Abstract
Significance: Activation of immune responses in plants is associated with a parallel burst of both reactive oxygen intermediates (ROIs) and nitric oxide (NO). The mechanisms by which these small redox-active molecules are synthesized and their signaling functions are critical for plants to defend themselves against pathogen infection. Recent Advances: The synthesis of apoplastic ROIs by plants after pathogen recognition has long been attributed to membrane-bound NAPDH oxidases. However, the emerging data suggest a role for other enzymes in various subcellular locations in ROI production after defense activation. It is becoming widely appreciated that NO exerts its biochemical function through the S-nitrosylation of reactive cysteine thiols on target proteins, constituting a key post-translational modification. Recent evidence suggests that S-nitrosylation of specific defense-related proteins regulates their activity. Critical Issues: The source(s) of NO production after pathogen recognition remain(s) poorly understood. Some NO synthesis can be attributed to the activity of nitrate reductase but to date, no nitric oxide synthase (NOS) has been identified in higher plants. However, the signaling functions of S-nitrosylation are becoming more apparent and thus dissecting the molecular machinery underpinning this redox-based modification is vital to further our understanding of plant disease resistance. Future Directions: In addition to identifying new contributors to the oxidative burst, the discovery of an NOS in higher plants would significantly move the field forward. Since S-nitrosylation has now been confirmed to play various roles in immune signaling, this redox-based modification is a potential target to exploit for improving disease resistance in crop species. Antioxid. Redox Signal. 19, 990–997.
Introduction
In order to survive, plants must continuously resist attempted infection by a large variety of pathogens, but unlike animals they lack an adaptive immune system and do not possess dedicated immune cells. However, plants have evolved a plethora of strategies to prevent pathogen ingress, from simple physical barriers and preformed antimicrobial compounds to a battery of inducible defense mechanisms. Plant pathogens must first subvert the plant cell wall, a complex but rigid structure that after defense activation can be strengthened at sites of attempted infection by the deposition of the polysaccharide, callose. The plant immune system can be split into two branches (as reviewed in 19), with the first branch triggered at the plant cell membrane where transmembrane pattern recognition receptors recognize molecular signatures of pathogens known as microbial- or pathogen-associated molecular patterns (MAMPs or PAMPs). This results in a relatively weak immune response termed PAMP-triggered immunity (PTI). Some pathogens can then deliver effector molecules inside plant cells that suppress PTI; these can in some cases be recognized by plant resistance (R)-gene products triggering the second, more powerful response of the immune system, known as effector-triggered immunity. This second branch of immunity commonly uses NB-LRR proteins, named after their nucleotide binding (NB) and leucine-rich repeat (LRR) domains. These NB-LRRs are the largest class of proteins encoded by the R-genes of plants (11). Pathogen genes encoding effectors that are recognized by the plant R-gene products can then be termed avr-genes as their recognition results in their avirulence. After recognition of pathogens, plants can deploy a wide range of defense responses to protect themselves against pathogen invasion. To prevent pathogen growth, plants often trigger the hypersensitive response (HR), the programmed execution of plant cells at the site of attempted infection (10). The next phase of the immune response then transmits signals systemically throughout the plant, which protect distal tissues against potential secondary infections, a mechanism termed systemic acquired resistance (34).
Changes in cellular redox status are a prominent feature of immune responses in higher eukaryotes. In this context, a hallmark of plant defense activation is the rapid production of reactive oxygen intermediates (ROIs), known as the oxidative burst and is primarily characterized by the production of superoxide (O2−) and hydrogen peroxide (H2O2). In addition, nitric oxide (NO) levels are also dramatically increased during the HR, constituting a parallel nitrosative burst. ROIs and NO, either alone or in combination, have been proposed to function as key orchestrators of the different branches of plant immunity. Here, we review the most recent findings in both the synthesis and signaling of ROIs and NO, and their molecular roles in regulating plant immunity. Further, we discuss downstream signaling by NO focusing on the post-translational regulation of defense-related proteins.
Production of ROIs by NADPH Oxidases
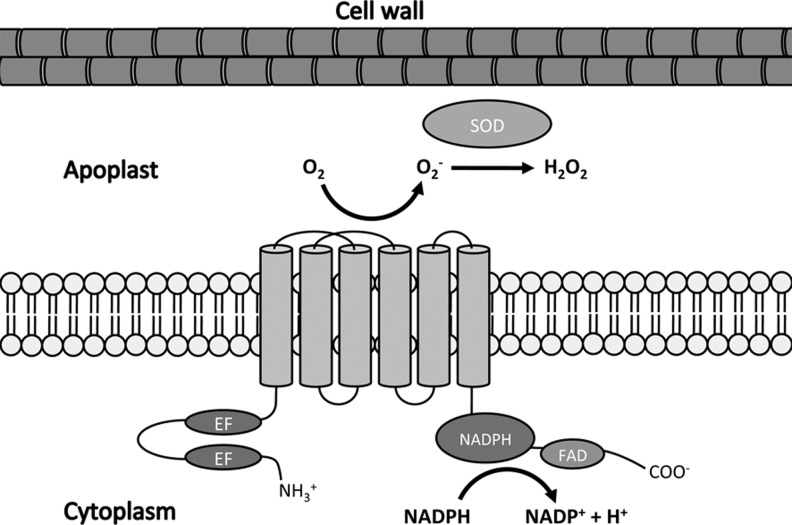
The production of apoplastic ROIs by plants after pathogen recognition has long been attributed to nicotinamide adenine dinucleotide phosphate (NADPH) oxidase enzymes related to those that perform the same role in mammalian phagocytes, termed respiratory burst oxidase homologues (RBOHs). The first report of a plant RBOH revealed that unlike its mammalian homologue gp91phox, it possesses additional potential regulatory EF-hand domains within an N-terminal extension (20). Like mammalian NADPH oxidases, plant RBOHs are transmembrane proteins that transfer electrons from intracellular NADPH across the plasma membrane before they are coupled to molecular oxygen in the apoplast to produce O2− and its subsequent dismutation product, H2O2 (Fig. 1). This dismutation reaction can ensue spontaneously but can be enhanced by the activity of superoxide dismutase. NADPH and the cofactor flavin adenine dinucleotide (FAD) both bind at regions within the C-terminal. The Arabidopsis genome contains ten predicted RBOH genes (RBOHA-RBOHJ) (42) and through genetic studies using loss-of-function mutants, RBOHD and RBOHF were shown to be the principle generators of ROIs after recognition of PAMPs and avirulent pathogens (43).
FIG. 1.
Schematic structure of a plant respiratory burst oxidase homologues (RBOH) protein. Plant RBOHs are located on the plasma membrane and synthesize reactive oxygen intermediates (ROIs) in the apoplast. Nicotinamide adenine dinucleotide phosphate (NADPH)- and flavin adenine dinucleotide (FAD)-binding domains are located at the C-terminal while the EF-hand domains are located near the N-terminal.
Since plants possess so many potential RBOH genes, this begs the question of whether these are functionally redundant or if there is in fact specificity for each RBOH to different cellular cues or requirements? The answer to this question remains largely enigmatic although recent work has begun to unravel these complex signaling networks. There is indeed evidence that RBOH proteins at least in part share redundancy as some phenotypes of atrbohD and atrbohF mutant plants, including reduced HR, are enhanced in the atrbohD atrbohF double mutant (43). Further study of these mutants surprisingly revealed that both RBOHD and RBOHF may actually suppress the spread of cell death during the HR in a lesion mimic mutant (lsd1) that exhibits “runaway” cell death in infected leaves (44). Most work on these enzymes has focused on their role in generation of ROIs after pathogen recognition and their effect on cell death during the HR. However, a recent study (7) investigated the effect of atrbohD and atrbohF mutations on defense-associated metabolic responses. The authors reported that both mutant lines showed similar resistance to Pseudomonas syringae pv. tomato DC3000 carrying the avirulence gene avrRpm1 but only atrbohF plants were more susceptible to the virulent form of the same pathogen. The Arabidopsis catalase 2 (cat2) mutant has increased intracellular H2O2 levels, which causes constitutive HR-like cell death and defense signaling when grown under long days in air (8), and these plants were used as a tool to uncover the effects of atrboh mutations on intracellular oxidative stress. This work revealed that RBOHF, but not RBOHD, was required to maintain various downstream responses emanating from high intracellular ROI levels in the cat2 mutant (7). These responses included the accumulation of salicylic acid (SA), an important signaling molecule essential for the establishment of disease resistance. Thus, although RBOHD and RBOHF have overlapping functions, there appears to be specificity in some instances. The complexity of these signaling networks makes it likely that the same set of proteins can induce different responses depending on the context of their requirement and multiple layers of regulation must exist for plants to fine-tune these both temporally and spatially.
Alternative Sources of ROIs
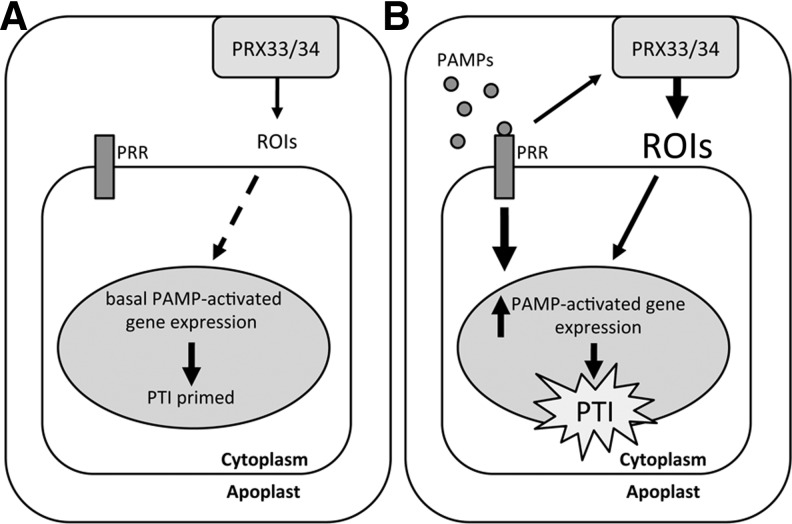
ROI synthesis by plants after pathogen recognition has also been attributed to a number of alternative mechanisms, including the activity of peroxidase enzymes. Transgenic Arabidopsis plants expressing an antisense cDNA against a French bean class III peroxidase were compromised in resistance against fungal and bacterial pathogens and showed a decreased oxidative burst, which was attributed to the silencing of two Arabidopsis peroxidase genes, PRX33 and PRX34 (4). A more recent study from the same laboratory (12) demonstrated that PRX33 and PRX34 knockdown Arabidopsis plants exhibit a reduced oxidative burst in response to PAMPs, including Flg22 and Elf26, synthetic peptides based on bacterial flagellin and elongation factor Tu, both well-established PAMPs recognized by plants. Further, these knockdown lines were impaired in the induction of several PAMP-activated genes after Flg22 and Elf26 treatment. Interestingly, these genes were also downregulated before PAMP treatment in the PRX knockdown lines compared to wild-type plants, suggesting that basal ROI production by PRX33 and/or PRX34 may constitutively prime plant defenses for attempted pathogen attack (12) (Fig. 2). Further evidence for peroxidases as a source of pathogen-induced ROIs emerged from a recent study in the moss Physcomitrella patens, where it was shown that Prx34 not only catalyzes ROI production but is also required for antifungal resistance (23). It must be noted that this peroxidase is unrelated to the previously mentioned Arabidopsis PRX33/34 (24) and the same gene nomenclature is purely coincidental.
FIG. 2.
A model for the role of PRX33/34 in PAMP-triggered immunity (PTI) signaling. (A) In unchallenged cells, PRX33/34 activity and presumably low-level apoplastic ROIs appear to maintain low-level expression of pathogen-associated molecular pattern (PAMP)-activated genes through an unknown mechanism. This possibly serves to pre-prime plant defenses for rapid activation after PAMP recognition. (B) Pathogen infection and PAMP recognition by pattern recognition receptors (PRRs) both activate intracellular signaling leading to PTI and also trigger the apoplastic production of ROIs by PRX33/34. This appears to positively regulate the expression of PAMP-activated genes and contributes to the establishment of PTI.
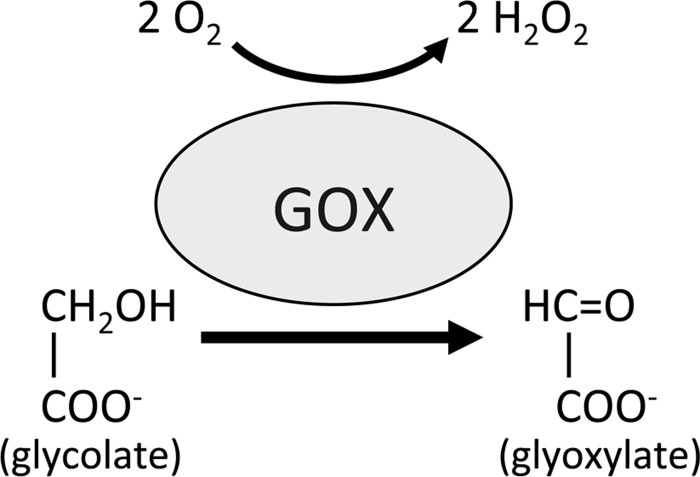
Another potential source of ROIs in plants is through the activity of glycolate oxidases (GOX). These enzymes are located in peroxisomes and catalyze the conversion of glycolate into glyoxylate during photorespiration, producing H2O2 in the process (Fig. 3). GOX has been proposed to be involved in disease resistance by various studies (5, 35, 41) although until recently its role has remained unclear. A recent study identified GOX in a screen for genes involved in non-host resistance (NHR) signaling in Nicotiana benthamiana (31). This work revealed that both GOX-silenced N. benthamiana and Arabidopsis gox mutant plants exhibit decreased NHR and gene-for-gene resistance. Further, these plants accumulated less H2O2 independent of NADPH oxidase activity and silencing RBOHD in gox plants did not affect their susceptibility to non-host pathogens. Thus, this study suggests that GOX is an important alternative source of ROIs during both NHR- and R-gene-mediated defense responses.
FIG. 3.
Hydrogen peroxide (H2O2) production by glycolate oxidase (GOX). Peroxisome-located GOX enzymes are a key component of photorespiration, catalyzing the conversion of glycolate to glyoxylate. This reaction consumes O2 and produces H2O2, which may have an important role in plant immunity.
Production of NO
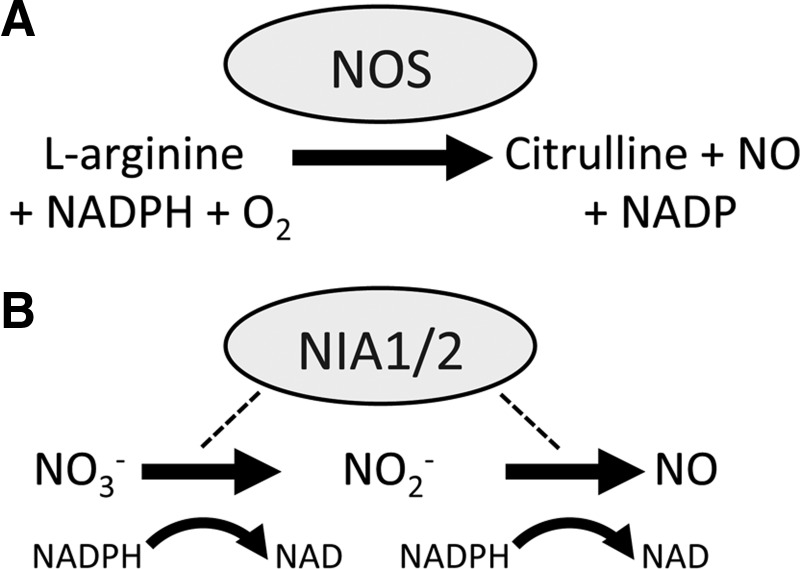
Despite accumulating evidence implicating a central role for NO in plant growth, development, and environmental responses, a nitric oxide synthase (NOS), structurally similar to those found in animals, has not been uncovered in higher plants. These NADP(H)-dependent enzymes catalyze the formation of NO and citrulline from l-arginine (Fig. 4A). It has been long understood that mammals possess three isoforms of NOS; neuronal-, endothelial-, (eNOS) and inducible-, the latter of which has been implicated in immune responses. A NOS similar in sequence and structure to human eNOS was recently identified in the single cell algae Ostreococcus tauri (16). The authors produced and purified recombinant protein from this predicted sequence and this was shown to exhibit catalytic kinetics similar to other NOS enzymes in vitro. Moreover, Escherichia coli cells expressing O. tauri NOS showed increased levels of NO and cell viability, demonstrating the NO-producing activity of this enzyme.
FIG. 4.
Possible routes of nitric oxide (NO) production in plants. (A) NO synthase (NOS) enzymes catalyze the formation of NO from l-arginine. (B) nitrate reductases catalyze the conversion of nitrate to nitrite (NO2−) but can also catalyze the formation of NO from NO2−.
While the hunt for an NOS in higher plants continues, other mechanisms for NO synthesis after pathogen recognition have been explored. A possible source of NO is through the action of cytosolic nitrate reductase (NR). The Arabidopsis genome encodes two of these genes, named NIA1 and NIA2, with NIA2 being responsible for the bulk of NR activity (47). These enzymes are involved in reducing nitrate, the main nitrogen source of plants, to nitrite (NO2−) in a NADPH-dependent fashion (Fig. 4B). However, NR appears to also be capable of producing NO in vitro and in vivo and this was unaffected by NOS inhibitors (30). The in vivo data from this study did rely on artificial chemical stimulation of NR and so may not be physiologically relevant. Further, the production of NO by NR is extremely inefficient and depends upon low O2 levels and high concentrations of NO2− (30). In the context of plant immunity, Arabidopsis nia1 and nia2 mutant plants, and wild-type plants treated with NR inhibitors might be impaired in NO production after recognition of toxins from the fungal pathogen Verticillium dahliae (36).
S-Nitrosoglutathione-Reductase and S-Nitrosylation
It is well established that NO levels increase in plants after pathogen infection but how does this small molecule convey its biochemical activity in plant cells? An emerging redox-based post-translational modification is S-nitrosylation, by which an NO moiety is coupled to a reactive cysteine thiol, forming an S-nitrosothiol (SNO). This form of protein modification was discovered in 1992 (38) and is analogous to the addition of a phosphate group during protein phosphorylation (45). In addition to being implicated in many cellular processes in animals (18), recent evidence has shown that S-nitrosylation is involved in the regulation of various important plant processes including plant defense-related gene expression and hypersensitive cell death (45).
Glutathione (GSH) is an antioxidant tripeptide that can be S-nitrosylated to form S-nitrosoglutathione (GSNO), a molecule thought to function as a reservoir of NO bioactivity. NO can be released from GSNO or can be directly transferred to a target by a process known as transnitrosylation (18). The enzyme responsible for controlling GSNO levels is known as GSNO-reductase (GSNOR) and reduces GSNO to oxidized GSH and NH3. While this enzyme is highly specific for GSNO, it indirectly controls the levels of protein SNOs, by turning over this natural NO donor (27). In Arabidopsis, loss of GSNOR function in the mutant line gsnor1-3 increases SNO levels and compromises R gene-mediated defense and NHR, while a gain-of-function mutant line gsnor1-1 showed enhanced resistance to virulent pathogens (15). This study provided evidence that SNO metabolism controlled by GSNOR had a key role in plant disease resistance. Further studies have since revealed that GSNOR appears to be a key regulator of cell death (9) and is also required for thermotolerance (22). Various studies discussed in the following section have also shown that S-nitrosylation of specific proteins is increased in gsnor1-3 mutant plants.
S-Nitrosylation of Plant Defense-Related Proteins
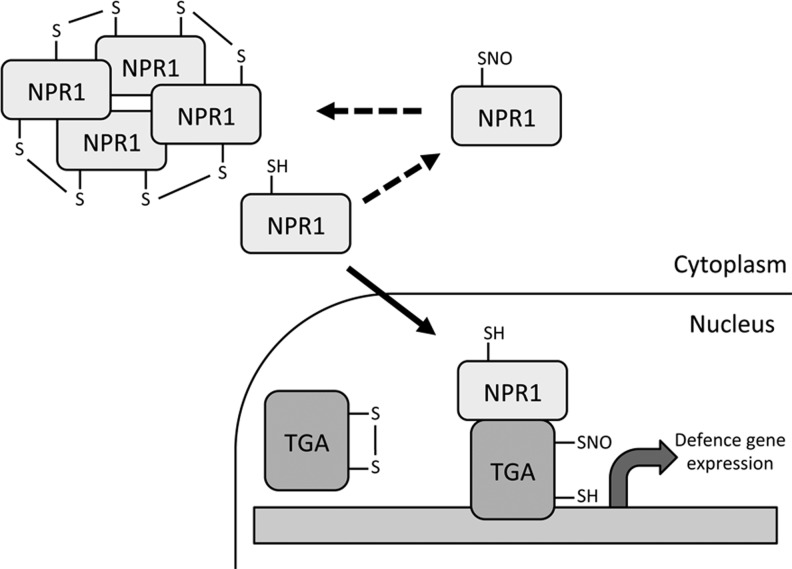
The defense-related transcription co-activator nonexpressor of pathogenesis-related genes 1 (NPR1) possesses many cysteine residues that form intermolecular disulfide bonds forming a high molecular-weight oligomer that is localized in the cytoplasm. After pathogen infection, these disulfide bonds are reduced allowing monomeric NPR1 to translocate to the nucleus where it exerts its co-activator function on many target genes (29). The complexity of this process was further revealed in a subsequent study where the authors showed that S-nitrosylation of NPR1 promoted its oligomerization and thus blunted its transcriptional activity (40). S-nitrosylation of NPR1 was increased in the gsnor1-3 mutant thus providing a link between high global SNO levels and the compromised disease resistance of this line. Cellular redox status not only affects NPR1 but also its binding partners TGA1 and TGA4. These transcription factors have been shown to contain intramolecular disulfide bridges at key cysteine residues that prevent NPR1 interaction (13). The authors of this study showed that reduction of these disulfide bonds allows NPR1-TGA1/4 interaction and NPR1-stimulated DNA binding of these transcription factors. A recent study revealed that these same cysteine residues can be S-nitrosylated and this may block further oxidative modification of TGA1/4, enhancing their DNA-binding capacity (26). There is now a wealth of evidence suggesting the NPR1-TGA transcriptional regulation system is under complex redox control with S-nitrosylation appearing to play a key role (37) (Fig. 5).
FIG. 5.
Redox-regulation of nonexpressor of pathogenesis-related genes 1 (NPR1) and TGA transcription factors. NPR1 exists as an oligomer in the cytoplasm, facilitated by intermolecular disulfide bonds. Upon pathogen attack these disulfides are reduced and monomeric NPR1 translocates to the nucleus where it interacts with TGA transcription factors and exerts its co-activator function on defense-related genes. S-nitrosylation of NPR1 promotes its oligomerization and thus inhibits its nuclear translocation. Formation of intramolecular disulfide bonds within TGAs prevent NPR1 binding, however, S-nitrosylation of TGAs may prevent these disulfides from forming, leading to increased defense gene expression.
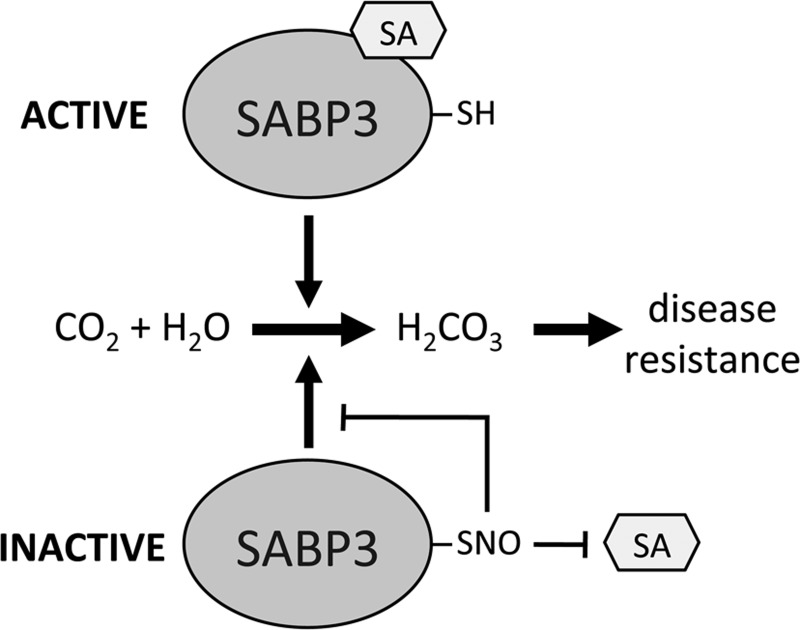
SA-binding protein 3 (SABP3) is a positive regulator of plant immunity that was identified as a target for S-nitrosylation (46). Similar to NPR1, S-nitrosylation of SABP3 is increased in the gsnor1-3 line. SABP3 possesses carbonic anhydrase activity that is required for expression of resistance in plants and this was shown to be inhibited by S-nitrosylation (Fig. 6). This raises a dilemma since a nitrosative burst and subsequent S-nitrosylation of proteins is associated with activation of plant defense responses, yet, S-nitrosylation of SABP3 inhibits its defensive activity. Interestingly, this could represent a negative feedback loop in plant defense signaling to regulate SA-mediated responses or could be a strategy employed by pathogens to suppress host immunity.
FIG. 6.
S-nitrosylation of salicylic acid-binding protein 3 (SABP3) inhibits its carbonic anydrase activity. SABP3 binds SA and possesses carbonic anydrase activity that is required for disease resistance. S-nitrosylation of SABP3 inhibits both SA binding and its carbonic anydrase activity.
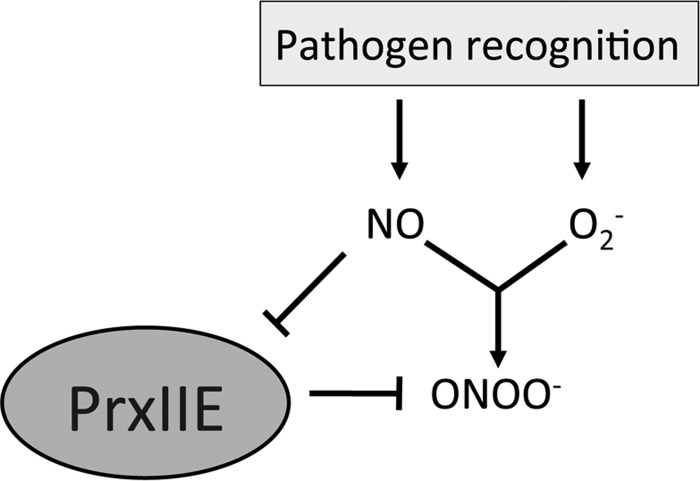
Peroxynitrite (ONOO−) is a reactive toxic agent formed by NO reacting with O2−. Peroxiredoxin II E (PrxIIE) is responsible for ONOO− detoxification in plants and S-nitrosylation of this enzyme was shown to inhibit this activity (33) (Fig. 7). ONOO− has been shown to induce programmed cell death (PCD) in animals (6) but not in plants, even in a PrxIIE mutant line (33). The S-nitrosylation of PrxIIE may serve as a regulatory mechanism that NO employs to control the levels of ONOO−, but whether this mechanism is implicated in PCD in plants remains an unanswered question. Caspases are animal proteins involved in triggering PCD in humans, and caspase-3 has been shown to be S-nitrosylated at its active-site cysteine in unstimulated human cell lines (28). This S-nitrosylation suppressed the cysteine protease activity of caspase-3, which is required for triggering PCD. Upon induction of the apoptotic pathway, SNO-caspase-3 levels decreased and this was associated with an increase in intracellular caspase activity and subsequent increased PCD. The Arabidopsis relative of caspase-3, metacaspase 9, has also been shown to be S-nitrosylated and similarly this suppresses its proteolytic activity (1); however, the biological relevance of this is not yet as well understood as in animals.
FIG. 7.
Peroxynitrite (ONOO−) detoxification by Peroxiredoxin II E (PrxIIE) is regulated by S-nitrosylation. NO and O2− can rapidly react to form toxic ONOO−, which is detoxified by PrxIIE. Pathogen infection triggers the parallel burst of NO and O2− and thus presumably increases the amount of cellular ONOO−. Further, increasing amounts of NO may promote S-nitrosylation of PrxIIE, which inhibits its activity leading to increased ONOO− levels.
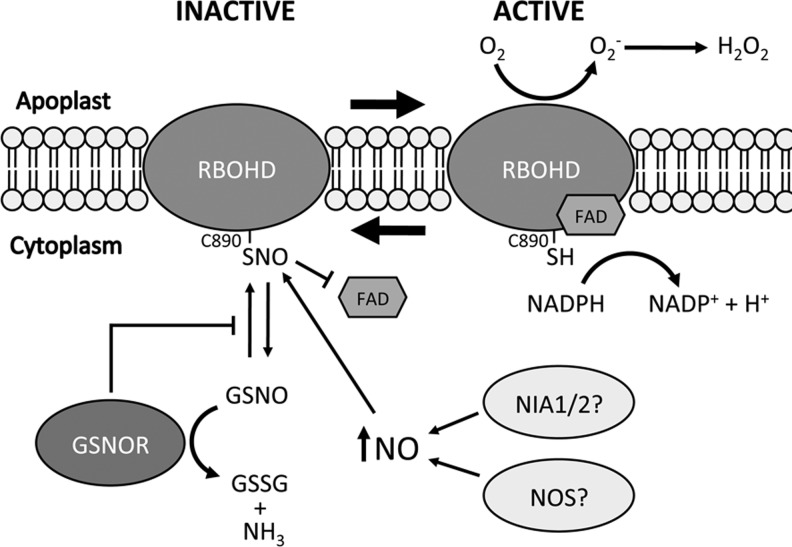
Recent work has identified Arabidopsis RBOHD as a target for S-nitrosylation during the plant immune response (48). The authors found that the production of ROIs after pathogen challenge was reduced in both gsnor1-3 and NO overproducing 1 mutant plants. Since both of these lines have elevated SNO levels it was hypothesized that S-nitrosylation of RBOH proteins could be regulating their ability to produce ROIs. Indeed, RBOHD was shown to be S-nitrosylated at Cys890 both in vitro and in vivo and this modification inhibited its NADPH oxidase activity. Computational modeling and experimental evidence showed that S-nitrosylation of Cys890 inhibited binding of the cofactor FAD, and this effect was abolished in a C890A mutant protein. Transgenic plants expressing a C890A mutant RBOHD showed greater pathogen-induced ROI accumulation and increased cell death during the HR. These findings suggest an interesting negative feedback loop by which the increasing NO levels during the HR promote S-nitrosylation of RBOHD at Cys890, attenuating its NADPH oxidase activity to prevent excessive ROI-induced cell death (Fig. 8). Importantly this cysteine reside is conserved from plants to humans and this study also showed that recombinant human and Drosophila melanogaster NADPH oxidases can be S-nitrosylated at this site in vitro, suggesting that this mechanism of inhibiting cell death may also operate in other complex eukaryotes.
FIG. 8.
S-nitrosylation of RBOHD inhibits FAD binding and its NADPH oxidase activity. S-nitrosylation of RBOHD at Cys890 blunts its NADPH oxidase activity by inhibiting the binding of the cofactor FAD. This may serve as a mechanism to attenuate cell death during immune responses through negative feedback signaling by increasing levels of NO. In gsnor1-3 mutant plants, RBOHD activity is reduced presumably due to its increased S-nitrosylation.
Identification of SNO-Proteins in Plants
The highly labile nature of the S–NO bond makes it a technically challenging post-translational modification to detect. Most work to date has relied on a technique known as the biotin-switch (17), where free cysteines are first blocked before SNOs are specifically reduced and “switched” with a biotin label to allow detection with antibodies against biotin or purification with streptavidin. Recent years have seen the emergence of proteome-wide screens for S-nitrosylated proteins in Arabidopsis (25), however, some of these have involved the application of an exogenous NO-donor and thus may not reflect the relevant in vivo catalogue of SNOs. Only one report to date exists describing the SNO-proteome of plants engaged in defense signaling (32) and this only uncovered 16 proteins involved in metabolism, antioxidant defense, and signaling. The fact that other proteins not identified in this study have been shown to be S-nitrosylated during defense signaling suggests that many other targets of this important modification are yet to be uncovered. A more recent study investigated the endogenous SNO-proteome under resting conditions and after salt stress (14) with only 53 proteins being identified as S-nitrosylation targets. Again, many proteins described in the previous section as being regulated by S-nitrosylation were not picked up in this screen although this could be expected as the plants in this study had not been challenged with a pathogen. Thus, a more complete assembly of the SNO-proteome after pathogen recognition and the subsequent nitrosative burst is desirable to identify more defense-related targets regulated by NO.
Denitrosylation by Thioredoxin Proteins in Plant Defense?
Although the cellular redox status and GSNOR activity govern global SNO levels, emerging data has revealed that NO can be specifically removed from proteins by enzymes with denitrosylating activity. Thioredoxin (TRX) proteins have been shown to reduce disulfide bonds and are of particular interest. Arabidopsis TRX-h5 (TRX5) has been shown to facilitate the reduction and subsequent monomerization of NPR1 oligomers upon SA-mediated plant defense activation leading to upregulation of defense genes (40). TRX5 has been implicated in plant immunity in response to victorin, produced by Victoria blight (39), and it is upregulated in response to avirulent bacterial pathogens (21). TRX proteins have recently emerged as enzymes with denitrosylating activity in human cells (2) and together with thioredoxin reductase (TRX-reductase) can remove SNO groups from target proteins. Interestingly, some proteins appear to be basally S-nitrosylated with stimulus-induced reduction/denitrosylation by TRX/TRX-reductase providing a cellular signal, while others are constitutively kept in their reduced/denitrosylated form by basal activity of TRX/TRX-reductase. A recent animal proteomic study revealed a list of SNO-proteins with diverse functions to be substrates for TRX/TRX-reductase (3) thus further enhancing the notion that denitrosylation of proteins by TRX may have important roles in cellular signaling (37).
Conclusions and Future Perspectives
A huge effort has been aimed at understanding plant NADPH oxidases and their role in producing the oxidative burst following pathogen recognition. However, the emerging data reveal that this class of enzyme may not provide the only source of ROIs, with new evidence implicating GOX and peroxidases in ROI synthesis after immune activation. It is likely that further contributors to the oxidative burst will be identified in the coming years. In addition to new players in the oxidative burst, the discovery of an NOS in higher plants would significantly move the field forward, although the current data suggest it may be of a structure different from the well-established mammalian NOSs. It is now appreciated that NO exerts its biological effect through the S-nitrosylation of reactive cysteine thiols in proteins and thus it is important to identify new targets of this key post-translational modification. Moreover, recent findings have confirmed that S-nitrosylation is an important regulatory mechanism in immune signaling. Thus, this redox-based modification is a potential target to exploit in breeding or plant design approaches to generate improved disease-resistant crops. Further, as dysregulation of S-nitrosylation is found to underpin the development of an increasing number of human diseases, the molecular machinery associated with this redox modification could represent an attractive drug target for future therapeutics.
Abbreviations Used
- avr
avirulence
- cat2
catalase 2
- eNOS
endothelial nitric oxide synthase
- FAD
flavin adenine dinucleotide
- GOX
glycolate oxidase
- GSH
glutathione
- GSNO
S-nitrosoglutathione
- GSNOR
GSNO-reductase
- H2O2
hydrogen peroxide
- HR
hypersensitive response
- lsd1
lesion simulating disease
- MAMP
microbial-associated molecular pattern
- NADPH
nicotinamide adenine dinucleotide phosphate
- NB-LRR
nucleotide-binding leucine-rich repeat
- NHR
non-host resistance
- NO
nitric oxide
- NO2−
nitrite
- NO3−
nitrate
- NOS
nitric oxide synthase
- NPR1
nonexpressor of pathogenesis-related genes 1
- NR
nitrate reductase
- O2−
superoxide
- ONOO−
peroxynitrite
- PAMP
pathogen-associated molecular pattern
- PCD
programmed cell death
- PRR
pattern recognition receptor
- PrxIIE
Peroxiredoxin II E
- PTI
PAMP-triggered immunity
- R
resistance
- RBOH
respiratory burst oxidase homologue
- ROIs
reactive oxygen intermediates
- SA
salicylic acid
- SNO
S-nitrosothiol
- SOD
superoxide dismutase
- TRX
thioredoxin
Acknowledgments
M.J.S. was the recipient of a Biotechnology and Biological Sciences Research Council (BBSRC) studentship, and work in the lab of G.J.L. is supported by BBSRC grant BB/D0118091/1.
References
- 1.Belenghi B. Romero-Puertas MC. Vercammen D. Brackenier A. Inzé D. Delledonne M. Van Breusegem F. Metacaspase activity of Arabidopsis thaliana is regulated by S-nitrosylation of a critical cysteine residue. J Biol Chem. 2007;282:1352–1358. doi: 10.1074/jbc.M608931200. [DOI] [PubMed] [Google Scholar]
- 2.Benhar M. Forrester MT. Hess DT. Stamler JS. Regulated protein denitrosylation by cytosolic and mitochondrial thioredoxins. Science. 2008;320:1050–1054. doi: 10.1126/science.1158265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benhar M. Thompson JW. Moseley MA. Stamler JS. Identification of S-nitrosylated targets of thioredoxin using a quantitative proteomic approach. Biochemistry. 2010;49:6963–6969. doi: 10.1021/bi100619k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bindschedler LV. Dewdney J. Blee KA. Stone JM. Asai T. Plotnikov J. Denoux C. Hayes T. Gerrish C. Davies DR. Ausubel FM. Bolwell GP. Peroxidase-dependent apoplastic oxidative burst in Arabidopsis required for pathogen resistance. Plant J. 2006;47:851–863. doi: 10.1111/j.1365-313X.2006.02837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bohman S. Wang M. Dixelius C. Arabidopsis thaliana-derived resistance against Leptosphaeria maculans in a Brassica napus genomic background. Theor Appl Genet. 2002;105:498–504. doi: 10.1007/s00122-002-0885-5. [DOI] [PubMed] [Google Scholar]
- 6.Bonfoco E. Krainc D. Ankarcrona M. Nicotera P. Lipton SA. Apoptosis and necrosis: two distinct events induced, respectively, by mild and intense insults with N-methyl-D-aspartate or nitric oxide/superoxide in cortical cell cultures. Proc Natl Acad Sci U S A. 1995;92:7162–7166. doi: 10.1073/pnas.92.16.7162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaouch S. Queval G. Noctor G. AtRbohF is a crucial modulator of defence-associated metabolism and a key actor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 2012;69:613–627. doi: 10.1111/j.1365-313X.2011.04816.x. [DOI] [PubMed] [Google Scholar]
- 8.Chaouch S. Queval G. Vanderauwera S. Mhamdi A. Vandorpe M. Langlois-Meurinne M. Van Breusegem F. Saindrenan P. Noctor G. Peroxisomal hydrogen peroxide is coupled to biotic defense responses by ISOCHORISMATE SYNTHASE1 in a daylength-related manner. Plant Physiol. 2010;153:1692–1705. doi: 10.1104/pp.110.153957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen R. Sun S. Wang C. Li Y. Liang Y. An F. Li C. Dong H. Yang X. Zhang J. Zuo J. The Arabidopsis PARAQUAT RESISTANT2 gene encodes an S-nitrosoglutathione reductase that is a key regulator of cell death. Cell Res. 2009;19:1377–1387. doi: 10.1038/cr.2009.117. [DOI] [PubMed] [Google Scholar]
- 10.Dangl JL. Dietrich RA. Richberg MH. Death don't have no mercy: cell death programs in plant-microbe interactions. Plant Cell. 1996;8:1793–1807. doi: 10.1105/tpc.8.10.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dangl JL. Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 12.Daudi A. Cheng Z. O'Brien JA. Mammarella N. Khan S. Ausubel FM. Bolwell GP. The apoplastic oxidative burst peroxidase in Arabidopsis is a major component of pattern-triggered immunity. Plant Cell. 2012;24:275–287. doi: 10.1105/tpc.111.093039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Després C. Chubak C. Rochon A. Clark R. Bethune T. Desveaux D. Fobert PR. The Arabidopsis NPR1 disease resistance protein is a novel cofactor that confers redox regulation of DNA binding activity to the basic domain/leucine zipper transcription factor TGA1. Plant Cell. 2003;15:2181–2191. doi: 10.1105/tpc.012849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fares A. Rossignol M. Peltier JB. Proteomics investigation of endogenous S-nitrosylation in Arabidopsis. Biochem Biophys Res Commun. 2011;416:331–336. doi: 10.1016/j.bbrc.2011.11.036. [DOI] [PubMed] [Google Scholar]
- 15.Feechan A. Kwon E. Yun BW. Wang Y. Pallas JA. Loake GJ. A central role for S-nitrosothiols in plant disease resistance. Proc Natl Acad Sci U S A. 2005;102:8054–8059. doi: 10.1073/pnas.0501456102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Foresi N. Correa-Aragunde N. Parisi G. Calo G. Salerno G. Lamattina L. Characterization of a nitric oxide synthase from the plant kingdom: NO generation from the green alga Ostreococcus tauri is light irradiance and growth phase dependent. Plant Cell. 2010;22:3816–3830. doi: 10.1105/tpc.109.073510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forrester MT. Foster MW. Benhar M. Stamler JS. Detection of protein S-nitrosylation with the biotin-switch technique. Free Radic Biol Med. 2009;46:119–126. doi: 10.1016/j.freeradbiomed.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hess DT. Matsumoto A. Kim SO. Marshall HE. Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–166. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 19.Jones JD. Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 20.Keller T. Damude HG. Werner D. Doerner P. Dixon RA. Lamb C. A plant homolog of the neutrophil NADPH oxidase gp91phox subunit gene encodes a plasma membrane protein with Ca2+ binding motifs. Plant Cell. 1998;10:255–266. doi: 10.1105/tpc.10.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Laloi C. Mestres-Ortega D. Marco Y. Meyer Y. Reichheld JP. The Arabidopsis cytosolic thioredoxin h5 gene induction by oxidative stress and its W-box-mediated response to pathogen elicitor. Plant Physiol. 2004;134:1006–1016. doi: 10.1104/pp.103.035782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee U. Wie C. Fernandez BO. Feelisch M. Vierling E. Modulation of nitrosative stress by S-nitrosoglutathione reductase is critical for thermotolerance and plant growth in Arabidopsis. Plant Cell. 2008;20:786–802. doi: 10.1105/tpc.107.052647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lehtonen MT. Akita M. Frank W. Reski R. Valkonen JP. Involvement of a class III peroxidase and the mitochondrial protein TSPO in oxidative burst upon treatment of moss plants with a fungal elicitor. Mol Plant Microbe Interact. 2012;25:363–371. doi: 10.1094/MPMI-10-11-0265. [DOI] [PubMed] [Google Scholar]
- 24.Lehtonen MT. Akita M. Kalkkinen N. Ahola-Iivarinen E. Rönnholm G. Somervuo P. Thelander M. Valkonen JP. Quickly-released peroxidase of moss in defense against fungal invaders. New Phytol. 2009;183:432–443. doi: 10.1111/j.1469-8137.2009.02864.x. [DOI] [PubMed] [Google Scholar]
- 25.Lindermayr C. Saalbach G. Durner J. Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 2005;137:921–930. doi: 10.1104/pp.104.058719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lindermayr C. Sell S. Müller B. Leister D. Durner J. Redox regulation of the NPR1-TGA1 system of Arabidopsis thaliana by nitric oxide. Plant Cell. 2010;22:2894–2907. doi: 10.1105/tpc.109.066464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu L. Hausladen A. Zeng M. Que L. Heitman J. Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature. 2001;410:490–494. doi: 10.1038/35068596. [DOI] [PubMed] [Google Scholar]
- 28.Mannick JB. Hausladen A. Liu L. Hess DT. Zeng M. Miao QX. Kane LS. Gow AJ. Stamler JS. Fas-induced caspase denitrosylation. Science. 1999;284:651–654. doi: 10.1126/science.284.5414.651. [DOI] [PubMed] [Google Scholar]
- 29.Mou Z. Fan W. Dong X. Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell. 2003;113:935–944. doi: 10.1016/s0092-8674(03)00429-x. [DOI] [PubMed] [Google Scholar]
- 30.Rockel P. Strube F. Rockel A. Wildt J. Kaiser WM. Regulation of nitric oxide (NO) production by plant nitrate reductase in vivo and in vitro. J Exp Bot. 2002;53:103–110. [PubMed] [Google Scholar]
- 31.Rojas CM. Senthil-Kumar M. Wang K. Ryu CM. Kaundal A. Mysore KS. Glycolate oxidase modulates reactive oxygen species-mediated signal transduction during nonhost resistance in nicotiana benthamiana and Arabidopsis. Plant Cell. 2012;24:336–352. doi: 10.1105/tpc.111.093245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Romero-Puertas MC. Campostrini N. Mattè A. Righetti PG. Perazzolli M. Zolla L. Roepstorff P. Delledonne M. Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics. 2008;8:1459–1469. doi: 10.1002/pmic.200700536. [DOI] [PubMed] [Google Scholar]
- 33.Romero-Puertas MC. Laxa M. Mattè A. Zaninotto F. Finkemeier I. Jones AM. Perazzolli M. Vandelle E. Dietz KJ. Delledonne M. S-nitrosylation of peroxiredoxin II E promotes peroxynitrite-mediated tyrosine nitration. Plant Cell. 2007;19:4120–4130. doi: 10.1105/tpc.107.055061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ross AF. Systemic acquired resistance induced by localized virus infections in plants. Virology. 1961;14:340–358. doi: 10.1016/0042-6822(61)90319-1. [DOI] [PubMed] [Google Scholar]
- 35.Schäfer P. Hückelhoven R. Kogel KH. The white barley mutant albostrians shows a supersusceptible but symptomless interaction phenotype with the hemibiotrophic fungus Bipolaris sorokiniana. Mol Plant Microbe Interact. 2004;17:366–373. doi: 10.1094/MPMI.2004.17.4.366. [DOI] [PubMed] [Google Scholar]
- 36.Shi FM. Li YZ. Verticillium dahliae toxins-induced nitric oxide production in Arabidopsis is major dependent on nitrate reductase. BMB Rep. 2008;41:79–85. doi: 10.5483/bmbrep.2008.41.1.079. [DOI] [PubMed] [Google Scholar]
- 37.Spoel SH. Loake GJ. Redox-based protein modifications: the missing link in plant immune signalling. Curr Opin Plant Biol. 2011;14:358–364. doi: 10.1016/j.pbi.2011.03.007. [DOI] [PubMed] [Google Scholar]
- 38.Stamler JS. Simon DI. Osborne JA. Mullins ME. Jaraki O. Michel T. Singel DJ. Loscalzo J. S-nitrosylation of proteins with nitric oxide: synthesis and characterization of biologically active compounds. Proc Natl Acad Sci U S A. 1992;89:444–448. doi: 10.1073/pnas.89.1.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sweat TA. Wolpert TJ. Thioredoxin h5 is required for victorin sensitivity mediated by a CC-NBS-LRR gene in Arabidopsis. Plant Cell. 2007;19:673–687. doi: 10.1105/tpc.106.047563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tada Y. Spoel SH. Pajerowska-Mukhtar K. Mou Z. Song J. Wang C. Zuo J. Dong X. Plant immunity requires conformational changes of NPR1 via S-nitrosylation and thioredoxins. Science. 2008;321:952–956. doi: 10.1126/science.1156970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taler D. Galperin M. Benjamin I. Cohen Y. Kenigsbuch D. Plant eR genes that encode photorespiratory enzymes confer resistance against disease. Plant Cell. 2004;16:172–184. doi: 10.1105/tpc.016352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Torres MA. Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 43.Torres MA. Dangl JL. Jones JD. Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc Natl Acad Sci U S A. 2002;99:517–522. doi: 10.1073/pnas.012452499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Torres MA. Jones JD. Dangl JL. Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat Genet. 2005;37:1130–1134. doi: 10.1038/ng1639. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y. Yun BW. Kwon E. Hong JK. Yoon J. Loake GJ. S-nitrosylation: an emerging redox-based post-translational modification in plants. J Exp Bot. 2006;57:1777–1784. doi: 10.1093/jxb/erj211. [DOI] [PubMed] [Google Scholar]
- 46.Wang YQ. Feechan A. Yun BW. Shafiei R. Hofmann A. Taylor P. Xue P. Yang FQ. Xie ZS. Pallas JA. Chu CC. Loake GJ. S-nitrosylation of AtSABP3 antagonizes the expression of plant immunity. J Biol Chem. 2009;284:2131–2137. doi: 10.1074/jbc.M806782200. [DOI] [PubMed] [Google Scholar]
- 47.Wilkinson JQ. Crawford NM. Identification of the Arabidopsis CHL3 gene as the nitrate reductase structural gene NIA2. Plant Cell. 1991;3:461–471. doi: 10.1105/tpc.3.5.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yun BW. Feechan A. Yin M. Saidi NB. Le Bihan T. Yu M. Moore JW. Kang JG. Kwon E. Spoel SH. Pallas JA. Loake GJ. S-nitrosylation of NADPH oxidase regulates cell death in plant immunity. Nature. 2011;478:264–268. doi: 10.1038/nature10427. [DOI] [PubMed] [Google Scholar]