Activating mutations in FLT3 (Fms-like tyrosine kinase 3) by internal tandem duplication (ITD) mutations are found in approximately 30% of patients with acute myeloid leukemia (AML) and are associated with poor outcome in this patient population. Numerous FLT3 inhibitors have been tested for the treatment of AML, but these inhibitors have shown variable responses that were attributed to heterogeneity in AML and mechanistic differences associated with inhibitory mechanism employed by these inhibitors.1 Nevertheless, many patients showed a reduction in blast counts and hematological improvements, but these remissions were not durable that questioned the validity of FLT3 as a therapeutic target in AML. To address this, an elegant translational study designed by Neil Shah's group has identified the AC220 resistant mutations in all FLT3-ITD relapsed patients that definitively demonstrated the validity of FLT3-ITD as a therapeutic target in human AML.2 Furthermore, the emergence of resistant mutations has been reported from the relapsed AML patients with FLT3-ITD treated with PKC4123 and sorafenib.4 In addition, an in vitro resistant screening identified mutation at gatekeeper residue conferred cross-resistance to all known FLT3 inhibitors.5 These observations suggest that secondary mutations conferring resistance in the kinase domain will pose a significant clinical challenge, which prompted us to identify new inhibitors against the FLT3 resistant variants.
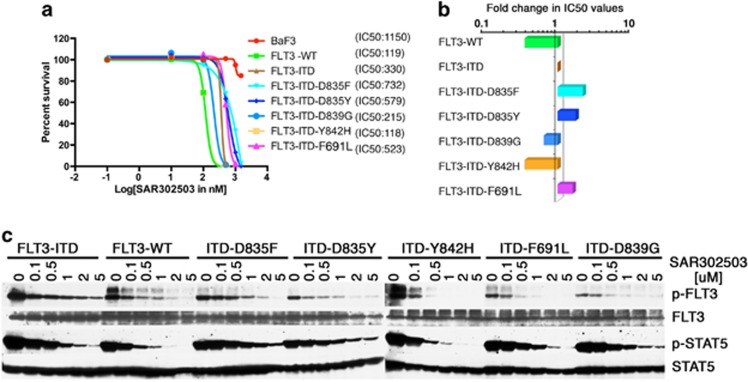
SAR302503 is a rationally designed small-molecule ATP-competitive inhibitor of JAK2, and it had been shown to have a high degree of kinase selectivity for JAK2 and FLT3 in in vitro kinase assays.6, 7 Given its equal potency toward FLT3 prompted us to test its efficacy against FLT3-ITD variants resistant to AC220. SAR302503 inhibited proliferation of BAF3 cells expressing FLT3 wild type and FLT3-ITD, with IC50 values of 119 and 330 nM, respectively (Figure 1a), whereas parental BAF3 cells and BAF3-JAK2-V617F cells were inhibited at IC50 values of ∼1100 and 600 nM, respectively (Supplementary Figure 1a). In accordance to this, western blotting of phospho-STAT5 and phospho-FLT3 showed reduced phosphorylation at concentrations that parallel the concentrations required to inhibit cell proliferation (Figure 1c). These observations suggest that SAR302503 is more selective to FLT3-WT>FLT3-ITD>JAK2-V617F, and it can be exploited for therapeutic targeting of FLT3-ITD in AML (Supplementary Figure 1a). Next, we analyzed the activity of SAR302503 against five different kinase domain variants of FLT3-ITD that had been shown to confer resistance against AC220. The growth of BAF3 cells expressing resistant variants of FLT3-ITD was inhibited completely within the range of 800 nM (Figure 1a). Interestingly, two FLT3-ITD variants (D839G and Y842H) were found to be hypersensitive to the drug (Figures 1a and b) while mutations at gatekeeper residue F691L and D835F/Y showed only twofold resistance to the drug (Figure 1b). Next, we performed in vitro resistant screening as described previously,8 to identify patterns of drug resistance using cells expressing FLT3-ITD, FLT3-ITD-F691L and FLT3-ITD-D835Y at 3 μM (Cmax value in human6, 9) of SAR302503. Surprisingly, we could not see the emergence of resistant clones against SAR302503 at this concentration (Supplementary Figure 1b). To rule out any off-target effect, we performed screening of BAF3 cells expressing Tel-JAK3 (having a cellular IC50 value of 2300 nm) at 2000 and 3000 nm of SAR302503, which showed growth of innumerable colonies (Supplementary Figure 1b). These observations suggest that the SAR302503 is a potent inhibitor of FLT3-ITD and is very effective against AC220 resistant variants and prevents the emergence of resistant clones.
Figure 1.
SAR302503 can efficiently inhibit the proliferation of native and AC220-resistant variants of FLT3-ITD. (a) Dose–response sigmoidal curve showing the cellular proliferation of BAF3 cells expressing FLT3 and its kinase domain AC220 resistant variants at different concentrations of SAR302503. IC50 values for each cell lines are indicated in parenthesis. (b) Bar graph showing relative fold difference in the IC50 values of FLT3 mutants normalized to native FLT3-ITD. (c) Immunoblot analysis showing the kinase activity of FLT3 and its kinase domain drug resistant variants against SAR302503 at different concentrations. Blots were probed using anti-phospho-FLT3, anti-phospho-STAT5, anti-FLT3 and anti-STAT5 antibody on the crude lysates from BAF3 cells expressing FLT3 and its drug resistant variants. Please note the reduced autophosphorylation in variants ITD-D835F/Y, ITD-F691L and ITD-D839G, whereas ITD-D842H showing increased autophosphorylation in untreated samples as compared with native FLT3-ITD.
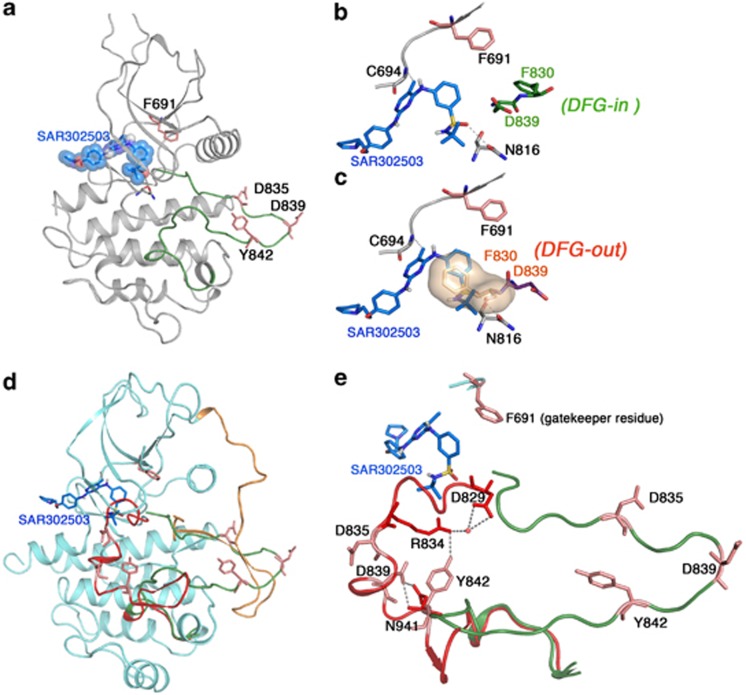
To understand the structural basis of SAR302503-mediated inhibition of FLT3 kinase, we constructed a model of the SAR302503-FLT3 kinase structure using a combination of homology modeling and induced-fit docking.10 We used the crystal structure of FLT3 crystallized in an inactive conformation and with related inhibitor−kinase complexes: for example, ABL complexed with nilotinib in closed and inactive conformation (PDB:3CS9); Ppy-A bound to the active conformation of ABL-T315I (PDB:2Z60); and JAK2 bound to isoquinoline in active conformation (PDB:2B7A). Our in silico analysis predicts that SAR302503 binds to an open and enzymatically active conformation of FLT3, in which residue Phe 830 of the DFG motif is displaced to accommodate the benzensulfonamide ring of the inhibitor (Figure 2). In this model, the pyrimidine ring of SAR302503 makes hydrophobic contacts with Leu 616 and Leu 818. Similar to other ATP-competitive inhibitors, SAR302503 anchors to the ATP site by two hydrogen bonds with residues Cys 694 from the kinase hinge region and Asn 816 from the catalytic loop (Figures 2a and b). Our model suggests that the enhanced sensitivity of SAR302503 for the variants D839G and Y842H is due to the destabilization of the inactive conformation by these mutations that in effect would stabilize the open and active conformation to which the drug preferentially binds, thus conferring hypersensitive response (Figures 2d and e). Given the lack of direct interaction of SAR302503 with resistant variants F691L and D835F/Y, our model suggests that these mutations would destabilize the open and active conformation rather than inactivating the kinase, therefore having sufficient catalytic activity to support cellular transformation. In support of this model, a substitution of Leucine for Phe 691 will weaken the hydrophobic spine that in effect will destabilize the active conformation and may weaken the kinase.11 However, substitution of Asp 835 with phenylalanine will destabilize the activation loop from both active and inactive conformation suggesting that it is stabilizing an intermediate conformational state.12 In inactive conformation, Asp 835 interacts with Ser 838 to stabilize the activation loop while it interacts with Gln 667 to stabilize the activation loop in open and active state, a similar interaction has been observed for the stabilization of ABL kinase in active conformation. In support, western blotting of FLT-3 autophosphorylation of these mutants, F691L and D835F/Y, from the untreated total cell extracts showed reduced autophosphorylation of FLT3 in comparison with FLT3-ITD (Figure 1c), thus supporting the notion that these mutations confer resistance by destabilizing the active state rather than direct steric hindrance to the drug.
Figure 2.
SAR302503 binds to ATP site, an active conformation of the FLT3 kinase. (a) Ribbon depiction of a structural model of FLT3-SAR302503 using the coordinates of the inactive FLT3 kinase (PDB:3CS9), ABL (PDB:2Z60) and JAK2 (PDB:2B7A). Activation loop is shown as green and extended conformation during active state. (b) A close-up view of the active site of the FLT3 kinase showing the interaction of SAR302503 by hydrogen bonding with Cys 694 and Asn 816. Further, SAR302503 can bind only in DFG-in conformational state (residues shown in green sticks), in which phenylalanine of the DFG motif is aligned in such a way that it coordinates with hydrophobic spine residues to stabilize the active state, a common feature in all kinases suggesting that SAR302503 can only bind to active state and thus can be classified as a type I inhibitor. (c) Active site showing the steric clash of the benzene sulfonamide group of SAR302503 with Phe 830 in DGF-out conformation (F830 from the DFG motif shown in surface). (d) Structure of autoinhibited FLT3 shown in ribbon with modeled activation loops in active conformation (shown in green) and in inactive conformation (shown in red). The juxtamembrane domain (orange) nearly spans the length of the kinase molecule and seemingly displaces the activation loop from the active state to favor inactive conformation. Amino-acid residues that were mutated to confer drug resistance are shown as sticks. (e) Conformation of activation loops in active (green) and inactive (red) states showing interaction of Y842 with D829 (DFG motif) through R834 and D839 with N934 to stabilize the inactive conformation, and thus mutation at these sites may favor or stabilize active state that will be more sensitive to inhibition by type I inhibitors. In support of this model, variant Y842H showing increased autophosphorylation in untreated sample, whereas D839G did not show significant change suggesting that it only destabilizes the inactive conformation and may be unable to stabilize the fully active conformation as we have observed in the Y842H. Nonetheless, it supports that mutant D839G stabilizes a conformation favoring efficient SAR302503 binding that reflects in hypersensitive response. The hypersensitive response of variant D839G is less than Y842H suggesting that the later is stabilizing a fully active state that binds the drug more efficiently. Nevertheless, these speculations require detail structural studies for better understanding and developing next-generation FLT3 inhibitors.
A recent study has shown the efficacy of Ponatinib on AC220 resistant mutations in F691L/I variant. However, it is ineffective against compound mutations and variants from the activation loop (D835F/Y), suggesting that these variants will become significant clinical challenge.13 Our study demonstrates that the SAR302503 is potently active against these resistant variants and prevents the emergence of resistant clones at Cmax (3 μM) of SAR302503.6, 9 Given the preclinical success of SAR302503 as a well-tolerated oral agent for the treatment of polycythemia vera and Myelofibrosis, its selectivity against tyrosine-kinase inhibitor (TKI) resistant variants of FLT3-ITD supports the clinical evaluation of SAR302503 in TKI-treated as well as TKI naive FLT3-ITD+ AML patients. To that end, SAR302503 may represent an effective first-line TKI therapy in AML patients. In addition, a lack of in vitro resistance against SAR302503 suggests that it will be more effective in managing the clinical resistance.
Acknowledgments
We are thankful to George Daley and Gary Gilliland for providing the FLT3 and FLT3-ITD retroviral constructs. This study was supported by grants to MA from the National Cancer Institutes at NIH (1RO1CA155091) and the Leukemia Research Foundation. MA is a recipient of the V-Scholar award from the V-Foundation.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Leung AY, Man CH, Kwong YL. FLT3 inhibition: a moving and evolving target in acute myeloid leukaemia. Leukemia. 27:260–268. doi: 10.1038/leu.2012.195. [DOI] [PubMed] [Google Scholar]
- Smith CC, Wang Q, Chin CS, Salerno S, Damon LE, Levis MJ, et al. Validation of ITD mutations in FLT3 as a therapeutic target in human acute myeloid leukaemia. Nature. 485:260–263. doi: 10.1038/nature11016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidel F, Solem FK, Breitenbuecher F, Lipka DB, Kasper S, Thiede MH, et al. Clinical resistance to the kinase inhibitor PKC412 in acute myeloid leukemia by mutation of Asn-676 in the FLT3 tyrosine kinase domain. Blood. 2006;107:293–300. doi: 10.1182/blood-2005-06-2469. [DOI] [PubMed] [Google Scholar]
- Man CH, Fung TK, Ho C, Han HH, Chow HC, Ma AC, et al. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood. 119:5133–5143. doi: 10.1182/blood-2011-06-363960. [DOI] [PubMed] [Google Scholar]
- Williams AB, Nguyen B, Li L, Brown P, Levis M, Leahy D, et al. Mutations of FLT3/ITD confer resistance to multiple tyrosine kinase inhibitors. Leukemia. 2013;27:48–55. doi: 10.1038/leu.2012.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernig G, Kharas MG, Okabe R, Moore SA, Leeman DS, Cullen DE, et al. Efficacy of TG101348, a selective JAK2 inhibitor, in treatment of a murine model of JAK2V617F-induced polycythemia vera. Cancer Cell. 2008;13:311–320. doi: 10.1016/j.ccr.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Hood J, Lasho T, Levine RL, Martin MB, Noronha G, et al. TG101209, a small molecule JAK2-selective kinase inhibitor potently inhibits myeloproliferative disorder-associated JAK2V617F and MPLW515L/K mutations. Leukemia. 2007;21:1658–1668. doi: 10.1038/sj.leu.2404750. [DOI] [PubMed] [Google Scholar]
- Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112:831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- Pardanani A, Gotlib JR, Jamieson C, Cortes JE, Talpaz M, Stone RM, et al. Safety and efficacy of TG101348, a selective JAK2 inhibitor, in myelofibrosis. J Clin Oncol. 29:789–796. doi: 10.1200/JCO.2010.32.8021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman W, Day T, Jacobson MP, Friesner RA, Farid R. Novel procedure for modeling ligand/receptor induced fit effects. J Med Chem. 2006;49:534–553. doi: 10.1021/jm050540c. [DOI] [PubMed] [Google Scholar]
- Azam M, Seeliger MA, Gray NS, Kuriyan J, Daley GQ. Activation of tyrosine kinases by mutation of the gatekeeper threonine. Nat Struct Mol Biol. 2008;15:1109–1118. doi: 10.1038/nsmb.1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azam M, Nardi V, Shakespeare WC, Metcalf CA, 3rd, Bohacek RS, Wang Y, et al. Activity of dual SRC-ABL inhibitors highlights the role of BCR/ABL kinase dynamics in drug resistance. Proc Natl Acad Sci USA. 2006;103:9244–9249. doi: 10.1073/pnas.0600001103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catherine C., Smith EAL, Zhu Xiaotian, Lin KimberlyC., Stewart WhitneyK., Lauren E Damon, et al. Activity of ponatinib against clinically-relevant AC220-resistant kinase domain mutants of FLT3-ITD. Blood. 2013;121:3165–3171. doi: 10.1182/blood-2012-07-442871. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.