Abstract
A keratinolytic enzyme secreted by Aspergillus flavus K-03 cultured in feather meal basal medium (FMBM) containing 2% (w/v) chicken feather was purified and characterized. Keratinolytic enzyme secretion was the maximal at day 16 of the incubation period at pH 8 and 28℃. No relationship was detected between enzyme yield and increase of fungal biomass. The fraction obtained at 80% ammonium sulfate saturation showed 2.39-fold purification and was further purified by gel filtration in Sephadex G-100 followed by ion exchange chromatography on DEAE-Sephadex A-50, yielding an active protein peak showing 11.53-fold purification. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and zymograms indicated that the purified keratinase is a monomeric enzyme with 31 kDa molecular weight. The extracellular keratinase of A. flavus was active in a board range of pH (7~10) and temperature (30℃~70℃) profiles with the optimal for keratinase activity at pH 8 and 45℃. The keratinase activity was totally inhibited by protease inhibitors such as phenylmethylsulfonyl fluoride (PMSF), iodoacetic acid, and ethylenediaminetetraacetate (EDTA) while no reduction of activity by the addition of dithiothreitol (DTT) was observed. N-terminal amino acid sequences were up to 80% homologous with the fungal subtilisins produced by Fusarium culmorum. Therefore, on the basis of these characteristics, the keratinase of A. flavus K-03 is determined to be subtilisins-like.
Keywords: Aspergillus flavus, Keratinase, Subtilisins, Serine protease
Keratin occurs in nature mainly in the form of hair, feathers, wool, horn, and nails and is the structural component of vertebrate skin. Arising as a waste product in a variety of ways, keratin is an insoluble protein and a component of epidermal and skeletal tissues (Bradbury, 1973). Because of a higher degree of cross-linking by disulphide bridges, hydrogen bonds and hydrophobic interactions, keratin is poorly susceptible to degradation and poorly susceptible to digestion by most common peptidases such as trypsin, pepsin, and papin (Bockel et al., 1995; Cohlberg, 1993). In particular, feathers constitute troublesome waste products, being mainly composed of hard β-keratin, and are produced in large quantities in commercial poultry processing plants. Therefore, their industrial utilization is economically and environmentally important (Onifade and Abu, 1998). Currently, a considerable portion of feather waste is utilized as a dietary protein supplement for animal feed (Bhargava and O'Neil, 1975), because of their low digestibility, invariable protein quality, and nutrient bioavailability (Papadopoulos et al., 1986).
To make better use of feather waste, pre-digestion of feathers with microorganisms has been proposed. Therefore, utilizing poultry feathers as a fermentation substrate in conjunction with a keratin-degrading microorganism or enzymatic biodegradation may be a better alternative to improve nutritional value of poultry feathers and reduce environmental waste. Recently, several extracellular keratinases, including Chrysosporium keratinophilium (Dozie et al., 1994), C. georgiae (El-Naghy et al., 1998), Aspergillus fumigatus (Do et al., 2004; Santos et al., 1996), and airborne dematophytes fungi (Marchisio et al., 1992), have been reported to be functional for the microbial and fungal invasion of skin and skim formation. As such, screening of non-pathogenic common fungi for producing keratinase is very important, since keratinase has potential applicability to the pharmaceutical and leather industries. We isolated the fungus Aspergillus flavus, which was selected as a prospective producer of keratinolytic enzyme (Kim, 2003). Here we report on the purification and characterization of an extracellular keratinolytic enzyme produced by A. flavus strain K-03.
Materials and Methods
Microorganism and culture conditions
A. flavus K-03, which was isolated from a poultry farm in South Korea (Kim, 2003), was maintained by serial passages in a potato-dextrose agar (PDA; Difco, Sparks, MD, USA) medium and was cultivated in 500 ml Erlenmeyer flasks, holding 200 ml of feather meal basal medium (FMBM) (Kim, 2003) containing 10 g of glucose, 0.025 g of MgSO4·7H2O, 0.025 g of CaCl2, 0.015 g of FeSO4·7H2O, 0.005 g of ZnSO4·7H2O per liter. The pH was adjusted to 8.0. Chicken feather (20 g/l) was used as carbon, nitrogen, and sulfur sources. Each flask was inoculated with 2 ml of a spore suspension (2 × 106 spores/ml) prepared from 7-day old subcultures of the fungus.
Protein determination
The protein content was determined by the method of Bradford (1976) using Bio-Rad assay reagent (Munich, Germany) and bovine serum albumin as a standard.
Keratinase activity
Keratinase activity was determined using a spectrophotometer according to the method of Letourneau et al. (1998). An aliquot of centrifuged culture broth containing 5 µg of protein sample was centrifuged and added to one-milliliter reaction mixture composed of 30 mg keratin azure (Sigma, St. Louis, MO, USA) in 50 mM Tris-HCl buffer (pH 8.0). The reaction was carried out at 28℃ for 1 h. The reaction mixture was constantly agitated at 120 rpm. After 1 h, the reaction was stopped by placing the tubes in an ice bath. The reaction mixture was then centrifuged at 10,000 ×g for 10 min. An assay without any sample was performed under the same conditions described above and used as a blank. All assays were done by measuring the absorbance at 595 nm, the amount of enzyme required to cause an increase of 0.001 in A595 nm per min.
Enzyme purification
2 l-Erlenmeyer flasks containing 1 l of FMBM were inoculated with a fungal suspension and incubated for 16 days at 28℃. The fungal culture broth was then centrifuged and the supernatants were filtrated successively through filter paper no. 2 (Whatmann Inc., Maidstone, UK) and 0.2 µm-pore-size filters (Millipore Corp., Billerica, MA, USA). In order to prevent enzyme autolysis, all procedures were carried out at 4℃. To purify keratinase, ammonium sulfate was added to the culture filtrate to 80% saturation. Each precipitate was dissolved in a certain amount of distilled water and dialyzed against distilled water. The precipitate resulting from ammonium sulfate fractionation at 80% saturation was suspended in 30 ml of 50 mM Tris-HCl buffer (pH 8.0) and dialyzed for 24 h at 4℃ in 2 l of 50 mM Tris-HCl buffer after dialysis. A portion (10 ml) was then loaded on a glass column (2.5 cm × 50 cm) packed with Sephadex G-75 (Sigma, St. Louis, USA) and equilibrated with 400 ml of 50 mM Tris-HCl buffer at pH 8.0. Protein was eluted with 50 mM Tris-HCl buffer (pH 8.0). Enzyme activity and protein content in each fraction were measured. Fractions showing high protein content and keratinase activity were collected and applied to an ion-exchange chromatography diethyaminoethyl (DEAE)-Sephadex A-50 column (2.5 cm × 50 cm) previously equilibrated with 200 ml of 50 mM Tris-HCl buffer (pH 8.0). Bound protein was eluted with 50 mM Tris-HCl buffer, followed by a linear gradient of NaCl (0~1 M NaCl equilibration buffer) at a flow rate of 60 ml/h. 5 ml-active fractions were pooled and protein content and keratinase activity for each fraction were monitored. Fractions exhibiting high keratinase activity were collected and concentrated by an ultrafiltration system with a 10 kDa-cutoff membrane (Ultra-15; Amicon Inc., Danvers, MA, USA). The retained material was applied to a Pharmacia HiLoad 16/60 Superdex 200 column equilibrated with 20 mM Tris-HCl buffer (pH 8.0) containing 0.15 M NaCl at 0.5 ml/min. The purified enzyme was stored in aliquots at -20℃ until use. Protein elution was monitored by measuring the absorbance at 590 nm.
Polyacrylamide gel electrophoresis
The purified enzyme was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a discontinuous buffer system (Laemmli, 1970). Separation gel (15%, w/v) and stacking gel (5%, w/v) were used. Proteins were stained by the silver staining method described by Merril et al. (1983) and the electrophoretically migration of the keratinolytic enzyme was compared with a broad range protein markers (Sigma, St. Louis, MO, USA).
Zymograms
SDS-PAGE 15% polyacryamide containing 10 mg/ml keratin was performed. Proteins and their active were visualized using Commassie brilliant blue R-250 (Sigma, St. Louis, MO, USA).
NH2-terminal amino acid sequence of keratinase
After electrophoresis, the purified enzyme was transferred to an Immobilon-p transfer membrane (Nihon Millipore Co., Tokyo, Japan) using the electoroblotting method (Kojima et al., 2006). The N-terminal amino acid sequence of the purified keratinase was determined with a protein sequencer (Model 476A, Applied Biosystems, Foster City, CA, USA). The sequences obtained were compared to sequences in the Swiss-Prot/TrEMBL database using the BLASTp algorithm (http://www.expasy.org/tools/blast/).
Effects of pH and temperature on enzyme activity
The optimal temperature for the purified enzyme activity was investigated by performing the enzyme reaction at different temperatures from 20℃ to 80℃ in 50 mM Tris-HCl buffer (pH 8.0). To determine the optimal pH, keratinolytic activity was assayed using different buffers, and the aliquots of the enzyme were adjusted from pH 3~12 using 50 mM buffers, i.e., citric acid/Na2HPO4 for pH 3~6, NaHPO4/Na2HPO4 for pH 6~8, Tris-HCl for pH 7~9, glycine/NaOH for pH 9~11, and NaHCO3/NaOH for 11~12. Each aliquot was incubated at 28℃ for 1 h and the remaining activity was assayed as previously mentioned.
Effects of protease inhibitors, metal ions, and reducing agents on keratinase activity
To investigate the effects of different protein inhibitors, metal ions, and reducing agents on keratinase activity, the purified enzyme solution was pre-incubated in 50 mM Tris-HCl buffer (pH 9.0) for 1 h at 40℃ with different reagents. Keratinase activity was determined as a percentage of residual activity relative to control. The enzyme was treated with the following protease inhibitors and metal ions: phenymethansesulphonyl fluoride (PMSF; 1 mM), pefabloc SC (1 mM), idoacetate (1 mM, ethylendiaminetetraacetate (EDTA; 1 mM), dithiothreitol (DTT; 1 mM), β-mercaptoethanol (1.1%; v/v), pepstatin A (3 µg/ml), and Ca2+, Mg2+, Mn2+, Cu2+, Fe2+, Hg2+, Zn2+ (1 mM each).
Results and Discussion
Growth and keratinase activity
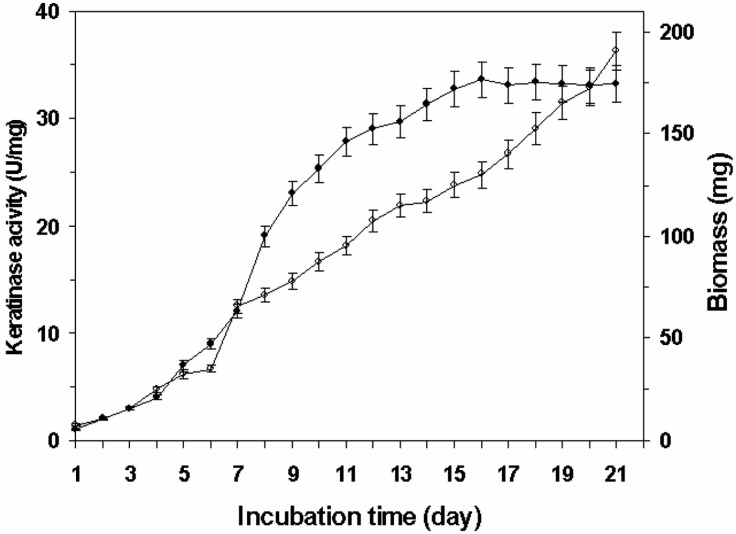
The time course for the production of extracellular keratinase activity in FMBM containing 2% (w/v) of feathers is shown in Fig. 1. The kinetics of enzyme synthesis demonstrated that production of the enzyme reached a maximum at day 16, and thereafter remained stable until day 21. In addition, the biomass increased steadily with incubation time. Similar production kinetics of keratinase has been reported for fungi such as Endothia parastica (Melzer and Boland, 1999), Trichophyton simii (Singh, 1997), Malbranchea gypsea (Singh, 1998), and Trichophyton vanbreuseghemii (Moallaei et al., 2006). In A. fumigatus (Do et al., 2004) and A. oryzae (Jousson et al., 2004), the rate of keratinase production reached a maximum concentration after 21 days of incubation and the concentration of extracellular keratinase produced by Lysobacter NCIMB 9497 (Allpress et al., 2002) was maximal after 29 days of growth.
Fig. 1.
Keratinase activity (•) and biomass (○) of A. flavus cultured in feather meal basal medium (FMBM) containing 2% (w/v) feather for 21 days at 28 ± 0.5℃, pH 8.0 and 120 rpm.
Purification of keratinase
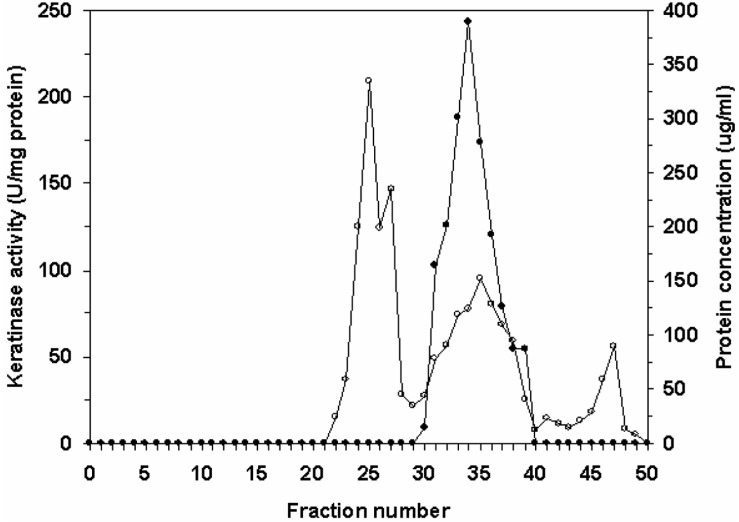
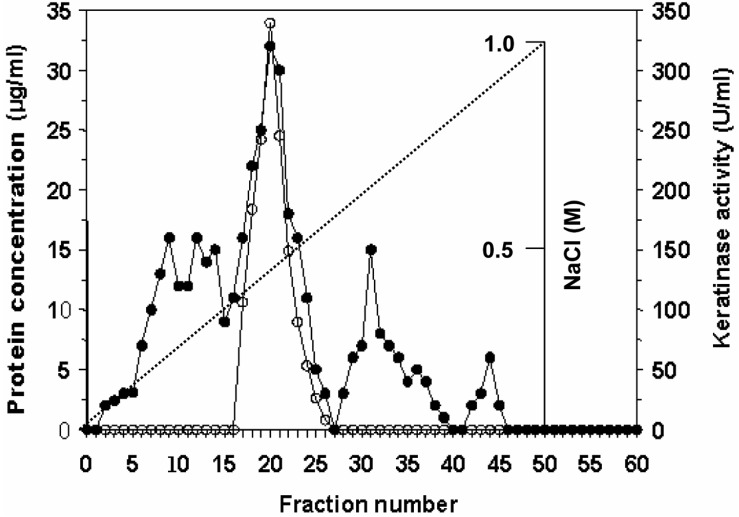
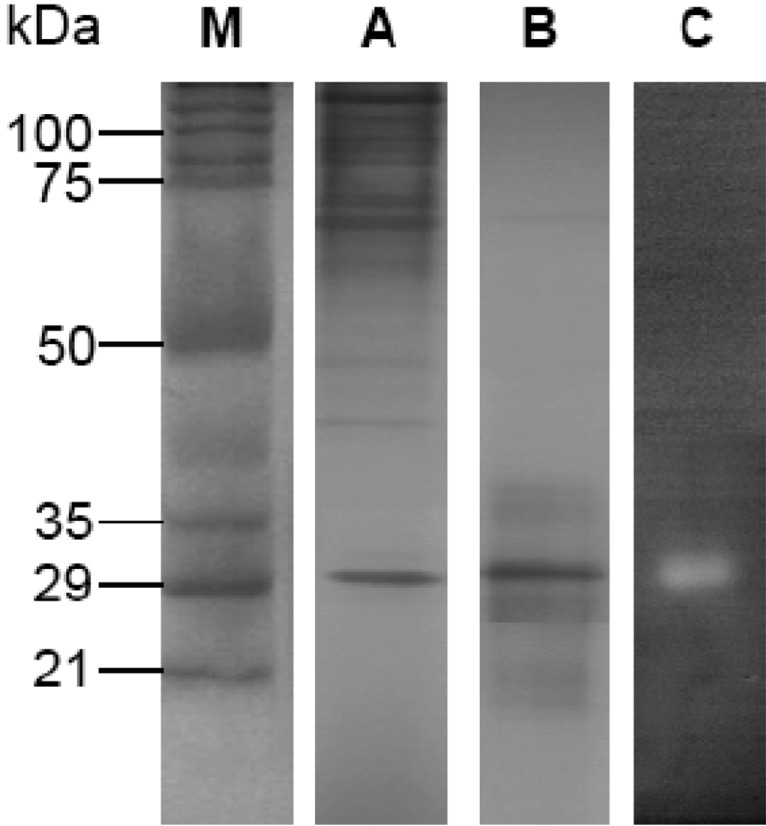
The fungus was cultivated for 16 days for all subsequent experiments. Purification of the keratinase was then undertaken. The crude enzyme, which was concentrated by centrifugation and precipitation with 80% saturation of ammonium sulfate, was dialyzed and subjected to gel filtration on a Sephadex G-75 column. The elution profiles for keratinase and protein from the Sephadex G-75 column are shown in Fig. 2. Three protein peaks were obtained. The second peak shows the highest specific keratinase activity (243.2 U/mg of protein). The most active fractions (numbers 30-39) from the Sephadex G-75 column were pooled and further purified by DEAE-Sephadex A-50 column chromatography (Fig. 3). Enzyme detection in the eluate revealed a second activity peak with high keratinase activity, corresponding to 0.4M NaCl, which superimposed with the third protein peak. The purification steps are summarized in Table 1. An overall recovery of 11.53-fold with a recovery of 32.36% and a specific activity of 316.15 U/mg protein were obtained. Analysis by SDS-PAGE revealed a single protein band (Fig. 4).
Fig. 2.
Gel filtration of Sephadex-75 of crude enzyme fraction precipitated with 80% ammonium sulfate. Symbols are (•) keratinase activity and (○) protein concentration.
Fig. 3.
Elution profile from A. flavus DEAE-Sephadex A-50. Symbols are (○) keratinase activity and (•) protein concentration.
Table 1.
Purification of the keratinase produced by A. flavus K-03

Fig. 4.
SDS-PAGE gel electrophoresis and zymograms of purified keratinase (5 µg) from A. flavus. Symbols are (M) molecular weight marker, (A) crud enzyme (10 µg), (B) purified keratinase after DEAE chromatography, and (C) zymograms of the purified keratinase.
Characterization of purified keratinase
The molecular mass of keratinase of A. flavus was estimated to be 31 kDa by both SDS-PAGE gel electrophoresis and zymograms (Fig. 4). In the case of the zymograms, only one band showed keratinolytic activity (Fig. 4, lane C). The enzyme appears to be a monomer. The molecular weight is in the range of other reported keratinases, between 16 kDa (Page and Stock, 1974) and 440 kDa (Yu et al., 1971). Most of these keratinases also have a molecular weight of around 30 kDa (Kang et al., 2005; Page and Stock, 1974). Gradisar et al. (2005) purified keratinases for non-pathogenic fungi, 33 kDa for Paecilomyces marquandii, 30 kDa for Doratomyces microsporus, and 22 kDa for A. flavus keratinase. All three keratinases are serine protease, as are the majority of keratinases.
NH2-terminal amino acid sequences
Table 3 lists the corresponding amino-acid sequences of several homologous fungal proteases. The NH2-terminal amino acid arrangement of the enzyme from the fungus A. flavus K-03 showed high homology with a substilisin-like protease produced by Fusarium culmorm (Pekkarinen et al., 2002), the sequences of which are 80% identical to those of A. flavus strain K-03. The proteases from several fungal species such as Cephalosporium cremonium, Trichoderma virens, T. hamatum, A. fumigatus, A. viridinutans, A. niger, and A. oryzae contained sequences that showed 44-76% homology. Subtilisin-like proteases from Microsporum canis (Descamps et al., 2002), A. niger (Jarai and Buxton, 1994), and A. oryzae (Tatsumi et al., 1991) have been detected, but their mechanisms have not been established.
Table 3.
Partial amino-acid sequence of fungal alkaline proteases that show homology with the A. flavus enzyme
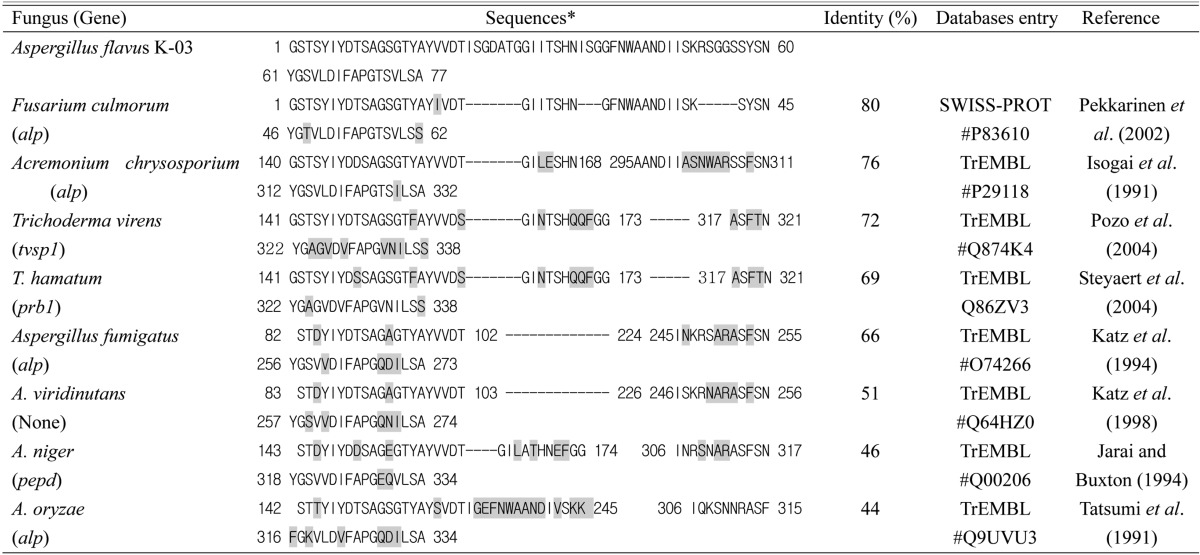
*Differential residues are indicated with shadows and missing peptides with dashes.
There was less homology in the NH2-terminal aminoacid arrangement between the purified keratinase and the proteases purified from A. fumigatus, A. viridinutans, A. niger, and A. oryzae (Table 3). Interestingly, most of the peptide sequences from C. cremonium, T. virens, and T. hamatum were generally conserved.
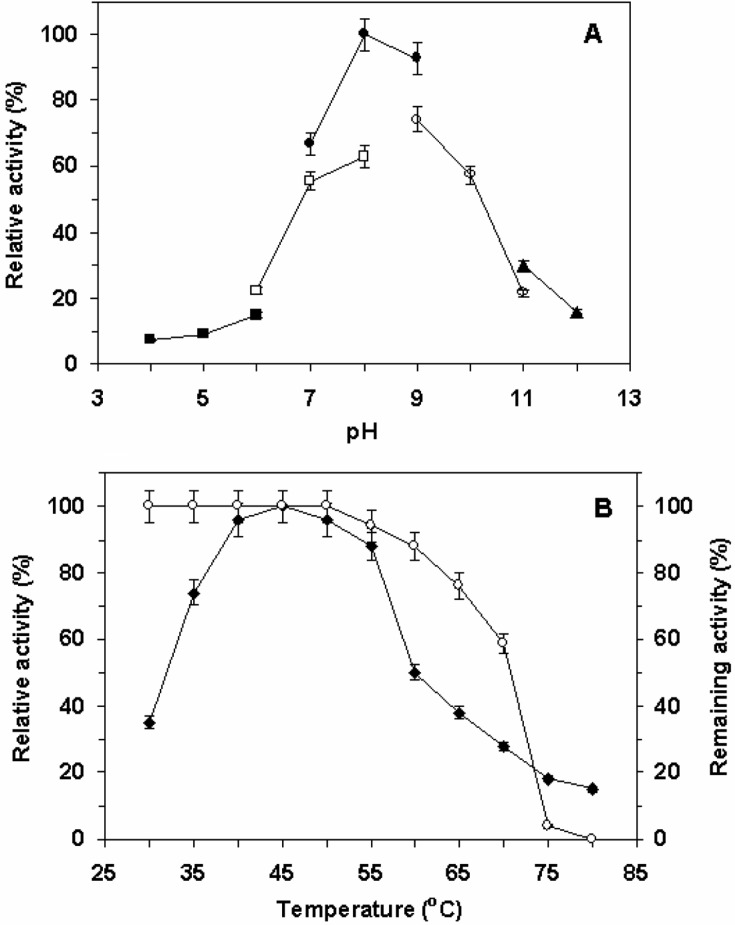
Effect of pH and temperature on keratinase
The optimal pH for keratinolytic activity was 8.0 (Fig. 5A). The enzyme activities declined rapidly at a pH higher than 8.0. 70% and 45% of the maximal enzyme activity were observed at pH 9.0 and 10.0, respectively. The enzyme was active even at pH 12.0. The optimal temperature for keratinase activity was determined by varying the reaction temperatures, between 30℃ and 80℃, at pH 8.0. The optimal temperature was 45℃ for keratinase activity, but above 65℃ the activity sharply decreased, as shown in Fig. 5B. The enzyme exhibited 30% and 15% of the maximum activity at 70℃ and 80℃, respectively. Thermal stability of the keratinase was investigated by heating the purified enzyme for 15 min at different temperatures in 100 mM Tris-HCl (pH 8.0). The enzyme was very stable up to 50℃ followed by a rapid loss of activity above 70℃. The enzyme retained more than 90% and 15% of its activity at 60℃ and 70℃, respectively. However, the enzyme was completely inactivated at 80℃. The activity and stability of the enzyme were higher than salt-tolerant keratinase from Aspergillus sp. FC-10 (Su and Lee, 2001). Su and Lee (2001) reported 80% inactivation of alkaline keratinase activity during incubation at 60℃. Nevertheless, the alkaline serine-type keratinase from A. terreus (Chakarabarti et al., 2000) showed maximum activity at pH 8.5 and 60℃. The enzyme retained about 60% at 70℃ whereas at 80℃ it retained only 20% of its keratinolytic activity. At 90℃, it was completely inactive.
Fig. 5.
(A) Effect of pH on keratinase activity. The pH profiles were determined in different buffers by varying pH values at 60℃. (▪), citrate buffer (pH 4~6); (□), NaHPO4/Na2HPO4 buffer (pH 6~8); (•), Tris-HCl buffer (pH 7~9); (○), glycine/NaOH buffer (pH 9~11); (▴), NaHCO3/NaOH buffer (pH 11~12). (B) Effect of temperature on the keratinase activity (♦) and on the thermal stability (○) of the purified keratinase. The enzyme was incubated in 100 mM Tris-HCl buffer (pH 8.0) at different temperatures for 15 min and the residual activity was determined as described in Materials and Methods.
Similar keratinase activities with purified keratinase from the fungal strain K-03 at temperatures in excess of 70℃ have been described only for Chrysosporium keratinophilus (Dozie et al., 1994), A. fumigatus (Santos et al., 1996), and Fervidobacterium pennivorans (Kim et al., 2004). Most of the other keratinases from bacteria and fungi (Cheng et al., 1995; Gradisar et al., 2005; Santos et al., 1996; Singh, 1997) are active at alkaline pH but show optimal activity at roughly 40℃, which is lower than the optimum temperature of the enzyme from the fungus A. flavus K-03.
Effect of metal ions and reagents on keratinase
The effects of various metals and reagents on the activity of purified keratinase from A. flavus K-03 are summarized in Table 2. The activity of the keratinase was reduced by serine-, metallo-, and cysteine-protease inhibitors (Table 2). Divalent metal ions such as Mn2+, Fe2+, and Hg2+ in concentrations up to 1 mM caused a slight activation of the extracellular keratinase (10%~20%). The activity was slightly reduced by Ca2+, Mg2+, Cu2+, and Zn2+ (5%~10%). These results, taken as a whole, suggest that the purified extracellular alkaline keratinase may be a metallo-like protease. In some reports (Kwak et al., 2004; Markaryan et al., 1994; Monod et al., 1993), A. fumigatus was also found to produce an extracellular Zn-metallo-like protease, representing up to 30% of the total protease activity, although the isoelectoric point of the enzyme purified by Monod et al. (1993) is 5.5. In a previous study (Kim, 2003), a serine type protease was purified from A. niger. However, the alkaline keratinase produced by A. flavus strain K-03 is clearly distinguished from the protease of A. niger, having characteristics of a serine-type protease. The extracellular keratinase of A. flavus strain K-03 was totally inhibited in the presence of phenylmethylsulfonyl fluoride (PMSF), iodoacetic acid, and ethylenediaminetetraacetate (EDTA). Since complete inhibition by PMSF could not be made reversible by the addition of dithiothreitol (DTT), the enzyme cannot be classified as a cysteine protease. The enzyme belongs to serine-type proteases.
Table 2.
Effect of metal ions, inhibitors, and reducing agents on the activity of keratinase purified from A. flavus K-03
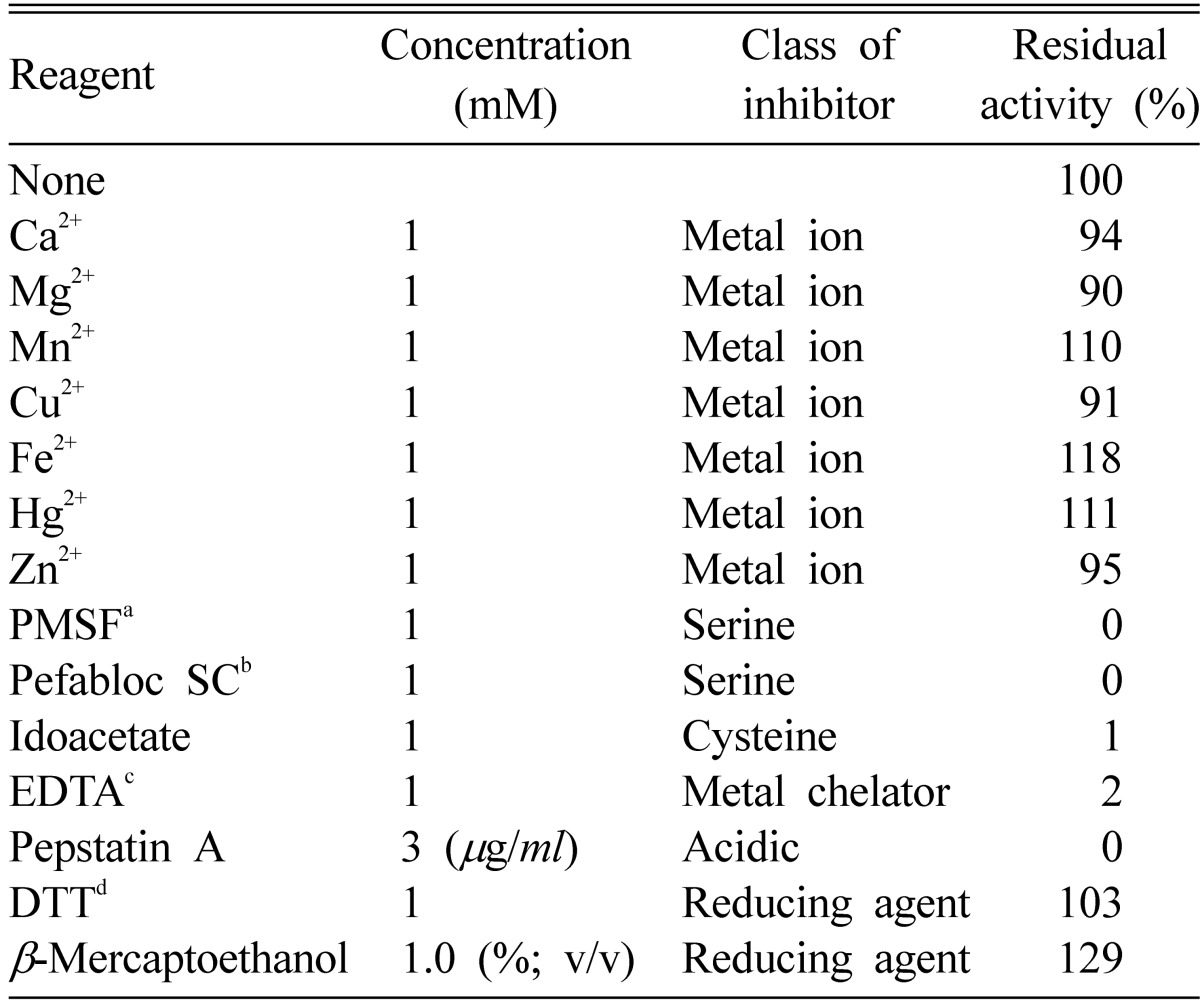
aPMSF; phenymethanesulphonyl fluoride.
bSigma-Aldrich, St. Louis, MO, USA.
cEDTA; ethylenediaminetetraacetate.
dDTT; dithiothreitol.
Therefore, the keratinase of A. flavus may have potential use in the detergent industry and should be of interest to leather industries. It also has considerable biotechnological potential for the processing of poultry feather waste and in microbiological keratin hydrolysates for feed use.
Acknowledgement
This study was supported by a Korea Research Foundation Grant (KRF-2006-J00702), for which the authors are thankful.
References
- 1.Allpress JD, Mountain G, Gowland PC. Production, purification and characterization of an extracellular keratinase from Lysobacter NCIMB 9497. Lett Appl Microbiol. 2002;34:337–342. doi: 10.1046/j.1472-765x.2002.01093.x. [DOI] [PubMed] [Google Scholar]
- 2.Bhargava K, O'Neil J. Composition and utilization of poultry by-product and hydrolyzed feather meal in broiled diets. Poultry Sci. 1975;54:1511–1518. [Google Scholar]
- 3.Bockel S, Diamy AM, Ricard A. Optical diagnostics of active species in N-2 microwave flowing postdischarges. Surface Coatings Technol. 1995;74-5:474–478. [Google Scholar]
- 4.Bradbury JH. The structure and chemistry of keratin fibers. Adv Protein Chem. 1973;27:111–211. doi: 10.1016/s0065-3233(08)60447-7. [DOI] [PubMed] [Google Scholar]
- 5.Bradford MM. A rapid and sensitive method for the quntitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti SK, Matsumura N, Ranu RS. Purification and characterization of an extracellular alkaline serine protease from Aspergillus terreus (IJIRA 62) Curr Microbiol. 2000;40:239–244. doi: 10.1007/s002849910048. [DOI] [PubMed] [Google Scholar]
- 7.Cheng SW, Hu HM, Shen SW, Takagi H, Asano M, Tsai YC. Production and characterization of keratinase of a feather-degrading Bacillus licheniformis PWD-1. Biosci Biotechnol Biochem. 1995;59:2239–2243. doi: 10.1271/bbb.59.2239. [DOI] [PubMed] [Google Scholar]
- 8.Cohlberg JA. Textbook error-the structure of alpha-keratin. Trends Biochem Sci. 1993;18:360–362. doi: 10.1016/0968-0004(93)90087-4. [DOI] [PubMed] [Google Scholar]
- 9.Descamps F, Brouta F, Monod M, Zaugg C, Baar D, Losson B, Mignon B. Isolation of a Microsporum canis gene family encoding three subtilisin-like proteases expressed in vivo. J Invest Dermatol. 2002;119:830–835. doi: 10.1046/j.1523-1747.2002.01784.x. [DOI] [PubMed] [Google Scholar]
- 10.Do JH, Anderson MJ, Denning DW, Bornberg-Bauer E. Inference of Aspergillus fumigatus pathway by computational genome anaysis: tricarboxylic acid cycle (TCA) and glyoxylate shunt. J Microbiol Biotechnol. 2004;14:74–80. [Google Scholar]
- 11.Dozie INS, Okeke CN, Unaeze NC. A thermostable, alkaline-active, keratinolytic proteinase from Chrysosporium keratinophilum. World J Microbiol Biotechnol. 1994;10:563–567. doi: 10.1007/BF00367668. [DOI] [PubMed] [Google Scholar]
- 12.El-Naghy MA, El-Ktatny MS, Fadl-Allah EM, Nazeer WW. Degradation of chicken feathers by Chrysosporium georgiae. Mycopathologia. 1998;143:77–84. doi: 10.1023/a:1006953910743. [DOI] [PubMed] [Google Scholar]
- 13.Gradisar H, Friedrich J, Krizaj I, Jerala R. Similarities and specificities of fungal keratinolytic proteases: Comparison of keratinases of Paecilomyces marquandii and Doratomyces microsporus to some known proteases. Appl Environ Microbiol. 2005;71:3420–3426. doi: 10.1128/AEM.71.7.3420-3426.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Isogai T, Fukagawa M, Kojo H, Kohsaka M, Aoki H, Imanaka H. Cloning and nucleotide sequences of the complementary and genomic DNAs for the alkaline protease from Acremonium chrysogenum. Agric Biol Chem. 1991;55:471–477. [PubMed] [Google Scholar]
- 15.Jarai G, Buxton FP. Cloning and characterization of the pepD gene of Aspergillus niger which codes for a subtilisin-like protease. Gene. 1994;139:51–57. doi: 10.1016/0378-1119(94)90522-3. [DOI] [PubMed] [Google Scholar]
- 16.Jousson O, Lechenne B, Bontems O, Capoccia S, Mignon B, Barblan J, Quadroni M, Monod M. Multiplication of an ancestral gene encoding secreted fungalysin preceded species differentiation in the dermatophytes Trichophyton and Microsporum. Microbiology. 2004;150:301–310. doi: 10.1099/mic.0.26690-0. [DOI] [PubMed] [Google Scholar]
- 17.Kang SW, Hong SI, Kim SW. Identification of Aspergillus strain with antifungal activity against Phytophthora species. J Microbiol Biotechnol. 2005;15:227–233. [Google Scholar]
- 18.Katz ME, Rice RN, Cheetham BF. Isolation and characterization of an Aspergillus nidulans gene encoding an alkaline protease. Gene. 1994;150:287–292. doi: 10.1016/0378-1119(94)90439-1. [DOI] [PubMed] [Google Scholar]
- 19.Katz ME, Dougall AM, Weeks K, Cheetham BF. Extreme DNA sequence variation in isolates of Aspergillus fumigatus. FEMS Immunol Med Microbiol. 1998;20:283–288. doi: 10.1111/j.1574-695X.1998.tb01138.x. [DOI] [PubMed] [Google Scholar]
- 20.Kim JD. Keratinolytic activity of five Aspergillus species isolated from poultry farm soil in Korea. Mycobiology. 2003;31:157–161. [Google Scholar]
- 21.Kim JS, Kluskens LD, de Vos WM, Huber R, van der Oost J. Crystal structure of fervidolysin from Fervidobacterium pennivorans, a keratinolytic enzyme related to subtilisin. J Mol Biol. 2004;335:787–797. doi: 10.1016/j.jmb.2003.11.006. [DOI] [PubMed] [Google Scholar]
- 22.Kojima M, Kanai M, Tominaga M, Kitazume S, Inoue A, Horikoshi K. Isolation and characterization of a feather-degrading enzyme from Bacillus pseudofirmus FA30-01. Extremophiles. 2006;10:229–235. doi: 10.1007/s00792-005-0491-y. [DOI] [PubMed] [Google Scholar]
- 23.Kwak BY, Kwon BJ, Kweon CH, Shon DH. Detection of Aspergillus, Penicillium, and Fusarium species by sandwich enzyme-linked immunosorbent assay using mixed monoclonal antibodies. J Microbiol Biotechnol. 2004;14:385–389. [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Letourneau F, Soussotte V, Bressollier P, Branland P, Verneuil B. Keratinolytic activity of Streptomyces sp SK1-02: a new isolated strain. Lett Appl Microbiol. 1998;26:77–80. doi: 10.1046/j.1472-765x.1998.00281.x. [DOI] [PubMed] [Google Scholar]
- 26.Filipello Marchisio V, Cassinelli C, Tullio V, Piscozzi A. Outdoor airborne dermatophytes and related fungi-survey in Turin (Italy) Mycoses. 1992;35:251–257. doi: 10.1111/j.1439-0507.1992.tb00857.x. [DOI] [PubMed] [Google Scholar]
- 27.Markaryan A, Morozova I, Yu HS, Kolattukudy PE. Purification and characterization of an elastinolytic metalloprotease from Aspergillus fumigatus and immunoelectron microscopic evidence of secretion of this enzyme by the fungus invading the murine lung. Infect Immun. 1994;62:2149–2157. doi: 10.1128/iai.62.6.2149-2157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Melzer MS, Boland GJ. CHV3-type dsRNAs and the GH2 genotype in a population of Cryphonectria parasitica in Ontario. Can J Plant Pathol. 1999;21:248–255. [Google Scholar]
- 29.Merril C, Goldman D, Van Keuren M. Silver staining methods for polyacrylamide gel electrophoresis. In: Merril C, Goldman D, Van Keuren M, editors. Methods in Enzymology. Netherlands: Academic Press Inc.; 1983. pp. 230–239. [DOI] [PubMed] [Google Scholar]
- 30.Moallaei H, Zaini F, Larcher G, Beucher B, Bouchara JP. Partial purification and characterization of a 37 kDa extracellular proteinase from Trichophyton vanbreuseghemii. Mycopathologia. 2006;161:369–375. doi: 10.1007/s11046-006-0019-8. [DOI] [PubMed] [Google Scholar]
- 31.Monod M, Paris S, Sanglard D, Jatonogay K, Bille J, Latge JP. Isolation and characterization of a secreted metalloprotease of Aspergillus fumigatus. Infect Immun. 1993;61:4099–4104. doi: 10.1128/iai.61.10.4099-4104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Onifade AA, Abu OA. Productive response of rabbits to supplemental copper in a diet based on tropical feedstuffs. J Appl Anim Res. 1998;13:129–135. [Google Scholar]
- 33.Page WJ, Stock JJ. Phosphate-mediated alteration of the Microsporum gypseum germination protease specificity for substrate: enhanced keratinase activity. J Bacteriol. 1974;117:422–431. doi: 10.1128/jb.117.2.422-431.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papadopoulos MC, El-Boushy AR, Roodbeen AE, Ketelaara EH. Effects of processing time and moisure content on amino acid composition and nitrogen characteristics of feather meal. Anim Feed Sci Technol. 1986;14:279–290. [Google Scholar]
- 35.Pekkarinen AI, Jones BL, Niku-Paavola ML. Purification and properties of an alkaline proteinase of Fusarium culmorum. Eur J Biochem. 2002;269:798–807. doi: 10.1046/j.0014-2956.2001.02697.x. [DOI] [PubMed] [Google Scholar]
- 36.Pozo MJ, Baek JM, Garcia JM, Kenerley CM. Functional analysis of tvsp1, a serine protease-encoding gene in the biocontrol agent Trichoderma virens. Fungal Genet Biol. 2004;41:336–348. doi: 10.1016/j.fgb.2003.11.002. [DOI] [PubMed] [Google Scholar]
- 37.Santos RMDB, Firmino AAP, deSa CM, Felix CR. Keratinolytic activity of Aspergillus fumigatus Fresenius. Curr Microbiol. 1996;33:364–370. doi: 10.1007/s002849900129. [DOI] [PubMed] [Google Scholar]
- 38.Singh CJ. Characterization of an extracellular keratinase of Trichophyton simii and its role in keratin degradation. Mycopathologia. 1997;137:13–16. doi: 10.1023/a:1006844201399. [DOI] [PubMed] [Google Scholar]
- 39.Singh CJ. Exocellular proteases of Malbranchea gypsea and their role in keratin deterioration. Mycopathologia. 1998;143:147–150. doi: 10.1023/a:1006968600404. [DOI] [PubMed] [Google Scholar]
- 40.Steyaert JM, Stewart A, Ridgway HJ. Co-expression of two genes, a chitinase (chit42) and proteinase (prb1), implicated in mycoparasitism by Trichoderma hamatum. Mycologia. 2004;96:1245–1252. [PubMed] [Google Scholar]
- 41.Su NW, Lee MH. Purification and characterization of a novel salt-tolerant protease from Aspergillus sp FC-10, soy sauce koji mold. J Ind Microbiol Biotechnol. 2001;26:253–258. doi: 10.1038/sj.jim.7000129. [DOI] [PubMed] [Google Scholar]
- 42.Tatsumi H, Ogawa Y, Murakami S, Ishida Y, Murakami K, Masaki A, Kawabe H, Arimura H, Nakano E, Motai H. Isolation and characterization of the alkaline protease gene of Aspergillus oryzae. Agric Biol Chem. 1991;55:2807–2811. [PubMed] [Google Scholar]
- 43.Yu RJ, Harmon SR, Grappel SF, Blank F. Two cell-bound keratinases of Trichophyton mentagrophytes. J Invest Dermatol. 1971;56:27–32. doi: 10.1111/1523-1747.ep12291869. [DOI] [PubMed] [Google Scholar]