Abstract
Pine tree death caused by pine wood nematode (PWN) involves phoretic relationships between PWN and its vector Japanese pine sawyer beetle (JPS). In an effort to understand the diversity of fungi involved in PWN life cycle, a total of 176 fungal isolates were collected from PWNs, adults and larvae of JPS, PWN-diseased Japanese black pine that was cut down in 2005 at Jinju, Korea. Based on microscopic observation and colony morphology, and sequence analysis of the ITS rDNA, the fungal isolates were identified at the level of genus. Three genera including Mucor, Ophiostoma, and Penicillium were identified from PWN. Two genera of Ophiostoma and Penicillium were discovered from JPS larvae. From JPS adult beetles, nine genera of Aspergillus, Gibberella, Hypocrea, Irpex, Leptosphaeria, Ophiostoma, Penicillium, and Plectosphaerella and unknown basidiomycetes were found. Ten genera from PWN-infected wood were confirmed as Bionectria, Botrytis, Camarops, Fusarium, Hypocrea, Nectrtia, Mucor, Ophiostoma, Penicillium, and Trichoderma. Penicillium and Ophiostoma were commonly distributed on PWN and its vector and host. This is first report of the fungi associated with PWN and its vector and host in Korea.
Keywords: ITS rDNA, Japanese black pine, Japanese pine sawyer, Pine wood nematode, Pine wood nematode-associated fungi
Pine wilt disease caused by the pine wood nematode (PWN, Bursaphelenchus xylophilus) was first reported from Japan in the beginning of 20 century. Since its occurrence in Japan, the nematode disease has been reported from other Asian countries such as China and Taiwan, and North America such as USA and Mexico. Recently, it has been detected in Portugal of Europe (Han et al., 2003; Mota and Vieria, 2004). The pine wood nematode disease also has been brought killing a lot of pine trees in Korea. Since its first finding at Busan area in 1988, PWN has been spread to Southern and middle regions of Korean peninsula.
It has been known that pine tree death by the nematode disease involves other biological agents that have symbiotic relationships with the nematodes. Japanese pine sawyer beetle (JPS, Monochamus alternatus) is known as the vector of the pine wood nematode transmission, and fungi associated with JPS are known as food resources of JPS (Donald et al., 2003). The main agents of pine wilt disease, pine wood nematode infects inside of pine wood after feeding pine shoot by its vector JPS and propagates in vascular tissues. The propagation of pine wood nematodes in vascular area hinders nutrients and water transportation in the infected tree and results in the symptom of pine tree wilt (Mamiya, 1980, 1985; Ishida et al., 1993; Ichihara et al., 2000).
In Korea, it has not been known that which fungal species are associated with PWN's ecosystem. To better understand the role of fungi in the disease biology of pine wood nematode it is prerequisite to investigate the mycoflora associated with PWN and its vector JPS and host. In the present paper we first report in Korea the isolation and identification of fungi present in PWN, JPS (adults and larvae) and PWN-infected Japanese black pine wood.
For the isolation of fungi, four PWN-diseased Japanese black pines were cut down and felled into logs (150 cm in length, 15 cm in diameter) and small discs (5 cm in length, 15 cm in diameter) in October of 2005 at Jinju, Kyungsangnamdo, Korea. To sample fungi from the PWN-infected pine wood, ten discs were made into tiny wood chips. One hundred chip pieces were surface-sterilized with 0.5% chlorine lax solution for 2 min, washed twice with sterile water, and air dried. The dried wood chip pieces were placed onto culture media below described. PWN was isolated using Baermann funnel method from the infected pine wood. The purified nematodes were washed several times with sterile distilled water before they were used for fungal isolation. To sample JPS larvae and adults, the felled logs were placed in a storage cage for PWN research in Southern Forest Research Center of Korean Forest Research Institute in Jinju. In the storage cage the eggs of JPS laid inside the PWN-diseased logs were allowed to hatch, and larvae to develop to pupas and adults, and adult beetles to emerge from the logs. In early May of 2006, ten living JPS larvae were captured from inside of 4 PWN-infected pine logs by breaking and carving out the tree stems. In late June of 2006, ten JPS adult beetles emerged from the stored logs were captured. For the isolation of fungi, the captured JPS (adults and larvae) and the purified PWN (about two thousands) were ground in sterile mortar and pestle with sterile water under aseptic conditions, and the resulting ground extracts were diluted and spread onto different culture media. Malt Extracts Agar (BD Science, USA), Potato Dextrose Agar (BD Science, USA) and Oxoid Malt Extracts Agar (Oxoid, England) were used as culture media with and without supplement of anti-bacterial agents and cycloheximide (100 µg/ml). To induce mycelia growth the inoculated culture media were incubated for 3~7 days at 20℃ at aerobic conditions. Mycelial tips grown on the culture media were transferred to new media plates. Fungal identification was performed based on the microscopic observation of colony properties and characteristics of microstructures referring books and online tools for fungal taxonomy and identification (De Hoog et al., 2000; Dugan, 2006; Jacobs and Wingfield, 2001; www.botany.utoronto.ca/ResearchLabs/MallochLab/Malloch/Moulds/Moulds.html) using a phase contrast light microscope (Karl Zeiss, Axioskop 40, Germany) and a dissect microscope (Olympus, SZ2-ILST, Japan). When the morphological characteristics were not confirmed, we carried out DNA sequencing analysis of the ITS rDNA region. For the amplification of ITS rDNA region, genomic DNA was prepared from purely isolated fungi using the method described Kim et al. (1999). To amplify the ITS rDNA region in fungal genome, PCR were performed in a Gene Amp-950 thermal cycler (ABI, USA) using universal primer pairs, ITS1-ITS4 (White et al., 1990). All the PCR reaction conditions were set according to the method described by Kim et al. (1999). The amplified DNA products were sequenced on Applied Biosystems ABI 373 DNA sequencer. Both strands of the PCR-amplified DNA fragments were sequenced using the PRISM Ready Reaction DyeDeoxy termination cycle sequencing kit and DNA sequences were determined in an Applied Biosystems ABI 373 DNA sequencer. The determined nucleotide sequences were searched for homologous ITS rDNA sequences of fungi in Blast-N on GenBank database (http://www.ncbi.nlm.nih.gov/BLAST/).
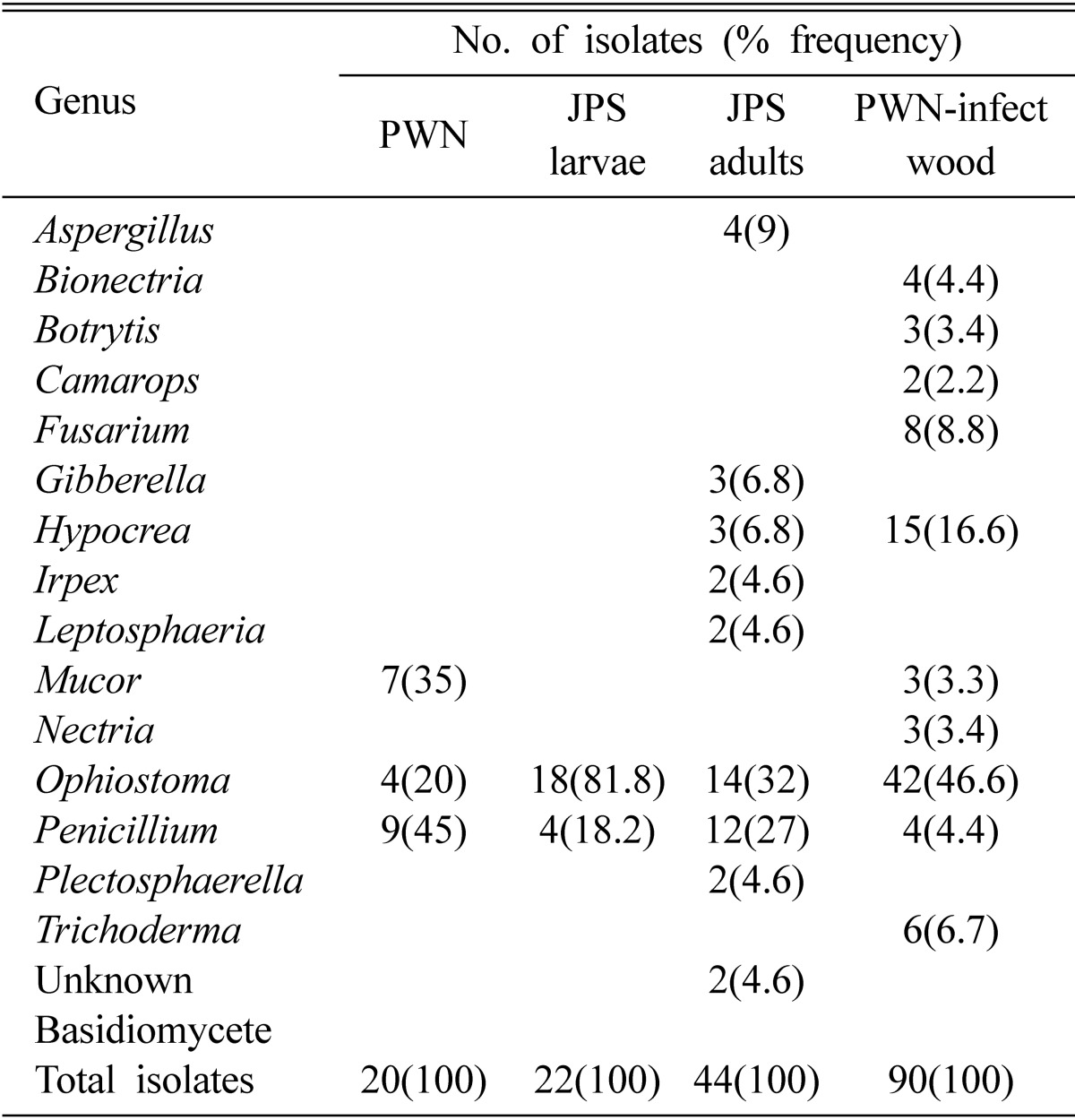
A total of 176 fungal isolates were obtained from the sampling results (Table 1). Among these fungal isolates, 20 isolates were isolated and purely cultured from PWN, 22 and 44 isolates from JPS larvae and adults, respectively, and 90 isolates from PWN-infested wood chips. From these isolates obtained we could identify 15 genera of fungi. From PWN, Mucor, Ophiostoma, and Penicillium were identified. The frequencies of isolation of these fungi were 35, 20, and 45%, respectively. From JPS larvae, Ophiostoma and Penicillium were discovered with frequency of 81.8 and 18.2%, respectively. It is noticeable that Ophiostoma has been isolated with very high frequency from JSP larvae. From JPS adults, more numbers of fungal genera were identified. These identified fungi were nine genera of Aspergillus, Gibberella, Hypocrea, Irpex, Leptosphaeria, Ophiostoma, Penicillium, and Plectosphaerella and unknown basidiomycetes. In the fungi from JPS adult beetles, Ophiostoma and Penicillium were the major groups of fungi. Except for these two genera all other genera showed less than 7% of isolation frequency. Ophiostoma showed the highest isolation frequency (32%). Regarding on the fungi from PWN-infected wood, ten genera were confirmed as Bionectria, Botrytis, Camarops, Fusarium, Hypocrea, Nectrtia, Mucor, Ophiostoma, Penicillium, and Trichoderma. Among these genera, Ophiostoma also was isolated with the highest frequency (46.6%). The remained genera showed less than 10% of isolation frequency except for Hypocrea (16.6%).
Table 1.
Fungi isolated from pine wood nematode (PNW), Japanese pine sawyer (JPS) larvae and adults, and PWN-infected Japanese black pine wood
Consequently, we could find from the results in Table 1 that 15 different genera were identified from the 176 fungal isolates. Among the 15 genera identified, Penicillium and Ophiostoma were isolated with high frequency and commonly distributed on PWN and its vector and host. Thus, we consider these fungal groups may do important roles in the PWN life cycle. Ophiostomatoid fungi including Ophiostma and Ceratocystis have been reported from PWN-infected wood in Japan and USA (Donald et al., 2005). These fungi have been considered as nutrient sources of PWN-vectoring beetles. Thus, our results of the isolation of Ophiostoma fungi are agreed with those of Japan and USA. Ye et al. (1995) isolated Pestalotiopsis, Fusarium, Ceratocystis, Colletotrichum, Alternaria, Sordaria, Chaetomium, Nigrospora, Phomopsis, Curvularia, Monochaetia, Trichoderma, Rhizopus, Penicillium, Aspergillus and more than ten other genera of unidentified fungi from Pinus in China. They also found that PWN could multiply on PDA grown with Ceraocystis but not with Penicillium.
In general, less number of fungal genus names was found from PWN and JPS larvae than from JPS adults and PWN-infected wood. There were differences in the fungal diversity between the fungi isolated from JPS larvae and the fungi from JPS adults. It seems that at the stage of JPS larva the fungal diversity is limited compared to JPS adult stage. Further investigation is needed to answer the question that why more fungal diversity are present at the stage of adults in JPS. Some of the fungal genera from PWN-infected wood were not found in the list of fungal genus from the JPS beetles. They are considered as the group of fungi that are not associated with PWN.
In conclusion, in this study, we first reported in Korea that diversity of fungi isolated from PWN, its vector JPS larvae and adults, and its infested host trees. Various fungi were found to be associated with pine tree disease caused by PWN. We could detect that the wood-staining group Ophiostoma and green mold group fungus Penicillium were commonly present as major groups in PWN itself and its associated environments. At present, PWN-related research is not in easy situation in Korea. Because of clear cut and burning of the PWN-infected trees on sites by a control strategy and of prohibition of the movement of the PWN-infected trees by a law, the obtaining of PWN-infected tree samples is extremely limited. Thus, we could not able to obtain PWN-infected tree samples largely enough. Although sample sizes are limited, the results of present study provide quite valuable basic information for the fungi associated with PWN disease research in Korea. We are currently analyzing the obtained fungi in this study at the level of species including their physiological and biochemical properties. These efforts will lead to further understand the roles of fungi in PWN biology and pine death-related ecology.
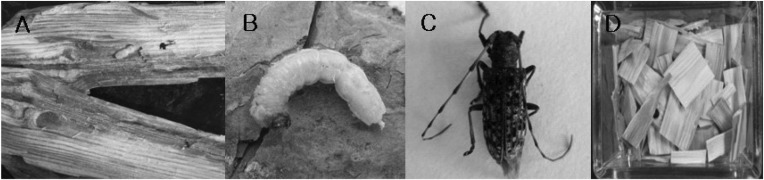
Fig. 1.
Examples of samples of pine wood infested by pine wood nematode (PNW), Japanese pine sawyer larva (B) and adult beetle (C), and Japanese black pine wood chips (D).
Acknowledgement
This study was supported by a Korea Forest Research Institute Grant for the General and Integrated Research Project for Pine Wilt Disease Control in the Republic of Korea.
References
- 1.Donald PA, Stamps WT, Linit MJ. Pine wilt disease in APSnet plant disease lessons. St. Paul, MN, USA: The American Phytopathological Society; 2003. [Google Scholar]
- 2.Dugan FM. The identification of fungi. Minnesota, USA: APS; 2006. [Google Scholar]
- 3.Han ZM, Hong YD, Zhao BG. A study on pathogenicity of bacteria carried by pine wood nematodes. J Phytopathol. 2003;151:683–689. [Google Scholar]
- 4.Ichihara Y, Fukuda K, Suzuki K. Early symptom development and histological changes associated with migration of Bursaphelenchus xylophilus in seedling tissues of Pinus thunbergii. Plant Disease. 2000;84:675–680. doi: 10.1094/PDIS.2000.84.6.675. [DOI] [PubMed] [Google Scholar]
- 5.Ishida K, Hogetsu T, Fukuda K, Suzuki K. Cortical responses in Japanese black pine to attack by the pine wood nematode. Can J Bot. 1993;71:1399–1405. [Google Scholar]
- 6.Jacobs K, Wingfield MJ. Leptographium species. Tree pathogens, insect associates and agents of blue stain. St. Paul, Minnesota: American Phytopathological Society; 2001. p. 207. [Google Scholar]
- 7.Kim SH, Uzunovic A, Brueil C. Rapid detection of Ophiostoma piceae and O. quercus in stained wood by PCR. Appl Environ Microbiol. 1999;65:287–290. doi: 10.1128/aem.65.1.287-290.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mamiya Y. Inoculation of the first year pine (Pinus densiflora) seedlings with Bursaphelenchus lignicolus and the histopathology of diseased seedlings. Jap J For Soc. 1980;62:176–183. [Google Scholar]
- 9.Mamiya Y. Initial pathological changes and disease development in pine trees induced by the pine wood nematode, Bursaphelenchus xylophilus. Ann Phytopathol Soc Japan. 1985;51:546–555. [Google Scholar]
- 10.Mota M, Vieira P. The pinewood nematode, Bursaphelenchus xylophilus. Proceedings of an international workshop; Nematology Monographs & Perspectives; August 20-22, 2001; University of Évora, Portugal. Leiden, The Netherlands: Brill Academic Publishers; 2004. p. 291. [Google Scholar]
- 11.White TJ, Bruns T, Lee S, Taylor J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Snindky JJ, White TJ, editors. PCR protocols: a guide to methods and applications. San Diego, California: Academic Press Inc.; 1990. pp. 315–322. [Google Scholar]
- 12.Ye W, Zhang Q, Hong S, Zhu D. Studies on fungi associated with Bursaphelenchus xylophilus on Pinus massoniana in Shenzhen, China; International Symposium on Pine Wilt Disease Caused by the Pine Wood Nematode; 31 October - 5 November 1995; Beijing, China. 1995. [Google Scholar]