Abstract
Significance: Cytochromes b561 (CYB561s) constitute a family of trans-membrane (TM), di-heme proteins, occurring in a variety of organs and cell types, in plants and animals, and using ascorbate (ASC) as an electron donor. CYB561s function as monodehydroascorbate reductase, regenerating ASC, and as Fe3+-reductases, providing reduced iron for TM transport. A CYB561-core domain is also associated with dopamine β-monooxygenase redox domains (DOMON) in ubiquitous CYBDOM proteins. In plants, CYBDOMs form large protein families. Physiological functions supported by CYB561s and CYBDOMs include stress defense, cell wall modifications, iron metabolism, tumor suppression, and various neurological processes, including memory retention. CYB561s, therefore, significantly broaden our view on the physiological roles of ASC. Recent Advances: The ubiquitous nature of CYB561s is only recently being recognized. Significant advances have been made through the study of recombinant CYB561s, revealing structural and functional properties of a unique “two-heme four-helix” protein configuration. In addition, the DOMON domains of CYBDOMs are suggested to contain another heme b. Critical Issues: New CYB561 proteins are still being identified, and there is a need to provide an insight and overview on the various roles of these proteins and their structural properties. Future Directions: Mutant studies will reveal in greater detail the mechanisms by which CYB561s and CYBDOMs participate in cell metabolism in plants and animals. Moreover, the availability of efficient heterologous expression systems should allow protein crystallization, more detailed (atomic-level) structural information, and insights into the intra-molecular mechanism of electron transport. Antioxid. Redox Signal. 19, 1026–1035.
Introduction
The impact of oxidation-reduction (redox) reactions on many fundamental cellular processes is increasingly recognized. An intricate network of redox proteins and low-molecular-weight electron donors and acceptors control redox homeostasis in many cell types. Ascorbate (ASC) is an essential component of this redox network in plants and animals, mainly as an electron donor in enzymatic reactions, including H2O2 scavenging by ASC peroxidase (APX) (19). However, ASC is also a principal electron donor to a family of di-heme, trans-membrane (TM) proteins, cytochromes b561 (CYB561s), catalyzing TM electron transport (TMET). CYB561s occur in a wide range of animal and plant phyla, and are generally encoded by small gene families. Their name is derived from the characteristic α-band absorption maximum (reduced minus oxidized).
CYB561 members are implicated in physiological processes, including plant ‘front-line’ (apoplastic) defense; iron uptake; ASC regeneration in neuroendocrine tissues; and tumor growth. CYB561s are also of interest because of their particular biochemical (structure-function) properties. They are composed of six TM α-helices, the central four of which make up the ‘CYB561-core,’ coordinating two heme b molecules with four conserved His. The core domain harbors the intra-molecular electron transfer, and, intriguingly, is a building block in other proteins, often in association with other redox domains such as the N-terminal domain of dopamine β-monooxygenase (DOMON). Together, the CYB561 protein family expands our horizon on the role of ASC in cell metabolism.
Physiological Functions and Biological Relevance
ASC recycling
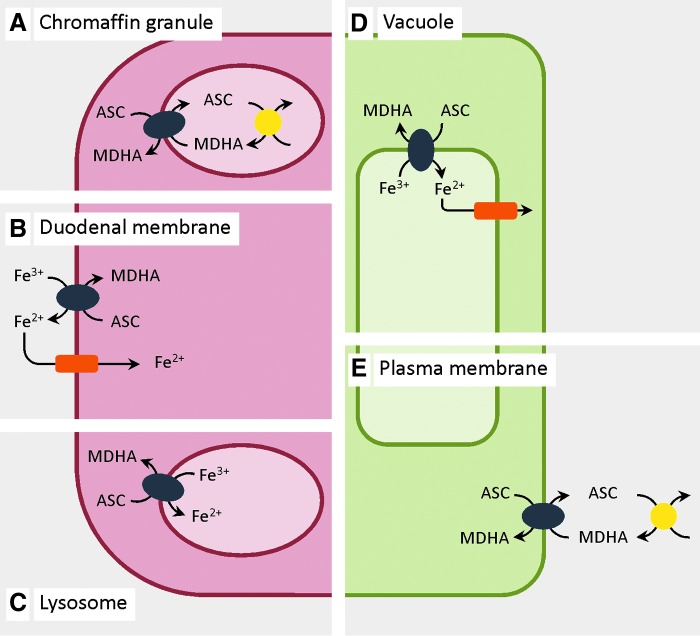
Some CYB561s support ASC regeneration, through one-electron reduction of partially oxidized ASC, monodehydroascorbate (MDHA) to ASC, using electrons provided by ASC, on ‘the other’ side of the membrane. In animals, such TMET activity is catalyzed by the chromaffin granule CYB561 (CGCytb/CYB561A1, for systematic nomenclature, see Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/ars), the first CYB561 identified (18). Chromaffin granules harbor dopamine synthesis, involving ASC-dependent dopamine β-hydroxylation, generating MDHA. A similar ASC-regenerating function is required in neuropeptide storage vesicles to sustain ASC-dependent amidated peptide biosynthesis. The essential role of CYB561s in this process, and its impact on neurological functions, was recently illustrated in the Drosophila mutant nemy, where decreased expression of a CYB561 homologue resulted in reduced memory retention (24). The ‘electron shuttle’ activity of CGCytb/CYB561A1, between ASC and membrane-impermeable ferricyanide or MDHA, was demonstrated in native chromaffin granule ‘ghost’ vesicles and reconstituted membrane systems (11, 44) (Fig. 1A). Further support on the role of CYB561s in ASC regeneration is found in erythrocytes, where intracellular ASC reduces extracellular MDHA (54, 60).
FIG. 1.
Subcellular localization and orientation of CYB561 family members in plant and animal cells. CYB561s (blue ovals) catalyze trans-membrane electron transport (TMET) through membranes, separating cytoplasm and acid compartments (secretory vesicles, vacuole, and extracellular matrix). Localization, orientation, and electron donor/acceptor couples are based on experimental evidence (on native and recombinant proteins), and ‘most plausible’ interpretations (see text). Ascorbate (ASC) appears a principal electron donor for all CYB561s, whereas both monodehydroascorbate (MDHA) and Fe3+ may act as electron acceptors. Animal CYB561s: (A) Chromaffin granules: CGCytb/CYB561A1 in chromaffin granule vesicle membranes, involved in catecholamine synthesis operating in concert with ASC-dependent oxidases (yellow circles); (B) duodenal DCytb/CYB561A2 in the duodenal cell membrane, reducing intestinal Fe3+-chelates before uptake through Fe2+-transporters (orange rectangles); (C) lysosomal membrane LCytb/CYB561A3. Plant CYB561: (D) plant tonoplast TCytb/CYB561B1 may reduce vacuolar Fe3+ for transport to the cytoplasm; (E) (putative) plant plasma membrane CYB561B acting in concert with extracellular oxidases (e.g., ASC oxidase). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Work on plant membranes revealed the presence of ASC-reducible, di-heme-like b-type cytochromes, with similar biophysical properties as bovine CGCytb/CYB561A1, in several species (4, 5, 22). A Phaseolus vulgaris CYB561, presumably plasma membrane (PM)-located, mediated TMET in purified, ASC-loaded, vesicles (2). Extracellular ASC provides antioxidant defense against gaseous oxidants (ozone, NOx), and CYB561 could be involved in apoplastic ASC regeneration (Fig. 1E). The Arabidopsis genome reveals four genes encoding six-TM CYB561s with an average of around 30% sequence identity to CGCytb/CYB561A1 (3), but it is not yet clear which of the gene products is involved in PM TMET.
Fe3+-reductase activity and iron homeostasis
A new CYB561 member was identified in the cell membrane of mouse duodenum epithelial cells, in response to hypoxia and iron deficiency (39). Mouse duodenal DCytb/CYB561A2 reduces intestinal Fe3+ before uptake (Fig. 1B). ASC is an electron donor to mouse DCytb/CYB561A2, at least in vitro (54). The elevated expression of DCytb/CYB561A2 in mice with systemic iron overload (hemochromatosis) provides additional support for a role in iron uptake (23).
ASC-mediated TM Fe3+-reductase activity was confirmed by complementation of Δfre1Δfre2 Fe3+-reductase-deficient yeast lines. Reduction of extracellular Fe3+-chelates in transformed yeast cells, expressing genes for three mouse and one Arabidopsis CYB561 protein (CGCytb/CYB561A1, DCytb/CYB561A2; LCytb/CYB561A3; TCytb/CYB561B1, respectively), was demonstrated (10, 54). For CGCytb/CYB561A1, the TM Fe3+-reductase activity was also demonstrated in ASC-loaded proteo-liposomes (11). These experiments suggest that, at least in vitro, all CYB561s may have Fe3+-reductase activity. Therefore, the actual in vivo activity of CYB561s, that is, Fe3+ reduction versus MDHA reduction, is probably dependent on substrate availability and biological context.
Free Fe3+ would readily be reduced by ASC, if occurring in the same compartment, and Fe2+ catalyzes free radical generation through Fenton-type reactions. Separation of Fe3+ and ASC, along with CYB561 catalyzing TMET, thus provides efficient control over ASC-mediated Fe3+ reduction.
Arabidopsis TCytb/CYB561B1, used in the yeast Fe3+-reduction assays, is localized in the tonoplast (20, 50) (Fig. 1D). Given that the Arabidopsis TCytb/CYB561B1 ASC-binding (electron acceptor) site is predicted on the cytoplasmic protein side, this suggests that this CYB561 could reduce Fe3+ in the vacuolar lumen, supporting transport to the cytoplasm. Vacuolar Fe2+-transporters (NRAMP3, NRAMP4), which may act in conjunction with TCytb/CYB561B1, have been identified (27). Possibly plant vacuoles store iron for remobilization when metabolic needs exceed supply. ASC-dependent CYB561-mediated Fe3+-reductase activity in plants may complement the well-characterized NADH-dependent activity of the FRO proteins (27).
Other physiological functions
In addition to a role in ASC and Fe metabolism, CYB561 members may be involved in other physiological processes. A watermelon PM CYB561B is up-regulated under drought and high-light stress and was proposed to constitute a route for thermal dissipation of excess light energy by functional interaction with apoplastic ASC oxidase (43) (Fig. 1E). In humans, a CYB561 (TSCytb/CYB561D2/101F6) is encoded in chromosomal region 3p21.3 and is related to suppression of tumor cell growth. Treating lung cancer cells with TSCytb/CYB561D2 plasmid-DNA nanoparticles and ASC, synergistically suppressed lung cancer cell development (46). For yet other CYB561s, limited functional information is still available. For example, mouse LCytb/CYB561A3 is located in lysosomal membranes in various tissues and is possibly associated specifically with macrophages (64) (Fig. 1C). However, its physiological impact is not yet clear. Lysosomal iron pools play a role in oxidative stress control (30), which could involve LCytb/CYB561A3-Fe3+-reductase activity. Finally, one should consider that other electron donors and acceptors (than ASC and MDHA/Fe3+) may still be identified, linking CYB561s to yet other physiological processes.
Tissue- and subcellular-distribution
CGCytb/CYB561A1 was first identified in catecholamine-synthesizing (chromaffin) vesicles, and was, consistently, demonstrated in various neuroendocrine tissues (53). It is now clear that plant and animal CYB561s occur in a wide range of organs, tissues, and cell types. Remarkably specific cell-type localizations are sometimes observed, but not yet understood, such as testicular Sertoli cells for mouse LCytb/CYB561A3 (64). Some animal organs (e.g., lung, spleen, brain, and duodenum) contain multiple CYB561 isoforms. However, in these cases, the proteins are apparently not located in identical cell types or membrane types. In Arabidopsis, CYB561 isoforms show some, but not complete, overlapping organ distribution patterns, and CYB561s occur in different membrane types (52, 62). Therefore, CYB561 isoform expression does not appear to be redundant. In a recently isolated Arabidopsis homozygous KO of TCytb/CYB561B1, transcription of at least three other CYB561 isoforms was not enhanced, supporting this notion (unpublished).
The subcellular membranes containing CYB561 consistently are membranes separating the cytoplasm and an acidic compartment, including acidic extracellular environments in plants and animals (Fig. 1). This pH gradient is most likely maintained by the activity of H+-transporting ATPases, and is probably important in CYB561 TMET function.
CYB561 Structure and Structure-Function Relationship
The CYB561 protein family, in plants and animals, was identified on the basis of sequence comparison to the amino acid sequence of the bovine chromaffin granule CGCytb/CYB561A1 (59) (Fig. 2). Until today, this is also the only native CYB561 protein for which biochemical and biophysical measurements are available. Biochemical and structural information of other plant and animal CYB561s is derived from recombinant proteins, expressed in yeast (Saccharomyces cerevisiae, Pichia pastoris), insect (Sf9) cells, and Escherichia coli (8, 11, 15, 20, 33, 35–37, 41, 45, 54). CYB561s are membrane-spanning proteins that are generally accepted to contain six TM α-helix domains and two pairs of His residues, arranged on four consecutive TM helices, coordinating two b-type hemes, located on each side of the membrane and accepting electrons from ASC (Figs. 2 and 3). Crystal-structure-level 3D atomic information is not (yet) available for any CYB561. However, 3D atomic resolution models were calculated for human CGCytb/CYB561A1, and Arabidopsis TCytb/CYB561B1 to support data analysis (6) (Fig. 4).
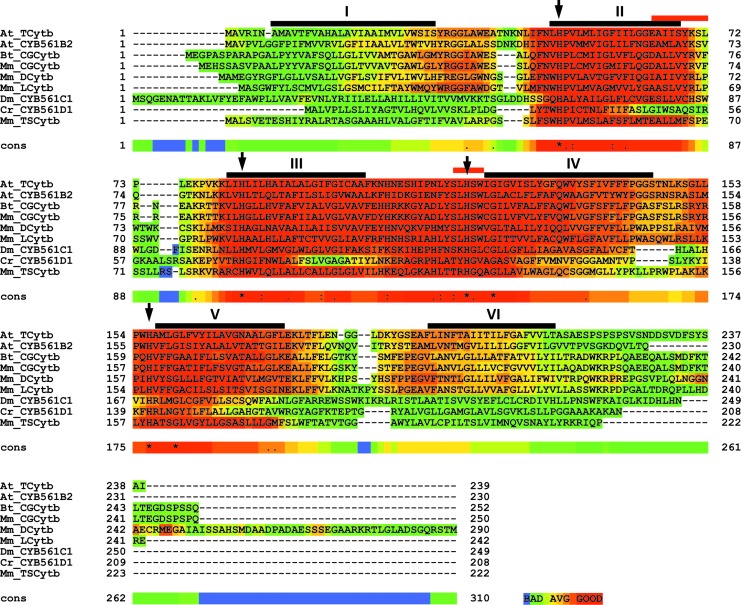
FIG. 2.
Multiple alignment of CYB561s, showing TM domains, conserved residues, and substrate-binding domains. ClustalW2.1 alignment of selected CYB561 sequences, representing isoforms from most evolutionary clusters (59) (see Fig. 5), showing TM domains (I–VI, black bars), conserved His residues (arrows), and putative substrate interacting sites (red bars). Sequences are as follows: (i) Arabidopsis thaliana tonoplast CYB561 (At_TCytb/CYB561B1; Q8L856); (ii) At_CYB561B2 (Q9SWS1); (iii) Bostaurus chromaffin granule CYB561 (Bt_CGCytb/CYB561A1; P10897); (iv) Mus musculus chromaffin granule CYB561 (Mm_CGCytb/CYB561A1; Q60720); (v) M. musculus duodenal CYB561 (Mm_DCytb/CYB561A2; Q925G2); (vi) M. musculus lysosomal CYB561 (Mm_LCytb/CYB561A3; Q6P1H1); (vii) Drosophila melanogaster CYTB561 (Dm_CYB561C1; Q9W4U9); (viii) Chlamydomonas reinhardtii CYB561 (Cr_CYB561E1; XP_001702111.1); and (ix) M. musculus tumor suppressor CYB561(Mm_TSCytb/CYB561D2; Q9WUE3). Color coding of sequence similarity is performed using the T-Coffee alignment evaluation (17). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 3.
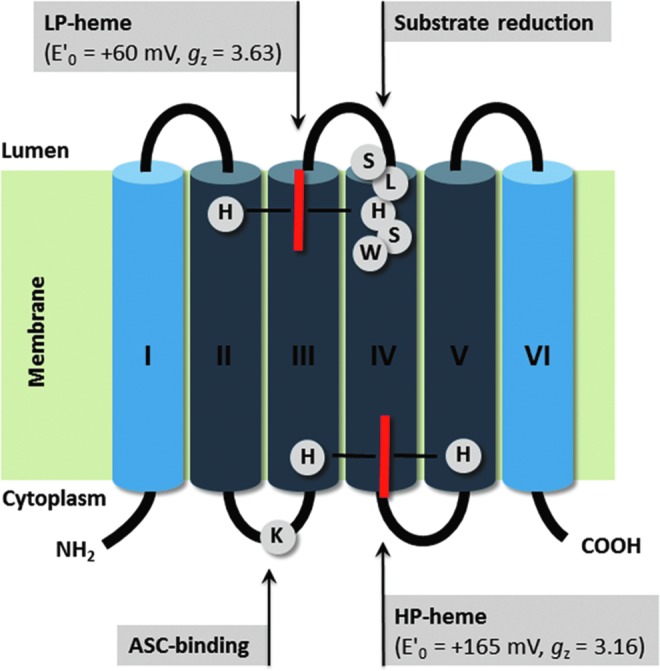
Schematic 2D structure of the Arabidopsis TCytb/CYB561B1 TM organization, including hemes, heme coordination, and putative substrate binding sites. CYB561s are composed of six TM domains (I-VI), with two heme-b molecules (red bars), coordinated by four conserved His on TM helices II-V. The high potential (HP)-heme is located on the cytoplasmic protein side, accepting electrons from ASC (or other electron donors). The heme coordination and orientation is confirmed through electron paramagnetic resonance studies on site-directed-mutant proteins. Also shown is a Lys residue, possibly involved in substrate (ASC) binding, and a well-conserved putative substrate-binding site (SLHSW). Dark blue: CYB561 four helix core domain. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
FIG. 4.
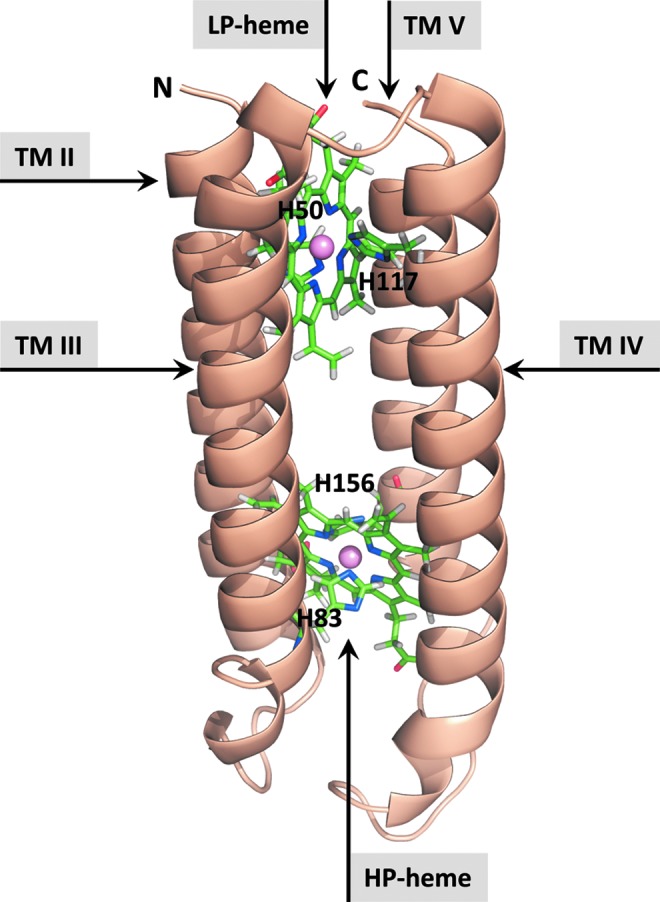
Calculated 3D atomic structure model of the Arabidopsis TCytb/CYB561B1-core domain. Based on extensive CYB561s sequence analysis, structural constraints were identified to predict 3D structural models, using molecular modeling tools. Counterclockwise helix topologies are shown, including the HP- and low-potential (LP) hemes (ball-and-stick). Balls represent the Fe-atoms [modified after (6)]. To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
Midpoint redox potentials
Redox titration results consistently reveal the presence of two one-electron redox centers (b-type hemes), with midpoint redox potential (E0′) values ranging from +80 to +190 mV for the ‘high potential’ (HP) heme, and −20 to +60 mV for the ‘low potential’ (LP) heme, (15, 35, 56). Among CYB561 isoforms, there is a relatively consistent difference of about 100 mV between the E0′ of the heme centers, supporting intra-molecular electron transfer mechanisms. Recently, it was demonstrated by electron paramagnetic resonance (EPR) and site-directed mutagenesis that the HP-heme is located on the cytoplasmic side of the TM proteins (16).
ASC reducibility
Reducibility by ASC is recognized as a particular CYB561 feature, generally reaching reduction levels of 50% to 90% (relative to Na-dithionite [DTH]). Why 100% reduction levels are not reached, even at high ASC concentrations, is not yet fully understood, in part because the ASC and/or DTH reduction mechanism is not yet fully unraveled. The midpoint redox potentials of the electron transfer reactions involving ASC are around +320 mV (MDHA/ASC) and −200 mV [dehydroascorbate (DHA)/MDHA], and +60 mV for the DHA/ASC redox couple (25). Since the MDHA concentration in the cell is orders of magnitude lower than that of ASC (due to disproportionation), the electron transfer from ASC to CYB561 is favored under physiological conditions. Thermodynamic evidence supports the idea that CYB561-mediated TMET may be coupled to a local protonation/de-protonation cycle (42, 44).
Direct molecular-binding interactions between ASC and CYB561 have not been measured, but steady-state and fast kinetic measurements on plant and animal CYB561s suggest the existence of two distinct ASC-‘affinity’ sites, with approximate values of 0.01 mM and 1 mM, for CGCytb/CYB561A1 (7, 11, 28). These parameters are not real binding constants but rather characterize the interaction between ASC and CYB561, resulting in the reduction of the HP and LP-hemes. The effect of diethylpyrocarbonate on ASC reduction kinetics also supports this hypothesis (58). A comparison of conserved sequence regions in the CYB561 core led to the tentative assignment of two substrate binding sites located on the cytoplasmic and luminal side of the protein (47, 59) (Fig. 2).
Spectrum analysis of mouse and bovine CGCytb/CYB561A1 and of TSCytb/CYB561D2 revealed the presence of two distinct, split α-bands in the spectrum of ASC-reduced proteins, which is consistent with the presence of two heme pockets (9, 12, 28). In Arabidopsis TCytb/CYB561B1 and mouse TSCytb/CYB561D2, singular value decomposition analysis identified two distinct b-type heme spectra that could be assigned to the two CYB561 hemes (12, 16). The property of split α-bands points to the fact that each heme is located in an anisotropic electrostatic field, originating from the distribution of charged amino acid residues around the heme pockets. Given the limited primary structure similarity between CYB561 isoforms, it is not surprising that the fine structure of the split α-band slightly differs, not only for the two hemes but also between isoforms (9, 12, 15, 16).
Other reducing agents
Common reductants such as NAD(P)H and reduced glutathione (GSH) are poor electron donors to CYB561s. Reduction by NAD(P)H of various CYB561 isoforms never exceeded 5% (relative to DTH) (12, 57). GSH does not reduce CYB561, but other thiol reagents, in particular dihydrolipoic acid (DHLA), are almost as efficient as ASC in reducing mouse CGCytb/CYB561A1 and TSCytb/CYB561D2; however, with only one apparent, high affinity, binding site (12). In addition, the Arabidopsis TCytb/CYB561B1 was reduced by DHLA, although with much lower sensitivity and biphasic concentration dependence (12). Thus, it is possible that DHLA plays a different role in CYB561 reduction in plants and animals. In cells, DHLA is present mostly in its lipoamine form, tethered to conserved Lys residues on lipoyl-accepting protein domains (13). It may, therefore, be worth to investigate lipoyl-domain-containing proteins as potential CYB561 redox partners. In addition, dithiothreitol, and its stereoisomer, dithioerythritol, reduce Arabidopsis TCytb/CYB561B and mouse TSCytb/CYB561E (12, and unpublished).
Site-directed mutagenesis
Important structure-function information is derived from targeted mutations in the highly conserved heme-coordinating His residues of the CYB561-core (Fig. 2 and 3). For example, detailed EPR studies of Arabidopsis TCytb/CYB561B1 mutant proteins revealed that the HP-heme center is located on the cytosolic side of the protein (16), opposite to that previously suggested (56, 58). Western blot analysis and spectroscopy demonstrated that mutation of His residues coordinating the LP-heme (intra-vesicular-side) of Arabidopsis TCytb/CYB561B1, and mouse CGCytb/CYB561A1, resulted in nearly undetectable protein levels. In contrast, mutations in the HP-heme-coordinating residues hardly affected CYB561 expression, but resulted in altered ASC-reduction kinetics and reduced heme content (11, 28, 34). Somewhat contradicting results were obtained with His mutations in human DCytb/CYB561A2, and need further investigation (37, 45).
Positively charged residues near the hemes may facilitate the interaction with the ASC and MDHA anions, as is, for example, the case in the ASC-mediated reduction of soybean APX (14, 31). Lys and Arg residues are frequently found in the loop regions between the TM helices (Fig. 2), and an Arg residue (R72 in mouse CGCytb/CYB561A1), located near the HP-heme, is fully conserved among CYB561A members (59). Mutagenesis of R72 resulted in the loss of the ‘high affinity’ ASC reduction, supporting its role in substrate binding and suggesting that the high-affinity ASC-binding site was close to the HP heme (11). Arabidopsis TCytb/CYB561B1 contains no Arg in the vicinity of the heme, but still displays two distinct affinity sites (7). Its role is possibly replaced by a conserved Lys (K80) residue. This idea is supported by mutagenesis of K83 in maize CYB561B1, which resulted in altered midpoint redox potentials and ASC-reduction kinetics (41).
Mutations in other conserved and partially conserved residues of mouse CYB561s had variable effects on TMET activity and ASC reduction (34, 41, 45, 54). Consistent with the putative location of the electron-accepting and -donating sides, mutations in loop regions on the intra-vesicular side severely decreased LCytb/CYB561A3 TM Fe3+-reductase activity, but mutations on the cytoplasmic side had comparatively less effect (54).
A key function of CYB561s is TMET, through sequential reduction and oxidation of the HP and LP-hemes. Conserved residues presumably located in TM domains (e.g., Q131 in mouse LCytb/CYB561A3/) have been identified, and mutation resulted in strongly decreased Fe3+-reductase activity (54). This suggests that electron transfer my involve oxidation/reduction of intramolecular amino acids. On the other hand, electron transfer along chains of prosthetic groups is well established, as long as the “edge-to-edge” distance between groups is smaller than 14 Å (15, 48).
CYB561-Domain-Containing Proteins
CYBDOM
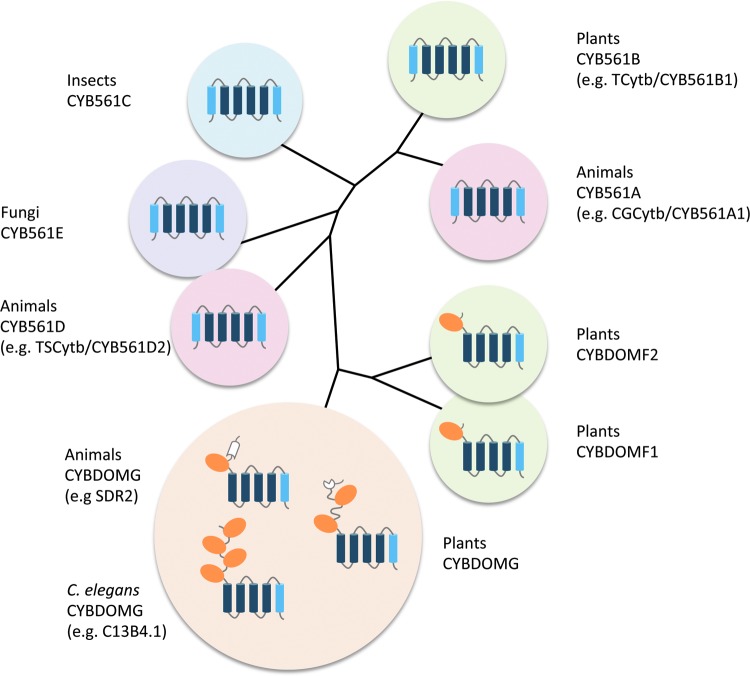
Identification of a CYB561-core domain at the C-terminus of a protein named stromal cell-derived receptor 2 (SDR2) provided evidence that CYB561s could be part of multi-domain proteins (49) (Fig. 5). Bio-informatic analyses demonstrated that most commonly a single CYB561-core follows a DOMON-domain (CYBDOM), but some proteins contain up to four DOMONs in a row (e.g., C13B4.1 of C. elegans; Fig. 5) (1, 26, 49, 51).
FIG. 5.
Evolutionary relationship between CYB561s and CYB561-core-containing CYBDOM proteins. CYB561 and CYBDOM proteins, cluster in various subgroups. The CYB561-core domain (four TM helices containing four conserved His and two hemes in bis-His coordination) is dark blue (see also Fig. 3). In the CYBDOM protein family, the CYB561 domain constituted by a CYB561-core plus the C-terminal α-helix of CYB561s is combined with one or more dopamine β-monooxygenase N-terminal domain (DOMON) domains (brown ovals). Cluster identifications are based on (59), except that clusters D and E were exchanged, for nomenclature reasons (see Supplementary Table S1). Each CYB561 cluster contains sequences from only animals or plants or insects or fungi, with the notable exception of Chlamydomonas reinhardtii CYB561 (XP_001702111.1, see also Supplementary Table S1) that clusters with the ‘fungal’ cluster CYB561E. CYBDOMF is divided into F1 and F2 clusters with diverging DOMON sequences. CYBDOMG includes sequences with one DOMON plus a reelin domain (white), sequences with four DOMONs, and plant sequences with two DOMONs and a DM13 domain (white). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
The DOMON-domain refers to the N-terminal domain of dopamine β-monooxygenase (or dopamine[beta]-hydroxylase, DBH); (1, 49), catalyzing the reduction of dopamine to norepinephrine in chromaffin granules. In the process, ASC is oxidized to MDHA, which may be regenerated by CYB561 (Fig. 1A). The DOMON-domain is widespread in nature, often in association with redox domains such as the Cu2+-monoxygenase domain in DBH or CYB561 in CYBDOMs, and/or with adhesion modules (e.g., reelin-domain in SDR2) (Fig. 5) (26). In DBH, this module is probably involved in tetramerisation (29), but not in catalysis or catecholamine binding (61).
CYBDOM sequences have been identified in the genome of animals, fungi, plants, and prokaryotes (26). Eukaryotic CYBDOMs were classified in two major groups. The ‘F’-group, including plant proteins with single CYB561- and DOMON-domains, and the ‘G’-group, including SDR2-related sequences (from animals or plants), sometimes with multiple DOMON domains (59) (Fig. 5). G-group CYBDOMs often also contain the reelin domain (animals) or DM13 domain (plants) (26, 51).
It is now clear that plants contain larger CYBDOM gene families than do other organisms (51, 59). For example, Arabidopsis contains at least ten CYBDOM-containing genes: eight belonging to the F-group, one to the G-group, and a divergent gene that could not easily be classified (51). In addition, Arabidopsis and other plant genomes contain CYBDOM-related genes with either no DOMON or no CYB561-domain (e.g., see section on heme-b binding). On the contrary, SDR2 is the only CYBDOM of the human genome, and single SDR2 orthologous genes are found in mouse and fly genomes, while C. elegans contains four G-group CYBDOMs (59).
All of these CYBDOMs contain a canonical CYB561-core, with four fully conserved His, at the N-terminus of four consecutive TM α-helices, plus an additional C-terminal α-helix, corresponding to the sixth α-helix of CYB561s (Fig. 5). In spite of these strictly conserved features, CYB561s and CYBDOMs have highly divergent sequences (59).
A common feature of CYBDOM sequences, at least in eukaryotes, is the presence of a signal sequence directing these proteins to the secretory pathway. Consistently, proteomic studies in plants have identified CYBDOM proteins on the PM (55), in some cases in association with lipid rafts (32). In tobacco roots expressing a chimeric CYBDOM-GFP from soybean, fluorescence was clearly associated with the PM (A. Costa and P. Trost, unpublished). In mammals, SDR2 is also believed to reside in the PM (61).
Topology predictions show that the C-terminus of CYBDOMs (corresponding to the C-terminus of CYB561s) is cytosolic and, consequently, the N-terminus of the five-α-helices CYB561-domain, connecting to the DOMON domain(s), is extracellular. Indeed, DOMON domains are commonly extracellular and either heme- or sugar-binding domains (26). The heme-binding DOMON domain of cellobiose dehydrogenase (CDH) of ligninolytic fungi plays a role in the attack to the lignocellulosic extracellular matrix of plants (63). Sugar-binding DOMON domains are also found in bacterial extracellular cellulases (26), overall suggesting a role of DOMON-containing proteins in modifying extracellular polymers (see section on Why CYBDOMs?).
Fe3+-reductase activity
The CYB561-core in CYBDOMs contains all necessary elements to catalyze TMET. Recombinant fly and mouse SDR2 demonstrate Fe3+-reductase activity in Xenopus oocytes (61). SDR2 is, therefore, considered a TM reductase and was renamed ferric chelate reductase 1 (e.g., in UniProt). Whether Fe3+-reductase activity can be extended to other CYBDOMs is unknown. Since the CYB561-domain can catalyze by itself the TM reduction of Fe3+-chelates, the role of the DOMON domain in SDR2 redox activity is uncertain. However, DOMON domains are most likely b-type cytochromes and may form a redox chain with CYB561 domains.
Heme-b binding in the DOMON domain
The DOMON-containing CDH of wood-degrading and phytopathogenic fungi is an extracellular flavo-cytochrome that participates in early events of lignocellulose degradation (38). The 3D structure of the DOMON-domain of CDH shows heme binding through an uncommon His-Met coordination (21) (Fig. 6). The heme is tightly fitted into a pocket of the concave inner β-sheet of the overall β-sandwich structure that is typical of the DOMON domain (26). Interestingly, both heme-axial ligands of Phanerochaete chrysosporium CDH (M65, H163) are conserved in CYBDOM sequences, strongly suggesting that DOMON-domains of CYBDOMs may also bind a heme b. These residues are conserved neither in the DOMON of DBH (that bind no hemes) nor in DOMON proteins that bind a sugar in place of the heme (26).
FIG. 6.

DOMON domain of P. chrysosporium cellobiose dehydrogenase (CDH), showing the uncommon b-type heme coordination by a Met-His pair. The DOMON domains of CYBDOM proteins are predicted to have the same 3D-fold based on two antiparallel β-sheets forming a β-sandwich structure as the DOMON domain of CDH (26). Although the sequence similarity between CYBDOM and CDH-DOMONs is limited, the pair of heme-coordinating residues Met65 and His163 (P. chrysosporium numbering) is strictly conserved. The structure was drawn from pdb file 1d7b (21). Heme and side chains of heme-coordinating residues are represented in sticks with the following color codes C (green), N (blue), O (red), and S (yellow). To see this illustration in color, the reader is referred to the web version of this article at www.liebertpub.com/ars
A close relative to CYBDOMs is the plant protein AIR12, which is composed of a single DOMON domain, including the conserved Met-His pair that in P. chrysosporium is involved in heme ligation (21, 51). AIR12 is attached to the external leaflet of plant PMs via a GPI anchor and was experimentally demonstrated to bind a single heme b (51). The peculiar electron paramagnetic resonance spectrum of AIR12 is consistent with the Met-His heme-coordination displayed by CDH. AIR12 is the only CYBDOM-like DOMON-domain for which heme binding was experimentally demonstrated. Arabidopsis contains a single AIR12 gene, and AIR12-knock out mutants are resistant to the necrotrophic pathogen Botrytis cinerea (R. Barbaro, A. Costa, F. Sicilia, P. Trost, unpublished). Whether the function of AIR12 depends on the interaction with a CYB561 isoform (reminiscent of CYBDOM proteins) remains to be demonstrated.
Why CYBDOMs?
The architecture of CYBDOM proteins, with a heme b-binding DOMON-domain linked to a five-α-helix CYB561-domain, suggests an ‘electron wire,’ extending well beyond the PM surface. Asking how the function of CYBDOMs may be related to their redox activity and architecture is particularly challenging in plants, where CYBDOMs are often coded by ten genes or more.
P. chrysosporium lines, defective in the DOMON-enzyme CDH, fail to colonize wood. It has been proposed that CDH may participate in lignin degradation (38, 63). In this view, CDH, extending into the cell wall, reduces an Fe3+-chelate (e.g., Fe3+-oxalate) with cellobiose being the reductant. In the presence of hydrogen peroxide (possibly produced by the flavo-domain of CDH itself), the reduced iron could generate hydroxyl radicals through Fenton chemistry, which may attack cell wall polymers, including lignin. CYBDOMs may well be involved in a similar reaction. The CYB561-core could provide cytoplasmic reducing equivalents to the extracellular heme(s), supporting hydroxyl radical-based modifications of cell wall polymers at a safe distance from the PM. As cell wall-loosening agents in plants, hydroxyl radicals are involved in fundamental processes such as for example, seed germination and cell elongation (40). In this context, CYBDOMs may constitute a redox bridge between cytosolic ASC and cell wall sites where hydroxyl radicals may be locally produced by DOMON-catalyzed Fenton chemistry, similar to the DOMON-containing CDH of basidiomycetes.
Supplementary Material
Abbreviations Used
- APX
ascorbate peroxidase
- ASC
ascorbate
- CDH
cellobiose dehydrogenase
- DHA
dehydroascorbate
- DHLA
dihydrolipoic acid
- DOMON
dopamine β-monooxygenase N-terminal domain
- HP
high potential
- LP
low potential
- MDHA
monodehydroascorbate
- PM
plasma membrane
- TM
trans-membrane
- TMET
trans-membrane electron transport
- SDR2
stromal cell-derived receptor 2
Acknowledgments
This work was supported by the Flemish Fund for Scientific Research, FWO (grant G.0118.07N to H.A.), by the Hungary-Romania Cross Border Cooperation Program of the EU (HURO/0901/219; to A.B.), and by the Italian Ministery of University and Research (PRIN2009 to PT and RB). The help of Dr. Tibor Pali (Biological Research Center, Szeged, Hungary) with the composition of Fig. 3, and of the HUGO Gene Nomenclature Committee, are gratefully acknowledged.
References
- 1.Aravind L. Domon: an ancient extracellular domain in dopamine beta-monooxygenase and other proteins. Trends Biochem Sci. 2001;26:524–526. doi: 10.1016/s0968-0004(01)01924-7. [DOI] [PubMed] [Google Scholar]
- 2.Asard H. Horemans N. Caubergs RJ. Transmembrane electron transport in ascorbate-loaded plasma membrane vesicles from higher plants involves a b-type cytochrome. FEBS Lett. 1992;306:143–146. doi: 10.1016/0014-5793(92)80986-q. [DOI] [PubMed] [Google Scholar]
- 3.Asard H. Terol-Alcayde J. Preger V. Del Favero J. Verelst W. Sparla F. Pérez-Alonso M. Trost P. Arabidopsis thaliana sequence analysis confirms the presence of cyt b-561 in plants: evidence for a novel protein family. Plant Physiol Biochem. 2000;38:905–912. [Google Scholar]
- 4.Asard H. Venken M. Caubergs R. Reijnders W. Oltmann FL. De Greef JA. b-Type cytochromes in higher plant plasma membranes. Plant Physiol. 1989;90:1077–1083. doi: 10.1104/pp.90.3.1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Askerlund P. Larsson C. Widell S. Cytochromes of plant plasma membranes. Characterization by absorbance difference spectroscopy and redox titration. Physiol Plant. 1989;76:123–134. [Google Scholar]
- 6.Bashtovyy D. Bérczi A. Asard H. Pali T. Structure prediction for the di-heme cytochrome b561 protein family. Protoplasma. 2003;221:31–40. doi: 10.1007/s00709-002-0065-0. [DOI] [PubMed] [Google Scholar]
- 7.Bérczi A. Asard H. Characterization of an ascorbate-reducible cytochrome b561 by site directed mutagenesis. Acta Biol Szeged. 2006;50:55–59. [Google Scholar]
- 8.Bérczi A. Asard H. Expression and purification of the recombinant mouse tumor suppressor cytochrome b561 protein. Acta Biol Szeged. 2008;52:257–265. [Google Scholar]
- 9.Bérczi A. Desmet F. Van Doorslaer S. Asard H. Spectral characterization of the recombinant mouse tumor suppressor 101F6 protein. Eur Biophys J. 2010;39:1129–1142. doi: 10.1007/s00249-009-0564-4. [DOI] [PubMed] [Google Scholar]
- 10.Bérczi A. Su D. Asard H. An Arabidopsis cytochrome b561 with trans-membrane ferrireductase capability. FEBS Lett. 2007;581:1505–1508. doi: 10.1016/j.febslet.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 11.Bérczi A. Su D. Lakshminarasimhan M. Vargas A. Asard H. Heterologous expression and site-directed mutagenesis of an ascorbate-reducible cytochrome b561. Arch Biochem Biophys. 2005;443:82–92. doi: 10.1016/j.abb.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 12.Bérczi A. Zimanyi L. Asard H. Dihydrolipoic acid reduces cytochrome b561 proteins. Eur Biophys J. 2012 doi: 10.1007/s00249-012-0812-x. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 13.Biewenga GP. Haenen GRMM. Bast A. Packer L. An overview of lipoate chemistry. In: Fuchs J, editor; Zimmer G, editor. Lipoic Acid in Health and Disease. New York: Marcell Dekker; 1997. pp. 1–32. [Google Scholar]
- 14.Bursey EH. Poulos TL. Two substrate binding sites in ascorbate peroxidase: the role of arginine 172. Biochemistry. 2000;39:7374–7379. doi: 10.1021/bi000446s. [DOI] [PubMed] [Google Scholar]
- 15.Cenacchi L. Busch M. Schleidt PG. Muller FG. Stumpp TV. Mantele W. Trost P. Lancaster CR. Heterologous production and characterisation of two distinct dihaem-containing membrane integral cytochrome b(561) enzymes from Arabidopsis thaliana in Pichia pastoris and Escherichia coli cells. Biochim Biophys Acta. 2012;1818:679–688. doi: 10.1016/j.bbamem.2011.10.030. [DOI] [PubMed] [Google Scholar]
- 16.Desmet F. Bérczi A. Zimanyi L. Asard H. Van Doorslaer S. Axial ligation of the high-potential heme center in an Arabidopsis cytochrome b561. FEBS Lett. 2011;585:545–548. doi: 10.1016/j.febslet.2011.01.006. [DOI] [PubMed] [Google Scholar]
- 17.Di Tommaso P. Moretti S. Xenarios I. Orobitg M. Montanyola A. Chang JM. Taly JF. Notredame C. T-Coffee: a web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res. 2011;39:W13–W17. doi: 10.1093/nar/gkr245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flatmark T. Terland O. Cytochrome b 561 of the bovine adrenal chromaffin granules. A high potential b-type cytochrome. Biochim Biophys Acta. 1971;253:487–491. doi: 10.1016/0005-2728(71)90052-1. [DOI] [PubMed] [Google Scholar]
- 19.Foyer CH. Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiol. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Griesen D. Su D. Bérczi A. Asard H. Localization of an ascorbate-reducible cytochrome b561 in the plant tonoplast. Plant Physiol. 2004;134:726–734. doi: 10.1104/pp.103.032359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hallberg BM. Bergfors T. Backbro K. Pettersson G. Henriksson G. Divne C. A new scaffold for binding haem in the cytochrome domain of the extracellular flavocytochrome cellobiose dehydrogenase. Structure. 2000;8:79–88. doi: 10.1016/s0969-2126(00)00082-4. [DOI] [PubMed] [Google Scholar]
- 22.Hendry GA. Houghton JD. Jones OT. The cytochromes in microsomal fractions of germinating mung beans. Biochem J. 1981;194:743–751. doi: 10.1042/bj1940743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herrmann T. Muckenthaler M. van der Hoeven F. Brennan K. Gehrke SG. Hubert N. Sergi C. Grone HJ. Kaiser I. Gosch I. Volkmann M. Riedel HD. Hentze MW. Stewart AF. Stremmel W. Iron overload in adult Hfe-deficient mice independent of changes in the steady-state expression of the duodenal iron transporters DMT1 and Ireg1/ferroportin. J Mol Med (Berl) 2004;82:39–48. doi: 10.1007/s00109-003-0508-x. [DOI] [PubMed] [Google Scholar]
- 24.Iliadi KG. Avivi A. Iliadi NN. Knight D. Korol AB. Nevo E. Taylor P. Moran MF. Kamyshev NG. Boulianne GL. nemy encodes a cytochrome b561 that is required for Drosophila learning and memory. Proc Natl Acad Sci U S A. 2008;105:19986–19991. doi: 10.1073/pnas.0810698105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iyanagi T. Yamazaki I. Anan KF. One-electron oxidation-reduction properties of ascorbic acid. Biochim Biophys Acta. 1985;806:255–261. [Google Scholar]
- 26.Iyer LM. Anantharaman V. Aravind L. The DOMON domains are involved in heme and sugar recognition. Bioinformatics. 2007;23:2660–2664. doi: 10.1093/bioinformatics/btm411. [DOI] [PubMed] [Google Scholar]
- 27.Jeong J. Guerinot ML. Homing in on iron homeostasis in plants. Trends Plant Sci. 2009;14:280–285. doi: 10.1016/j.tplants.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 28.Kamensky Y. Liu W. Tsai AL. Kulmacz RJ. Palmer G. Axial ligation and stoichiometry of heme centers in adrenal cytochrome b561. Biochemistry. 2007;46:8647–8658. doi: 10.1021/bi700054g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kapoor A. Shandilya M. Kundu S. Structural insight of dopamine beta-hydroxylase, a drug target for complex traits, and functional significance of exonic single nucleotide polymorphisms. PLoS One. 2011;6:e26509. doi: 10.1371/journal.pone.0026509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kurz T. Gustafsson B. Brunk UT. Intralysosomal iron chelation protects against oxidative stress-induced cellular damage. FEBS J. 2006;273:3106–3117. doi: 10.1111/j.1742-4658.2006.05321.x. [DOI] [PubMed] [Google Scholar]
- 31.Lad L. Mewies M. Raven EL. Substrate binding and catalytic mechanism in ascorbate peroxidase: evidence for two ascorbate binding sites. Biochemistry. 2002;41:13774–13781. doi: 10.1021/bi0261591. [DOI] [PubMed] [Google Scholar]
- 32.Lefebvre B. Furt F. Hartmann MA. Michaelson LV. Carde JP. Sargueil-Boiron F. Rossignol M. Napier JA. Cullimore J. Bessoule JJ. Mongrand S. Characterization of lipid rafts from Medicago truncatula root plasma membranes: a proteomic study reveals the presence of a raft-associated redox system. Plant Physiol. 2007;144:402–418. doi: 10.1104/pp.106.094102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu W. Kamensky Y. Kakkar R. Foley E. Kulmacz RJ. Palmer G. Purification and characterization of bovine adrenal cytochrome b561 expressed in insect and yeast cell systems. Protein Expr Purif. 2005;40:429–439. doi: 10.1016/j.pep.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 34.Liu W. Rogge CE. da Silva GF. Shinkarev VP. Tsai AL. Kamensky Y. Palmer G. Kulmacz RJ. His92 and His110 selectively affect different heme centers of adrenal cytochrome b(561) Biochim Biophys Acta. 2008;1777:1218–1228. doi: 10.1016/j.bbabio.2008.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu W. Rogge CE. Kamensky Y. Tsai AL. Kulmacz RJ. Development of a bacterial system for high yield expression of fully functional adrenal cytochrome b561. Protein Expr Purif. 2007;56:145–152. doi: 10.1016/j.pep.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 36.Liu W. Wu G. Tsai AL. Kulmacz RJ. High-yield production, purification and characterization of functional human duodenal cytochrome b in an Escherichia coli system. Protein Expr Purif. 2011;79:115–121. doi: 10.1016/j.pep.2011.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludwiczek S. Rosell FI. Ludwiczek ML. Mauk AG. Recombinant expression and initial characterization of the putative human enteric ferric reductase Dcytb. Biochemistry. 2008;47:753–761. doi: 10.1021/bi701793a. [DOI] [PubMed] [Google Scholar]
- 38.Ludwig R. Harreither W. Tasca F. Gorton L. Cellobiose dehydrogenase: a versatile catalyst for electrochemical applications. Chem Phys Chem. 2010;11:2674–2697. doi: 10.1002/cphc.201000216. [DOI] [PubMed] [Google Scholar]
- 39.McKie AT. Barrow D. Latunde-Dada GO. Rolfs A. Sager G. Mudaly E. Mudaly M. Richardson C. Barlow D. Bomford A. Peters TJ. Raja KB. Shirali S. Hediger MA. Farzaneh F. Simpson RJ. An iron-regulated ferric reductase associated with the absorption of dietary iron. Science. 2001;291:1755–1759. doi: 10.1126/science.1057206. [DOI] [PubMed] [Google Scholar]
- 40.Muller K. Linkies A. Vreeburg RA. Fry SC. Krieger-Liszkay A. Leubner-Metzger G. In vivo cell wall loosening by hydroxyl radicals during cress seed germination and elongation growth. Plant Physiol. 2009;150:1855–1865. doi: 10.1104/pp.109.139204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakanishi N. Rahman MM. Sakamoto Y. Takigami T. Kobayashi K. Hori H. Hase T. Park SY. Tsubaki M. Importance of the conserved lysine 83 residue of Zea mays cytochrome b(561) for ascorbate-specific transmembrane electron transfer as revealed by site-directed mutagenesis studies. Biochemistry. 2009;48:10665–10678. doi: 10.1021/bi9010682. [DOI] [PubMed] [Google Scholar]
- 42.Nakanishi N. Takeuchi F. Tsubaki M. Histidine cycle mechanism for the concerted proton/electron transfer from ascorbate to the cytosolic haem b centre of cytochrome b561: a unique machinery for the biological transmembrane electron transfer. J Biochem. 2007;142:553–560. doi: 10.1093/jb/mvm181. [DOI] [PubMed] [Google Scholar]
- 43.Nanasato Y. Akashi K. Yokota A. Co-expression of cytochrome b561 and ascorbate oxidase in leaves of wild watermelon under drought and high light conditions. Plant Cell Physiol. 2005;46:1515–1524. doi: 10.1093/pcp/pci164. [DOI] [PubMed] [Google Scholar]
- 44.Njus D. Kelley PM. The secretory-vesicle ascorbate-regenerating system: a chain of concerted H+/e(-)-transfer reactions. Biochim Biophys Acta. 1993;1144:235–248. doi: 10.1016/0005-2728(93)90108-r. [DOI] [PubMed] [Google Scholar]
- 45.Oakhill JS. Marritt SJ. Gareta EG. Cammack R. McKie AT. Functional characterization of human duodenal cytochrome b (Cybrd1): redox properties in relation to iron and ascorbate metabolism. Biochim Biophys Acta. 2008;1777:260–268. doi: 10.1016/j.bbabio.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 46.Ohtani S. Iwamaru A. Deng W. Ueda K. Wu G. Jayachandran G. Kondo S. Atkinson EN. Minna JD. Roth JA. Ji L. Tumor suppressor 101F6 and ascorbate synergistically and selectively inhibit non-small cell lung cancer growth by caspase-independent apoptosis and autophagy. Cancer Res. 2007;67:6293–6303. doi: 10.1158/0008-5472.CAN-06-3884. [DOI] [PubMed] [Google Scholar]
- 47.Okuyama E. Yamamoto R. Ichikawa Y. Tsubaki M. Structural basis for the electron transfer across the chromaffin vesicle membranes catalyzed by cytochrome b561: analyses of cDNA nucleotide sequences and visible absorption spectra. Biochim Biophys Acta. 1998;1383:269–278. doi: 10.1016/s0167-4838(97)00216-1. [DOI] [PubMed] [Google Scholar]
- 48.Page CC. Moser CC. Chen X. Dutton PL. Natural engineering principles of electron tunneling in biological oxidation-reduction. Nature. 1999;402:47–52. doi: 10.1038/46972. [DOI] [PubMed] [Google Scholar]
- 49.Ponting CP. Domain homologues of dopamine beta-hydroxylase and ferric reductase: roles for iron metabolism in neurodegenerative disorders? Hum Mol Genet. 2001;10:1853–1858. doi: 10.1093/hmg/10.17.1853. [DOI] [PubMed] [Google Scholar]
- 50.Preger V. Scagliarini S. Pupillo P. Trost P. Identification of an ascorbate-dependent cytochrome b of the tonoplast membrane sharing biochemical features with members of the cytochrome b561 family. Planta. 2005;220:365–375. doi: 10.1007/s00425-004-1360-0. [DOI] [PubMed] [Google Scholar]
- 51.Preger V. Tango N. Marchand C. Lemaire SD. Carbonera D. Di Valentin M. Costa A. Pupillo P. Trost P. Auxin-responsive genes AIR12 code for a new family of plasma membrane b-type cytochromes specific to flowering plants. Plant Physiol. 2009;150:606–620. doi: 10.1104/pp.109.139170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Provart NJ. University of Toronto; [Apr 1;2012 ]. 2011. Arabidopsis eFP Browser. [Google Scholar]
- 53.Pruss RM. Shepard EA. Cytochrome b561 can be detected in many neuroendocrine tissues using a specific monoclonal antibody. Neuroscience. 1987;22:149–157. doi: 10.1016/0306-4522(87)90205-3. [DOI] [PubMed] [Google Scholar]
- 54.Su D. Asard H. Three mammalian cytochromes b561 are ascorbate-dependent ferrireductases. FEBS J. 2006;273:3722–3734. doi: 10.1111/j.1742-4658.2006.05381.x. [DOI] [PubMed] [Google Scholar]
- 55.Sun Q. Zybailov B. Majeran W. Friso G. Olinares PD. van Wijk KJ. PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res. 2008;37(Database issue):D969–74. doi: 10.1093/nar/gkn654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Takeuchi F. Hori H. Obayashi E. Shiro Y. Tsubaki M. Properties of two distinct heme centers of cytochrome b561 from bovine chromaffin vesicles studied by EPR, resonance Raman, and ascorbate reduction assay. J Biochem. 2004;135:53–64. doi: 10.1093/jb/mvh006. [DOI] [PubMed] [Google Scholar]
- 57.Terland O. Flatmark T. Oxidoreductase activities of chromaffin granule ghosts isolated from the bovine adrenal medulla. Biochim Biophys Acta. 1980;597:318–330. doi: 10.1016/0005-2736(80)90109-1. [DOI] [PubMed] [Google Scholar]
- 58.Tsubaki M. Kobayashi K. Ichise T. Takeuchi F. Tagawa S. Diethyl pyrocarbonate modification abolishes fast electron accepting ability of cytochrome b561 from ascorbate but does not influence electron donation to monodehydroascorbate radical: identification of the modification sites by mass spectrometric analysis. Biochemistry. 2000;39:3276–3284. doi: 10.1021/bi991883d. [DOI] [PubMed] [Google Scholar]
- 59.Tsubaki M. Takeuchi F. Nakanishi N. Cytochrome b561 protein family: expanding roles and versatile transmembrane electron transfer abilities as predicted by a new classification system and protein sequence motif analyses. Biochim Biophys Acta. 2005;1753:174–190. doi: 10.1016/j.bbapap.2005.08.015. [DOI] [PubMed] [Google Scholar]
- 60.VanDuijn MM. Tijssen K. VanSteveninck J. Van Den Broek PJ. Van Der Zee J. Erythrocytes reduce extracellular ascorbate free radicals using intracellular ascorbate as an electron donor. J Biol Chem. 2000;275:27720–27725. doi: 10.1074/jbc.M910281199. [DOI] [PubMed] [Google Scholar]
- 61.Vargas JD. Herpers B. McKie AT. Gledhill S. McDonnell J. van den Heuvel M. Davies KE. Ponting CP. Stromal cell-derived receptor 2 and cytochrome b561 are functional ferric reductases. Biochim Biophys Acta. 2003;1651:116–123. doi: 10.1016/s1570-9639(03)00242-5. [DOI] [PubMed] [Google Scholar]
- 62.Verelst W. Asard H. Analysis of an Arabidopsis thaliana protein family, structurally related to cytochromes b561 and potentially involved in catecholamine biochemistry in plants. J Plant Physiol. 2004;161:175–181. doi: 10.1078/0176-1617-01064. [DOI] [PubMed] [Google Scholar]
- 63.Zamocky M. Ludwig R. Peterbauer C. Hallberg BM. Divne C. Nicholls P. Haltrich D. Cellobiose dehydrogenase—a flavocytochrome from wood-degrading, phytopathogenic and saprotropic fungi. Curr Protein Pept Sci. 2006;7:255–280. doi: 10.2174/138920306777452367. [DOI] [PubMed] [Google Scholar]
- 64.Zhang DL. Su D. Bérczi A. Vargas A. Asard H. An ascorbate-reducible cytochrome b561 is localized in macrophage lysosomes. Biochim Biophys Acta. 2006;1760:1903–1913. doi: 10.1016/j.bbagen.2006.07.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.