Abstract
Stored rice was collected from rice processing complexes of National Agricultural Cooperative Federation of 11 regions in Korea to evaluate the occurrence of fungi and bacteria and to identify the predominant fungi and bacteria to the genus levels. Most rice samples generally produced the higher levels of fungi and bacteria than white rice. The occurrence of fungi and bacteria varied in various locations of Korea. Among fungi observed, Aspergillus spp. and Penicillium spp. were dominant in the samples and Aspergillus spp. were observed more frequently than Penicillium spp. Predominant bacteria from rice and white rice samples tentatively belonged to the Genus Bacillus, Pectobacterium, Pantoea, and Microbacterium according to BIOLOG and FAME analyses. The results of this study showed that rice in Korea was contaminated in a relatively high level by two dominant storage fungi such as Aspergillus spp. and Penicillium spp. In addition, occurrence of mycotoxins in rice by the fungi could be possible and thus it is necessary to control the storage fungi.
Keywords: Aspergillus, Penicillium, Rice, Storage bacteria, Storage fungi
Rice (Oryza sativa L.) is a principle food grain in Korea and other countries. In Korea, rice is the main crop to control by governmental organizations in its cultivation, storage and processing. As the technology for cultivation was developed, the harvest of rice also could be increased. However, the change of food intake for consumers leads the decrease of consumption of rice every year (Oh, 2006). Consequently, harvested and stored rice has been increased and excessively supplied. In case of rice stored in RPC, the yearly harvested rice is regularly stored for 3 to 4 months from fall to winter as the shape of unpolished rice. To consider the storage of rice in individual home and markets, the expected storage period may be expanded up to 4 to 5 years (Oh, 2006). As the storage period would be longer, the physical, chemical, and biological properties will be changed. In inappropriate storage condition, the loss of quality and quantity of rice by contamination of microorganisms is not possible to avoid (Yoo et al., 1993). To minimize the loss of quality and quantity of crops due to microbial contamination, there are several methods such as the use of host resistance, cultural practices, and application of chemicals or biological agents to reduce the population of pests (Cottyn et al., 2001).
The pests associated with rice to be controlled include not only plant pathogens but also post-harvest fungi or bacteria. The genus Aspergillus and Penicillium are the well-known storage fungi (Filtenborg et al., 1996). They are ubiquitous and have ability to produce various kinds of mycotoxins, which are harmful to plants, animals, and humans (Park et al., 2005). For the control of storage fungi, the basic information on the population of naturally occurring fungi and bacteria is essential. Therefore, the objectives of this study were to evaluate the occurrence of fungi and bacteria in rice and white rice stored in rice processing complexes (RPC) of National Agricultural Cooperative Federation and to identify the predominant fungi and bacteria to the genus levels.
Rice and white rice samples were collected from 11 regions of Korea such as Andong, Anseong, Cheonan, Gimje, Gimpo, Gumi, Hamyang, Jinju, Naju, Namwon and Yeongdeok in 2005 and/or 2006. Since rice was stored in ton bags and stacked up to the ceiling of the bin in RPC, samplings were done with a sampler stabbing into bags. Temperature in RPC varied from 4℃ to room temperatures and moisture content of the rice and white rice samples was approximately 15%.
The samples (3 g) were finely ground with the analytical mill (IKA A11 basic, IKA® Works, Inc. Wilmington, USA). The ground rice was suspended in sterile distilled water and incubated for 50 minutes at 28℃ with shaking (120 rpm). After then, several dilutions of the rice suspension were made and smeared on different media for evaluation of fungi and bacteria. The treated media were then incubated in the dark at 28℃ for 2 and 5 days for bacteria and fungi, respectively. For assessing fungi in rice samples, dichloran 18% glycerol agar [(DG18), Dichloran glycerol 31.6 g, anhydrous glycerol 220 g, ZnSO4·7H2O 10 mg, CuSO4·5H2O 5 mg, chloramphenicol 50 mg in 1 l H2O] amended with 50 mg chlortetracycline was used. DG18 agar is selective for the fungi grown in dry conditions while inhibits the growth of yeast, bacteria and the mucoraceous fungi such as Rhizopus and Mucor species, which can disturb the normal growth of the storage fungi (Swan et al., 1998). Genus Aspergillus, Penicillium or other fungi on the medium were also determined under an optical microscope (Klich et al., 2002; Pitt et al., 1985).
For assessing bacteria, nutrient agar (NA) amended with 50 mg NaCl and 50 mg cycloheximide was used. The dominant and representative bacterial colonies on NA were isolated and inoculated on tryptic soy agar (TSA) containing 5% sheep blood (Difco, Detroit, MI, USA) for identification through BIOLOG analysis and incubated at 28℃ for 24 hr. The bacterial strains grown on TSA were harvested and suspended into 20 ml of Biolog buffer (0.4% sodium chloride, 0.03% pluronic F-68, 0.01% gellan gum) in test tubes. Then absorbance of the test tubes with bacterial strains was adjusted to 50% using percent transmittance. One hundred microliters of each of the bacterial suspensions in Biolog plates were incubated at 30℃ for 24 hr. Microlog GN Microplate System read the reaction data for metabolic fingerprinting with the Microlog 3 database release 4.01A (BIOLOG, Hayward, USA) at 24 hr. Along with BIOLOG analysis, the analysis of fatty acid methyl-esters (FAME) for the representative bacterial strains was conducted for identification to the genus level by gas chromatography (GC) using the MIDI system (Microbial Identification System, Inc., Newark, USA). Bacterial cells were harvested from colonies grown on TSA and then fatty acids were isolated from the cellular lipids by saponification. The fatty acids were methylated to increase volatility by forming the volatile methyl esters. These were extracted into the organic phase and automatically injected into a gas chromatograph for separation, identification, and quantification. The electronic signal of the GC was passed to the computer, which compared the fatty acid methyl ester composition of the tested bacteria to those of known bacteria in a stored database library to select the most similar species with the tested bacteria. The experiments were conducted with three replications and the data were analyzed with the Statistical Analysis System (SAS Institute, Cary, USA).
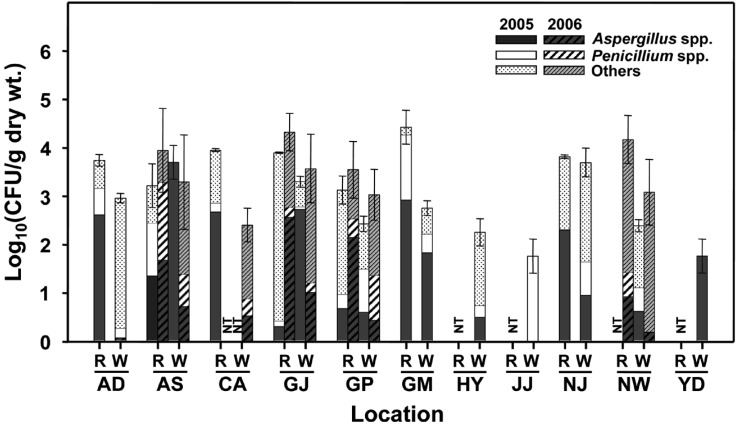
In fungal populations, most rice samples generally produced the higher levels of fungal populations [102 to 105 colony forming units (CFU) per gram dry weight] than that of white rice (101 to 104 CFU per gram dry weight) (Fig. 1). In addition, fungal populations also varied in locations. Among fungi observed, Aspergillus spp. and Penicillium spp. were dominant in the samples and Aspergillus spp. were observed more frequently than Penicillium spp. (Fig. 1). The unique characteristic of Aspergillus spp. for their identification was round spore-bearing structure on the tip of conidiophores, formation of 'T' or 'L' shape of connection between conidiophore and vegetative hyphae, so called the 'foot cell' (Klich et al., 2002). Approximately seven distinct species of Aspergillus were observed from the samples of rice and white rice. The genus Penicillium also has the distinctive characteristic of conidiophores with dense brush-like spore-bearing structures and colonies usually with bluish or greenish grey color and closed texture (Pitt, 1973). Although Penicillium spp. were more diverse relative to Aspergillus spp., they were difficult to identify to species level because of the lack of obvious distinguishing features as reported by Pitt (1973).
Fig. 1.
Populations of fungi occurred in the stored rice samples collected from 11 locations of Korea in 2005 and 2006. Different marks in each column indicate the percentage composition of Aspergillus spp., Penicillium spp., and other fungi of the sample. A bar on each columm indicates the standard deviation of the mean from three replications. CFU = colony forming unit; R = rice; W = white rice; NT = not tested; AD = Andong, AS = Anseong, CA = Cheonan, GJ = Gimje, GP = Gimpo, GM = Gumi, HY = Hamyang, JJ = Jinju, NJ = Naju, NW = Namwon, and YD = Yeongdeok.
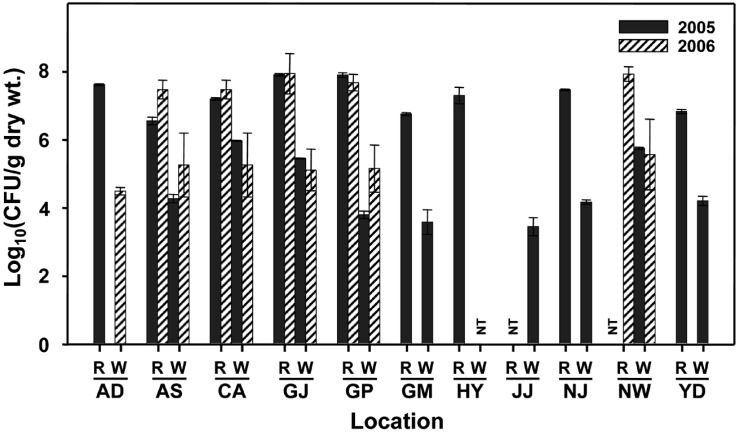
The populations of bacteria in the samples of 2005 and 2006 approximately ranged from 104 to 108 CFU per gram of rice and white rice (Fig. 2). Generally, bacterial populations in rice were higher (101 and 104 CFU per gram) than that of white rice from each location (Fig. 2). There were no distinctive differences of bacterial populations in samples between 2005 and 2006 while were differences among locations where samples were taken. The dominant bacteria were isolated and tested for their identification through BIOLOG and FAME analyses. According to the data of FAME analysis, bacterial strains were to be Bacillus cereus (similarity index = 0.284), Pectobacterium carotovorum (similarity index = 0.446), Pantoea ananatis (similarity index = 0.682), and Microbacterium lacticum (similarity index = 0.912) at the species level. Therefore, the bacterial strains were tentatively identified to be Genus Bacillus, Pectobacterium, Pantoea, and Microbacterium at the genera level. Bacillus spp. showed white, wide-spread colonies with irregular margins and rod cells. Bacillus mainly existed in rice and/or white rice samples from Anseong, Andong, Gimje, Namwon, and Naju. Especially they dominated in white rice samples from Gumi and Hamyang. Pectobacterium with yellow, wide spread colonies and Pantoea with distinct yellow, convex colonies have been detected in most samples regardless of locations. In addition, the species of Microbacterium was detected from rice and/or white rice samples of Andong, Gumi, and Naju. Previously, this bacterium has been reported to be causal agent of red stripe of rice by Kaku et al. (2000).
Fig. 2.
Populations of bacteria occurred in the stored rice samples collected from 11 locations of Korea in 2005 and 2006. A bar on each column indicates the standard deviation of the mean from three replications. CFU = colony forming unit; R = rice; W = white rice, NT = not tested; AD = Andong, AS = Anseong, CA = Cheonan, GJ = Gimje, GP = Gimpo, GM = Gumi, HY = Hamyang, JJ = Jinju, NJ = Naju, NW = Namwon, and YD = Yeongdeok.
The results of this study showed that rice in Korea was contaminated in a relatively high level by two dominant storage fungi such as Aspergillus spp. and Penicillium spp. In addition, occurrence of mycotoxin in rice by the fungi could be possible and thus it is necessary to control the storage fungi. Based on the basic information on population and diversity of bacteria, searching biologically effective bacteria as biocontrol agents from rice could be needed to reduce the fungal contamination. Further studies on current situation of mycotoxins occurring in rice in Korea by the fungus should be needed.
Acknowledgment
This study was carried out with the support of "Specific Joint Agricultural Research-Promoting Projects (Project No. 20050301-033-202-055-03-00 and 20060301-033-003-001-03-00)", RDA, Republic of Korea.
References
- 1.Cottyn B, Regalado E, Lanoot B, De Cleene M, Mew TW, Swings J. Bacterial populations associated with rice seed in the tropical environment. Phytopathology. 2001;91:282–292. doi: 10.1094/PHYTO.2001.91.3.282. [DOI] [PubMed] [Google Scholar]
- 2.Filtenborg O, Frisvad JC, Thrane U. Moulds in food spoilage. Int J Food Microbiol. 1996;33:85–102. doi: 10.1016/0168-1605(96)01153-1. [DOI] [PubMed] [Google Scholar]
- 3.Kaku H, Subandiyah S, Ochiai H. Red stripe of rice is caused by a bacterium Microbacterium sp. J Gen Plant Pathol. 2000;66:149–152. [Google Scholar]
- 4.Klich MA. Identification of common Aspergillus species. Utrecht, The Netherlands: Centraalbureau voor Schimmelcultures; 2002. [Google Scholar]
- 5.Oh KW. Food grain consumption survey report. Republic of Korea: National Statistical Office; 2006. [Google Scholar]
- 6.Park JW, Choi SY, Hwang HJ, Kim YB. Fungal mycoflora and mycotoxins in Korean polished rice destined for humans. Int J Food Microbiol. 2005;103:305–314. doi: 10.1016/j.ijfoodmicro.2005.02.001. [DOI] [PubMed] [Google Scholar]
- 7.Pitt JI. An appraisal of identification methods for Penicillium species: novel taxonomic criteria based on temperature and water relations. Mycologia. 1973;65:1135–1157. [PubMed] [Google Scholar]
- 8.Pitt JI. A laboratory guide to common Penicillium species. Australia: Food Science Australia; 1985. [Google Scholar]
- 9.Swan JRM, Crook B. Airborne microorganisms associated with grain handling. Ann Agric Environ Med. 1998;5:7–15. [PubMed] [Google Scholar]
- 10.Taligoola HK, Ismail MA, Chebon SK. Mycobiota associated with rice grains marketed in Uganda. J Biol Sci. 2004;4:271–278. [Google Scholar]
- 11.Yoo CC, Hwang HJ, Kim YB. Studies on fungal invasion of stored rice kernels. J Inst Biotechnol. 1993;5:48–53. [Google Scholar]