubMed] [Google Scholar]
- 59.Camps C, Buffa FM, Colella S, Moore J, Sotiriou C, Sheldon H, Harris AL, Gleadle JM, Ragoussis J. Hsa-miR-210 is induced by hypoxia and is an independent prognostic factor in breast cancer. Clin Cancer Res. 2008;14:1340–1348. doi: 10.1158/1078-0432.CCR-07-1755. [DOI] [PubMed] [Google Scholar]
- 60.Huang X, Ding L, Bennewith KL, Tong RT, Welford SM, Ang KK, Story M, Le QT, Giaccia AJ. Hypoxia-inducible mir-210 regulates normoxic gene expression involved in tumor initiation. Mol Cell. 2009;35:856–867. doi: 10.1016/j.molcel.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chan SY, Loscalzo J. MicroRNA-210: A unique and pleiotropic hypoxamir. Cell Cycle. 2010;9:1072–1083. doi: 10.4161/cc.9.6.11006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fasanaro P, Greco S, Lorenzi M, Pescatori M, Brioschi M, Kulshreshtha R, Banfi C, Stubbs A, Calin GA, Ivan M, Capogrossi MC, Martelli F. An integrated approach for experimental target identification of hypoxia-induced miR-210. J Biol Chem. 2009;284:35134–35143. doi: 10.1074/jbc.M109.052779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chan YC, Banerjee J, Choi SY, Sen CK. miR-210: The master hypoxamir. Microcirculation. 2012;19:215–223. doi: 10.1111/j.1549-8719.2011.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gorospe M, Tominaga K, Wu X, Fahling M, Ivan M. Post-transcriptional control of the hypoxic response by RNA-binding proteins and MicroRNAs. Front Mol Neurosci. 2011;4:7. doi: 10.3389/fnmol.2011.00007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Parikh VN, Jin RC, Rabello S, Gulbahce N, White K, Hale A, Cottrill KA, Shaik RS, Waxman AB, Zhang YY, Maron BA, Hartner JC, Fujiwara Y, Orkin SH, Haley KJ, Barabasi AL, Loscalzo J, Chan SY. MicroRNA-21 integrates pathogenic signaling to control pulmonary hypertension: Results of a network bioinformatics approach. Circulation. 2012;125:1520–1532. doi: 10.1161/CIRCULATIONAHA.111.060269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polytarchou C, Iliopoulos D, Hatziapostolou M, Kottakis F, Maroulakou I, Struhl K, Tsichlis PN. Akt2 regulates all akt isoforms and promotes resistance to hypoxia through induction of miR-21 upon oxygen deprivation. Cancer Res. 2011;71:4720–4731. doi: 10.1158/0008-5472.CAN-11-0365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Haque I, Banerjee S, Mehta S, De A, Majumder M, Mayo MS, Kambhampati S, Campbell DR, Banerjee SK. Cysteine-rich 61-connective tissue growth factor-nephroblastoma-overexpressed 5 (CCN5)/wnt-1-induced signaling protein-2 (WISP-2) regulates microRNA-10b via hypoxia-inducible factor-1alpha-TWIST signaling networks in human breast cancer cells. J Biol Chem. 2011;286:43475–43485. doi: 10.1074/jbc.M111.284158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Preis M, Gardner TB, Gordon SR, Pipas JM, Mackenzie TA, Klein EE, Longnecker DS, Gutmann EJ, Sempere LF, Korc M. MicroRNA-10b expression correlates with response to neoadjuvant therapy and survival in pancreatic ductal adenocarcinoma. Clin Cancer Res. 2011;17:5812–5821. doi: 10.1158/1078-0432.CCR-11-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 70.Klein U, Lia M, Crespo M, Siegel R, Shen Q, Mo T, Ambesi-Impiombato A, Califano A, Migliazza A, Bhagat G, Dalla-Favera R. The DLEU2/miR-15a/16–1 cluster controls B cell proliferation and its deletion leads to chronic lymphocytic leukemia. Cancer Cell. 2010;17:28–40. doi: 10.1016/j.ccr.2009.11.019. [DOI] [PubMed] [Google Scholar]
- 71.Chang CJ, Chao CH, Xia W, Yang JY, Xiong Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, Liu M, Chen CT, Yu D, Hung MC. p53 regulates epithelial-mesenchymal transition and stem cell properties through modulating miRNAs. Nat Cell Biol. 2011;13:317–323. doi: 10.1038/ncb2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim T, Veronese A, Pichiorri F, Lee TJ, Jeon YJ, Volinia S, Pineau P, Marchio A, Palatini J, Suh SS, Alder H, Liu CG, Dejean A, Croce CM. p53 regulates epithelial-mesenchymal transition through microRNAs targeting ZEB1 and ZEB2. J Exp Med. 2011;208:875–883. doi: 10.1084/jem.20110235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hermeking H. MicroRNAs in the p53 network: Micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 74.Mutharasan RK, Nagpal V, Ichikawa Y, Ardehali H. microRNA-210 is upregulated in hypoxic cardiomyocytes through akt- and p53-dependent pathways and exerts cytoprotective effects. Am J Physiol Heart Circ Physiol. 2011;301:H1519–30. doi: 10.1152/ajpheart.01080.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Boldin MP, Baltimore D. MicroRNAs, new effectors and regulators of NF-kappaB. Immunol Rev. 2012;246:205–220. doi: 10.1111/j.1600-065X.2011.01089.x. [DOI] [PubMed] [Google Scholar]
- 76.Boldin MP, Taganov KD, Rao DS, Yang L, Zhao JL, Kalwani M, Garcia-Flores Y, Luong M, Devrekanli A, Xu J, Sun G, Tay J, Linsley PS, Baltimore D. miR-146a is a significant brake on autoimmunity, myeloproliferation, and cancer in mice. J Exp Med. 2011;208:1189–1201. doi: 10.1084/jem.20101823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vigorito E, Perks KL, Abreu-Goodger C, Bunting S, Xiang Z, Kohlhaas S, Das PP, Miska EA, Rodriguez A, Bradley A, Smith KG, Rada C, Enright AJ, Toellner KM, Maclennan IC, Turner M. microRNA-155 regulates the generation of immunoglobulin class-switched plasma cells. Immunity. 2007;27:847–859. doi: 10.1016/j.immuni.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sheedy FJ, Palsson-McDermott E, Hennessy EJ, Martin C, O’Leary JJ, Ruan Q, Johnson DS, Chen Y, O’Neill LA. Negative regulation of TLR4 via targeting of the proinflammatory tumor suppressor PDCD4 by the microRNA miR-21. Nat Immunol. 2010;11:141–147. doi: 10.1038/ni.1828. [DOI] [PubMed] [Google Scholar]
- 79.Garzon R, Liu S, Fabbri M, Liu Z, Heaphy CE, Callegari E, Schwind S, Pang J, Yu J, Muthusamy N, Havelange V, Volinia S, Blum W, Rush LJ, Perrotti D, Andreeff M, Bloomfield CD, Byrd JC, Chan K, Wu LC, Croce CM, Marcucci G. MicroRNA-29b induces global DNA hypomethylation and tumor suppressor gene reexpression in acute myeloid leukemia by targeting directly DNMT3A and 3B and indirectly DNMT1. Blood. 2009;113:6411–6418. doi: 10.1182/blood-2008-07-170589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lujambio A, Portela A, Liz J, Melo SA, Rossi S, Spizzo R, Croce CM, Calin GA, Esteller M. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010;29:6390–6401. doi: 10.1038/onc.2010.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xiong L, Wang F, Huang X, Liu ZH, Zhao T, Wu LY, Wu K, Ding X, Liu S, Wu Y, Zhao Y, Zhu LL, Fan M. DNA demethylation regulates the expression of miR-210 in neural progenitor cells subjected to hypoxia. FEBS J. 2012;279:4318–4326. doi: 10.1111/febs.12021. [DOI] [PubMed] [Google Scholar]
- 82.Caruso P, MacLean MR, Khanin R, McClure J, Soon E, Southgate M, MacDonald RA, Greig JA, Robertson KE, Masson R, Denby L, Dempsie Y, Long L, Morrell NW, Baker AH. Dynamic changes in lung microRNA profiles during the development of pulmonary hypertension due to chronic hypoxia and monocrotaline. Arterioscler Thromb Vasc Biol. 2010;30:716–723. doi: 10.1161/ATVBAHA.109.202028. [DOI] [PubMed] [Google Scholar]
- 83.Ho JJ, Metcalf JL, Yan MS, Turgeon PJ, Wang JJ, Chalsev M, Petruzziello-Pellegrini TN, Tsui AK, He JZ, Dhamko H, Man HS, Robb GB, Teh BT, Ohh M, Marsden PA. Functional importance of dicer protein in the adaptive cellular response to hypoxia. J Biol Chem. 2012;287:29003–29020. doi: 10.1074/jbc.M112.373365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wu C, So J, Davis-Dusenbery BN, Qi HH, Bloch DB, Shi Y, Lagna G, Hata A. Hypoxia potentiates microRNA-mediated gene silencing through posttranslational modification of Argonaute2. Mol Cell Biol. 2011;31:4760–4774. doi: 10.1128/MCB.05776-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Eltzschig HK, Abdulla P, Hoffman E, Hamilton KE, Daniels D, Schonfeld C, Loffler M, Reyes G, Duszenko M, Karhausen J, Robinson A, Westerman KA, Coe IR, Colgan SP. HIF-1-dependent repression of equilibrative nucleoside transporter (ENT) in hypoxia. J Exp Med. 2005;202:1493–1505. doi: 10.1084/jem.20050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morote-Garcia JC, Rosenberger P, Kuhlicke J, Eltzschig HK. HIF-1-dependent repression of adenosine kinase attenuates hypoxia-induced vascular leak. Blood. 2008;111:5571–5580. doi: 10.1182/blood-2007-11-126763. [DOI] [PubMed] [Google Scholar]
- 87.Ibla JC, Khoury J, Kong T, Robinson A, Colgan SP. Transcriptional repression of na-K-2Cl cotransporter NKCC1 by hypoxia-inducible factor-1. Am J Physiol Cell Physiol. 2006;291:C282–C289. doi: 10.1152/ajpcell.00564.2005. [DOI] [PubMed] [Google Scholar]
- 88.Weigand JE, Boeckel JN, Gellert P, Dimmeler S. Hypoxia-induced alternative splicing in endothelial cells. PLoS One. 2012;7:e42697. doi: 10.1371/journal.pone.0042697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yan K, Liu P, Wu CA, Yang GD, Xu R, Guo QH, Huang JG, Zheng CC. Stress-induced alternative splicing provides a mechanism for the regulation of MicroRNA processing in arabidopsis thaliana. Mol Cell. 2012;48:421–531. doi: 10.1016/j.molcel.2012.08.032. [DOI] [PubMed] [Google Scholar]
- 90.Nevo-Caspi Y, Amariglio N, Rechavi G, Paret G. A-to-I RNA editing is induced upon hypoxia. Shock. 2011;35:585–589. doi: 10.1097/SHK.0b013e31820fe4b7. [DOI] [PubMed] [Google Scholar]
- 91.Choudhury Y, Tay FC, Lam DH, Sandanaraj E, Tang C, Ang BT, Wang S. Attenuated adenosine-to-inosine editing of microRNA-376a* promotes invasiveness of glioblastoma cells. J Clin Invest. 2012;122:4059–4076. doi: 10.1172/JCI62925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yu Z, Hecht NB. The DNA/RNA-binding protein, translin, binds microRNA122a and increases its in vivo stability. J Androl. 2008;29:572–579. doi: 10.2164/jandrol.108.005090. [DOI] [PubMed] [Google Scholar]
- 94.Wang J, Haubrock M, Cao KM, Hua X, Zhang CY, Wingender E, Li J. Regulatory coordination of clustered microRNAs based on microRNA-transcription factor regulatory network. BMC Syst Biol. 2011;5:199. doi: 10.1186/1752-0509-5-199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Piriyapongsa J, Jordan IK, Conley AB, Ronan T, Smalheiser NR. Transcription factor binding sites are highly enriched within microRNA precursor sequences. Biol Direct. 2011;6:61. doi: 10.1186/1745-6150-6-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Weir EK, Archer SL. The role of redox changes in oxygen sensing. Respir Physiol Neurobiol. 2010;174:182–191. doi: 10.1016/j.resp.2010.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Venkataraman S, Alimova I, Fan R, Harris P, Foreman N, Vibhakar R. MicroRNA 128a increases intracellular ROS level by targeting bmi-1 and inhibits medulloblastoma cancer cell growth by promoting senescence. PLoS One. 2010;5:e10748. doi: 10.1371/journal.pone.0010748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Zhang X, Ng WL, Wang P, Tian L, Werner E, Wang H, Doetsch P, Wang Y. MicroRNA-21 modulates the levels of reactive oxygen species by targeting SOD3 and TNFalpha. Cancer Res. 2012;72:4707–4713. doi: 10.1158/0008-5472.CAN-12-0639. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- 99.Xu X, Jia R, Zhou Y, Song X, Wang J, Qian G, Ge S, Fan X. Microarray-based analysis: Identification of hypoxia-regulated microRNAs in retinoblastoma cells. Int J Oncol. 2011;38:1385–1393. doi: 10.3892/ijo.2011.961. [DOI] [PubMed] [Google Scholar]
- 100.Thulasingam S, Massilamany C, Gangaplara A, Dai H, Yarbaeva S, Subramaniam S, Riethoven JJ, Eudy J, Lou M, Reddy J. miR-27b*, an oxidative stress-responsive microRNA modulates nuclear factor-kB pathway in RAW 264.7 cells. Mol Cell Biochem. 2011;352:181–188. doi: 10.1007/s11010-011-0752-2. [DOI] [PubMed] [Google Scholar]
- 101.Chandel NS, McClintock DS, Feliciano CE, Wood TM, Melendez JA, Rodriguez AM, Schumacker PT. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: A mechanism of O2 sensing. J Biol Chem. 2000;275:25130–25138. doi: 10.1074/jbc.M001914200. [DOI] [PubMed] [Google Scholar]
- 102.Schroedl C, McClintock DS, Budinger GR, Chandel NS. Hypoxic but not anoxic stabilization of HIF-1alpha requires mitochondrial reactive oxygen species. Am J Physiol Lung Cell Mol Physiol. 2002;283:L922–L931. doi: 10.1152/ajplung.00014.2002. [DOI] [PubMed] [Google Scholar]
- 103.Bonello S, Zahringer C, BelAiba RS, Djordjevic T, Hess J, Michiels C, Kietzmann T, Gorlach A. Reactive oxygen species activate the HIF-1alpha promoter via a functional NFkappaB site. Arterioscler Thromb Vasc Biol. 2007;27:755–761. doi: 10.1161/01.ATV.0000258979.92828.bc. [DOI] [PubMed] [Google Scholar]
- 104.Wiesen JL, Tomasi TB. Dicer is regulated by cellular stresses and interferons. Mol Immunol. 2009;46:1222–1228. doi: 10.1016/j.molimm.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ghosh AK, Shanafelt TD, Cimmino A, Taccioli C, Volinia S, Liu CG, Calin GA, Croce CM, Chan DA, Giaccia AJ, Secreto C, Wellik LE, Lee YK, Mukhopadhyay D, Kay NE. Aberrant regulation of pVHL levels by microRNA promotes the HIF/VEGF axis in CLL B cells. Blood. 2009;113:5568–5574. doi: 10.1182/blood-2008-10-185686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Gonsalves CS, Kalra VK. Hypoxia-mediated expression of 5-lipoxygenase-activating protein involves HIF-1alpha and NF-kappaB and microRNAs 135a and 199a-5p. J Immunol. 2010;184:3878–3888. doi: 10.4049/jimmunol.0902594. [DOI] [PubMed] [Google Scholar]
- 108.Jian X, Xiao-yan Z, Bin H, Yu-feng Z, Bo K, Zhi-nong W, Xin N. MiR-204 regulate cardiomyocyte autophagy induced by hypoxia-reoxygenation through LC3-II. Int J Cardiol. 2011;148:110–112. doi: 10.1016/j.ijcard.2011.01.029. [DOI] [PubMed] [Google Scholar]
- 109.Mouillet JF, Chu T, Nelson DM, Mishima T, Sadovsky Y. MiR-205 silences MED1 in hypoxic primary human trophoblasts. FASEB J. 2010;24:2030–2039. doi: 10.1096/fj.09-149724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Muth M, Hussein K, Jacobi C, Kreipe H, Bock O. Hypoxia-induced down-regulation of microRNA-449a/b impairs control over targeted SERPINE1 (PAI-1) mRNA - a mechanism involved in SERPINE1 (PAI-1) overexpression. J Transl Med. 2011;9:24. doi: 10.1186/1479-5876-9-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rane S, He M, Sayed D, Vashistha H, Malhotra A, Sadoshima J, Vatner DE, Vatner SF, Abdellatif M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ Res. 2009;104:879–886. doi: 10.1161/CIRCRESAHA.108.193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ye Y, Perez-Polo JR, Qian J, Birnbaum Y. The role of microRNA in modulating myocardial ischemia-reperfusion injury. Physiol Genomics. 2011;43:534–542. doi: 10.1152/physiolgenomics.00130.2010. [DOI] [PubMed] [Google Scholar]
- 113.Saito K, Kondo E, Matsushita M. MicroRNA 130 family regulates the hypoxia response signal through the P-body protein DDX6. Nucleic Acids Res. 2011;39:6086–6099. doi: 10.1093/nar/gkr194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cheng Y, Zhu P, Yang J, Liu X, Dong S, Wang X, Chun B, Zhuang J, Zhang C. Ischaemic preconditioning-regulated miR-21 protects heart against ischaemia/reperfusion injury via anti-apoptosis through its target PDCD4. Cardiovasc Res. 2010;87:431–439. doi: 10.1093/cvr/cvq082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Sarkar J, Gou D, Turaka P, Viktorova E, Ramchandran R, Raj JU. MicroRNA-21 plays a role in hypoxia-mediated pulmonary artery smooth muscle cell proliferation and migration. Am J Physiol Lung Cell Mol Physiol. 2010;299:L861–71. doi: 10.1152/ajplung.00201.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Sayed D, He M, Hong C, Gao S, Rane S, Yang Z, Abdellatif M. MicroRNA-21 is a downstream effector of AKT that mediates its antiapoptotic effects via suppression of fas ligand. J Biol Chem. 2010;285:20281–20290. doi: 10.1074/jbc.M110.109207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Suh JH, Choi E, Cha MJ, Song BW, Ham O, Lee SY, Yoon C, Lee CY, Park JH, Lee SH, Hwang KC. Up-regulation of miR-26a promotes apoptosis of hypoxic rat neonatal cardiomyocytes by repressing GSK-3beta protein expression. Biochem Biophys Res Commun. 2012;423:404–410. doi: 10.1016/j.bbrc.2012.05.138. [DOI] [PubMed] [Google Scholar]
- 118.Urbich C, Kaluza D, Fromel T, Knau A, Bennewitz K, Boon RA, Bonauer A, Doebele C, Boeckel JN, Hergenreider E, Zeiher AM, Kroll J, Fleming I, Dimmeler S. MicroRNA-27a/b controls endothelial cell repulsion and angiogenesis by targeting semaphorin 6A. Blood. 2012;119:1607–1616. doi: 10.1182/blood-2011-08-373886. [DOI] [PubMed] [Google Scholar]
- 119.Xiao J, Zhu X, He B, Zhang Y, Kang B, Wang Z, Ni X. MiR-204 regulates cardiomyocyte autophagy induced by ischemia-reperfusion through LC3-II. J Biomed Sci. 2011;18:35. doi: 10.1186/1423-0127-18-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Puissegur MP, Mazure NM, Bertero T, Pradelli L, Grosso S, Robbe-Sermesant K, Maurin T, Lebrigand K, Cardinaud B, Hofman V, Fourre S, Magnone V, Ricci JE, Pouyssegur J, Gounon P, Hofman P, Barbry P, Mari B. miR-210 is overexpressed in late stages of lung cancer and mediates mitochondrial alterations associated with modulation of HIF-1 activity. Cell Death Differ. 2011;18:465–478. doi: 10.1038/cdd.2010.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chen Z, Li Y, Zhang H, Huang P, Luthra R. Hypoxia-regulated microRNA-210 modulates mitochondrial function and decreases ISCU and COX10 expression. Oncogene. 2010;29:4362–4368. doi: 10.1038/onc.2010.193. [DOI] [PubMed] [Google Scholar]
- 122.Ivan M, Harris AL, Martelli F, Kulshreshtha R. Hypoxia response and microRNAs: No longer two separate worlds. J Cell Mol Med. 2008;12:1426–1431. doi: 10.1111/j.1582-4934.2008.00398.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Du R, Sun W, Xia L, Zhao A, Yu Y, Zhao L, Wang H, Huang C, Sun S. Hypoxia-induced down-regulation of microRNA-34a promotes EMT by targeting the notch signaling pathway in tubular epithelial cells. PLoS One. 2012;7:e30771. doi: 10.1371/journal.pone.0030771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Yang F, Zhang L, Wang F, Wang Y, Huo XS, Yin YX, Wang YQ, Zhang L, Sun SH. Modulation of the unfolded protein response is the core of microRNA-122-involved sensitivity to chemotherapy in hepatocellular carcinoma. Neoplasia. 2011;13:590–600. doi: 10.1593/neo.11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chan YC, Khanna S, Roy S, Sen CK. miR-200b targets ets-1 and is down-regulated by hypoxia to induce angiogenic response of endothelial cells. J Biol Chem. 2011;286:2047–2056. doi: 10.1074/jbc.M110.158790. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.