Abstract
Grape fruits with blue mold symptoms were collected from house storages in different locations in Korea and were investigated for their association with Penicillium species. A total of 12 isolates of Penicillium were isolated from the collected fruits. Based on morphological and cultural characteristics and β-tublin gene sequence data analysis, they were identified as P. bialowiezense, P. citrinum, P. echinulatum, P. expansum, P. solitum and unidentified Penicillium species. P. solitum was the predominant followed by P. expansum. P. bialowiezense and P. echinulatum were newly recorded in Korea. β-Tubulin gene sequences could be used to distinguish each species of Penicillium and the molecular groups were correlated well with the morphological species. The unidentified species was supposed to be a new species, not previously reported in literature.
Keywords: Blue mold, Colony characteristics, Grape, Morphological characteristics, Penicillium, β-Tubulin gene
During the storage period, fruits of grape may undergo various attacks from insects and microorganisms like fungi, yeasts and bacteria. Penicillium species are a major cause of deterioration and decay amongst a wide range of post harvest plant products, particularly fruits such as grape, oriental pear and citrus.
Penicillium of Korea are insufficiently known. The published Korean literature on the genus is scattered and fragmentary and pertains to about 60 species (Anonymous, 2004; Cho et al., 2005; Lee et al., 2003). Most of them are soil-borne and only 10 species have been reported to be associated with storage diseases of plant products (Anonymous, 2004; Kim et al., 2002; Oh et al., 1999).
Identification of Penicillium is not easy. It is a large genus, and many common species look alike to the uninitiated. At the same time there is a great deal of variability within the species, therefore, unambiguous identification of the species requires molecular identification (Guerche et al., 2004). Among the molecular tools available, β-tubulin gene has proven useful for identification of closely related Penicillium species (Kim et al., 2006; Seifert and Louis-Seize, 2000; Samson et al., 2004).
The objectives of this study were to clearly identify Penicillium isolates from blue mold of grapes, by using two approaches; the analysis of morphological and cultural characteristics and the analysis of their β-tubulin gene sequences.
Materials and Methods
Isolation
Grape fruits with blue mold were collected from store houses in Daejeon, Naju and Suwon in Korea during the period of December 2004 and February 2006. The conidia assumed to be Penicillium were picked up from blue molds of grapes and transferred to malt extract agar (MEA; malt extract 20 g, peptone 1.0 g, glucose 20 g, agar 20 g, distilled water 1 liter) and grown for 7 days at 25℃.
Culture
Isolates were inoculated onto Czapek yeast extract agar (CYA; K2HPO4 1.0 g, Czapek concentrate 10 mg, yeast extract 5 g, sucrose 30 g, agar 15 g, distilled water 1 liter) and malt extract agar (MEA) media in 3 points in a petridish. Colony appearance, exudates production, pigmentation and reverse coloration were assessed and colony diameters were measured and recorded after one week incubation at 25℃.
Morphological observations
Penicillium isolates were identified with the help of keys developed by Pitt (1979, 2000) and Frisvad and Samson (2004). Cultures were inoculated on CYA and 2% MEA media in three-points of 9 cm plastic Petri dishes. Petri dishes were incubated at 25℃ in a dark condition. The cultures were examined after 7 days of incubation. All morphological data were examined on cultures grown on 2% MEA for 7 days at 25℃. The examination and measurements of conidiophores and conidia were made from slide preparations stained with 3% KOH. Differential interference contrast microscopy was used for the observation and 30 units of each morphological character were measured.
DNA extraction and PCR amplification
Isolates were grown on liquid shake culture of potato dextrose broth medium for 3~4 days at 25℃. Mycelia were collected from the cultures by filtration and transferred to 1.5 ml tubes. These samples were frozen at -70℃ before use. DNA was extracted by the method of Cubero et al. (1999). For the amplification of β-tubulin gene, primers Bt2a (5'-GGTAACCAAATCGGTGCTGCTTTC-3') and Bt2b (5'-ACCCTCAGTGTAGTGACCCTTGGC-3') (Glass and Donaldson, 1995) were used. PCR mixture contained 0.5 pmol of each primer, 0.2 mM of dNTP's, 10 mM Tris-HCl, 50 mM KCl, 1.5 MgCl2, 2.5 U Taq polymerase and 15 ng of template DNA. PCR amplification was carried out in a i-cycler (BIO-RAD, USA) for 30 cycles of 94℃ for 40 s denaturing, 55℃ for 40 s annealing and 72℃ for 1 min extension. Initial denaturing at 94℃ was extended to 3 min and the final extension was at 72℃ for 15 min.
The PCR products were purified by using a Wizard PCR prep. kit (Promega, Madison, WI, USA). Purified double stranded PCR fragments were directly sequenced with BigDye terminator cycle sequencing kits (Applied Biosystems, Forster City, CA, USA) by following the manufacturer's instructions. Same primer sets with PCR amplification were used to sequence both DNA strands. The gel electrophoresis and data collection were performed on an ABI Prism 310 Genetic Analyzer (Applied Biosystems, Forster City, CA, USA).
Sequencing and phylogenetic analysis
The sequences were compared with β-tubulin gene sequence available in the GenBank by the BLAST search analysis. Sequences generated from materials in this study and retrieved from GenBank were initially aligned using the program CLUSTAL X (Thompson et al., 1997) and then the alignment was refined manually using the PHYDIT program version 3.0 (Chun, 1995; available at http://plaza.sun.ac.kr/~jchun/phydit).
Ambiguously aligned regions were excluded from the subsequent analyses. A neighbor-joining tree was reconstructed with Kimura's 2-parameter distance model (Kimura, 1980) using the PHYDIT 3.57c package (Felsenstein, 1995). The bootstrap analysis using 1000 replications were performed to assess the relative stability of the branches.
Results and Discussion
Grape fruits with blue mold were collected from different store houses in Korea. Twelve isolates were selected as Penicillium on the basis of their morphological structure. According to cultural and morphological characteristics, the Penicillium species were divided into 6 groups. On the basis of previous description by Pitt (1979, 2000) and Frisvad and Samson (2004), each group was identified as P. bialowiezense, P. citrinum, P. echinulatum, P. expansum, P. solitum and unidentified Penicillium sp. Taxonomic descriptions, photos of colonies and drawings of fungal structures of each species are given below.
Penicillium bialowiezense K. Zaleski, Figs. 1(A, B), 3(A) Bul. Int. Acad. Pol. Sci. Lett., Ser. B 1927: 462, 1927
Fig. 1.
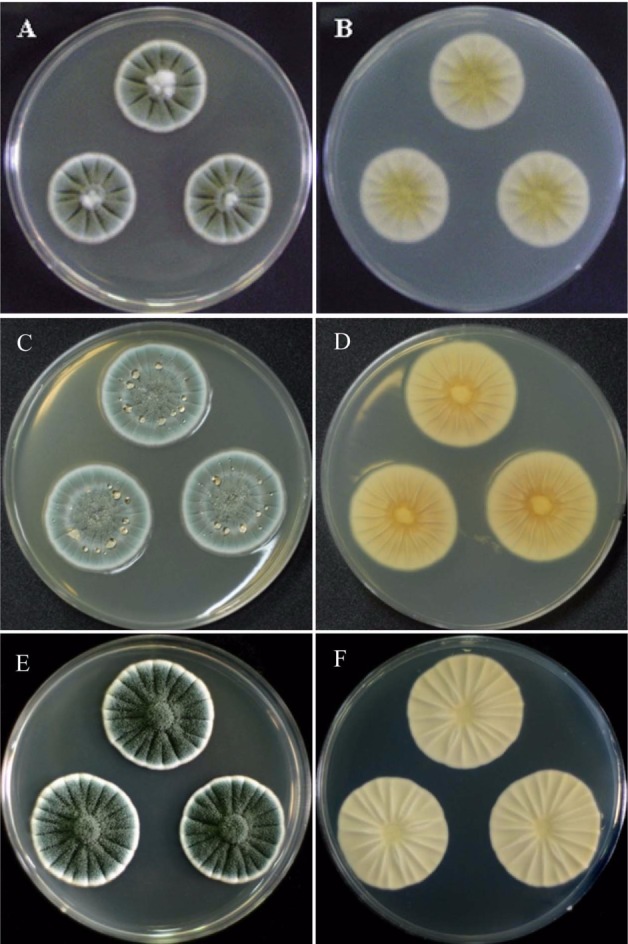
Penicillium colonies on CYA after 7 days of incubation: (A, B) P. bialowiezense; (C, D) P. citrinum; (E, F) P. echinulatum: (A, C, E) obverse; (B, D, F) reverse.
Fig. 3.
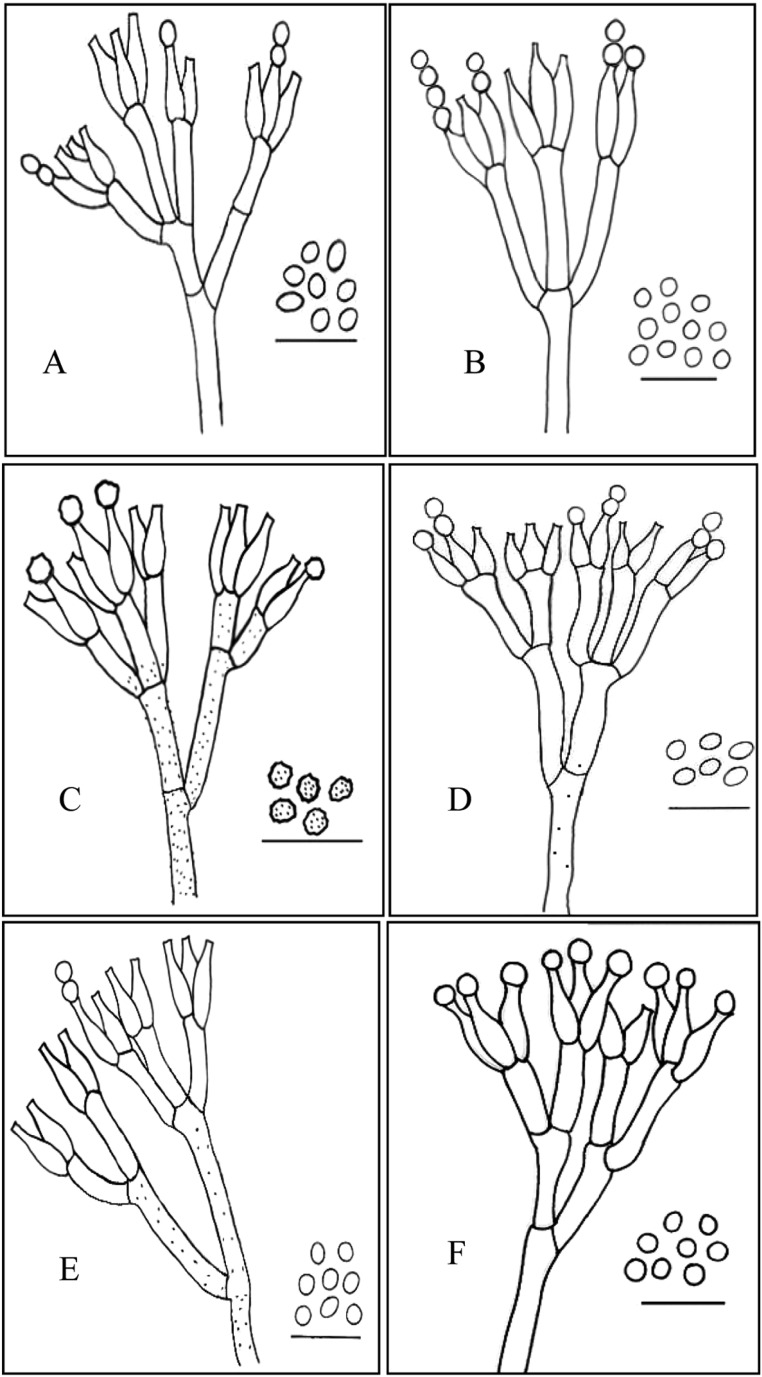
Conidiophores and conidia of (A) P. bialowiezense, (B) P. citrinum, (C) P. echinulatum, (D) P. expansum, (E) P. solitum and (F) unidentified Penicillium sp. Scale bar = 12 µm.
Colonies on CYA 20~28 mm diam, radily sulcate, surface texture vulutinous; conidiogenesis moderate to heavy, dark green to grey green; exudate often present as pale to reddish brown droplets; reverse beige to yellowish cream.
Colonies on MEA 15~23 mm diam, pale or slightly sulcate; surface texture velutinous, floccose in center; conidiogenesis light to moderate, pale green to pale yellow green; exudates absent; reverse pale yellow to yellow orange.
Conidiophores long, appressed, stipes 250~450 µm long, smooth-walled, terverticilate; rami 20~25 µm long; metulae cylindrical but apically inflated,, 10~15 µm long, phialides cylindrical with gradually tapering collula, 6~10 × 2.5~3 µm; conidia subglobose to ellipsoidal, finely roughened, 2.5~3.8 × 2~3 µm.
Isolate examined: CNU 060156.
Notes: Colony characteristics and micromorphology of the fungus agreed well with the description of P. bialowiezense (Frisvad and Samson, 2004). The species is most closely related to P. bravicompectum. P. bialowiezense has shorter and less wide stipes than P. bravicompectum which differentiate it from P. bravicompectum. The fungus has been reported from forest soil, wheat bread, grapes etc. in Denmark, Poland, Italy, Slovenia, Chile, Wyoming, USA, Canada, Saudi Arabia etc. (Frisvad and Samson, 2004).
Penicillium citrinum Thom, Figs. 1(C, D), 3(A) Bull. Bur. Anim. Ind. US Dep. Agric. 118: 61, 1910
Colonies on CYA 26~32 mm diam, radily sulcate, surface texture floccose in center and vulutinous at margins; conidiogenous light to moderate, greyish green; exudates clear, pale yellow or pale brown to rddish brown; reverse yellow, yellow brown to reddish brown.
Colonies on MEA 20~22 mm diam, plane or radially sulcate; surface texture floccose; conidiogenesis moderate to heavy, grey blue at the margin, elsewhere dull green; exudates absent; reverse pale brown to yellow brown.
Conidiophores arising from subsurface or surface hyphae, stipes 100~300 µm long, smooth walled, biverticilate with 3~5 divergent metulae in a whorl; metulae usually uniform, 15~20 × 2~5 µm, bearing 6~10 phialids; phialids ampulliform, 7~10 × 2~2.5 µm; conidia globose to subglobose, 2.5~3.0 µm diam, smooth-walled or finely rough-walled, produced in long columns.
Isolates examined: CNU 04245, CNU 04246.
Notes: The principle distinguishing character of this species is the penicillus, which consists of 3~5 divergent and usually vesiculate metulae, bearing long well defined columns of conidia (Pitt, 1979). Colonies on CYA at 25℃ were dominated by copious clear to yeallow or brown exudates at center; on MEA, growth was slow and dense, with heavy conidiogenesis (Pitt, 1979). This fungus has been reported from soils in Korea (Kim, 1979; Kim and Lee, 1980; Min et al., 1980).
Penicillium echinulatum Fassatiova, Figs. 1(E, F), 3(C) Acta Univ. Carol. Biol. 12: 326, 1977
Coloies on CYA 32~40 mm diam, deeply radially sulcate, dense, texture velutinous to weakly fasciculate, floccose in center; conidiogenesis heavy, dull green to dark green; exudate absent or clear droplets; reverse light bluish grey or cream coloured.
Colonies on MEA 20~30 mm diam, lightly radially sulcate, texture velutinous to granular, occasionally crustose centrally; conidiogenesis moderate to heavy, dull green to dark green; exudates clear; reverse yellow orange to orange brown.
Conidiophores borne from subsurface hyphae, singly or in fascicles, appressed, stipes 250~500 × 3.0~4.0 µm long, rough-walled, terverticillate; rami cylindrical 12~20 × 3.5~ 4.0 µm long; metulae cylindrical, 10~15 × 2~3 µm; phialides ampulliform tapering to a distinct column, 8~11 × 2~3 µm; conidia globose to subglobose, rough-walled, 3.5~4.5 µm.
Isolate examined: CNU 04225.
Notes: Colony characteristics and micromorphology of the fungus agreed well with the description of P. echinulatum (Frisvad and Samson, 2004). The species is closely related to P. solitum and P. discolor, but differs from P. solitum by producing rough-walled conidia and from P. discolor by inability to produce a diffusible red pigment (Frisvad and Samson, 2004). The fungus has been reported from food (margarine, chesse and other lipid contaiing substrate) in Japan, Denmark, Sweden, Norway, the Netherlands, Germany, France, Russia, USA, Wyoming, Thailand, South Africa and Canada (Frisvad and Samson, 2004). This is the first record of P. echinulatum from grape in Korea.
Penicillium expansum Link, Figs. 2(A, B), 3(D) Obs. Mycol. 1: 16, 1809
Fig. 2.
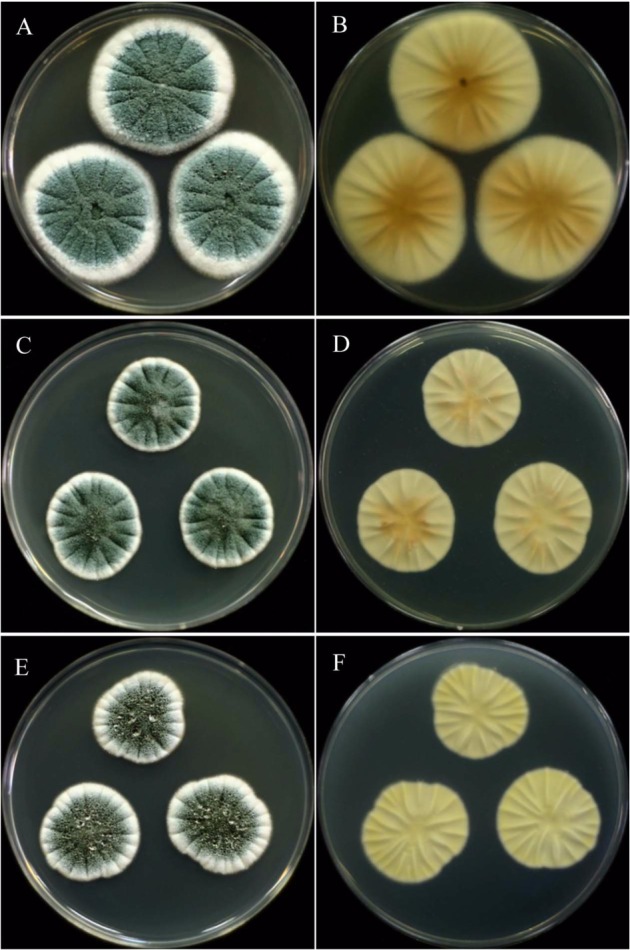
Penicillium colonies on CYA after 7 days of incubation: (A, B) P. expansum; (C, D) P. solitum; (E, F) unidentified Penicillium sp.: (A, C, E) obverse; (B, D, F) reverse.
Colonies on CYA 35~50 mm diam, radily sulcate, moderately deep, texture floccose to weakly fasciculate; conidiogesis moderate, blue green to moderate bluish green; exudate clear to pale orange brown; reverse light to vivid yellow.
Colonies on MEA 25~35 mm diam, plane, texture velutinose or coremial, sometimes becoming crustose; conidiogenesis heavy, yellow green to blue green; reverse pale to orange brown.
Conidiophores borne from subsurface or aerial hyphae, single or in fascicles, appressed, stipes usually smooth-walled, occasionally very finely rough-walled, terverticillate, less commonly biverticillate; rami cylindrical, 15~25 × 3~4 µm; metulae more or less cylindrical, 10~17 × 3~4 µm; phialids ampulliform, 9~12 × 2~4 µm; conidia ellipsoidal to subglobose, smooth-walled, 3~3.5 × 2.5~3.0 µm diam.
Isolates examined: CNU 04209, CNU 04210.
Notes: Colony characteristics and micromorphology of the fungus agreed well with the description of P. expansum (Frisvad and Samson, 2004). The species is similar to P. crustosum and P. viridicatum in colony morphology and colouration. The fungus, however, has ellipsoidal conidia and usually smooth stipes. The fungus has been reported from apples, pears and other pomaceous fruits, cherries, peaches etc. in Denmark, United Kingdom, Sweden, Norway and many other countries (Frisvad and Samson, 2004).
Penicillium solitum Wrestling, Figs. 2(C, D), 3(E) Ark. Bot. 11: 52, 1911
Colonies on CYA 20~30 mm diam, radily sulcate, surface texture vulutinous, less commonly fasciculate; conidiogenesis very heavy, dark bluish green to dark green; exudate often present, clear to light yellow droplets on the surface; reverse pale or cream to light beige.
Colonies on MEA 15~27 mm diam, radially sulcate; surface texture velutinous; conidiogenesis heavy, pale bluish green to greyish blue green; exudates rarely present, clear to light yellow; reverse light to moderate orange yellow.
Conidiophores arising from subsurface hyphae, stipes 200~400 µm long, rough-walled, terverticilate; rami cylindrical 10~18 µm long; metulae cylindrical, 10~15 µm long; phialides ampuliform, 9~11 × 2~3 µm; conidia globose to subglobose, less commonly broadly ellipsoidal, smooth to finely rough-walled, 3~4.5 µm.
Isolates examined: CNU 04211, CNU 04213, CNU 04214.
Notes: Colony characteristics and micromorphology of the fungus agreed well with the description of P. solitum (Frisvad and Samson, 2004). The species is most closely related to P. echinulatum and P. discolor, but differs from P. echinulatum by producing smooth-walled conidia and from P. discolor by inability to produce a diffusible red pigment (Frisvad and Samson, 2004). The fungus has been reported from apples, oriental pear, grape, citrus, processed meat in Japan, Denmark, Greenland, United Kingdom, Sweden, Norway, the Netherlands, Germany, France, Russia, USA and Canada (Frisvad and Samson, 2004).
Penicillium taxon, Figs. 2(E, F), 3(F)
Colonies on CYA 20~30 mm diam, radily sulcate, surface texture vulutinous; conidiogenesis moderate to heavy, green to dull green; exudate present as clear or colourless droplets on surface; reverse pale greenish yellow to greyish yellow green.
Colonies on MEA 15~25 mm diam, plane or lightly sulcate; surface texture velutinous; conidiogenesis light to moderate, pale green; exudates absent, reverse light yellow to vivid yellow.
Conidiophores arising from submerged or aerial hyphae, stipes 150~300 µm long, smooth or rough-walled, usually terverticilate, sometime biverticillate; rami cylindrical 15~25 µm long; metulae cylindrical, 8~14 µm long, phialids ampuliform, flask-shaped with short necks, 10~13 mm; conidia globose to subglobose, smooth-walled, 2.5~4.0 µm diam.
Isolate examined: CNU 060161.
Notes: This taxon can be distinguished from P. bialowiezense by its globose conidia.
Sequence analysis of β-tubulin gene
The partial β-tubulin gene from 12 isolates of Penicillium from blue mold fruits of grape was amplified. Amplication of β-tubulin gene with primers Bt2a and Bt2b yields fragments of approximately 500 bp. BLAST database searches were performed with partial β-tubulin gene as queries to reveal relationship to published sequences.
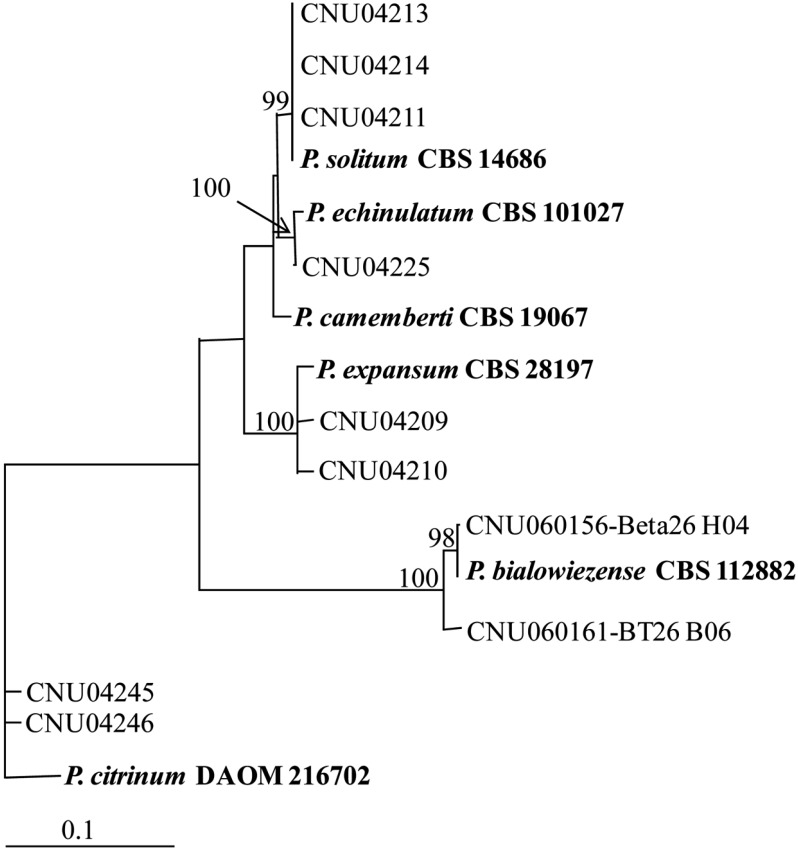
In a distance analysis with neighbor-joining method, sequence of isolates CNU 04211, CNU 04213 and CNU 04214 was 100% identical to that of P. solitum CBS 14686, with a bootstrap value of 99% (Fig. 4, Table 1). Sequence of CNU 04225 isolate was 100% identical to that of P. echinulatum CBS 19067, with a bootstrap value of 100%. Isolates CNU 04209 and CNU 04210 and P. expansum CBS 28197 were belonged to the same group. Sequence similarity among them was 100%, which was supported by a bootstrap value of 100%. β-tubulin sequence of CNU 060156 and CNU 060161 were 100% and 98% identical to P. bialowiezense CBS 112882, respectively. Sequence of isolates CNU 04245 and CNU 04246 was 99.5% identical to that of P. citrnum DAOM 216702 (Table 1). Sequence of CNU 04206 isolate matched well with that of P. camemberti CBS 19067 (Fig. 3, Table 1).
Fig. 4.
Neighbor-joining tree based on phylogenetic analysis of β-tubulin gene sequences. The number below each branch indicates bootstrap values of distance. The bootstrap values were obtained after a bootstrap test of 1000 replications. References, which were taken from GenBank database, are in bold type.
Table 1.
DNA similarity matrix for â-tubulin gene sequences of P. bialowiezense, P. citrinum, P. echinulatum, P. expansum, P. solitum and unidentified Penicillium species and their related species
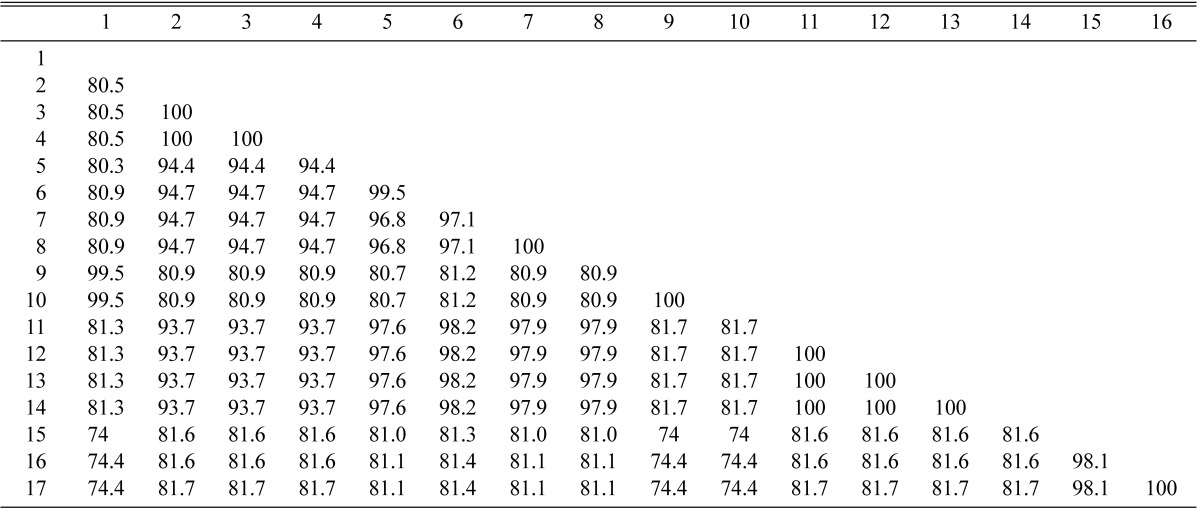
1. P. citrinum DAOM216702 (GenBank database); 2. P. expansum CBS28197 (GenBank database); 3. P. expansum CNU04209; 4. P. expansum CNU04210; 5. P. echinulatum 101027(GenBank database); 6. P. echinulatum CNU04225; 7. P. camemberti CBS19067 (GenBank database); 8. P. sp. CNU04206; 9. P. citrinum CNU04245; 10. P. citrinum CNU04246; 11. P. solitum CBS14686 (GenBank database); 12. P. solitum CNU04211; 13. P. solitum CNU04213; 14. P. solitum CNU04214; 15. P. bialowiezense CNU060161; 16. P. bialowiezense CNU060156; 17. P. bialowiezense CBS112882 (GenBank database).
The β-tubulin gene sequence has been used extensively to characterize a wide range of fungi, specially Penicillium. The amount of variation is suitable for studying phylogenetic relationship among closely related species of Penicillium (Samson et al., 2004) and filamentous fungi (Glass and Donaldson, 1995).
In this study, the phylogenetic tree inferred from the sequences of β-tubulin gene correlated well with the species that were defined by cultural and morphological characteristics except CNU 04206 and P. camemberti. The β-tubulin gene sequence evidenced that isolate CNU 04206 was identical to the GenBank sequence of P. camemberti CBS 19067 (Table 1), but morphological and cultural characteristics of our isolates did not agree with the description of P. camemberti (Frisbbad and Samson, 2004). The species was not identified and was supposed to a new species, not previously reported in literature.
Acknowledgement
This study was supported by a grant from the BioGreen 21 Program of Rural Development Administration, Republic of Korea.
References
- 1.Anonymous. List of plant diseases in Korea. 4th ed. The Korean Society of Plant Pathology; 2004. [Google Scholar]
- 2.Cho HS, Hong SB, Go SJ. First report of Penicillium brasilianum and P. dalear isolated form soil in Korea. Mycobiology. 2005;33:113–117. doi: 10.4489/MYCO.2005.33.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chun J. Computer-assisted classification and identification of actinomycetes. Newcastle upon Tyne, UK: University of Newcastle; 1995. PhD thesis. [Google Scholar]
- 4.Cubero OF, Crespo ANA, Fatehi F, Bridge PD. DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. Plant Syst Evol. 1999;216:243–249. [Google Scholar]
- 5.Felsenstein J. PHYLIP (Phylogeny Inference Package) Version 3.57C. Seattle, WA: University of Washington; 1995. [Google Scholar]
- 6.Frisvad JC, Samson RA. Phylogenetic taxonomy of Penicillium subgenus Penicillium. A guide to identification of food and air-borne terverticillate penicilla and their mycotoxins. Stud Mycol. 2004;49:1–173. [Google Scholar]
- 7.Glass NL, Donaldson GC. Development of primer sets designed for use with the PCR to amplify conserved gene from filamentous Ascomycetes. Appl Environ Microbiol. 1995;61:1323–1330. doi: 10.1128/aem.61.4.1323-1330.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerche SL, Garcia C, Darriet P, Dubourdieu D, Labarere J. Characterization of Penicillium species isolated from grape berries by their internal transcribed spacer (ITS) sequences and by gas chromatography-mass spectrometry analysis of geomsin production. Curr Microbiol. 2004;48:405–411. doi: 10.1007/s00284-003-4176-4. [DOI] [PubMed] [Google Scholar]
- 9.Kim JH, Lee WH, Cheong SS, Choi JS, Ryu J, Choi YG. Identification and chracteristics of Penicillium spp isolated from post-harvest decay of pear. Res Plant Dis. 2002;82:113–116. [Google Scholar]
- 10.Kim WK, Park MS, Hahm SS, Yu SH. Two new records of Penicillium with blue moldy bulbs of lily in Korea. Mycobiology. 2006;34:176–179. doi: 10.4489/MYCO.2006.34.4.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim KS. Soil-borne fungi of Phyllosticta reticulata forests soil in Korea (I) Korean J Mycol. 1979;7:91–116. [Google Scholar]
- 12.Kim KS, Lee JY. Soil-borne fungi of Phyllosticta reticulata foreats soil in Korea (II) Korean J Mycol. 1980;8:45–51. [Google Scholar]
- 13.Kimura M. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequence. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 14.Lee S, Hong SB, Kim CY. Contribution to the checklist of soil-inhabiting fungi in Korea. Mycobiology. 2003;31:9–18. [Google Scholar]
- 15.Min KH, Hong SW, Yokoyama T. Hyphomycetes from Korean soil. The genus Penicillium with a teleomorphic state Eupenicillium javanicum. Korean J Mycol. 1980;18:91–103. [Google Scholar]
- 16.Oh SY, Chung IM, Paik SB, Yu SH. Survey and control of the occurrence of mycotoxins from post-harvest fruits. 1. Mycotoxins produced by Penicillium isolates from apple, pear, citrus and grape. Plant Dis Agric. 1999;5:100–104. [Google Scholar]
- 17.Pitt JI. Distribution of ubiquinones in Penicillium and related genera. Mycol Res. 1991;95:705–711. [Google Scholar]
- 18.Pitt JI. The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London, UK: Academic press; 1979. p. 634. [Google Scholar]
- 19.Samson RA, Seifert KA, Kuijpers AFA, Houbraken JAMP, Frisvad JC. Phylogenetic analysis of Penicillium subgenus Penicillium using partial β-tublin sequences. Stud Mycol. 2004;49:175–200. [Google Scholar]
- 20.Seifert KA, Louis-Seize G. Integration of Modern Taxonomic Methods for Penicillium and Aspergillus Classification. Amsterdam, the Netherlands: Harwood Academic Publishers; 2000. Phylogeny and species concepts in the Penicillium aurantiogriseum complex as inferred from partial β-tubulin gene DNA sequences; pp. 189–198.pp. 510 [Google Scholar]
- 21.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTALX windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucl Acids Res. 1997;25:4876–4878. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]