Abstract
The glucocorticoid receptor (GR) functions to regulate a wide group of physiological processes through hormone inducible interaction with genomic loci and subsequent manipulation of the transcriptional output of target genes. Despite expression in a wide variety of tissues, the GR has diverse roles that are regulated tightly in a cell type specific manner. With the advent of whole genome approaches, the details of that diversity and the mechanisms regulating them are beginning to be elucidated. This review aims describe the recent advances detailing the role chromatin structure plays in dictating GR specificity.
Introduction
The glucocorticoid receptor (GR) is a member of the nuclear receptor superfamily of transcription factors that functions to control a wide array of physiological processes including proliferation, development, inflammation, and metabolic homeostasis (Sapolsky, Romero and Munck, 2000). Once activated by ligand, the receptor translocates to the nucleus and dimerizes on sequence specific response elements (GREs) (Mangelsdorf, Thummel, Beato et al., 1995). The receptor is able recruit accessory molecules that aid in preparing the target gene for transcriptional activation or repression (Lonard and O’Malley, 2012). Many of these accessory molecules, also termed co-regulators, act to modify the chromatin structure at the response element and within the promoter of target genes (Trotter and Archer, 2008, Wolf, Heitzer, Grubisha et al., 2008). It has become abundantly clear that biology uses that chromatin structure and the ability to modify it as an important step in regulating transcription factor function (Robyr and Wolffe, 1998).
DNA in eukaryotes is packaged into nucleosomes that act as the basic units of chromatin. Each nuclesome is made up of roughly 146 bp of DNA wrapped around a histone octamer containing two copies of histones H2A, H2B, H3, and H4 (Luger, Mader, Richmond et al., 1997). These nucleosomes are connected in an array by spans of linker DNA associated with histone H1 (Happel and Doenecke, 2009). The abundance of the histone H1 within this linker region has an important role in maintaining nucleosome positioning as well as defining the higher order structure of chromatin (Pennings, Meersseman and Bradbury, 1994, Bednar, Horowitz, Grigoryev et al., 1998). Both of these functions are critical to regulating transcription factor activity. The higher order chromatin structure can be manipulated from completely open or accessible in the “beads on a string” 10 nm fiber conformation to a closed or inaccessible structure (Vaquero, Loyola and Reinberg, 2003). The accessibility of the chromatin acts as a critical regulator of DNA-protein interactions (Li and Reinberg, 2011).
The role of chromatin in GR mediated transactivation has been intensely studied for many years using a number of model promoters, such as MMTV. Using these exogenous promoters, a model of GR activation has evolved in which the activated receptor acts to reorganize the chromatin architecture from an inactive to active state. The MMTV promoter, when introduced into a eukaryotic genome, assembles into an ordered array of nucleosomes (Richard-Foy and Hager, 1987). Recruitment of the receptor results in a number of changes in the chromatin structure. Most notably, the position of the nucleosome surrounding the GRE is altered increasing the accessibility of the promoter (Richard-Foy and Hager, 1987, Pina, Bruggemeier and Beato, 1990). This allows stable binding of other factors, such as NF-1 and Oct-1 which aid to regulate transcriptional output (Archer, Cordingley, Wolford et al., 1991, Bruggemeier, Kalff, Franke et al., 1991, Hebbar and Archer, 2007, Eisfeld, Candau, Truss et al., 1997). In this sense, the GR acts as a pioneering factor to aid other transcription factor binding. Together, a model was generated in which responsive regions are held in an inactive and inaccessible state that is remodeled by recruitment of GR and its accessory proteins to a transcriptionally permissive state. This review aims to update this model from recent work and novel technologies that have changed the perspective of GR and other nuclear receptor activities.
The Whole Genome as a Model System
The development of microarray technology altered the methodology of nuclear receptor research away from model promoters. This progression has only been strengthened with the advent of next-generation sequencing. Moving from model promoters to all promoters has changed the perspective in which we view transcriptional initiation. The initial work performed with tiled microarrays looking at nuclear receptor recruitment immediately altered the paradigm as the majority of binding sites were not identified in classically defined promoters (Carroll, Liu, Brodsky et al., 2005). This finding has been validated for many nuclear receptors including GR in multiple cell types (Wang, Li, Zhang et al., 2009, Welboren, van Driel, Janssen-Megens et al., 2009, So, Chaivorapol, Bolton et al., 2007, Nielsen, Pedersen, Hagenbeek et al., 2008, Reddy, Pauli, Sprouse et al., 2009, Pan, Kocherginsky and Conzen, 2011, John, Sabo, Thurman et al., 2011, Yu, Mayba, Lee et al., 2010, Polman, Welten, Bosch et al., 2012). The distribution of binding sites in the nucleus suggests that GR mediates much of its effects through long-range interactions with transcriptional start sites (TSS). In fact, one study showed that 70% of hormone regulated genes did not demonstrate GR binding within 10 kb of the TSS (Reddy et al., 2009). Another study described extremely distal binding regions to the FKBP5 TSS with strong enhancer activity (Makkonen, Kauhanen, Paakinaho et al., 2009). The genes Ciz1 and Lcn2 were shown to be regulated through a large chromatin loop of 30 kb (Hakim, John, Ling et al., 2009). These types of distal interactions are suggestive of an overall chromatin geography that allows the GR to influence transcription across great genomic distances and potentially different chromosomes.
The distribution of the GR binding sites and the role that distal binding elements have on transcriptional regulation has made the linkage between response elements and gene regulation cloudy. It is difficult to assign a specific binding event to a potential gene target on a genomic scale. In fact, only 50% of the genes that have the closest TSS to a GR binding site demonstrate a dexamethasone dependent transcriptional response (Yu et al., 2010). Nonetheless, it is clear that genes regulated by hormone are enriched for GR binding within 100kb (So et al., 2007, Yu et al., 2010). This linkage between binding and regulation is more evident if cell type specific binding is compared. Looking at binding events in the A549 cell line, genes regulated only in A549 cells had a much higher percentage (27%) of binding events when compared to genes only regulated in U20S cells (1.8%) (So et al., 2007). Together, these data have provided an alternative view to the classical model by which the receptor binds proximal promoters and activates genes. The majority of GR actions are clearly occurring in distal enhancer elements. This phenomenon is limiting the true power of these assays by hindering the ability to clearly associate recruitment to transcriptional responses. The guiding mechanisms by which these sites are connected to TSSs for gene regulation are not understood. A clue to this organization can be found in a study of the estrogen receptor which utilized CTCF binding to separate the genome into blocks and found that the receptor was more likely to regulate a gene within its block than an adjacent block (Chan and Song, 2008). This correlation was not dependent upon distance and suggests that the insulator behavior of CTCF elements may act to direct the receptor to its gene target. It appears that there is an organization to the genome dictated possibly by chromatin landmarks that link response elements to genes. Until this organization is better understood, the ability to link binding events to transcriptional regulation will be limited.
An interesting correlation in these global GR recruitment studies has shown genes that are induced by hormone are more associated with binding events than genes that are repressed (Reddy et al., 2009, Pan et al., 2011). The implication of these data suggests that many repressive events are not caused in cis by bound receptor, yet recent work has demonstrated the GR binds negative GREs (nGRE) that are abundant throughout the genome (Surjit, Ganti, Mukherji et al., 2011). Binding to these response elements results in an altered conformation represented by two GR monomers able to recruit repressor complex consisting of NCOR/SMRT and HDACs (Surjit et al., 2011, Hudson, Youn and Ortlund, 2013). Thus, strong evidence suggests the direct repression of genes by GR despite the lower abundance of ChIP peaks near repressed genes. One explanation may be the lower reported affinity for nGRE by GR results in lower ChIP signal that in many cases may not meet peak calling thresholds (Surjit et al., 2011). Clearly, more work is needed to clarify the role of nGREs and GR on a genomic level.
Chromatin is a Determinate in GR Recruitment
With the global cistrome of GR binding being defined now in several systems, the key mechanisms directing GR recruitment have been investigated. While the vast majority of binding sites (62–80%) contain sequences considered to be a classical GRE, it is clear the GR can be located at other regions either by tethering or through alternate recognition motifs (So et al., 2007, Reddy et al., 2009, John et al., 2011). Recent work has tried to define the molecular characteristics of true recruitment sites that may differentiate them from in silico GREs. In fact, the ChIP data has been fairly consistent in demonstrating that only a small percentage of possible binding sites are actually occupied and that the specific recruitment sites are cell type dependent.
Initial investigations into the mechanisms behind cell type specificity have concentrated on chromatin organization and structure. Work investigating the Ciz1/Lcn2 gene regulation identified a chromatin loop present prior to GR recruitment but only in cells that show an inducible response (Hakim et al., 2009). Global spacial interactions confirmed that cells are preset in the cellular organization and that hormone induction does not induce dramatic reorganization (Hakim, Sung, Voss et al., 2012). This phenomenon suggests that cells can program responsive regions through modification of the underlying chromatin structure. In fact, initial analysis of a 150kb region of the genome demonstrated recruitment of the receptor to constitutively DNase I hypersensitive sites (John, Sabo, Johnson et al., 2008). Further investigation has shown GR is preferentially recruited to sites of DNase I accessibility. Global analysis in a murine cell line indicated that 71% of occupancy sites are accessible prior to receptor recruitment (John et al., 2011). A strong correlation in human cell lines has also demonstrated a preference (69% of binding sites) for DNase I hypersensitivity (Reddy, Gertz, Crawford et al., 2012). Utilizing Formaldehyde Assisted Isolation of Regulatory Elements (FAIRE) as an alternate method of enriching for “open” chromatin structure gave a similar profile with 67% of GR binding sites demonstrating significantly higher signal as compared to background (Burd, Ward, Crusselle-Davis et al., 2012). Comparing different GRE sequences at binding sites to the DNase accessibility of those same sites demonstrated that roughly half of the GRE motifs had an absolute requirement for accessible chromatin (John et al., 2011). Thus, roughly 50% of response element sequences require accessible chromatin for GR binding. Interestingly, these accessible sites of GR recruitment appear to be cell type specific. The overlap between recruitment seen in a pituitary line and mammary line was only 11.4%, with cell type specific binding events only seen in cells with preexisting accessible chromatin. While chromatin accessibility may play an integral role in determining cell type specificity, recent work has also implicated it as a rheostat for ligand availability. This work identified classes of GR binding that are associated with a low abundance of ligand (hypersensitive) to high abundance of ligand (low sensitivity) (Reddy et al., 2012). While all classes of binding showed a preference for DNase I sensitive regions of the genome, ligand hypersensitive sites showed not only the strongest overlap with DNase I accessibility, but also the strongest signal of accessibility. Thus, chromatin structure can be utilized to fine tune the glucocorticoid response within cells.
These data put forth a model, by which GR is specifically recruited to accessible sites rather than the previous dogma of initiating a transition from closed to open chromatin. Nonetheless, there is still a clear chromatin remodeling event that occurs in chromatin structure upon GR recruitment, and that this activity is required for gene activation.
Pioneering Factors and Master Regulators
The role chromatin plays in defining the cellular response that a transcription factor has in a given cell type has led to an investigation into the mechanisms by which these underlying tracks of specificity are laid. The identification of FOXA1 as a critical regulator of estrogen receptor action reintroduced the idea of pioneering factors (Carroll, Meyer, Song et al., 2006, Lupien, Eeckhoute, Meyer et al., 2008, Eeckhoute, Carroll, Geistlinger et al., 2006). The idea of master regulators defining the cell specificity of transcription factors is not specific for nuclear receptors as TGF-β activity is controlled through a small set of cell type specific factors (Mullen, Orlando, Newman et al.). While the idea of pioneering factors was not new, previously the GR had been considered for that role specifically in regards to factors such as NF1 (Richard-Foy and Hager, 1987). However, the relationship of NF1 and GR has been shown to be much more reciprocal than a purely GR pioneering function (Hebbar and Archer, 2007). The recent data on estrogen receptor, androgen receptor, and other master regulatory networks has spurred the hunt within the genomic data sets for GR pioneering factors.
Our work utilizing FAIRE signal across GR binding sites was not able to single out another transcription factor that was uniquely correlated with chromatin structure. However, motif and transcription factor binding analysis did illustrate that GR binding occurred within hotspots of potential factor binding (Burd et al., 2012). This finding is also supported in alternate work investigating PPARγ signaling and showed similar hotspots of transcription factor binding, including GR (Siersbaek, Nielsen, John et al.). However, other analyses have identified potential factors that may influence the chromatin state prior to GR recruitment and thus act as pioneering factors to specify activity. Factors such as AP-1, AML1, and NFkB were shown to be associated with preprogrammed GR binding locations in a murine pituitary line while FoxA2 (HNF3), TAL1, and NF3 motifs were associated with preprogrammed sites in a murine mammary line (John et al., 2011). Only AP-1 was associated with inaccessible sites implicating the other factors in either the development or maintenance of the accessible chromatin state. Another analysis in human cells also demonstrated an overlap of GR with AP-1, FoxA1, FoxA2, and CREB1 (Reddy et al., 2012). Interestingly, CREB1 demonstrated a strong correlation with those sites that were most sensitive to glucocorticoids and overall had the most overlap with DNase I accessible regions. In a neuronal cell line, GR binding correlated with a number of transcription factor binding motifs that have not typically been associated with the receptor. These include Gabpa, Prxx2, Zfp81, Gata1, and Zbtb3 (Polman et al., 2012). The divergent factors identified in each cell type support the notion that cell type specificity can be dictated by a small group of regulators that may direct recruitment through manipulation of chromatin accessibility.
With the exception of a small set of these regulators, the role of these transcription factors in regulating chromatin has not been well studied. It is unclear if these binding partners truly act as pioneering factors or as coregulators, tethering mechanisms, or another transcriptionally cooperative relationship. Analysis of chromatin structure and GR binding following knockdown or deletion of these factors has not been extensively studied with the exception of recent work studying AP-1. That work demonstrated that sites in which GR and AP-1 sites overlap, GR binding was dependent upon an active form of fos. Moreover, both a dominant negative form of fos and knockdown of the protein showed a decrease in DNase I hypersensitivity at recruitment sites (Biddie, John, Sabo et al., 2011). It is unclear if this is true pioneering activity, which is typically identified with the capacity to bind inaccessible DNA and promote a chromatin transition, rather than an ability to maintain the already remodeled genomic region. Another factor, C/EBPβ has been shown to be present in adipocytes prior to chromatin remodeling during development and is required for formation of the transcription factor hotspots (Siersbaek et al., 2011). The forkhead family of proteins are known pioneering factors that have the ability to bind inaccessible chromatin and reorganize that structure (Cirillo, Lin, Cuesta et al., 2002). FoxA1 has been previously implicated in controlling the MMTV chromatin state and GR mediated activation (Holmqvist, Belikov, Zaret et al., 2005). However, the role of FoxA1 appears to be cooperative with GR in which both hormone and expression levels modulate the activities of both proteins in a reciprocal manner similar to the situation seen between GR and NF1 (Belikov, Holmqvist, Astrand et al., 2012, Belikov, Astrand and Wrange, 2009). Finally, an analysis of the crosstalk between NFKB and GR investigated receptor recruitment with and without tumor necrosing factor (TNF). A significant percentage of binding locations (12%) were only utilized in the presence of active NFKB (Rao, McCalman, Moulos et al., 2011). It is clear the GR has a cooperative relationship with a number of proteins in which the receptor may act as a pioneering factor as well as be influenced by the activity of those partners. In fact, evidence suggests that factor binding is not ordered in a sequential basis but allows for quick residence times in which the activity of each element can influence subsequent events (Voss, Schiltz, Sung et al., 2011).
Chromatin Remodeling Complexes
There is quite a body of research detailing the requirement of chromatin remodeling complexes, most notably SWI/SNF, in nuclear receptor and GR mediated gene transactivation (Fryer and Archer, 1998, Trotter and Archer, 2007). The core subunits of the SWI/SNF complex, either Brg1 or Brm, are able to utilized the energy from ATP hydrolysis to functionally reorder chromatin structure (Kwon, Imbalzano, Khavari et al., 1994). During GR activation, the complex is recruited to remodel chromatin structure and facilitates the chromatin structure required for additional protein loading such as NF1, OTFs, and TBP (Archer, Lefebvre, Wolford et al., 1992, Archer, 1993, Fletcher, Xiao, Mautino et al., 2002). These studies indicated the importance of this activity for GR to function as a pioneering factor, but the role of this complex in organizing pre-programmed binding sites has only recently been addressed.
To date, whole genome analysis of Brg1 on global chromatin architecture in regards to nuclear receptor recruitment has not been addressed. However, studies investigating the role of Brg1 in maintaining these accessible binding regions have been done utilizing several endogenous targets. Work utilizing an inducible dominant negative form of Brg-1 demonstrated instances of pre-existing DNase I accessible sites relying on the function SWI/SNF (John et al., 2008). However, it is unclear if this is a direct or indirect effect of SWI/SNF, as recruitment of the complex to those sites in the absence of hormone has yet to be done. A similar study investigating Brm knockdown in a human lung cancer cell line showed similar findings in that a subset of GR binding locations are SWI/SNF dependent both at the transcriptional level and receptor recruitment stage (Engel and Yamamoto, 2011). ChIP performed at those loci for Brm demonstrated a hormone inducible enrichment, but also suggested that basal SWI/SNF complexes may be associated throughout the genome. This concept would support a direct role of SWI/SNF in promoting a chromatin state prior to receptor recruitment. An alternate study investigating nucleosome occupancy by MNase digestion assays showed that hormone inducible genes have an increase in nucleosomes surrounding the TSS following GR activation. This increase as well as the maintenance of the low nucleosome occupancy prior to receptor activation was dependent upon SWI/SNF activity (Pham, Sims, Archer et al., 2011). The basal nucleosome phenomenon was not specific for GR regulated genes and may represent either a general function of the complex in maintaining chromatin structure or a secondary effect of a protein who relies on complex function for expression. However, investigating 21 GR recruitment sites, we only saw one site that had changes in basal FAIRE signal following inducible shRNA knockdown of Brg1 (Burd et al., 2012). Taken together, it appears that SWI/SNF may play a role in presetting chromatin structure at GR binding loci, but the details and mechanism remain unclear. While much of the work up to this point has concentrated on the identification of potential pioneering factors, reorganization of nucleosome positioning is usually associated with some type of ATPase dependent chromatin remodeling. It will be interesting to see in future studies what role SWI/SNF and/or other ATPase dependent chromatin remodeling complexes have in the activity of pioneering factors required for GR recruitment.
Chromatin Characterization of GR Binding Loci
The recent work has demonstrated that GR binding regions usually maintain an accessible state prior to receptor binding. It is less clear what that accessibility represents in terms of physical chromatin structure and nucleosome makeup. The human genome encodes for several variants of the histone proteins each of which can be extensively posttranslationally modified (review in (Campos and Reinberg, 2009)). The initial work characterizing recruitment sites in detail has only begun to characterize potential GR binding locations. Nonetheless, preliminary findings are beginning to elucidate some defining characteristics enriched at GR binding loci.
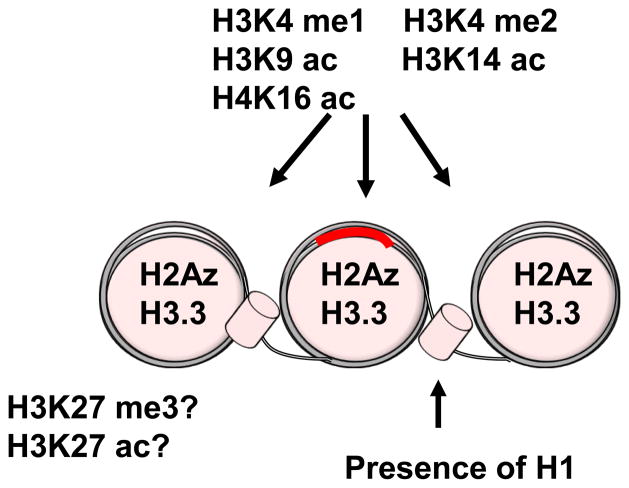
In vitro transcription work investigating the MMTV promoter has given some insights to the roles of NF1 and Oct1 in presetting the chromatin for GR binding. In the Xenopus oocyte system, the addition of NF1 and Oct1 increased H3K14, H3K9, and H4K16 acetylation and that acetylation of the promoter increased GR occupancy (Astrand, Belikov and Wrange, 2009). Furthermore, topography assays demonstrated a reduction in negative superhelicity illustrating an overall change in chromatin architecture. This preset chromatin state induced by NF1 and Oct1 did not result in loss of histone H1 which is normally involved in hormone dependent chromatin remodeling and may give a clue to the differences between a preset chromatin state and an active one. In this same system, an alternate pioneering factor, FoxA1, was able to promote H4K16 acetylation but not H3K14 or H3K9 (Belikov et al., 2012). These types of posttranslational modifications (PTMs) are not surprising as some have been associated with enhancer elements. Since the majority of GR binding sites occur outside classically defined promoters, enhancers may be the predominant dock for GR recruitment. Thus, they provide possibly the best target to understand chromatin structure underlying GR recruitment. In particular, monomethylation of H3K4 (Heintzman, Stuart, Hon et al., 2007, Birney, Stamatoyannopoulos, Dutta et al., 2007) is associated with distal enhancers. These enhancers may be in a poised or active state depending upon H3K27 status (Buecker and Wysocka, 2012). The trimethylated state indicates a poised enhancer and upon transcription factor binding is converted to an acetylated state which marks an active enhancer (Creyghton, Cheng, Welstead et al., 2010, Rada-Iglesias, Bajpai, Swigut et al., 2010). It is unclear if the preset, accessible GR binding locations are enriched for poised or possibly even active enhancing marks. Another potential mark that may characterize these sites is H3K4 dimethylation. For the androgen receptor, this mark is present at both enhancer and promoters in a distinct pattern occupying two nucleosomes flanking the receptor binding site (He, Meyer, Shin et al., 2010). It is unclear if this phenomenon applies to GR binding. In the end, there may be no unifying histone PTM that identifies a preset GR binding location, but potentially the combination of several marks will provide useful to discern the specificity of GR action (Figure 1).
Figure 1.
The makeup of an enhancer element is marked by many potential histone PTM and variants. The marks may be able to discern which elements are primed for GR recruitment. It is still unclear whether GR responsive enhancer elements are in the classical poised or active state (H3K27 status) prior to receptor binding.
There is also a potential role of the histone variants H2Az and H3.3 in defining GR binding sites (Jin, Zang, Wei et al., 2009). The H2Az variant is enriched at binding loci of both preset and de novo binding locations (John et al., 2008). Similarly, the variant is enriched in the androgen inducible PSA locus prior to receptor activation (Dryhurst, McMullen, Fazli et al., 2012). H2Az has been implicated in marking euchromatin boundaries and preventing the spread of heterochromatin (Meneghini, Wu and Madhani, 2003). In this capacity, the variant may act to maintain accessible regions at the majority GR binding regions that are distal from the other regulatory elements and may be surrounded by heterochromatic expanses. Following this line of thinking, H2Az has also been shown to overlap substantially with all three H3K4 methylation states as well as DNase I hypersensitive regions (Barski, Cuddapah, Cui et al., 2007). Together, these data provide strong support for an H2Az role in maintaining a poised chromatin state.
In addition to PTMs and histone variants, DNA methylation has also been implicated in regulating the GR responsive regions. Sites that have the most accessibility prior to receptor binding also have an enrichment for CpG elements (Wiench, John, Baek et al., 2011), while sites that require de novo remodeling by GR have low CpG density. Interestingly, the former set has a methylation pattern dependent upon cell type where accessibility is correlated with lower methylation levels. Furthermore, sparsely populated CpGs at de novo remodeling locations have higher methylation levels that are demethylated upon receptor recruitment. A potential role of DNA methylation in the regulation of GR and other transcription factor activity is not new (Becker, Ruppert and Schutz, 1987, Bhardwaj, Song, Beildeck et al., 2012, Kress, Thomassin and Grange, 2001, Kress, Thomassin and Grange, 2006), but recent work is helping define the mechanistic role it may play in regulating GR specificity.
Concluding Remarks
The importance of chromatin in dictating transcription factor actions is apparent, but the mechanisms of control are only beginning to be understood. Prior accessibility of the recruitment site correlates strongly with binding events in the presence of hormone and the role of pioneering factors in laying the tracks of that accessibility is becoming clearer. Yet, we still don’t understand the coordination of these factors to dictate cell type specificity. This control is clearly maintained by a combination of factors in a dynamic process that is limited by the current genomic techniques which only look at a snapshot of transcriptional activation. This is both true for the binding events measured by ChIP and the transcriptional analysis characterized by RNA-seq. These techniques also rely on bioinformatic processing of large sets of data that may miss less substantial changes but are nonetheless critical mediators of transcription regulation.
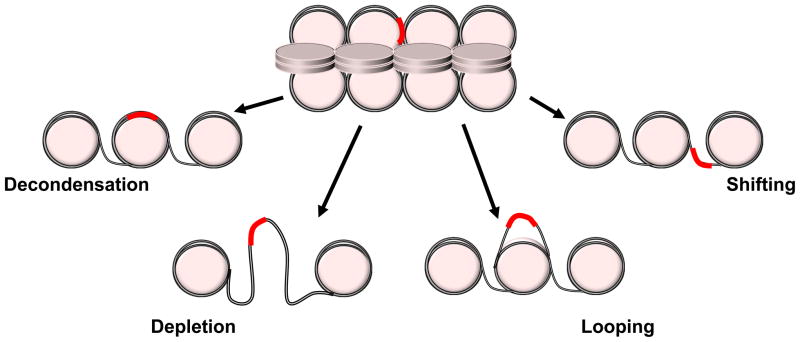
The progression of nuclear receptor study from model promoters to the genome has identified new concepts in how cells direct a limited number of transcription factors to potentiate a diverse set of transcriptional outcomes. Chromatin is an integral regulatory mechanism for defining genetic programs to stimuli such as steroid hormones. The development that the majority of GR binding regions are defined by accessible chromatin architecture prior to receptor recruitment certainly belies its previous role as a pioneering factor. Nonetheless, the majority of binding loci still undergo further chromatin remodeling following GR recruitment suggesting that our view of chromatin being “open” and “closed” is too constrained. Prior to recruitment, most sites must have some transitional state of accessibility to allow for GR to recognize its response element. This state can be controlled in a cell type specific manner, most notably by pioneering factors, to define precise genetic programs. However, in order for activation of transcription, at the majority of sites further remodeling is required and is thus both dependent upon pioneering factors while also acting as one. Furthermore, this receptor-permissive chromatin state has really only been characterized by some defining traits without a full understanding of the actual chromatin structure. These traits include an accessibility, certain histone post-translational modifications, and histone variants. Yet, questions about nucleosome occupancy and positioning are still debated. This accessibility may be nucleosome depletion, response element repositioning to the linker region, transient reorganization of the DNA wrapped around the nucleosome, or simply a reorganization of the tertiary structure of the chromatin structure (Figure 2). In fact, there may not be one unifying mechanism by which accessibility is managed and all or some of these possibilities could be utilized. However, understanding the specifics of the chromatin structure at accessible response sites will greatly aid in our ability to predict GR binding in silico.
Figure 2.
GR binding locations (red) are marked by accessibility prior to receptor interaction. The transition from an inactive to an active recruitment site could be explained by multiple transitions in chromatin structure.
One of the most surprising aspects of the recent GR studies is the relative distance of binding sites to TSS thus making it difficult to link response elements to potential target genes. This limitation has restricted the ability to apply mechanistic insight to the GR binding, histone modification, and chromatin accessibility maps. The critical goal of all of this research is to predict a genetic outcome based upon activation and recruitment of the receptor. At this point, those predictions are still largely guesswork. It is still unclear which binding regions are actually productive in generating a transcriptional response and which ones are simply parking lots for the receptor. In order to address these questions, the most important challenge ahead will be to understand the organization within the genome and link binding events with chromatin characteristics to better predict genetic outcome.
Highlights.
Glucocorticoid Receptor binding sites have a distinct chromatin architecture
Preset chromatin accessibility defines specificity
Glucocorticoid receptor relies on pioneer factors to preset binding sites
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sapolsky RM, Romero LM, Munck AU. How do glucocorticoids influence stress responses? Integrating permissive, suppressive, stimulatory, and preparative actions. Endocr Rev. 2000;21:55–89. doi: 10.1210/edrv.21.1.0389. [DOI] [PubMed] [Google Scholar]
- 2.Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–9. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lonard DM, O’Malley BW. Nuclear receptor coregulators: modulators of pathology and therapeutic targets. Nat Rev Endocrinol. 2012;8:598–604. doi: 10.1038/nrendo.2012.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Trotter KW, Archer TK. The BRG1 transcriptional coregulator. Nucl Recept Signal. 2008;6:e004. doi: 10.1621/nrs.06004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wolf IM, Heitzer MD, Grubisha M, DeFranco DB. Coactivators and nuclear receptor transactivation. J Cell Biochem. 2008;104:1580–6. doi: 10.1002/jcb.21755. [DOI] [PubMed] [Google Scholar]
- 6.Robyr D, Wolffe P. Hormone action and chromatin remodelling. Cell Mol Life Sci. 1998;54:113–24. doi: 10.1007/s000180050130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–60. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 8.Happel N, Doenecke D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene. 2009;431:1–12. doi: 10.1016/j.gene.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 9.Pennings S, Meersseman G, Bradbury EM. Linker histones H1 and H5 prevent the mobility of positioned nucleosomes. Proc Natl Acad Sci U S A. 1994;91:10275–9. doi: 10.1073/pnas.91.22.10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bednar J, Horowitz RA, Grigoryev SA, Carruthers LM, Hansen JC, Koster AJ, Woodcock CL. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci U S A. 1998;95:14173–8. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaquero A, Loyola A, Reinberg D. The constantly changing face of chromatin. Sci Aging Knowledge Environ. 2003;2003:RE4. doi: 10.1126/sageke.2003.14.re4. [DOI] [PubMed] [Google Scholar]
- 12.Li G, Reinberg D. Chromatin higher-order structures and gene regulation. Curr Opin Genet Dev. 2011;21:175–86. doi: 10.1016/j.gde.2011.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. Embo J. 1987;6:2321–8. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pina B, Bruggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–31. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 15.Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–98. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bruggemeier U, Kalff M, Franke S, Scheidereit C, Beato M. Ubiquitous transcription factor OTF-1 mediates induction of the MMTV promoter through synergistic interaction with hormone receptors. Cell. 1991;64:565–72. doi: 10.1016/0092-8674(91)90240-y. [DOI] [PubMed] [Google Scholar]
- 17.Hebbar PB, Archer TK. Chromatin-dependent cooperativity between site-specific transcription factors in vivo. J Biol Chem. 2007;282:8284–91. doi: 10.1074/jbc.M610554200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisfeld K, Candau R, Truss M, Beato M. Binding of NF1 to the MMTV promoter in nucleosomes: influence of rotational phasing, translational positioning and histone H1. Nucleic Acids Res. 1997;25:3733–42. doi: 10.1093/nar/25.18.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Janne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138:245–56. doi: 10.1016/j.cell.2009.04.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welboren WJ, van Driel MA, Janssen-Megens EM, van Heeringen SJ, Sweep FC, Span PN, Stunnenberg HG. ChIP-Seq of ERalpha and RNA polymerase II defines genes differentially responding to ligands. Embo J. 2009;28:1418–28. doi: 10.1038/emboj.2009.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, Stunnenberg HG. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–67. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reddy TE, Pauli F, Sprouse RO, Neff NF, Newberry KM, Garabedian MJ, Myers RM. Genomic determination of the glucocorticoid response reveals unexpected mechanisms of gene regulation. Genome Res. 2009;19:2163–71. doi: 10.1101/gr.097022.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pan D, Kocherginsky M, Conzen SD. Activation of the glucocorticoid receptor is associated with poor prognosis in estrogen receptor-negative breast cancer. Cancer Res. 2011;71:6360–70. doi: 10.1158/0008-5472.CAN-11-0362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.John S, Sabo PJ, Thurman RE, Sung MH, Biddie SC, Johnson TA, Hager GL, Stamatoyannopoulos JA. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu CY, Mayba O, Lee JV, Tran J, Harris C, Speed TP, Wang JC. Genome-wide analysis of glucocorticoid receptor binding regions in adipocytes reveal gene network involved in triglyceride homeostasis. PLoS One. 2010;5:e15188. doi: 10.1371/journal.pone.0015188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Polman JA, Welten JE, Bosch DS, de Jonge RT, Balog J, van der Maarel SM, de Kloet ER, Datson NA. A genome-wide signature of glucocorticoid receptor binding in neuronal PC12 cells. BMC Neurosci. 2012;13:118. doi: 10.1186/1471-2202-13-118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Makkonen H, Kauhanen M, Paakinaho V, Jaaskelainen T, Palvimo JJ. Long-range activation of FKBP51 transcription by the androgen receptor via distal intronic enhancers. Nucleic Acids Res. 2009;37:4135–48. doi: 10.1093/nar/gkp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hakim O, John S, Ling JQ, Biddie SC, Hoffman AR, Hager GL. Glucocorticoid receptor activation of the Ciz1-Lcn2 locus by long range interactions. J Biol Chem. 2009;284:6048–52. doi: 10.1074/jbc.C800212200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan CS, Song JS. CCCTC-binding factor confines the distal action of estrogen receptor. Cancer Res. 2008;68:9041–9. doi: 10.1158/0008-5472.CAN-08-2632. [DOI] [PubMed] [Google Scholar]
- 32.Surjit M, Ganti KP, Mukherji A, Ye T, Hua G, Metzger D, Li M, Chambon P. Widespread negative response elements mediate direct repression by agonist-liganded glucocorticoid receptor. Cell. 2011;145:224–41. doi: 10.1016/j.cell.2011.03.027. [DOI] [PubMed] [Google Scholar]
- 33.Hudson WH, Youn C, Ortlund EA. The structural basis of direct glucocorticoid-mediated transrepression. Nat Struct Mol Biol. 2013;20:53–58. doi: 10.1038/nsmb.2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hakim O, Sung MH, Voss TC, Splinter E, John S, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, de Laat W, Hager GL. Diverse gene reprogramming events occur in the same spatial clusters of distal regulatory elements. Genome Res. 2012;21:697–706. doi: 10.1101/gr.111153.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.John S, Sabo PJ, Johnson TA, Sung MH, Biddie SC, Lightman SL, Voss TC, Davis SR, Meltzer PS, Stamatoyannopoulos JA, Hager GL. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–24. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 36.Reddy TE, Gertz J, Crawford GE, Garabedian MJ, Myers RM. The hypersensitive glucocorticoid response specifically regulates period 1 and expression of circadian genes. Mol Cell Biol. 2012;32:3756–67. doi: 10.1128/MCB.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Burd CJ, Ward JM, Crusselle-Davis VJ, Kissling GE, Phadke D, Shah RR, Archer TK. Analysis of chromatin dynamics during glucocorticoid receptor activation. Mol Cell Biol. 2012;32:1805–17. doi: 10.1128/MCB.06206-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M. Genome-wide analysis of estrogen receptor binding sites. Nat Genet. 2006;38:1289–97. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- 39.Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M. FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell. 2008;132:958–70. doi: 10.1016/j.cell.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eeckhoute J, Carroll JS, Geistlinger TR, Torres-Arzayus MI, Brown M. A cell-type-specific transcriptional network required for estrogen regulation of cyclin D1 and cell cycle progression in breast cancer. Genes Dev. 2006;20:2513–26. doi: 10.1101/gad.1446006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mullen AC, Orlando DA, Newman JJ, Loven J, Kumar RM, Bilodeau S, Reddy J, Guenther MG, DeKoter RP, Young RA. Master transcription factors determine cell-type-specific responses to TGF-beta signaling. Cell. 2011;147:565–76. doi: 10.1016/j.cell.2011.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siersbaek R, Nielsen R, John S, Sung MH, Baek S, Loft A, Hager GL, Mandrup S. Extensive chromatin remodelling and establishment of transcription factor ‘hotspots’ during early adipogenesis. Embo J. 2011;30:1459–72. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biddie SC, John S, Sabo PJ, Thurman RE, Johnson TA, Schiltz RL, Miranda TB, Sung MH, Trump S, Lightman SL, Vinson C, Stamatoyannopoulos JA, Hager GL. Transcription factor AP1 potentiates chromatin accessibility and glucocorticoid receptor binding. Mol Cell. 2011;43:145–55. doi: 10.1016/j.molcel.2011.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–89. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- 45.Holmqvist PH, Belikov S, Zaret KS, Wrange O. FoxA1 binding to the MMTV LTR modulates chromatin structure and transcription. Exp Cell Res. 2005;304:593–603. doi: 10.1016/j.yexcr.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 46.Belikov S, Holmqvist PH, Astrand C, Wrange O. FoxA1 and glucocorticoid receptor crosstalk via histone H4K16 acetylation at a hormone regulated enhancer. Exp Cell Res. 2012;318:61–74. doi: 10.1016/j.yexcr.2011.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Belikov S, Astrand C, Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol. 2009;29:5413–25. doi: 10.1128/MCB.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rao NA, McCalman MT, Moulos P, Francoijs KJ, Chatziioannou A, Kolisis FN, Alexis MN, Mitsiou DJ, Stunnenberg HG. Coactivation of GR and NFKB alters the repertoire of their binding sites and target genes. Genome Res. 2011;21:1404–16. doi: 10.1101/gr.118042.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Voss TC, Schiltz RL, Sung MH, Yen PM, Stamatoyannopoulos JA, Biddie SC, Johnson TA, Miranda TB, John S, Hager GL. Dynamic exchange at regulatory elements during chromatin remodeling underlies assisted loading mechanism. Cell. 2011;146:544–54. doi: 10.1016/j.cell.2011.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 51.Trotter KW, Archer TK. Nuclear receptors and chromatin remodeling machinery. Mol Cell Endocrinol. 2007;265–266:162–7. doi: 10.1016/j.mce.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–81. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 53.Archer TK, Lefebvre P, Wolford RG, Hager GL. Transcription factor loading on the MMTV promoter: a bimodal mechanism for promoter activation. Science. 1992;255:1573–6. doi: 10.1126/science.1347958. [DOI] [PubMed] [Google Scholar]
- 54.Archer TK. Nucleosomes modulate access of transcription factor to the MMTV promoter in vivo and in vitro. Ann N Y Acad Sci. 1993;684:196–8. doi: 10.1111/j.1749-6632.1993.tb32282.x. [DOI] [PubMed] [Google Scholar]
- 55.Fletcher TM, Xiao N, Mautino G, Baumann CT, Wolford R, Warren BS, Hager GL. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–63. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Engel KB, Yamamoto KR. The glucocorticoid receptor and the coregulator Brm selectively modulate each other’s occupancy and activity in a gene-specific manner. Mol Cell Biol. 2011;31:3267–76. doi: 10.1128/MCB.05351-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pham CD, Sims HI, Archer TK, Schnitzler GR. Multiple distinct stimuli increase measured nucleosome occupancy around human promoters. PLoS One. 2011;6:e23490. doi: 10.1371/journal.pone.0023490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Campos EI, Reinberg D. Histones: annotating chromatin. Annu Rev Genet. 2009;43:559–99. doi: 10.1146/annurev.genet.032608.103928. [DOI] [PubMed] [Google Scholar]
- 59.Astrand C, Belikov S, Wrange O. Histone acetylation characterizes chromatin presetting by NF1 and Oct1 and enhances glucocorticoid receptor binding to the MMTV promoter. Exp Cell Res. 2009;315:2604–15. doi: 10.1016/j.yexcr.2009.05.012. [DOI] [PubMed] [Google Scholar]
- 60.Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, Wang W, Weng Z, Green RD, Crawford GE, Ren B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 61.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, Kuehn MS, Taylor CM, Neph S, Koch CM, Asthana S, Malhotra A, Adzhubei I, Greenbaum JA, Andrews RM, Flicek P, Boyle PJ, Cao H, Carter NP, Clelland GK, Davis S, Day N, Dhami P, Dillon SC, Dorschner MO, Fiegler H, Giresi PG, Goldy J, Hawrylycz M, Haydock A, Humbert R, James KD, Johnson BE, Johnson EM, Frum TT, Rosenzweig ER, Karnani N, Lee K, Lefebvre GC, Navas PA, Neri F, Parker SC, Sabo PJ, Sandstrom R, Shafer A, Vetrie D, Weaver M, Wilcox S, Yu M, Collins FS, Dekker J, Lieb JD, Tullius TD, Crawford GE, Sunyaev S, Noble WS, Dunham I, Denoeud F, Reymond A, Kapranov P, Rozowsky J, Zheng D, Castelo R, Frankish A, Harrow J, Ghosh S, Sandelin A, Hofacker IL, Baertsch R, Keefe D, Dike S, Cheng J, Hirsch HA, Sekinger EA, Lagarde J, Abril JF, Shahab A, Flamm C, Fried C, Hackermuller J, Hertel J, Lindemeyer M, Missal K, Tanzer A, Washietl S, Korbel J, Emanuelsson O, Pedersen JS, Holroyd N, Taylor R, Swarbreck D, Matthews N, Dickson MC, Thomas DJ, Weirauch MT, Gilbert J, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Buecker C, Wysocka J. Enhancers as information integration hubs in development: lessons from genomics. Trends Genet. 2012;28:276–84. doi: 10.1016/j.tig.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, Boyer LA, Young RA, Jaenisch R. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A. 2010;107:21931–6. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature. 2010;470:279–83. doi: 10.1038/nature09692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M, Mieczkowski P, Lieb JD, Zhao K, Brown M, Liu XS. Nucleosome dynamics define transcriptional enhancers. Nat Genet. 2010;42:343–7. doi: 10.1038/ng.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jin C, Zang C, Wei G, Cui K, Peng W, Zhao K, Felsenfeld G. H3.3/H2A.Z double variant-containing nucleosomes mark ‘nucleosome-free regions’ of active promoters and other regulatory regions. Nat Genet. 2009;41:941–5. doi: 10.1038/ng.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Dryhurst D, McMullen B, Fazli L, Rennie PS, Ausio J. Histone H2A.Z prepares the prostate specific antigen (PSA) gene for androgen receptor-mediated transcription and is upregulated in a model of prostate cancer progression. Cancer Lett. 2012;315:38–47. doi: 10.1016/j.canlet.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 68.Meneghini MD, Wu M, Madhani HD. Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell. 2003;112:725–36. doi: 10.1016/s0092-8674(03)00123-5. [DOI] [PubMed] [Google Scholar]
- 69.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 70.Wiench M, John S, Baek S, Johnson TA, Sung MH, Escobar T, Simmons CA, Pearce KH, Biddie SC, Sabo PJ, Thurman RE, Stamatoyannopoulos JA, Hager GL. DNA methylation status predicts cell type-specific enhancer activity. Embo J. 2011;30:3028–39. doi: 10.1038/emboj.2011.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becker PB, Ruppert S, Schutz G. Genomic footprinting reveals cell type-specific DNA binding of ubiquitous factors. Cell. 1987;51:435–43. doi: 10.1016/0092-8674(87)90639-8. [DOI] [PubMed] [Google Scholar]
- 72.Bhardwaj A, Song HW, Beildeck M, Kerkhofs S, Castoro R, Shanker S, De Gendt K, Suzuki K, Claessens F, Issa JP, Orgebin-Crist MC, Wilkinson MF. DNA demethylation-dependent AR recruitment and GATA factors drive Rhox5 homeobox gene transcription in the epididymis. Mol Endocrinol. 2012;26:538–49. doi: 10.1210/me.2011-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kress C, Thomassin H, Grange T. Local DNA demethylation in vertebrates: how could it be performed and targeted? FEBS Lett. 2001;494:135–40. doi: 10.1016/s0014-5793(01)02328-6. [DOI] [PubMed] [Google Scholar]
- 74.Kress C, Thomassin H, Grange T. Active cytosine demethylation triggered by a nuclear receptor involves DNA strand breaks. Proc Natl Acad Sci U S A. 2006;103:11112–7. doi: 10.1073/pnas.0601793103. [DOI] [PMC free article] [PubMed] [Google Scholar]