Abstract
Dynamic shape changes of the plasma membrane are fundamental to many processes ranging from morphogenesis and cell migration to phagocytosis and viral propagation. Here we demonstrate that Exo70, a component of the exocyst complex, induces tubular membrane invaginations towards the lumen of synthetic vesicles in vitro and generates protrusions on the surface of cells. Biochemical analyses using Exo70 mutants and independent molecular dynamics simulations based on Exo70 structure demonstrate that Exo70 generates negative membrane curvature through an oligomerization-based mechanism. In cells, the membrane-deformation function of Exo70 is required for protrusion formation and directional cell migration. Exo70 thus represents a membrane-bending protein that may couple actin dynamics and plasma membrane remodeling for morphogenesis.
Keywords: exocyst, Exo70, actin, cell migration, membrane curvature
INTRODUCTION
Cells can be shaped by proteins that directly bind to the plasma membrane and sense, induce, or stabilize its curvature (McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006; Frost et al., 2009). The BAR family of proteins form oligomeric crescent-shaped domains that abut the membrane through their concave surfaces (McMahon and Gallop, 2005; Frost et al., 2009). They induce tubular extensions from synthetic vesicles in vitro and, in cells, cause invaginations of the plasma membrane towards the cytoplasm during processes such as endocytosis. On the other hand, most I-BAR (inverse-BAR) proteins bind to the membrane through their convex surface, and induce membrane tubules towards the lumen of the vesicles in vitro and generate surface protrusions such as filopodia in cells (Mattila and Lappalainen, 2008; Scita et al., 2008; Zhao et al., 2011). The curvatures generated towards the cytoplasm by BAR proteins are often termed as “positive curvature”, whereas those generated in the form of surface protrusions by I-BAR proteins are termed as “negative curvature” (McMahon and Gallop, 2005; Zimmerberg and Kozlov, 2006).
The exocyst complex, consisting of Sec3, Sec5, Sec6, Sec8, Sec10, Sec15, Exo70 and Exo84, mediates the tethering of secretory vesicles at the plasma membrane for exocytosis and cell surface expansion (Hsu et al., 2004; Munson and Novick, 2006; He and Guo, 2009). The exocyst is implicated in many cellular processes such as epithelia formation, cytokinesis, and neurite branching. Recent evidence has demonstrated that the exocyst is involved in cell migration (Zuo et al., 2006; Rosse et al., 2006; Spiczka and Yeaman, 2008; Liu et al., 2009a; Thapa et al., 2012; Ren and Guo, 2012; Liu et al., 2012). In particular, the exocyst component Exo70 directly interacts with the Arpc1 subunit of the Arp2/3 complex and kinetically stimulates actin polymerization and branching at the leading edges of the migrating cells (Zuo et al., 2006; Liu et al., 2009a; Liu et al., 2012). Exo70 has a long rod-like structure that binds to PI(4,5)P2 through positively charged residues on its surface (Dong et al., 2005; Hamburger et al., 2006; Moore et al., 2007; Liu et al., 2007). Overexpression of Exo70, but not any other subunit of the exocyst complex, induces filopodia formation in cells independent of its function in exocytosis (Zuo et al., 2006).
In this study, combining biochemical and cell biological analyses and in silico molecular dynamics and mesoscale simulations, we demonstrate that Exo70 induces negative membrane curvature through an oligomerization-based scaffolding mechanism. In cells, membrane-deformation by Exo70 is required for the generation of membrane protrusions and cell migration. Our study not only reveals a function of Exo70 in membrane curvature induction, but also provides a mechanism that couples actin dynamics and membrane remodeling for morphogenesis and directional cell migration.
RESULTS
Exo70 induces membrane tubules towards the lumen of synthetic vesicles
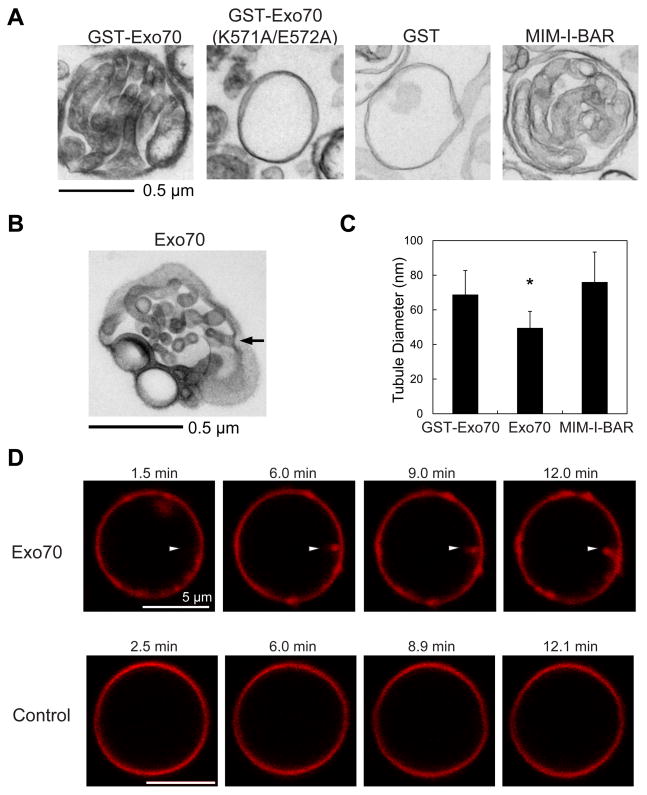
Exo70 was previously shown to bind to PI(4,5)P2 and associate with the cytosolic side of the plasma membrane (Liu et al., 2007; He et al., 2007). Moreover, overexpression of Exo70 induces membrane protrusions similar to the I-BAR proteins (Zuo et al., 2006; Mattila et al., 2007; Yang et al., 2009). We thus tested whether Exo70, like I-BAR, affects membrane morphology under the same condition described previously (Mattila et al., 2007). Synthetic large unilamellar vesicles (LUVs) containing 30% PI(4,5)P2 were incubated with recombinant Exo70 or the I-BAR domain of MIM. The morphology of these LUVs was then analyzed by transmission electron microscopy (EM). Strikingly, the LUVs incubated with GST-Exo70 displayed tubular invaginations that extended towards the lumen (Figure 1A). The average diameter of these membrane tubules was 68.8 ± 13.8 nm (Figure 1C). Exo70 without the GST tag also induced membrane invaginations (Figure 1B), albeit the average diameter of the tubules was smaller (49.5 ± 9.6 nm) (Figure 1C). The I-BAR domain of MIM induced membrane tubules that were slightly larger in diameter than those induced by Exo70 (76.1 ± 17.4 nm) (Figure 1A and 1C). As a negative control, GST had no effect on the LUVs, and the Exo70 (K571A/E572A) mutant, which is defective in PI(4,5)P2 binding (Liu et al., 2007), failed to induce invaginations (Figure 1A). Three-dimensional reconstruction of the deformed LUVs using a series of tilted images demonstrated the connectivity of the tubular invaginations with the exterior (see arrow in Figure 1B and Supplemental Movie 1a for tilted series). We have also performed the experiment using LUVs containing 5% PI(4,5)P2. Exo70 induced invaginations, albeit the number of tubules was smaller than samples using LUVs containing 30% PI(4,5)P2 (see Supplemental Movie 1b for tilted series).
Figure 1. Exo70 induces tubular invaginations on synthetic vesicles.
(A) Transmission EM showing that GST-Exo70 induced tubular invaginations towards the interior of the lumen of LUVs similar to the MIM I-BAR domain. GST, GST-Exo70(K571A/E572A) did not induce any membrane tubules. See EXPERIMENTAL PROCEDURES for details. Scale bar, 0.5 μm. (B) Untagged Exo70 induced membrane invaginations. The arrow shows a representative region where the tubular invagination was connected to the exterior. See also Supplemental Movie 1a for 3-D tomography of this LUV. (C) Comparison of the diameters of the membrane tubules induced by different proteins. Error bars represent standard deviation (SD). n=80; *, p<0.01. (D) Confocal microscopy of fluorescence-labeled Giant Unilamellar Vesicles (GUVs) incubated with Exo70 (upper panel) or buffer control (lower panel). Sequential frames show the inward growth of a tubule (arrowhead) from a GUV incubated with Exo70. Scale bar, 5 μm.
In addition to using LUVs and EM, we also investigated membrane tubulation induced by Exo70 using giant unilamellar vesicles (GUVs) containing 5% PI(4,5)P2 and fluorescence-labeled lipids. The large size of GUVs and the fluorescence labeling allow the direct visualization of membrane deformation in real-time. As shown in Figure 1D, addition of Exo70 induced inward growth of membrane tubules from these GUVs, which was not observed in the absence of Exo70.
Oligomerization of Exo70
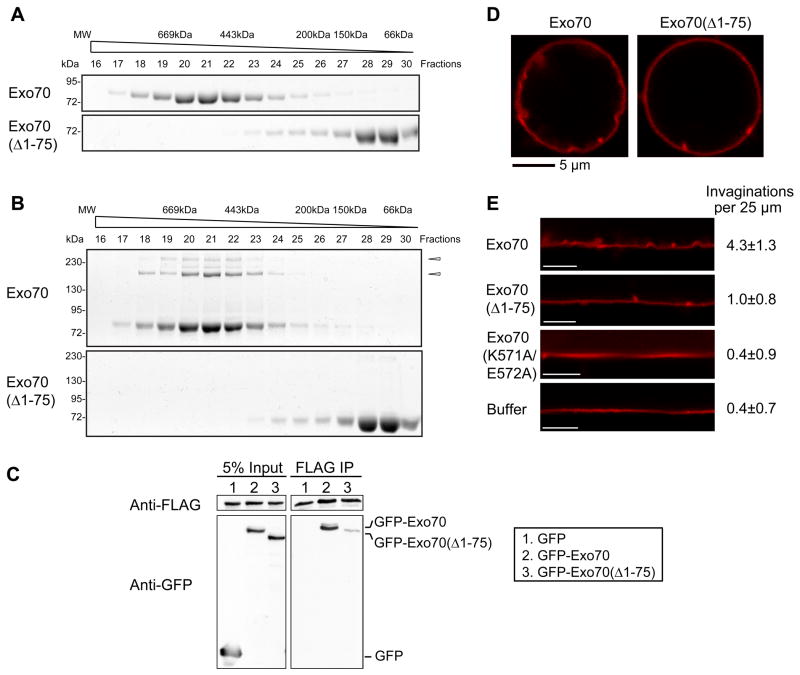
BAR proteins usually deform membrane by forming α-helical dimers or oligomers that scaffold the negatively charged membranes (McMahon and Gallop, 2005; Frost et al., 2009; Zhao et al., 2011). We therefore examined the potential oligomerization of recombinant Exo70 by gel-filtration chromatography. The full-length Exo70 was distributed in high-molecular weight (MW) fractions (Figure 2A), suggesting that Exo70 formed oligomers. Sequence analysis by COILS program (Lupas et al., 1991) predicts that the N-terminus of Exo70 has a high probability to form a coiled-coil structure, which may participate in Exo70 oligomerization. We found that Exo70(Δ1–75), which lacks the N-terminal 75 amino acids, was distributed in low-MW chromatographic fractions. To further confirm the oligomerization of Exo70, we treated the fractions with the cysteine cross-linker BM(PEG)2. A range of oligomeric species was detected in Exo70 but not Exo70(Δ1–75) samples (Figure 2B), suggesting that the high-MW fractions of Exo70 were indeed formed by oligomerization rather than due to the elongated shape of the protein. In addition to the experiments using recombinant proteins, we also detected the self-association of Exo70 in HEK293T cells by immunoprecipitation (Figure 2C).
Figure 2. Oligomerization of Exo70 is required for membrane deformation.
(A) Recombinant Exo70 proteins were loaded onto a Superdex 200 10/300GL column and eluted fractions were subjected to SDS-PAGE. Exo70, but not Exo70(Δ1–75), was eluted in high MW fractions. (B) Gel-filtration fractions were lightly treated with cross-linker BM(PEG)2, which links proteins in vicinity. High MW species were observed in Exo70 but not in Exo70(Δ1–75) fractions. Arrowheads: Exo70 oligomers. (C) Lysates of HEK293T cell expressing Exo70-FLAG together with GFP, GFP-Exo70, or GFP-Exo70(Δ1–75) were incubated with anti-FLAG (M2) beads. The inputs and bound proteins were analyzed by Western blotting using anti-FLAG (upper panels) and anti-GFP monoclonal antibodies (lower panels). Exo70-FLAG was able to pull down GFP-Exo70, but to a much lesser extent, GFP-Exo70(Δ1–75). (D) Exo70 or Exo70(Δ1–75) was incubated with GUVs and subjected to confocal microscopy. Exo70(Δ1–75) induced much fewer invaginations than Exo70. (E) The contours of GUV membranes were linearized. The numbers of invaginations per 25 μm were calculated (average ± SD). Exo70(Δ1–75) and Exo70(K571A/E572A) induced much fewer invaginations than the full-length Exo70 (n=10; p<0.01). Scale bar, 5 μm.
The ability of Exo70(Δ1–75) to deform membrane was examined using fluorescence-labeled GUVs (Figure 2D and 2E). Exo70(Δ1–75) induced much fewer invaginations than the full-length Exo70. In addition, the Exo70(K571A/E572A) mutant, which does not bind to the membrane, failed to induce invaginations, similar to the buffer control (Figure 2E). These results suggest that Exo70 employs a scaffolding mechanism to deform the membrane, which requires both membrane binding and protein oligomerization.
Molecular dynamics and mesoscale simulations on Exo70-induced membrane deformation
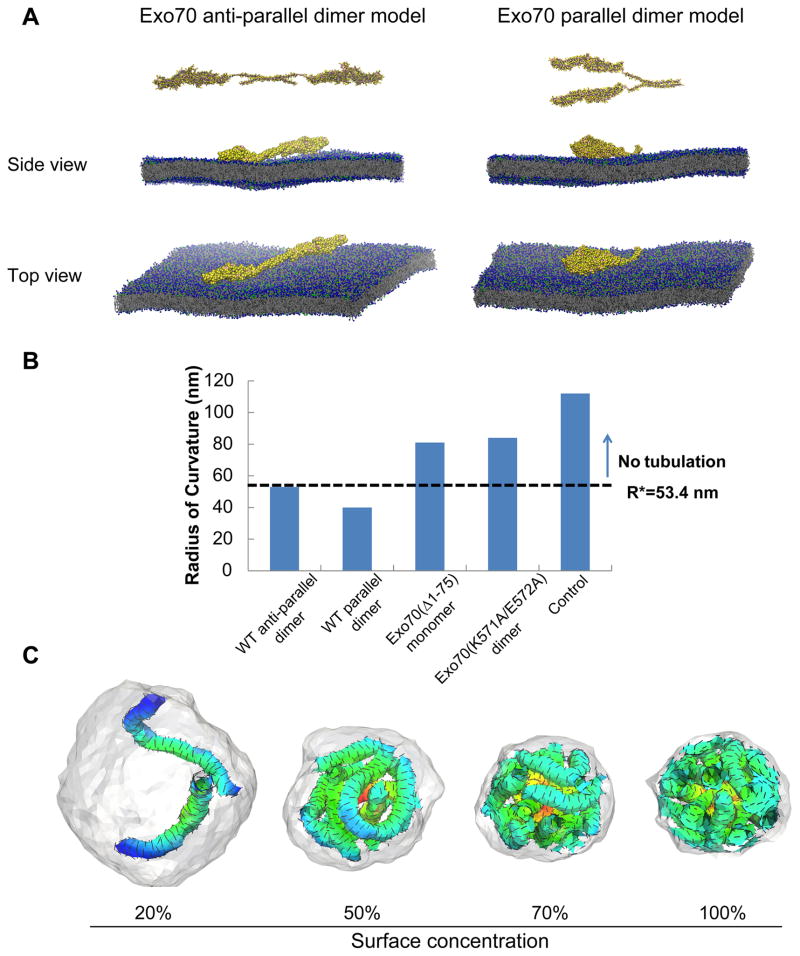
Molecular dynamics simulations were previously used to study the BAR domain-induced positive curvature and resolve at the molecular level the mechanisms, by which BAR domains sculpt the lipid bilayers (Humphrey et al., 1996; Arkhipov et al., 2008; Yin et al., 2009; Ayton and Voth, 2009). Recently, coarse-grained molecular dynamics (CGMD) simulations were employed to study the molecular interactions of ENTH domain oligomerization and association with membranes leading to the stabilization of tubular membrane geometries (Lai et al., 2012). Here we employed similar approaches to investigate the interaction between Exo70 and the membrane. We test whether the minimal oligomerized state (namely a dimer) shows enhanced curvature induction in comparison to the monomer. If the enhancement is present at the dimer level, this effect is expected to be accentuated further in the case of higher-order oligomers. The crystal structure of mouse Exo70 (amino acid 85–653) features a 170 × 35 Å rod composed of α-helical bundles slightly curved in the middle (Hamburger et al., 2006; Moore et al., 2007); and the N-terminal, non-crystalized region is predicted to be a coiled-coil structure. Based on the structural information, coarse-grained Exo70 anti-parallel and parallel dimer models were constructed using MODELLER and relaxed using all-atom molecular dynamics simulations, and then attached to an equilibrated bilayer containing 12,800 lipids with a 4:1 DOPC:DOPS ratio (Figure 3A). The system was simulated with a standard MARTINI model with time step of 40 fs up to 300 ns in real-time (see Supplemental Movie 2a-b). In addition, we simulated Exo70(Δ1–75) monomer, Exo70(K571A/E572A) mutant dimers and no protein control in the system. We have characterized the induced curvature as well as the fluctuations associated with the curvature according to a statistical procedure (see Supplemental Experimental Procedures), in which we fit frame-by-frame a two-dimensional Gaussian function to the height of the bilayer mid-plane (Supplemental Movies 3a-e). These fits characterize the dynamic negative-curvature and anisotropy associated with dimples induced by Exo70 on the bilayer in each conformation; the anisotropy is particularly pronounced in the wild-type dimers. We computed the average maximum induced curvature for each system, the extent of the curvature field (and the associated anisotropy by defining the standard deviation of the Gaussian fit to the mid-plane deformation along the major and minor axes), as well as the estimate of the deformation energy to create the equivalent membrane deformation induced by the protein (see Supplemental Figure 1A-C and associated text in Supplemental Experimental Procedures). These estimates for a single unit (i.e. protein monomer or protein dimer) interacting with a patch of a bilayer indicate that both anti-parallel and parallel dimers induce significantly more negative curvature than Exo70(Δ1–75) monomer, Exo70(K571A/E572A) mutant dimer, and control simulations of a bare membrane. The results are presented in Figure 3B in terms of R, the radius of curvature estimates (here R = 〈2Hmax〉−1). Analysis of the protein and lipid conformations in our simulations revealed that the induction of the curvature was mediated by the positively charged residues throughout the surface of the Exo70 dimers that interact with the negatively charged phospholipids, which suggests that curvature induction requires a scaffold of at least two linked Exo70 monomers that act in concert to remodel the associated lipid bilayer from its native planar state; a representation of the electrostatic map depicting this charge complementarity between the protein and the lipid interfaces is shown in Supplemental Figure 1D and E.
Figure 3. Molecular dynamics and mesoscale simulations demonstrate that Exo70 induces negative membrane curvature.
(A) Molecular dynamics simulation of modeled Exo70 anti-parallel (left) and parallel (right) dimers on a DOPC/DOPS bilayer. Top and side views of the simulation snapshots show negative curvature formed in both directions. All snapshots were rendered with VMD (Humphrey et al., 1996). See also Supplemental Movie 2 for a more comprehensive view and Supplemental Movie 3 for analysis of negative curvature recorded in the simulations. (B) Comparison of the radius of curvature estimates (here R = 〈2Hmax〉−1) for different Exo70 variants on the membrane. Dashed line at 53.4 nm represents the threshold above which no inward membrane tubulation was detected in mesoscale simulations. (C) Snapshots from mesoscale simulations of inward growing tubules for model Exo70 domains at different surface concentration (from 20% to 100%); a0=14 nm, H0||=−1.0/a0 and εLL =1kBT. The extent of tubulation is Exo70 concentration-dependent. Exo70 proteins on the outer surface of the membrane are not shown in order to enhance the visibility of the tubules. See also Supplemental Figure 1.
Mesoscale simulations have been carried out in the field to investigate endocytic vesicle formation in yeast and mammalian cells, which provide important mechanistic insights of protein-lipid interplays during endocytosis (Liu et al., 2006; Liu et al., 2009b; Agrawal et al., 2010). More recently, an extension of this approach along with concepts from the theory of self-assembly in liquid crystalline phases has been employed to study the nucleation of tubular morphologies (Ramakrishnan et al., 2010). Using this new approach, we performed Monte Carlo simulations starting from a closed vesicle with Exo70 dimers (Figure 3C). The equations and parameter settings are explained in detail in Supplemental Experimental Procedures. We have examined the collective effect of negative curvature-inducing fields on membrane morphology. In our model, the membrane surface is triangulated and each vertex can have up to one field vector of strength H0||; we note that each field vector on a vertex can represent the collective effect of multiple (but a fixed number of) proteins acting on the membrane area associated with that vertex. The average length of a link between two adjacent vertices is 1.3a0, where a0 is the intrinsic length of the triangulated membrane lattice. We set the value of a0 to be 14 nm by requiring the span of the field vector (1.3a0) in the mesoscale model to be consistent with the extent of the long-axis of the curvature field in the CGMD model for the protein (see σy values in Supplemental Figure 1C), i.e. 1.3a0=σy. We explore surface concentrations of the field vector (i.e. percentage of vertices at which a curvature field vector resides) in the range 20% to 100% and field vector strength in the range H0|| = −0.4/a0 to −1.0/a0 to find parameter regimes which stabilize inward tubulation. Figure 3C depicts typical membrane morphologies when H0|| = −1.0/a0 and surface concentration of curvature fields from 20% to 100%. At 50% and 70% surface concentration, the resulting tubular morphologies are remarkably similar to the experimentally observed morphologies depicted in the electron micrograph of Figure 1A. The extent of tubulation depends on Exo70 concentration: inward tubules were observed to form at low field vector surface concentration (20%); as the concentration increased, the number of tubules increased. Our simulations also indicated that below a field vector strength of |H0|| |< |−0.6/a0|, the inward membrane tubulation was absent. Based on the tubular morphology in our simulation, we performed geometric analyses of individual tubules to find that the average tubule diameter was 2.6a0=36 nm at H0|| =−1.0/a0 and 3.4a0=48 nm for H0|| =−0.6/a0. The threshold protein density required to induce tubular invaginations is shown in Supplemental Figure 1F; see also associated text in Supplemental Experimental Procedures for the estimation of the threshold.
All together, the simulations suggest that Exo70 generates pronounced negative membrane curvature even at the dimer-level. Estimates of the surface density of proteins required for stabilization of inward tubulation reveals that only the wild type dimers are able to induce sufficient curvature to meet the estimated threshold and the monomer as well as the lipid binding mutant fails to meet the threshold criteria.
Exo70 induces actin-free membrane protrusions in cells
In cells, overexpression of membrane-deforming proteins often results in the formation of numerous tubules in the cytoplasm (for BAR proteins such as amphiphysin) (Takei et al., 1999; Peter et al., 2004) or protrusions at the cell surface (for I-BAR proteins such as IRSp53) (Mattila et al., 2007). The expression of these BAR proteins at non-physiological levels in cells had served to test their role in generating membrane curvatures in vivo. Exo70 overexpression was previously shown to induce filopodial protrusions (Zuo et al., 2006). The protrusions on the cell surface are topologically equivalent to the inward membrane tubules observed in the synthetic vesicles shown above. However, filopodial protrusions on the cell surface also involve filamentous actin bundles (Mattila and Lappalainen, 2008). Therefore, a careful examination of these Exo70-induced protrusions is needed to better differentiate the contributions from actin and curvature induction, respectively.
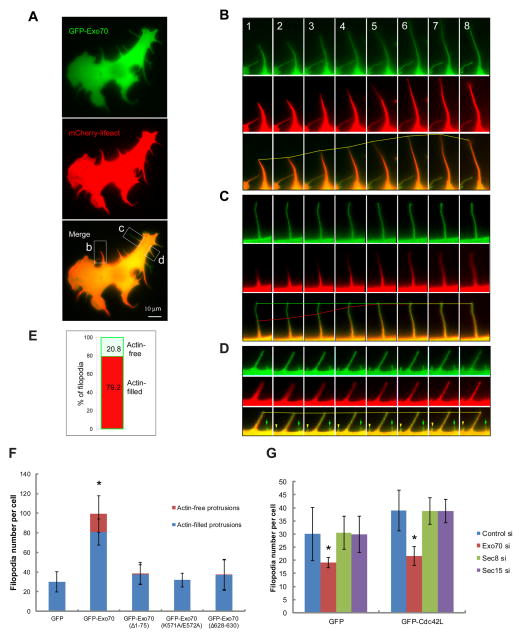
Using time-lapse fluorescence microscopy, we examined the dynamics of Exo70 and F-actin in filopodial protrusions induced by over-expressing GFP-Exo70 in B16F1 cells (Figure 4). GFP-Exo70 and F-actin (labeled by mCherry-Lifeact) frequently co-distributed along the entire length of filopodia and displayed coordinated dynamics (Figure 4B and 4D). Interestingly, there was a substantial fraction (20.8%) of GFP-Exo70-positive filopodia that were devoid of F-actin (Figure 4C-E). This phenotype was also observed previously with the overexpression of I-BAR domains, where ~10% actin-free protrusions were found (Yang et al., 2009). We observed that approximately 5% of the protrusions were first positive only with GFP-Exo70, and later filled with F-actin (Figure 4C and 4D). We have also performed line scan analysis of the GFP-Exo70-induced protrusions (Supplemental Figure 2A-C). For each protrusion, we measured the fluorescence intensity of GFP-Exo70 and actin signals along the length of protrusion, and estimated the Full Width at Half Maximum (FWHM) by Gaussian fitting using Matlab (Mathworks). The ratio between GFP-Exo70 and actin is above 1 in all protrusions analyzed (n=20), usually higher in the distal region of the protrusions, suggesting that the GFP-Exo70 signal is outside actin signal, especially at the tips. In addition to GFP-tagged Exo70, we found that the monomeric RFP-Ruby-tagged Exo70 induced membrane protrusions in a similar manner (Supplemental Figure 2D). Therefore the generation of the observed protrusions was unlikely an artifact of GFP dimerization. While the wild-type GFP-Exo70 stimulated protrusion formation, the Exo70 mutants, Exo70(Δ1–75) (defective in oligomerization), Exo70(K571A/E572A) (defective in membrane association) and Exo70(Δ628–630) (defective in binding the Arp2/3 complex) (Zuo et al., 2006; Liu et al., 2012) lost their ability to stimulate protrusions. Furthermore, in these cells, actin-free protrusions were barely detectable in the existing filopodia (Figure 4F, Supplemental Figure 2E).
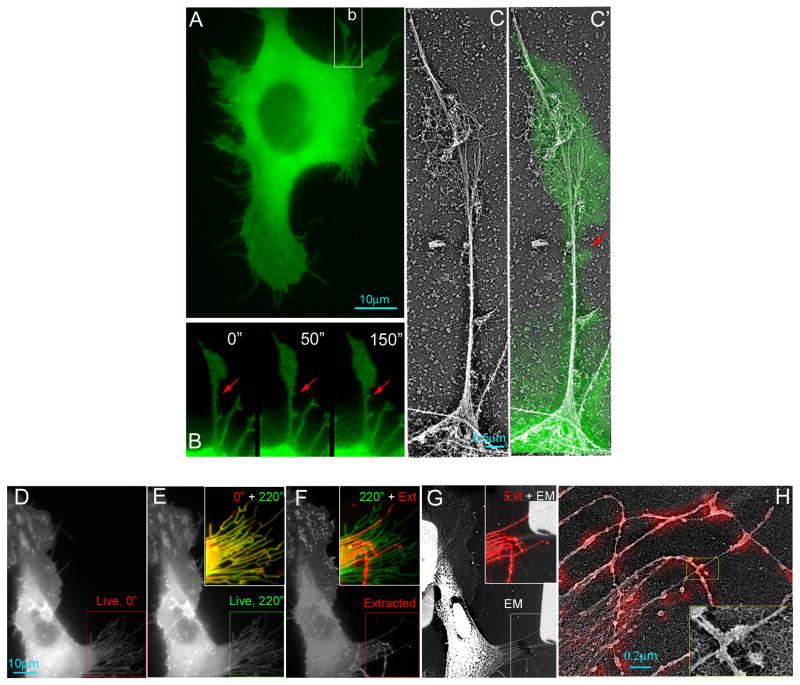
Figure 4. Characterization of Exo70-induced membrane protrusions on the cell surface.
(A) Overview of a B16F1 cell co-expressing GFP-Exo70 and mCherry-Lifeact (labeling F-actin). (B-D) Time-lapse microscopy showing dynamics of individual filopodia in Box b, c, d, of (A). Time interval between frames is 42 seconds. The lines mark the most distal points of GFP-Exo70 (green) and actin (red) signals in the filopodia over time; the yellow lines indicate the coincidence of these signals. (B) For a regular filopodium, the dynamics of GFP-Exo70 and F-actin were indistinguishable. (C) While GFP-Exo70 signal was stationary, F-actin lagged behind and gradually filled the filopodium. (D) GFP-Exo70 and F-actin co-localized in the long stationary filopodium, whereas the nascent GFP-Exo70-positive filopodium (green arrow) was actin-free. In the same field, a new membrane protrusion (yellow arrowhead) was emerging and gradually filled with F-actin. (E) The percentage of filopodia completely filled with F-actin (red) and lacking F-actin (blue) in the interior. A total of 1687 filopodia in 31 cells were counted. (F) B16F1 cells expressing GFP-Exo70 variants were stained for F-actin. GFP-Exo70 induced significantly more filopodia than GFP or GFP-Exo70 mutants. In addition, approximately 20% of the GFP-Exo70-induced membrane protrusions were actin-free. Much lower percentages of the protrusions in cells expressing GFP or GFP-Exo70 mutants (see text for description of the mutants) were actin-free. Error bars, SD. n=10; *, p<0.01. See also Supplemental Figure 2E. (G) B16F1 cells treated with siRNA targeting Exo70, Sec8 or Sec15 were transfected with GFP or GFP-Cdc42L. The expression of GFP-Cdc42L induced more filopodia comparing to GFP cells. The number of filopodia was significantly reduced upon Exo70 knockdown. Knockdown of Sec8 or Sec15 did not affect the filopodia formation in GFP-Cdc42L or GFP cells. Error bars, SD. n=10; *, p<0.01. See also Supplemental Figure 3B for Western blot showing the knockdown efficiency.
To examine the ultrastructure of actin in GFP-Exo70 induced protrusions, we performed platinum replica EM. For filopodia that did contain actin, there were fewer (2–4, usually more than 15 in normal filopodia, Lewis and Bridgman, 1992) or loosely bundled actin filaments (Supplemental Figure 2F-I). To further confirm the above observation, correlative time-lapse fluorescence and platinum replica EM were performed on GFP-Exo70 expressing cells. We found that many filopodial protrusions were either empty or contained very few linear actin filaments (Figure 5). Furthermore, after detergent extraction, more than 50% of these protrusions were completely dissolved (~20%) or lost their distal regions (~30%), suggesting the absence of actin in the detergent-sensitive regions (Figure 5D-H). Our observation that Exo70 induces membrane protrusions devoid of actin is consistent with the in vitro finding of tubule generation inside the lumen of synthetic vesicles (Figure 1) as they are topologically equivalent in the direction of tubulation (“negative curvature”).
Figure 5. Dynamics and ultrastructures of Exo70-induced membrane protrusions.
(A-C) Correlative fluorescence and platinum replica EM showing the organization of actin in protrusive structures in a GFP-Exo70-expressing cell. (A) Overview of a B16F1 cell expressing GFP-Exo70. Scale bar, 10 μm. (B) Frames from the time-lapse sequence showing dynamics of a protruding filopodium (corresponding to box b in A). Correlative EM of region B is shown in C or with overlaid GFP-Exo70 fluorescence (green) (C′). The red arrows show a newly generated membrane protrusion that was devoid of actin. (D-H) Correlative fluorescence and platinum replica EM of a GFP-Exo70-expressing cell shown by GFP fluorescence (D-F) or EM (G) at the beginning (D) and the end (E and F) of the 220 sec time-lapse sequence and after detergent extraction (“Ext”, with 1% Triton X-100 in PEM buffer containing 2 μM phalloidin without 2% PEG) (F, G). Inset in E is an overlay of boxed regions from D and E, showing that most of filopodial protrusions marked by GFP-Exo70 was barely changed. Inset in F is an overlay of boxed regions from E and F showing that half of membrane protrusions were removed by detergent extraction, while others remained (shown in red), which contained actin filaments as shown by EM (inset in G). (H) Correlative EM showing that each detergent-resistant filopodia contained only 2–4 actin filaments, which are superimposed with GFP-Exo70 signals (colored red).
To test whether the ability of Exo70 to stimulate membrane protrusion depends on the other members of the exocyst complex, we examined the filopodial protrusions in B16F1 cells with Sec8 and Sec15 (two other components of the exocyst complex) knocked down by RNAi. GFP-Exo70 induced numerous protrusions in these knockdown cells similar to the control cells, with approximately 20% of the protrusions actin-free (Supplemental Figure 3A). The constitutively active form of Cdc42, Cdc42(Q61L), stimulated filopodia formation in B16F1 cells (Zuo et al., 2006; Figure 4G). Knockdown of Exo70 reduced the number of filopodia induced by Cdc42(Q61L), whereas knockdown of Sec8 or Sec15 had no effect, further suggesting that Exo70 is specifically required for filopodia induction (Figure 4G; Supplemental Figure 3B). In cells, higher percentage of endogenous Exo70 was localized to the plasma membrane comparing to Exo84 and Sec10 (Supplemental Figure 3C-D). By gel filtration, we found that the endogenous Exo70 exists in a free pool in addition to the holo-complex (Supplemental Figure 3E). Taken together, it is possible that Exo70 induces membrane protrusions independent of the other exocyst components.
Exo70 mediates leading edge protrusion and directional cell migration
Membrane protrusions at the leading edge are important for directional cell migration (Pollard and Borisy, 2003; Mattila and Lappalainen, 2008; Ridley, 2011). A branched actin network mediated by the Arp2/3 complex “pushes” the plasma membrane to generate surface protrusions, which is the physical step that initiates directional cell migration (Pollard and Borisy, 2003; Ridley, 2011). Recently, accumulating evidence suggests an important role of membrane curvature induction for protrusion formation (Bereiter-Hahn, 2005; Suetsugu et al., 2006; Mattila et al., 2007; Ji et al., 2008; Yang et al., 2009; Guerrier et al., 2009). Since Exo70, like I-BAR domains (Suetsugu et al., 2006; Mattila et al., 2007; Yang et al., 2009) and srGAP2 F-BAR (Guerrier et al., 2009), can induce negative membrane curvature, it is possible that Exo70 uses its membrane-deforming activity for leading edge protrusion and directional cell migration.
We tested this hypothesis by taking advantage of the Exo70 mutants defective in deforming membrane in vitro. The Exo70(K571A/E572A) and Exo70(Δ1–75) mutants were selected because they maintained their ability to bind to other exocyst components and to interact with Arpc1 (Supplemental Figure 4A-B). In addition, in our exocytosis assays using VSV-G-ts45, Exo70(Δ1–75) was able to rescue the exocytosis block in Exo70 knockdown cells similar to the wild-type Exo70 (Supplemental Figure 4C-E). Exo70 mutants did not induce filopodial protrusions in B16F1 cells (Figure 4F, Supplemental Figure 2E), consistent with in vitro (Figure 2) and in silico (Figure 3) results.
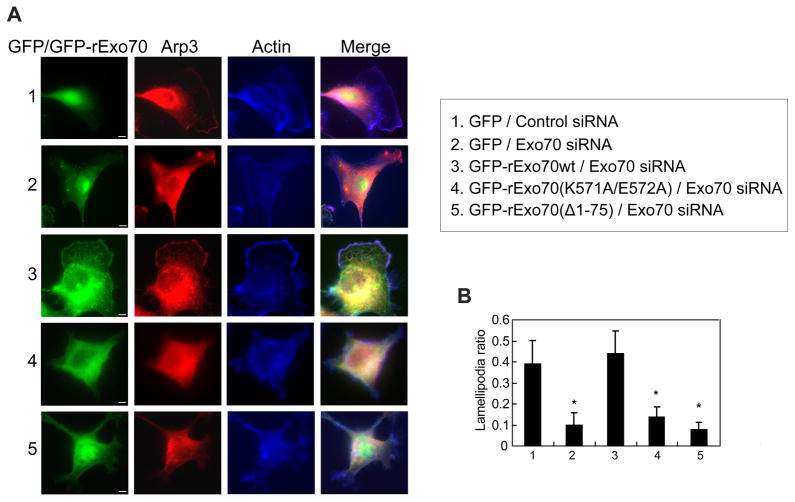
Human MDA-MB-231 cell lines stably expressing GFP-tagged rat Exo70 mutants Exo70(K571A/E572A) and Exo70(Δ1–75) were generated. The endogenous Exo70 was then knocked down by siRNA in these cells. The levels of knockdown were confirmed by Western blotting (Supplemental Figure 5A). The Exo70 knockdown cells expressing Exo70 membrane-deformation mutants were dramatically defective in their ability to form lamellipodial protrusions as revealed by Arp3 and F-actin staining (Figure 6A). The “lamellipodia ratio”, defined as the length of the leading edge divided by the total cell perimeter, was significantly lower in the knockdown cells expressing Exo70 mutants (Figure 6B). Exo70 was shown to stimulate the Arp2/3 complex-mediated actin polymerization (Liu et al., 2012). The problem in lamellipodia formation in the Exo70(Δ1–75) cells was, however, unlikely due to a defect in actin dynamics, as the lysates from these cells had comparable stimulatory effect as those from wild type Exo70-expressing cells on actin polymerization in our pyrene actin assay (Supplemental Figure 4F).
Figure 6. The function of Exo70 in membrane deformation is required for lamellipodia formation.
(A) MDA-MB-231 cells expressing indicated siRNA and GFP-tagged rat Exo70 variants were stained for Arp3 (red) and F-actin (blue) to detect lamellipodia. Control siRNA-treated cells (Panel 1) had clear and extended lamellipodia. In Exo70-knockdown cells (Panel 2), the formation of lamellipodia was impaired. Expression of GFP-rExo70 (Panel 3) rescued lamellipodia formation, whereas the expression of Exo70 mutants failed to rescue the defect (Panel 4–5). Scale bar, 5 μm. (B) The ratios between the length of the lamellipodia and the total cell perimeters (“lamellipodia ratio”) were compared for each group. The mutant groups have lower lamellipodia ratios. Error bars, SD. n=25; *, p<0.01.
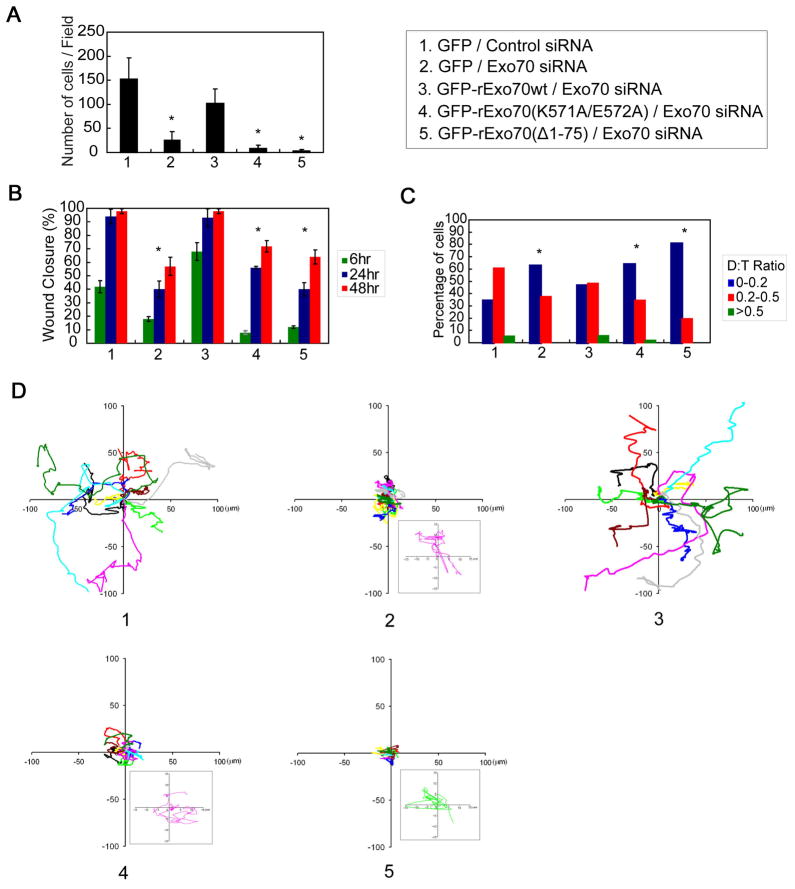
The migratory properties of these cells were also examined by three different assays. In the transwell assay, much fewer Exo70 knockdown cells expressing the Exo70 mutants migrated to the lower chamber, as compared to the control cells (Figure 7A). Similarly, in a wound-healing assay, it took the Exo70 mutant cells longer time to close the wounds (Figure 7B and Supplemental Figure 5B). Finally, by tracking the trajectories of individual cells over time, we found that cells expressing Exo70 mutants were not only slower in migration, but also less capable of maintaining the direction of their movement (Figure 7C and 7D). The directional persistence, quantified as the ratio of the net displacement to the total migration distance (“D:T ratio”), was much smaller in cells expressing Exo70 mutants (Figure 7C). Taken together, these results suggest that the membrane-deforming activity of Exo70 plays an important role in membrane protrusion formation and directional cell migration.
Figure 7. The function of Exo70 in membrane deformation is required for directional cell migration.
(A) Transwell assays were performed using cells described in Figure 6. The bars indicate the average number of migrated cells per field for each group. Error bars, SD. n=3; *, p<0.01. (B) Wound-healing assays were performed. The percentage of wound closure (closure distance over initial opening) after 6, 24, and 48 hours of migration was calculated for each group. Error bars, SD. n=3; *, p<0.01. See also Supplemental Figure 5B for wound healing images. (C) Single cell movement was tracked using time-lapse microscopy. Directional persistence of individual cells was calculated as D:T ratio (see text). The percentage of cells with different D:T ratios (0–0.2; 0.2–0.5; >0.5) is indicated. n=60; *, p<0.01. (D) The trajectories of 10 representative cells at 5 min intervals during a 400-min migration period are presented for each group. The origins of each track were superimposed at position (0, 0). The boxed regions in corresponding treatments showed 7× magnified migration track from one representative cell in each group, which indicates that the cells were not stationary.
DISCUSSION
Combining biochemical and cell biological approaches and in silico molecular simulations, our study reveals a function for Exo70 in the generation of negative membrane curvature in vitro. In cells, overexpression of Exo70 induces filopodial protrusions, many of which are actin-free at the tip, supporting the role of Exo70 in membrane bending. Furthermore, cells with inhibition of Exo70, or expressing Exo70 mutants deficient in curvature induction, are defective in generating filopodia and lamellipodia, and fail in directional migration.
Phospholipid-binding and protein oligomerization-mediated scaffolding is one of the major mechanisms for membrane curvature induction (McMahon and Gallop, 2005; Frost et al., 2009; Mim et al., 2012; Boucrot et al, 2012). By sequence alignment, Exo70 does not belong to any of the BAR domain families. On the other hand, the rod-like structure of Exo70 and its oligomerization may bestow its overall resemblance to the BAR proteins. Through the characterization of Exo70 mutants, we demonstrate that both phospholipid-binding and oligomerization are needed for Exo70 to induce membrane curvature. In addition to the biochemistry and cell biology experiments, we investigate the molecular mechanism of Exo70-induced membrane deformation by molecular dynamics simulations. Based on the known Exo70 sequence and its crystal structure, our analysis of the protein-membrane interactions predicts the ability of dimeric Exo70 to induce negative curvature through positively charged residues on its surface that interact with phospholipids. Moreover, mesoscale Monte Carlo simulations demonstrate that Exo70 generates membrane tubules on liposomes with remarkable similarities to the EM micrographs obtained from our in vitro membrane tubulation assay. The simulations of protein-membrane interactions at the atomic scale and membrane tubulation at the mesoscale further suggest an oligomerization-mediated mechanism for Exo70-induced membrane curvature induction.
Membrane curvature induction is often coupled with cytoskeletal activities (for review, see Doherty and McMahon, 2008; Scita et al., 2008). Exo70 was previously shown to directly interact with and kinetically stimulate the Arp2/3-complex-mediated actin polymerization and branching (Zuo et al., 2006; Liu et al., 2012). The function of Exo70 on actin dynamics can be coordinated with its ability to induce membrane curvature during such processes as morphogenesis and cell migration. It is thought in the field that a branched actin network mediated by the Arp2/3 complex “pushes” the plasma membrane for cells to protrude. Recent findings on I-BAR and F-BAR proteins led to the appreciation of membrane curvature in protrusion formation (Mattila et al., 2007; Mattila and Lappalainen, 2008; Yang et al., 2009; Guerrier et al., 2009). It is plausible that Exo70, like the above-mentioned proteins, uses its membrane-deforming activity in leading edge protrusion and directional cell migration. The ability of Exo70 to induce negative membrane curvature may create a space at the leading edge to accommodate active actin polymerization and branching mediated by the Arp2/3 complex. Exo70 may thus couple membrane remodeling and actin dynamics for effective protrusion formation and cell migration.
In this study, we find that overexpression of Exo70 can induce actin-free filopodial protrusions, further supporting the role of Exo70 in membrane curvature induction in cells. Similar phenotypes have been observed with the overexpression of I-BAR domains (Suetsugu et al., 2006; Yang et al., 2009) and srGAP2 F-BAR (Guerrier et al., 2009) that are capable of inducing negative membrane curvatures. The “actin-free” filopodial protrusions observed in these studies are consistent with the emerging view that membrane curvature induction plays a role in protrusion formation (Scita et al., 2008; Mattila and Lappalainen, 2008). The actin networks in lamellipodia during cell migration are mostly branched filaments generated by the Arp2/3 complex (Pollard and Borisy, 2003), whereas those in filopodia are bundled filaments (Mattila and Lappalainen, 2008). Unlike in lamellipodia, the Arp2/3 complex in the distal tips of filopodia is less prevailing (Svitkina et al., 1999). As such, in filopodia, Exo70 cannot effectively exert its stimulatory effect on actin branch formation as it does in lamellipodia (Liu et al., 2012). The filopodial tips devoid of actin in the Exo70-overexpressing cells may reflect that the membrane-deforming activity of Exo70 surpasses its ability to stimulate actin in those regions.
The function of Exo70 in membrane deformation may be independent of the exocyst complex. None of the other exocyst subunits can induce membrane protrusions in cells upon overexpression. Exo70 is able to induce protrusions even when other exocyst subunits are knocked down. Inhibition of Exo70, but not any other exocyst components, affects membrane protrusion formation in cells with or without Cdc42 induction. Exo70 has a clear enrichment in the plasma membrane comparing to other subunits. Furthermore, the endogenous Exo70 exists in free pools as well as being part of the exocyst holo-complex; similar observation has recently been made in yeast cells (Morgera et al., 2012). It is possible that the free pool of Exo70 exerts its function in membrane curvature induction during protrusion formation.
In summary, our studies reveal that Exo70, through an oligomerization-based scaffolding mechanism, generates negative curvature on the plasma membrane. In addition to cell migration, Exo70 and other members of the exocyst have been shown to function in such cellular processes as primary ciliogenesis, viral progression, neuritis growth and branching, all of which involve plasma membrane remodeling. It will be interesting to test whether the function of Exo70 in membrane deformation reported here also applies to these processes. Moreover, as Exo70 is regulated by small GTPases and kinases (Inoue et al., 2003; Ren and Guo, 2012), it will be interesting to investigate how the function of Exo70 is spatially and temporally controlled in various cellular contexts to fulfill different physiological functions.
EXPERIMENTAL PROCEDURES
Detailed information on plasmids, antibodies, cell culture, protein purification, immunoprecipitation and in vitro binding assays, additional information on molecular dynamics simulations and mesoscale simulations, VSV-G trafficking assay, and pyrene actin assay is described in the Supplemental Experimental Procedures.
Statistics
Statistical analyses of the data were all performed using two-tailed Student’s t-test. Sample size (n) and error estimates such as standard deviation (SD) or standard error (s.e.m) are indicated in the text or figure legends.
LUV preparation and thin-section electron microscopy
PI(4,5)P2(L-α-phosphatidylinositol-4,5-bisphosphate; porcine brain triammonium salt) and DOPC (1,2-dioleoyl-sn-glycero-3-phosphocholine) were purchased from Avanti Polar Lipids. LUVs were prepared using a protocol adapted from Mattila et al, 2007. PI(4,5)P2 and DOPC were mixed at a molar ratio of 30:70, or PI(4,5)P2: DOPS: DOPE: DOPC were mixed at a molar ratio of 5:15:10:70, dried with nitrogen stream, and hydrated in 20 mM Hepes-KOH, pH 7.5, and 100 mM NaCl to a total lipid concentration of 0.5mM. The mixed lipids were subjected to five cycles of freeze-thaw and extrusion through 1 μm filters using a mini-extruder. The concentrations of LUVs and proteins were chosen based on previous studies (Peter et al., 2004; Liu, et al., 2007; Mattila et al., 2007). 100 μM LUVs were incubated with 2 μM recombinant proteins at room temperature for 1 hour, fixed with 2% glutaraldehyde and 0.1% tannic acid in 0.1M Na-cacodylate buffer for 1hour at 4°C. Samples were then sedimented (17,000 g, 30 min at 4°C) and post-fixed in 1% OsO4 for 1 hour at 4°C, followed by en bloc staining with 2% uranyl acetate for 1 hour at 4°C, dehydration and Spurr embedding. 100 nm sections were prepared and stained with uranyl acetate and lead citrate. All samples were visualized using a JEM 1011 transmission electron microscope (JEOL USA) operated at 100 kV. For tilt series, specimen was tilted ±20º at 5º intervals. Images were captured by ORIUS 835.10W CCD camera (Gatan). The diameters of the membrane tubules inside the LUVs were measured by ImageJ. For 3-D reconstruction, images were aligned and reconstituted with TomoJ plugin in ImageJ.
GUV preparation and confocal microscopy
POPC (1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine), DOPS (1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-L-serine, sodium salt) and DSPE-Bio-PEG2000 (1,2-distearoyl-sn-glycero-3-phosphoethanolamine-N-[biotinyl(polyethyleneglycol)-200], ammonium salt) were purchased from Avanti Polar Lipids and Texas Red-DHPE (Texas Red-1,2-dihexadecanoyl-sn-glycero-3-phosphoethanolamine) from Invitrogen. GUVs were prepared from a lipid mixture containing POPC, POPS, PI(4,5)P2, Texas Red-DHPE and DSPE-Bio-PEG2000 (molar ratio 80:15:5:0.3:0.3) by electroformation (Angelova et al., 1986; Mathivet et al., 1996), as described (Tian and Baumgart, 2009), except that the sucrose concentration was 350 mM to osmotically balance the solutions.
GUVs were introduced into protein solutions within polypropylene tubes at room temperature at a final protein concentration of 5 μM. Aliquots were transferred between glass coverslips separated by Parafilm. GUV images were collected using a confocal fluorescence microscope (Tian and Baumgart, 2009), with 543 nm laser excitation of Texas Red-DHPE. The contours of the GUVs were transformed to a co-ordinate system using the Polar Transformation plugin in ImageJ.
Gel filtration
200 μl (600 μg) recombinant Exo70 or Exo70(Δ1–75) were loaded onto a Superdex 200 10/300GL column and eluted with PBS containing 1mM DTT at a flow rate of 0.5 ml/min. 500 μl fractions were collected. 20 μl of each fraction was subjected to 8% SDS-PAGE and stained with SimplyBlue SafeStain (Invitrogen). The column was calibrated using molecular weight markers (MW-GF-1000, Sigma). For cross-linking, fractions were treated with 40nM BM(PEG)2 (Thermo Scientific) for 1 hour at room temperature prior to SDS-PGAE. Exo70 has 4 cysteines and BM(PEG)2 has a spacer arm of 14.7Å.
Gel filtration of MDA-MB-231 cell lysates was performed using a Superdex 200 10/300GL column in a buffer containing 20 mM HEPES, pH 7.4, 1 % NP-40, 100 mM KCl, 5 mM MgCl2, 1 mM NaF, 1 mM DTT, 1mM PMSF. Fractions were collected and analyzed by Western blotting.
Molecular dynamics (MD) simulations
A coarse-grained Exo70 model was constructed according to the crystal structure in the protein data bank (pdb code 2PFT) (Moore et al., 2007). Structures for the unresolved loops and alpha-helical N-terminus were constructed with MODELLER (Sali and Blundell, 1993). The resulting structure was relaxed using a short, 10 ns all-atom simulation under the CHARMM27 force field in the GROMACS molecular dynamics simulation package (Bjelkmar et al., 2010). A representative snapshot was then coarse-grained for use in the MARTINI force field (Monticelli et al., 2008). To maintain secondary structure, elastic bonds were applied to all backbone beads within 0.5 – 0.9 nm with a force of 500 kJ/mol-nm2, except for the N-terminal tail, for which we used a much weaker 5 kJ/mol-nm2 force constant for residues 1–20 and 50 kJ/mol-nm2 for residues 21–90.
Wild type Exo70 was modeled in a parallel or anti-parallel dimer conformation while the Exo70(K571A/E572A) mutant was modeled in an anti-parallel conformation and compared to the corresponding wild type control. Exo70(Δ1–75) model was effectively a monomer as the dimerization domain is considerably shortened. Each modeled Exo70 variant was attached to an equilibrated bilayer containing 12,800 lipids with a 4:1 DOPC:DOPS ratio. The charge (−1) on DOPS partly serves as a surrogate for the higher charge (−4) on PIP2 at a lower concentration. The systems were minimized and simulated with a standard MARTINI time step of 40 fs for 100 ns. Due to the smoothed energy landscape in the MARTINI force field, this corresponds to a real time of 3–4 times longer. The Berendsen thermostat and barostat maintained a temperature of 310K and semi-isotropic pressure coupling of 1.0 bar both parallel and perpendicular to the bilayer plane with coupling time constants of 1.0 and 0.2 ps, respectively. Simulations were solvated with 600,000 water beads, corresponding to 4 water molecules each, and counter-ions were added at a physiological concentration of 150 mM. All other parameters were set per the MARTINI force field specifications (Monticelli et al., 2008). All simulations were performed on supercomputing platforms at the Texas Advanced Computing Center (TACC). The coarse-grained MD (CGMD) simulations of the protein-membrane system were run on parallel architectures with 48–46 processors.
Live cell imaging
B16F1 cells were co-transfected with GFP-Exo70 and mCherry-Lifeact. The cells were re-plated the next day, and transferred into phenol red–free L-15 medium supplemented with 10% FBS and kept on the microscope stage at 37ºC during observation. Light microscopy was performed using an Eclipse TE2000-U inverted microscope (Nikon) equipped with Planapo 100× 1.3 NA objective and Cascade 512B CCD camera (Photometrics) driven by Metamorph imaging software (Molecular Devices). For quantification purposes, filopodia with F-actin-free tips were defined as filopodia lacking detectable mCherry-Lifeact fluorescence signal within a distance of 0.6μm or more from the tip of the GFP-Exo70 signal in the same filopodium.
Electron microscopy of B16F1 cells
Cells for platinum replica EM were processed as described previously (Svitkina and Borisy, 2006; Svitkina, 2007). Briefly, cells were extracted for 5 min at room temperature with 1% Triton X-100 in PEM buffer containing 2 μM phalloidin with or without 2% PEG (MW 35,000), followed by fixation with 2% glutaraldehyde in 0.1 M Na-cacodylate (pH 7.3).
Correlative EM was performed as described using cells growing on marked coverslips (Svitkina and Borisy, 2006; Svitkina, 2007). Samples were analyzed using a JEM 1011 transmission electron microscope (JEOL USA) operated at 100 kV. Images were captured by an ORIUS 835.10W CCD camera (Gatan) and presented in inverted contrast.
Fluorescence microscopy
MDA-MB-231 and B16F1 cells with different treatments were grown on coverslips, washed with phosphate-buffered saline (PBS), fixed in 4% formaldehyde at room temperature for 12 min, washed, permeabilized for 5 min with PBST (PBS-Tween), and blocked for 10 min with 2% bovine serum albumin in PBST. To quantify lamellipodial protrusion, MDA-MB-231 cells were co-stained with an anti-Arp3 polyclonal antibody and Alexa Fluor 350 phalloidin (Invitrogen/Molecular Probes) for fluorescence observation using Leica DMI 6000B inverted microscope equipped with a DFC350 FX camera and a 63× objective. Images were processed with Adobe Photoshop (Adobe Systems, version 7.0). “Lamellipodia ratio” was defined as the perimeter occupied by the lamellipodia to the whole cell perimeter as measured by ImageJ 1.73v software. To quantify the number of filopodia, B16F1 cells were stained with Alexa Fluor 594 phalloidin (Invitrogen/Molecular Probes). To quantify the localization of different exocyst subunits at the protruding plasma membrane, B16F1 cells were co-stained with anti-Exo70 monolocal antibody and anti-Exo84 or Sec10 polyclonal antibody. The fluorescence intensities at the protruding plasma membrane and the whole cell were measured using ImageJ.
Wound healing assay
MDA-MB-231 cells were seeded in 6-well plates (~2×106 cells per well) for 24 hours. When the cells grew to confluence, scratches were introduced using a sterile 20-μl pipette tip. Cells were washed with PBS and refreshed with regular growth medium. Cells were imaged after wounding at 0, 6, 24 and 48 hours by phase contrast microscopy using Leica DMI 6000B inverted microscope equipped with a DFC350 FX camera and a 10× objective. The percentage of wound closure was calculated by dividing the wound distance by the initial distance.
Transwell migration assay
MDA-MB-231 cells (1 ×105 cells per well) with different treatments were seeded in the upper chamber of a transwell filter with 6.5-mm diameter, 8-μm pore size membrane (Corning Inc.) in DMEM without serum. Cells were allowed to migrate across the filter for 24 hours at 37°C towards the lower side of the chamber containing DMEM plus 10% FBS. Cells on the upper side of the filter were removed with cotton swabs, and migrated cells on the lower side of the chamber were fixed and stained with Giemsa stain. The average number of migrated cells in five randomly chosen fields per transwell insert was counted to quantify the extent of migration.
Tracking individual cell migration by time-lapse microscopy
MDA-MB-231 cells were plated on 35-mm glass bottom dishes (MatTek Corporation) at a density of 4×104 cells in Leibovitz’s (L-15) medium (GIBCO) and placed in a temperature-controlled chamber. The movement of individual cells was traced with an inverted microscope (Eclipse TE2000-U, Nikon) using a 4× objective. Phase time-lapse recording started 24 hours after plating and images were collected at 5-min intervals over 15 hours. The motility parameters, including migration path, net distance and rate, were obtained from time-lapse movies using Metamorph imaging software. The migration paths were shown as graphs using the Excel (Microsoft). The directional persistence was calculated as a ratio of the net displacement and the total length of migration paths (the “D:T ratio”) during a 400-min period. 60 cells were analyzed for each treatment. Each set of experiments was performed in duplicate.
Supplementary Material
Highlights.
Exo70 induces negative curvature on synthetic vesicles and modeled membranes
Exo70 employs an oligomerization-based scaffolding mechanism to deform membrane
Exo70 induces actin-free filopodia, independent of the other exocyst subunits
Membrane-deformation by Exo70 is required for lamellipodia formation and cell migration
Acknowledgments
We thank Drs. Roberto Dominguez, Pekka Lappalainen, Hongxia Zhao, Shu-Chan Hsu, Charles Yeaman, Bing He for reagents and helpful discussions. We also thank Ms. Jian Zhang for her technical assistance. The work is supported by grants from National Institutes of Health to W.G. (GM085146). Y.Z. is supported by an American Heart Association pre-doctoral fellowship. The JEOL 1011 transmission electron microscope was supported by the NIH Shared Instrumentation Grant RR-22482 to T.S. Supercomputing resources to perform large scale simulations were available through an XSEDE allocation grant.
Footnotes
AUTHOR CONTRIBUTIONS
Y.Z., J.L. and W.G. conceived the project and designed experiments; Y.Z., J.L., C.Y., B.R.C., X.X. and T.S. designed and/or carried out some of the experiments; R.P.B, N.R., and R.R. carried out the molecular dynamics and mesoscale simulations. Y.Z., J.L. and W.G. analyzed the data and wrote the manuscript. Y.Z. and J.L. contributed equally to this work. The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Agrawal NJ, Nukpezah J, Radhakrishnan R. Minimal mesoscale model for protein-mediated vesiculation in clathrin-dependent endocytosis. PLoS Comput Biol. 2010;6:e1000926. doi: 10.1371/journal.pcbi.1000926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelova MI, Dimitrov DS. Liposome electroformation. Faraday Discuss Chem Soc. 1986;81:303–311. [Google Scholar]
- Arkhipov A, Yin Y, Schulten K. Four-scale description of membrane sculpting by BAR domains. Biophys J. 2008;95:2806–2821. doi: 10.1529/biophysj.108.132563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayton GS, Voth GA. Systematic multiscale simulation of membrane protein systems. Curr Opin Struct Biol. 2009;19:138– 144. doi: 10.1016/j.sbi.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bereiter-Hahn J. Mechanics of crawling cells. Med Eng Phys. 2005;27:743–753. doi: 10.1016/j.medengphy.2005.04.021. [DOI] [PubMed] [Google Scholar]
- Bjelkmar P, Larsson P, Cuendet MA, Hess B, Lindahl E. Implementation of the CHARMM force field in GROMACS: Analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J Chem Theory Comput. 2010;6:459–466. doi: 10.1021/ct900549r. [DOI] [PubMed] [Google Scholar]
- Boucrot E, Pick A, Çamdere G, Liska N, Evergren E, McMahon HT, Kozlov MM. Cell membrane fission is promoted by insertion of amphipathic helices and is restricted by crescent BAR domains. Cell. 2012;149:124–136. doi: 10.1016/j.cell.2012.01.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mediation, modulation, and consequences of membrane-cytoskeleton interactions. Annu Rev Biophys. 2008;37:65–95. doi: 10.1146/annurev.biophys.37.032807.125912. [DOI] [PubMed] [Google Scholar]
- Dong G, Hutagalung AH, Fu C, Novick P, Reinisch KM. The structures of exocyst subunit Exo70p and the Exo84p C-terminal domains reveal a common motif. Nat Struct Mol Biol. 2005;12:1094–1100. doi: 10.1038/nsmb1017. [DOI] [PubMed] [Google Scholar]
- Frost A, Unger VM, De Camilli P. The BAR Domain Superfamily: Membrane-Molding Macromolecules. Cell. 2009;137:191–196. doi: 10.1016/j.cell.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrier S, Coutinho-Budd J, Sassa T, Gresset A, Jordan NV, Chen K, Jin WL, Frost A, Polleux F. The F-BAR domain of srGAP2 induces membrane protrusions required for neuronal migration and morphogenesis. Cell. 2009;138:990–1004. doi: 10.1016/j.cell.2009.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger ZA, Hamburger AE, West AP, Jr, Weis WI. Crystal structure of the S.cerevisiae exocyst component Exo70p. J Mol Biol. 2006;356:9–21. doi: 10.1016/j.jmb.2005.09.099. [DOI] [PubMed] [Google Scholar]
- He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Xi F, Zhang X, Zhang J, Guo W. Exo70 interacts with phospholipids and mediates the targeting of the exocyst to the plasma membrane. EMBO J. 2007;26:4053–4065. doi: 10.1038/sj.emboj.7601834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu S-C, TerBush D, Abraham M, Guo W. The exocyst complex in polarized exocytosis. Inter Rev Cytology (2004) 2004;233:243–265. doi: 10.1016/S0074-7696(04)33006-8. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Inoue M, Chang L, Hwang J, Chiang SH, Saltiel AR. The exocyst complex is required for targeting of Glut4 to the plasma membrane by insulin. Nature. 2003;422:629–633. doi: 10.1038/nature01533. [DOI] [PubMed] [Google Scholar]
- Ji L, Lim J, Danuser G. Fluctuations of intracellular forces during cell protrusion. Nat Cell Biol. 2008;10:1393–1400. doi: 10.1038/ncb1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CL, Jao CC, Lyman E, Gallop JL, Peter BJ, McMahon HT, Langen R, Voth GA. Membrane binding and self-association of the epsin N-terminal homology domain. J Mol Biol. 2012;423:800–817. doi: 10.1016/j.jmb.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Kaksonen M, Drubin DG, Oster G. Endocytic vesicle scission by lipid phase boundary forces. Proc Natl Acad Sci USA. 2006;103:10277–10282. doi: 10.1073/pnas.0601045103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zuo X, Yue P, Guo W. Phosphatidylinositol 4,5-bisphosphate mediates the targeting of the exocyst to the plasma membrane for exocytosis in mammalian cells. Mol Biol Cell. 2007;18:4483–4492. doi: 10.1091/mbc.E07-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yue P, Artym VV, Mueller SC, Guo W. The role of the exocyst in matrix metalloproteinase secretion and actin dynamics during tumor cell invadopodia formation. Mol Biol Cell. 2009;20:3763–3771. doi: 10.1091/mbc.E08-09-0967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Sun Y, Drubin DG, Oster GF. The Mechanochemistry of Endocytosis. PLoS Biol. 2009;7:e1000204. doi: 10.1371/journal.pbio.1000204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Zhao Y, Sun Y, He B, Yang C, Svitkina T, Goldman YE, Guo W. Exo70 Stimulates the Arp2/3 Complex for Lamellipodia Formation and Directional Cell Migration. Curr Biol. 2012 doi: 10.1016/j.cub.2012.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- Mathivet L, Cribier S, Devaux PF. Shape change and physical properties of giant phospholipid vesicles prepared in the presence of an AC electric field. Biophys J. 1996;70:1112–1121. doi: 10.1016/S0006-3495(96)79693-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Pykalainen A, Saarikangas J, Paavilainen VO, Vihinen H, Jokitalo E, Lappalainen P. Missing-in-metastasis and IRSp53 deform PI(4,5)P2-rich membranes by an inverse BAR domain-like mechanism. J Cell Biol. 2007;176:953–964. doi: 10.1083/jcb.200609176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattila PK, Lappalainen P. Filopodia: molecular architecture and cellular functions. Nat Rev Mol Cell Biol. 2008;9:446–454. doi: 10.1038/nrm2406. [DOI] [PubMed] [Google Scholar]
- McMahon HT, Gallop JL. Membrane curvature and mechanisms of dynamic cell membrane remodelling. Nature. 2005;438:590–596. doi: 10.1038/nature04396. [DOI] [PubMed] [Google Scholar]
- Mim C, Cui H, Gawronski-Salerno JA, Frost A, Lyman E, Voth GA, Unger VM. Cell Structural Basis of Membrane Bending by the N-BAR Protein Endophilin. Cell. 2012;149:137–145. doi: 10.1016/j.cell.2012.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. The MARTINI coarse-grained force field: Extension to proteins. J Chem Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- Moore BA, Robinson HH, Xu Z. The crystal structure of mouse Exo70 reveals unique features of the mammalian exocyst. J Mol Biol. 2007;371:410–421. doi: 10.1016/j.jmb.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M, Novick P. The exocyst defrocked, a framework of rods revealed. Nat Struct Mol Biol. 2006;13:577–581. doi: 10.1038/nsmb1097. [DOI] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJ, Evans PR, McMahon HT. BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science. 2004;303:495–499. doi: 10.1126/science.1092586. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan N, Sunil Kumar PB, Ipsen JH. Monte Carlo simulations of fluid vesicles with in-plane orientational ordering. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;81:041922. doi: 10.1103/PhysRevE.81.041922. [DOI] [PubMed] [Google Scholar]
- Ren J, Guo W. ERK1/2 Regulate exocytosis through direct phosphorylation of the exocyst component Exo70. Dev Cell. 2012;22:967–978. doi: 10.1016/j.devcel.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ridley AJ. Life at the leading edge. Cell. 2011;145:1012–1022. doi: 10.1016/j.cell.2011.06.010. [DOI] [PubMed] [Google Scholar]
- Rosse C, Hatzoglou A, Parrini MC, White MA, Chavrier P, Camonis J. RalB mobilizes the exocyst to drive cell migration. Mol Cell Biol. 2006;26:727–734. doi: 10.1128/MCB.26.2.727-734.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Scita G, Confalonieri S, Lappalainen P, Suetsugu S. IRSp53: crossing the road of membrane and actin dynamics in the formation of membrane protrusions. Trends Cell Biol. 2008;18:52–60. doi: 10.1016/j.tcb.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Spiczka KS, Yeaman C. Ral-regulated interaction between Sec5 and paxillin targets Exocyst to focal complexes during cell migration. J Cell Sci. 2008;121:2880–2891. doi: 10.1242/jcs.031641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetsugu S, Murayama K, Sakamoto A, Hanawa-Suetsugu K, Seto A, Oikawa T, Mishima C, Shirouzu M, Takenawa T, Yokoyama S. The RAC binding domain/IRSp53-MIM homology domain of IRSp53 induces RAC-dependent membrane deformation. J Biol Chem. 2006;281:35347–35358. doi: 10.1074/jbc.M606814200. [DOI] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Arp2/3 complex and actin depolymerizing factor/cofilin in dendritic organization and treadmilling of actin filament array in lamellipodia. J Cell Biol. 1999;145:1009–1026. doi: 10.1083/jcb.145.5.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkina TM, Borisy GG. Cell Biology: A Laboratory Handbook. 3. 2006. Correlative light and electron microscopy studies of cytoskeletal dynamics; pp. 277–285. [Google Scholar]
- Svitkina TM. Electron microscopic analysis of the leading edge in migrating cells. Methods Cell Biol. 2007;79:295–319. doi: 10.1016/S0091-679X(06)79012-4. [DOI] [PubMed] [Google Scholar]
- Takei K, Slepnev VI, Haucke V, De Camilli P. Functional partnership between amphiphysin and dynamin in clathrin-mediated endocytosis. Nat Cell Biol. 1999;1:33–39. doi: 10.1038/9004. [DOI] [PubMed] [Google Scholar]
- Thapa N, Sun Y, Schramp M, Choi S, Ling K, Anderson RA. Phosphoinositide signaling regulates the exocyst complex and polarized integrin trafficking in directionally migrating cells. Dev Cell. 2012;22:116–130. doi: 10.1016/j.devcel.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian A, Baumgart T. Sorting of lipids and proteins in membrane curvature gradients. Biophys J. 2009;96:2676–2688. doi: 10.1016/j.bpj.2008.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C, Hoelzle M, Disanza A, Scita G, Svitkina T. Coordination of membrane and actin cytoskeleton dynamics during filopodia protrusion. PLoS One. 2009;4:e5678. doi: 10.1371/journal.pone.0005678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Arkhipov A, Schulten K. Simulations of membrane tubulation by lattices of amphiphysin N-BAR domains. Structure. 2009;17:882–892. doi: 10.1016/j.str.2009.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Pykalainen A, Lappalainen P. I-BAR domain proteins: linking actin and plasma membrane dynamics. Curr Opin Cell Biol. 2011;23:14–21. doi: 10.1016/j.ceb.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Zimmerberg J, Kozlov MM. How proteins produce cellular membrane curvature. Nat Rev Mol Cell Biol. 2006;7:9–19. doi: 10.1038/nrm1784. [DOI] [PubMed] [Google Scholar]
- Zuo X, Zhang J, Zhang Y, Hsu SC, Zhou D, Guo W. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat Cell Biol. 2006;8:1383–1388. doi: 10.1038/ncb1505. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.