Abstract
Thirty seven species of Fusarium were evaluated for their ability of producing extracellular enzymes using chromogenic medium containing substrates such as starch, cellobiose, CM-cellulose, xylan, and pectin. Among the tested species Fusarium mesoamericanum, F. graminearum, F. asiaticum, and F. acuminatum showed high β-glucosidase acitivity. Xylanase activity was strongly detected in F. proliferatum and F. oxysporum. Strong pectinase activity was also found in F. oxysporum and F. proliferatum. Amylase activity was apparent in F. oxysporum. No clear activity in cellulase was found from all the Fusarium species tested.
Keywords: Chromogenic media, Extracellular enzymes, Fusarium
Fungi can produce diverse extracellular enzymes those are used to break down complex polysaccharides into simple sugars to be assimilated and used for growth and reproduction. These enzymes are useful for environmental and industrial applications such as food processing, brewery, biofuels, bioremediation etc. For those applications, exploration of new sources of useful fungal extracellular enzymes is demanded. The discomycete Fusarium is one of well-known fungi in agriculture and forestry as problematic plant pathogens and mycotoxin contaminating agents that can affect human and animal health (Booth, 1977; Nelson et al., 1983). In fact, Fusarium is a large genus of filamentous fungi, and most of Fusarium species are harmless saprobes and relatively abundant members of the soil microbial community (Domsch et al., 1980; Nwanma et al., 1993). This ecological habitat of the fungus implies that Fusarium would be a useful resource of extracellular enzymes. However, information on the ability of producing enzymes in Fusarium spp. is rarely reported.
Culture collection is one of great resources where we can find new sources of extracellular enzymes. Currently diverse species of Fusarium are deposited and preserved in domestic fungal culture collections. The present study was carried out to find potent new sources of valuable fungal enzymes from Fusarium species. For this purpose, the ability of producing enzymes is evaluated with various Fusarium species using the plate screening methods with chromogenic substrates (Lee and Lee, 1997). This method is relatively straightforward and simply applicable tools for specific detection of polysaccharide degrading microorganisms (Castro et al., 1995).
A total of 37 Fusarium species were obtained from Korean Agricultural Culture Collection (KACC, Suwon, Korea) and tested for their ability of extracellular enzyme activities in chromogenic media. The obtained fungi were maintained on potato dextrose agar (Difco, USA). Preculturing of the fungi were made either on PDA or on 2% malt extract agar (Difco, USA) at 25℃ for 5 days. For the observation of fungal extracellular enzyme activity, the precultures were transferred onto the media containing each of 0.5% CM-cellulose (Sigma, USA), D-cellobiose (Sigma, USA), starch from potato (Sigma, USA), polygalacturonic acid (MP Biomedical, France), xylan from oat spelts (Sigma, USA) as enzymatic carbon source, 0.1% yeast nitrogen base (Difco, USA) as its fundamental nitrogen source, and 1.5% agar powder. 0.5% Congo red dyes (Teather and Wood, 1982) was used for chromogenic reaction due to its better performance in extracellular enzyme activity detection with various fungal species (Yoon et al., 2007). After 5 days of culturing at 25℃, we could observe clear zone formed by reaction between enzyme and chromogenic substrates. For the clear zone measurement, cultured Petri plates were upside placed on white light box and photographed. The degree of extracellular enzyme activity was evaluated based on the size of formed clear zone. Clear zone size is divided into three groups. The first is strong activity group (defined as S at Table 1) that shows 2.5~4 cm of clear zone size. The second is moderate activity group (defined as M at Table 1) that shows 1~2.4 cm of clear zone size. The third is weak activity group that forms less than 1 cm of clear zone size and/or doesn't form clear zone (defined as N at Table 1). Mycelial growth was also measured simultaneously with the measurement of clear zone.
Table 1.
The ability of producing extracellular enzymes in Fusarium spp.
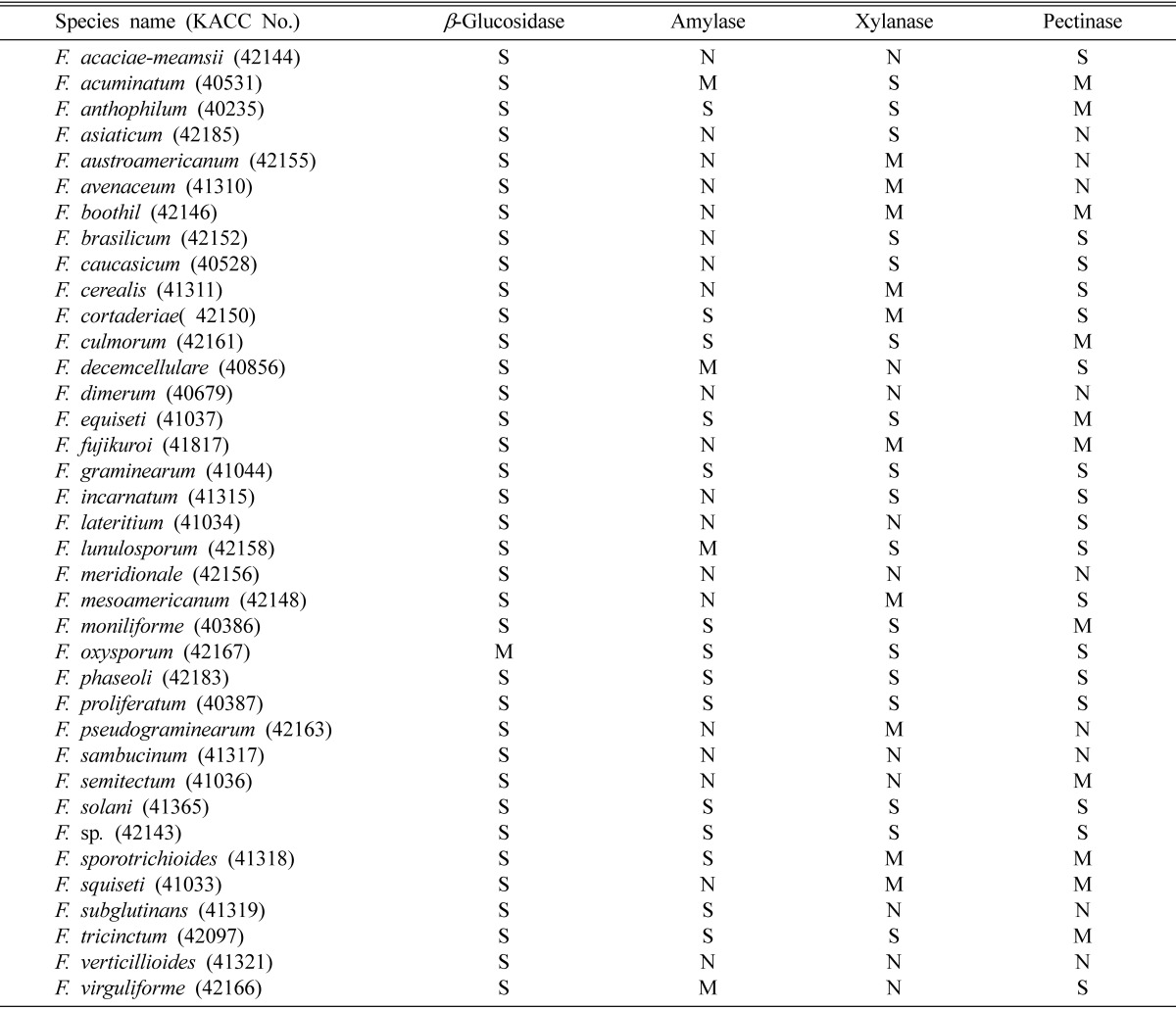
S: strong activity, M: moderate activity and N: weak or no activity.
The results of activity detection of four different extracellular enzymes in 37 Fusarium species are given at Table 1. β-glucosidase, amylase, pectinase, and xylanase were detected but no cellulase was detected in the 37 species. Thirty-six of 37 Fusarium species tested showed strong activity of β-glucosidase, the enzyme that degrades D-cellobiose to glucose. Thus, the production of this enzyme looks popular in Fusarium. An example of this enzyme detection is in Fig. 1. Among these 36 species, strong activity was much apparent in F. mesoamericanum, F. graminearum, F. asiaticum, and F. acuminatum. In amylase detection, fourteen of 37 species showed strong activity. The strongest activity was detected in F. oxysporum. Amylase is the enzyme which degrades amylose in starch polymers. Fig. 2 shows few examples of amylase detection on chromogenic media. Regarding xylanase detection, strong activity was found in 17 of 37 species. F. proliferatum and F. oxysporum produced strongest activity among the 17 species of strong activity pool. The patterns of colony growth and clear zone formed on chromogenic plates are shown in Fig. 3. Strong activity of pectinase (Fig. 4) was also detected from 17 of 37 species listed at Table 1. The strongest pectinase activity was also apparent in F. oxysporum and F. proliferatum.
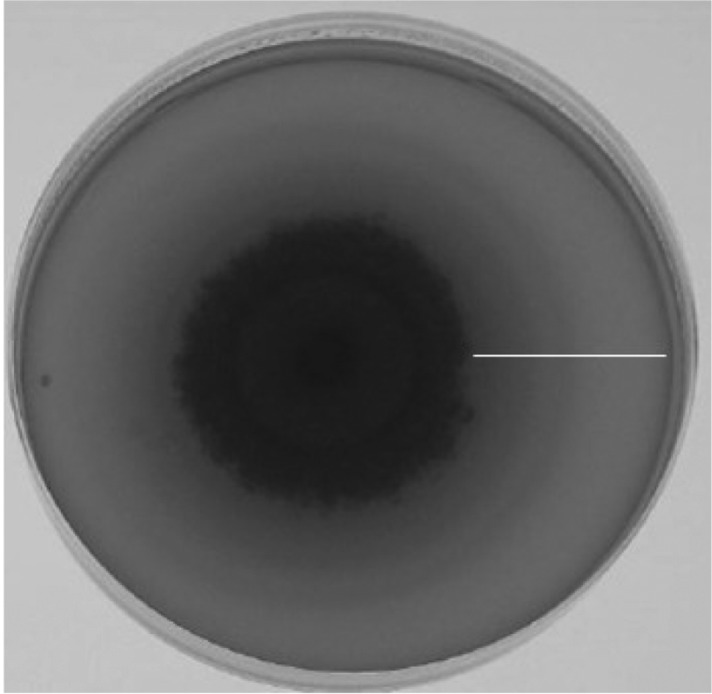
Fig. 1.
An example of strong β-glucosidase activity shown chromogenic media by a Fusarium species. Bar indicates clear zone.
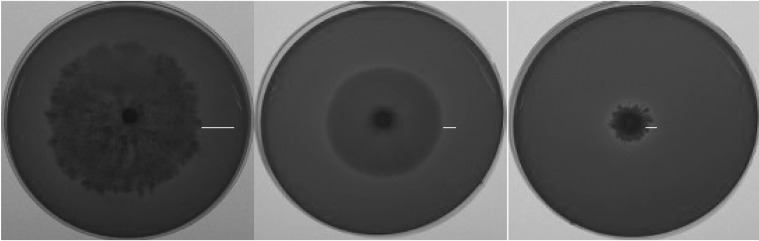
Fig. 2.
Examples of amylase activities shown on chromogenic media by different Fusarium species. Bar indicates clear zones.
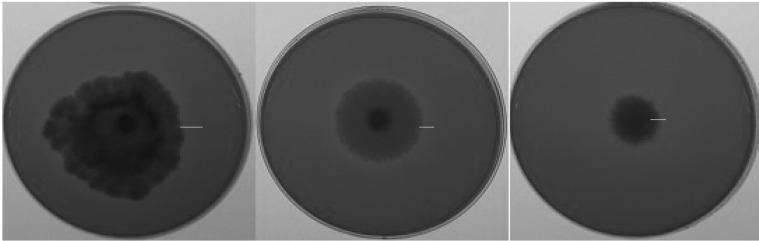
Fig. 3.
Examples of xylanse activities shown on chromogenic media by different Fusarium species. Bar indicates clear zones.
Fig. 4.
Examples of pectinase activities shown on chromogenic media by different Fusarium species. Bar indicates clear zones.
When we compared each Fusarium culture for enzyme production ability, only F. graminearum, F. phaseoli, F. proliferatum, F. solani, and F. sp. showed that they have the strong ability of producing β-glucosidase, amylase, pectinase, and xylanase. On the contrary, F. dimerum, F. meridionale, F. sambucinum, and F. verticillioides showed the least ability of extracellular enzyme production. These Fusarium species showed weak or no activity of amylase, pectinase, and xylanase.
Yoon et al. (2007) successfully detected cellulase in F. solani using the same plate assay methods based on chrormogenic reaction. In this study we could not detect cellulase not only from F. solani but also from other Fusarium species. Considering that F. solani strains used for this study and that of Yoon's are different, the discrepancy in the results of cellulase detection in F. solani is likely from difference in strain properties. Since we could not detect cellulase from any of the species listed at Table 1, cellulose is not common extracellular enzyme in Fusarium.
Overall, we have found that chromogenic media is also useful for the detection of extracellular β-glucosidase, amylase, pectinase, and xylanase in Fusarium spp. There were differences in the production of these enzymes among the tested 37 Fusarium species. Since most of the cultures used in this study are Korean origin, the results of our work provide would provide basic information on the biochemical properties of the Fusarium species deposited in KACC. Based on information of Table 1 that F. oxysporum produces strong pectinase activity and on information of the enzyme gene in GenBank database, Yoon (2007) easily cloned a gene encoding one of pectinases from F. oxysporum using PCR-based cloning approach. Since protein expression technology using protein expression vectors is developing, Yoon's results show that the use of DNA database and the fungal culture that demonstrated the ability of producing enzyme activity is synergistic for the work of a target enzyme and its gene. Thus, the finding of five Fusarium species that show strong activities in β-glucosidase, amylase, pectinase, and xylanase implies that these cultures deserve to further look to explore their potent values in industrial application.
Acknowledgment
This study was supported by a grant (project no:20060401034815) of BioGreen21 program from the Rural Development of Administration in Korea.
References
- 1.Booth C. Fusarium Laboratory Guide to the Identification of the Major Species. Kew: Commonweath Mycological Institute; 1977. p. 55. [Google Scholar]
- 2.Castro GR, Baigori MD, Sineriz F. A plate technique for screening of inulin degrading microorganisms. J Microbiol Methods. 1995;22:51–56. [Google Scholar]
- 3.Domsch KH, Gams W, Anderson TH. Compendium of soil fungi. London: Academic Press; 1980. [Google Scholar]
- 4.Lee ST, Lee JJ. Insoluble dye substrate for screening and assay of xylan-degrading enzymes. J Microbiol Methods. 1997;29:1–5. [Google Scholar]
- 5.Nelson PE, Tousson TA, Marasas WFO. Fusarium species. An illustrated manual for identification. London: Pennsylvania State Univ Press; 1983. [Google Scholar]
- 6.Onyike NBN, Nelson PE. The distribution of Fusarium species in soils planted to millet and sorghum in Lesotho, Nigeria and Zimbabwe. Mycopathologia. 1993;121:105–114. [Google Scholar]
- 7.Teather RM, Wood PJ. Use of congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from bovine rumen. Appl Environ Microbiol. 1982;43:777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoon JH. Study on the detection of extracellular enzyme activities in Korean isolates of Colletotrichum, Fusarium, Penicillium, and Trichoderma. Cheonan, Korea: Dankook University; 2007. MSc. Thesis. [Google Scholar]
- 9.Yoon JH, Park JE, Suh DY, Hong SB, Ko SJ. Comparison of dyes for easy detection of extracellulase in fungi. Mycobiology. 2007;35:21–24. doi: 10.4489/MYCO.2007.35.1.021. [DOI] [PMC free article] [PubMed] [Google Scholar]