Abstract
Overexpression of the classical homeobox transcription factor HOXC6 is frequent in prostate cancers and correlates with adverse clinical parameters. Since surprisingly many HOXC6 target genes are downregulated in prostate cancer, it has been posited that oncogenic effects of HOXC6 in prostate cancer may be unmasked by concurrent epigenetic downregulation of target genes exerting tumor suppressive effects. To test this hypothesis, we have studied the expression of three HOXC6 target genes, CNTN1 (encoding a cell adhesion protein), DKK3 and WIF1 (encoding WNT growth factor antagonists) as well as DNA methylation of DKK3 and WIF1. HOXC6 upregulation and association with poor prognosis were confirmed in our tissue series. The three target genes were each significantly downregulated in cancer tissues and expression of each one correlated inversely with that of HOXC6. Cases with lower WIF1 expression showed significantly earlier recurrence (p = 0.021), whereas no statistical significance was reached for CNTN1 and DKK3. Hypermethylation of DKK3 or WIF1 gene promoters was observed in a subset of cancers with downregulated expression, but was often weak. Our data support the hypothesis that HOXC6 target genes exerting tumor-suppressive effects are epigenetically downregulated in prostate cancer, but DNA methylation appears to follow or bolster rather than to cause their transcriptional inactivation.
Keywords: prostate cancer, epigenetic silencing, DNA methylation, homeobox transcription factors, WNT signaling
1. Introduction
Prostate cancer is distinguished by a profusion of epigenetic alterations, which include consistent hypermethylation of several genes, frequent hypermethylation of numerous others, genome-wide hypomethylation of repeat sequences, changes in histone modifications and altered expression of chromatin regulatory factors [1,2]. Some of these epigenetic changes, notably hypermethylation of genes like GSTP1, appear to be associated with earlier stages of tumor development, whereas others are rather associated with tumor progression. The latter changes include hypermethylation of additional genes, hypomethylation of retroelements and overexpression of the histone methyltransferase EZH2. Accordingly, there is considerable interest in exploiting epigenetic changes associated with early development for the detection of prostate cancer on the one hand and alterations associated with progression for classification, molecular staging and prognostic purposes on the other hand [3,4].
Among the prominent targets of epigenetic alterations in human cancers are classical HOX genes encoding transcription factors regulating cell fate and differentiation [5,6]. These genes are located in four clusters. Whereas normal prostate expresses predominantly posterior genes from the A and B clusters, genes from the C and D clusters become activated in cancer tissues [7-12]. Both the causes and consequences of cancer-associated changes in HOX gene expression are insufficiently understood. Nevertheless, it is now well established that individual classical HOX genes can act as oncogenes or tumor suppressors in various human cancers [6,13].
In the prostate, specifically, there is convincing evidence for an oncogenic function of HOXC6. Its mRNA and protein have been found to be strongly overexpressed in many prostate cancers compared to their low expression in benign tissues [7,10,14,15]. The degree of HOXC overexpression parallels several clinical parameters of tumor progression, including Gleason scores [10,14,16]. Analyses of genes affected by HoxC6 knockout in murine prostates or by up- or downregulation of HOXC6 in human prostatic cells identified targets in the WNT and Notch signaling pathways as well as BMP7, FGFR2 and PDGFRA [15]. These target genes are upregulated by HOXC6 and could plausibly mediate its effects on prostate cancer progression and metastasis. Curiously, however, about half of the genes positively regulated by HOXC6 in experimental models are actually downregulated in prostate cancers, including three genes encoding inhibitors of WNT signaling, WIF1, DKK3 and SFRP1 [15]. Moreno [17] has proposed an elegant explanation for this apparent discrepancy. According to this hypothesis, HOXC6 can activate both target genes promoting and preventing prostate cancer progression, but epigenetic inactivation of its tumor-suppressive targets would restrict its effect to cancer-promoting genes. Indeed, all three WNT inhibitor genes have been reported to be downregulated or hypermethylated in prostate cancer [18-26]. Therefore, DNA methylation of these genes may prevent their activation by HOXC6. However, this hypothesis has not been investigated explicitly by studying expression of HOXC6 together with methylation and expression of these target genes in prostatic tissue samples.
Here we report an expression analysis of HOXC6 and three of its target genes in a well-characterized series of prostate cancer tissues. Our data confirm the reported correlation of HOXC6 expression with clinical parameters of prostate cancer progression. As predicted, WIF1 and DKK3 downregulation was related to HOXC6 overexpression. Both genes were hypermethylated in some prostate cancer samples, but their hypermethylation was not well correlated with downregulation. Likewise, a third HOXC6 target gene, CNTN1, was concordantly downregulated. Taken together, our data suggest that downregulation of HOXC6 target genes are often accompanied by DNA methylation but can occur independently of this epigenetic modification.
2. Results and Discussion
2.1. Expression of HOXC6 in Prostate Cancer Tissues
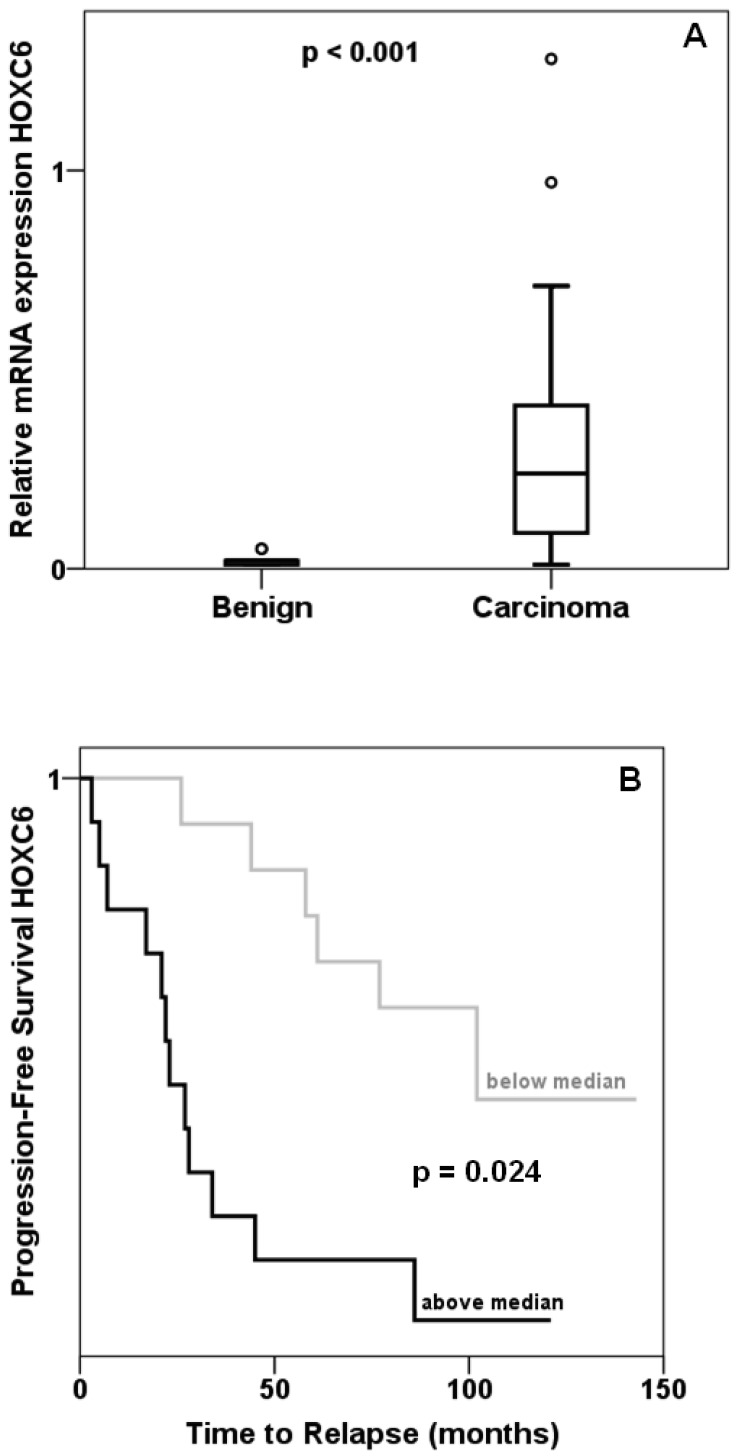
Expression of HOXC6 was determined by quantitative RT-PCR in 45 prostate cancer and 13 benign tissues collected from prostatectomies. The majority of cancer tissues displayed—often grossly— elevated levels of HOXC6 mRNA resulting in an overall highly significant difference compared to benign tissues (Figure 1A). As reported by others, cancers with high HOXC6 expression had significantly higher T stage, had more often spread to lymph nodes and were assigned higher Gleason scores. Expression of MKI67 encoding the proliferation marker Ki67 was likewise enhanced in these cases (Mann-Whitney test: p = 0.004). Cancers with above median HOXC6 expression recurred significantly (log-rank p = 0.024) earlier than cancers with below median expression (Figure 1B). These data confirm previous reports on frequent HOXC6 overexpression in prostate cancer [7,10,14,15] and the association of increasing HOXC6 overexpression with adverse clinical parameters.
Figure 1.
Expression of HOXC6 in prostate cancer. (A) Expression of HOXC6 mRNA as measured by quantitative RT-PCR in 45 prostate carcinoma and 13 benign prostate tissues; (B) Kaplan-Meier analysis of effect of HOXC6 expression on biochemical recurrence.
2.2. Expression of Presumed HOXC6 Target Genes in Prostate Cancer Tissues
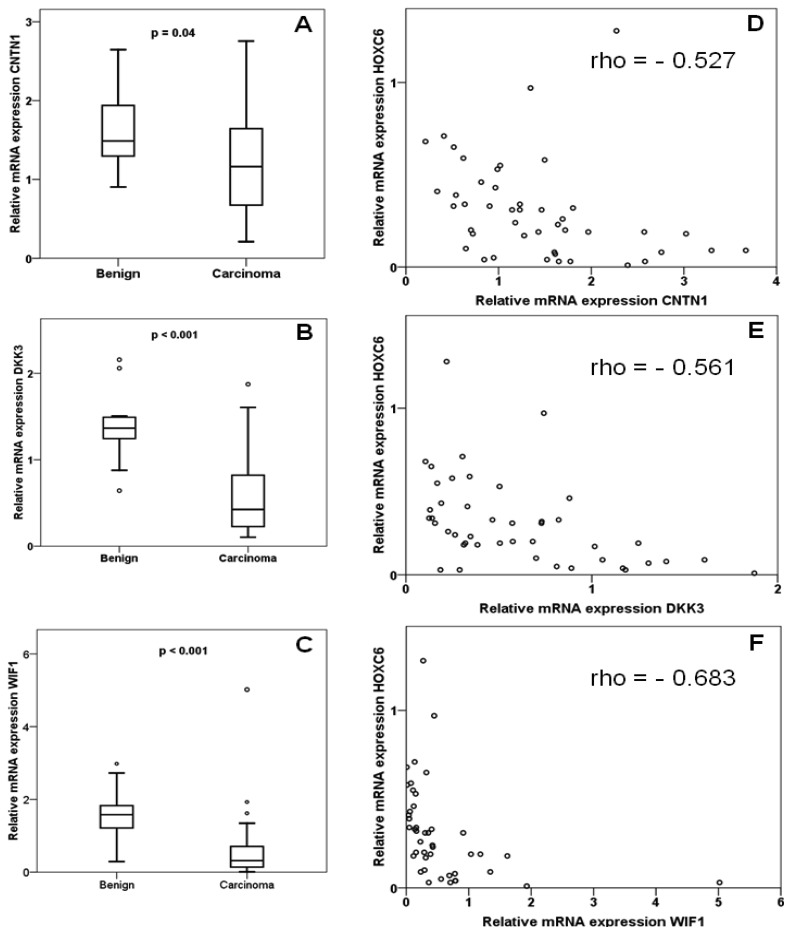
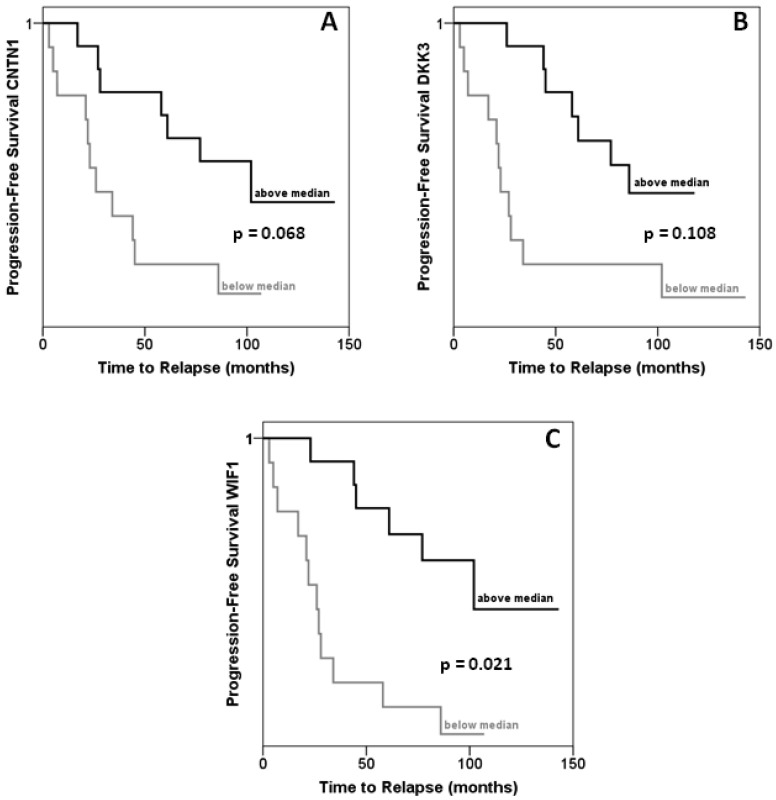
In the same set of samples, expression of DKK3, WIF1 and CNTN1 was observed to be significantly decreased (Figures 2A–C). Expression of each gene correlated inversely with that of HOXC6 in a statistically significant (each p < 0.001) manner (Figures 2D–F). Accordingly, expression of each target gene correlated significantly positively with that of each other, with Spearman rho coefficients between 0.4 and 0.6. Cases with lower than median expression of each target gene, CNTN1, DKK3 or WIF1, showed earlier recurrence, but the association was only significant at the p < 0.05 level for WIF1 (Figures 3A–C). In addition, low WIF1 expression was significantly associated with lymph node involvement (p = 0.036) and higher Gleason scores (p = 0.026), but not with tumor stage (pT2 vs. pT3). Expression of CNTN1 or DKK3 was not significantly associated with any histopathological parameter in our series.
Figure 2.
Expression of HOXC6 target genes in prostate cancer. (A) Expression of CNTN1 mRNA as measured by quantitative RT-PCR in 45 prostate carcinoma and 13 benign prostate tissues; (B) Expression of DKK3 mRNA in the same set of tissues; (C) Expression of WIF1 mRNA in the same set of tissues; (D–F) Plots of CNTN1, DKK3 and WIF1 expression against HOXC6 expression. In each case, there was a highly significant (p < 0.001) inverse correlation.
Figure 3.
Relation of HOXC6 target gene expression to prostate cancer recurrence. Kaplan-Meier analysis of relation of CNTN1 (A), DKK3 (B) and WIF1 (C) expression to progression-free survival, measured as biochemical recurrence. Samples were stratified by median for each gene.
Our measurements confirm the reported downregulation of the WNT factor antagonists DKK3 and WIF1 in prostate cancer [18,21-23,25,26]. In addition, our data hint at an association of stronger WIF1 downregulation with worse prognosis. Most importantly, our study demonstrates for the first time explicitly that expression of certain target genes is inversely correlated with overexpression of HOXC6. This is also the first report on CNTN1 in prostate cancer. The gene encodes a member of the contactin family which serves as a membrane receptor for chondroitin sulfate and regulates receptor tyrosine phosphatases. The function of contactin 1 has mainly been studied in neuronal and glial cells, where it regulates cell-cell and cell-substrate adhesion. Two studies in lung cancers and gliomas suggest that this function may also be relevant for cancer cell invasion and metastasis [27,28]. Investigations on the function of contactin 1 in prostate cancer and of the epigenetic regulation of its complex gene might therefore be rewarding.
2.3. Methylation of Presumed HOXC6 Target Genes in Prostate Cancer Tissues
Methylation of DKK3 and WIF1 promoter CpG-islands was analyzed by methylation-specific PCR as described in previous publications [21,29]. Methylation of DKK3 was observed in 16 of the 92 carcinoma samples, but not in benign controls. It was significantly more frequent in cases with lymph node involvement and significantly less frequent in cases with Gleason score < 6 (χ2 test, p < 0.05). However, expression of DKK3 was not significantly different between carcinoma samples with or without hypermethylation. WIF1 methylation was more prevalent, being detectable in 31 of 92 carcinoma samples, but also in eight of 17 benign controls. The latter finding may relate to previous findings [18] suggesting that WIF1 downregulation may commence at early stages of prostate cancer. As for DKK3, WIF1 hypermethylation and the extent of downregulation of expression were not significantly related to each other. Because the bands obtained in the WIF1 MS-PCR assay with the methylated-specific primers were often weak (except in LNCaP cells used as a positive control), bisulfite sequencing was conducted across the region interrogated by the assay in prostate tissue samples and controls (Figure 4). The analyzed part of the WIF1 CpG-island was completely unmethylated in blood leukocytes, but was quite densely methylated in the prostate cancer cell line LNCaP which lacks WIF1 expression. In contrast, only occasional sites were methylated in the expressing 22Rv1 line. In all benign and carcinoma tissues, only patchy and weak methylation was found, as suggested by the results of the MS-PCR assay. Of note, we have previously shown that in this set of prostate cancer tissues GSTP1, EPB41L3 and several other genes are each hypermethylated at frequencies of 60%–80%, often displaying dense methylation [19,30]. Thus, DKK3 and WIF1 hypermethylation was much less widespread than downregulation and was often weak if it occurred.
Figure 4.
WIF1 methylation in prostate cancer. Bisulfite sequencing analysis of WIF1 promoter methylation in blood leukocytes, prostate cancer cell lines with high (22Rv1) and low (LNCaP) expression, benign and carcinoma prostate tissues. Each line represents one cloned PCR product, each circle represents one CpG site. Dark circles indicate methylated and light circles unmethylated sites. Some sites at the 3′-end of the sequence were difficult to read and are labeled by x.
3. Experimental Section
3.1. Patients and Tissue Samples
High quality RNA was prepared from 13 normal prostate tissues from cancer-carrying prostates and 45 carcinomas from patients aged 59–74 years undergoing total prostatectomy as described [19]. According to the IUAC 2007 TNM classification, tumors were staged as pT2 in 20, pT3 in 23 and pT4 in 2 cases. A Gleason score of 7 was detected in 26 tumors, < 7 in 13 tumors and > 7 in 6 tumors. None of the patients had distant metastases, but 11 cancers had spread to local lymph nodes. High quality DNA was available from 92 cancer tissues encompassing the specimens used for RNA analysis. Of these, 43 were staged as pT2 and 49 as pT3 or pT4. Sixteen patients had lymph node metastases, but none distant metastases. Each 27 carcinomas were assigned a Gleason score > 7 or < 7 and 38 a score of 7. The median follow-up period was 98 months. The study was approved by the ethics committee of the Heinrich Heine University medical faculty.
3.2. RNA Extraction and Quantitative RT-PCR
Total RNA was isolated from preconfluent cells using the RNeasy ® Mini Kit (Qiagen, Hilden, Germany). Two μg RNA were reversed transcribed using SuperscriptII (Invitrogen, Karlsruhe, Germany) with oligo-dT primers according to the manufacturer's protocol. Quantitative real-time PCR for CNTN1, DKK3, WIF1, and the reference gene TBP was performed using SYBR-Green reaction mix (Qiagen) with 0.4 μM of each primer in an ABI Prism 7900HT instrument (Applied Biosystems, Darmstadt, Germany) with the following conditions: activation at 95 °C for 15 minutes followed by 45 cycles of denaturation at 94 °C for 15 s, elongation at 72 °C for 30 s and measuring at 88 °C for 15 s. Amplification lasted 30 s with temperatures at 55 °C for CNTN1, 57 °C for DKK3 and WIF1, 61 °C for TBP. The quality of the amplification was assured by a melting curve established by incubation at 95 °C, 60 °C and 99 °C for 15 s each. HOXC6 expression was measured by Taqman assays Hs00171690 mL (Applied Biosystems). Duplicate measurements gave < 10% difference. For each gene, a standard curve was constructed using a reference cell line with high expression and expression in the samples was expressed relative to this standard. The same procedure was performed for the TBP control, to which the measurements were then adjusted.
3.3. DNA Extraction and Methylation Analyses
High-quality DNA was extracted from tissues and prostate carcinoma cell lines as described [19]. The EZ DNA Methylation Kit (Zymo Research, Orange, CA, USA) was used for bisulfite conversion. PCRs were performed in a 50 μL reaction mixture consisting of 1 × buffer, 150 μM dNTPs, 15 pmol of each primer, 1 U Hotstar Taq polymerase, water and 2 μL bisulfite-converted DNA each. The following program was used: initial Taq activation at 94 °C for 15 min, followed denaturation at 95 °C for 30 s, annealing for 30 s, elongation at 72 °C for 45 s, with a final elongation for 10 min. MS-PCR amplification was performed for DKK3 unmethylated/methylated for 36/38 cycles with annealing at 61/65 °C, for WIF1 unmethylated/methylated MS-PCR amplification was performed for 34/36 cycles with annealing at 53 °C for both genes. Fully methylated and completely methylated controls were carried in each reaction as described [19]. For WIF1 bisulfite sequencing PCR was conducted for 35 cycles with annealing at 60 °C. PCR products were examined by agarose gel electrophoresis. For bisulfite sequencing PCR products were cloned into the E. coli TOPO 10 vector (TOPO TA Cloning Kit, Invitrogen). Four clones from each sample were sequenced using standard methods. Primer sequences are compiled in Table 1.
Table 1.
Primers Used.
| Primer names | Forward 5′ → 3′ | Reverse 5′ → 3′ | Product size (bp) | Reference |
|---|---|---|---|---|
| DKK3-MS-PCR (meth.) | GGG GCG GGC GGC GGG GC | ACA TCT CCG CTC TAC GCC CG | 120 | [29] |
| DKK3-MS-PCR (unmeth.) | TTA GGG GTG GGT GGT GGG GT | CTA CAT CTC CAC TCT ACA CCC A | 125 | [29] |
| DKK3-qRT-PCR | TTG CCA GCT TCC AGT ACA CC | TGC AGT GAC CCC AGA CAC A | 105 | self-designed |
| WIF1-MS-PCR (meth.) | CGT TTT ATT GGG CGT ATC GT | ACT AAC GCG AAC GAA ATA CGA | 145 | [29] |
| WIF1-MS-PCR (unmeth.) | GGG TGT TTT ATT GGG TGT ATT GT | AAA AAA ACT AAC ACA AAC AAA ATA CAA AC | 154 | [29] |
| WIF1-qRT-PCR | TAA TGG AGG GAC CTG TTT CTA CC | CCA TTT CGA CAG GGT TGT G | 102 | self-designed |
| WIF1-Bisulfite sequencing-PCR | GTT TTA GGG GTT TTT GAG TGT T | CAA CTC CCT CAA CCA AAA CTA | 463 | [31] |
3.4. Statistical Methods
Statistical calculations were performed using SPSS 19.0.
4. Conclusions
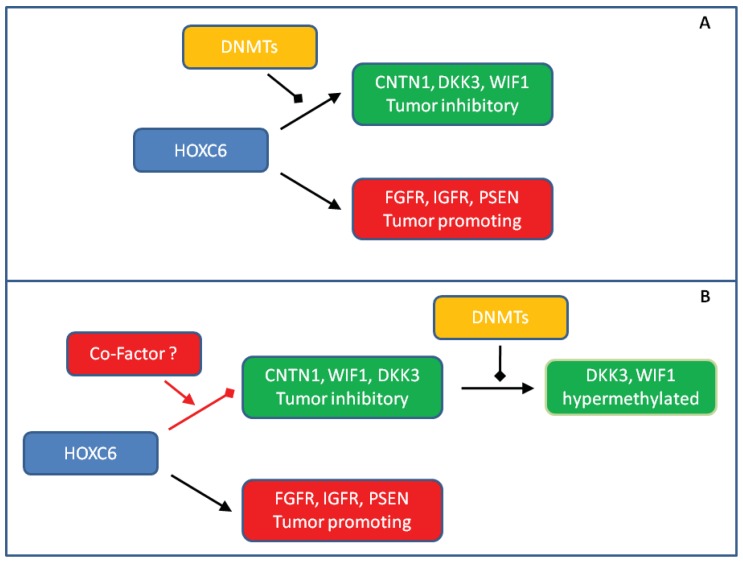
We observed striking HOXC6 overexpression in our small, but well-characterized prostate cancer tissue series. Our findings underscore that the previously described strong association of HOXC6 overexpression with adverse clinical parameters [7,10,14,15] is robust and deserves to be explored for the purposes of developing prognostic and molecular staging biomarkers. Our confirmation of these relationships accentuates the question of how HOXC6 contributes to oncogenesis and tumor progression in the prostate. Obviously, the answer is not trivial. Since HOXC6 is well-established as a transcription activator, it is most likely to act by regulating gene expression. Indeed, a range of positively regulated target genes have been defined by various experimental approaches. As reviewed by Moreno [17], among them are plausible candidates for promoting oncogenesis in the prostate, e.g., through WNT, BMP, FGF and NOTCH signaling pathways. However, it seems paradoxical that several genes activated by HOXC6 in experimental settings would be expected to counteract tumorigenesis in vivo, especially antagonists of WNT signaling like DKK3 and WIF1 [22,26,32]. These have indeed been reported to be downregulated or hypermethylated in prostate cancer in other studies [16,21,23]. One plausible explanation for the paradox is that genes antagonizing tumor development become hypermethylated in prostate by independent mechanisms and are no longer accessible to transcriptional activation by HOXC6 (Figure 5A).
Figure 5.
Hypotheses on the function of DNA methylation in inactivation of HOXC6 target genes in prostate cancer. (A) initial hypothesis based on ref. [14]; (B) hypothesis modified to explain the results of our study.
Several results from the present study argue against this hypothesis in its most simple form. First, although we observed DKK3 and WIF1 hypermethylation in some cases, it was relatively weak and transcriptional downregulation was more generalized. Of note, we have previously found that hypermethylation of another HOXC6 target, SFRP1, is also relatively rare in our tissue series, whereas around 80% of the cases harbor hypermethylated GSTP1, as expected [19]. According results have been published by others [25]. Second, hypermethylation was not well correlated with transcriptional downregulation and was also observed in many benign adjacent tissues in the case of WIF1. These findings argue that DNA methylation is not the primary cause of transcriptional silencing of these genes, but may rather follow and bolster their initial downregulation by other epigenetic mechanisms (Figure 5B).
Third, we found the strongest downregulation of CNTN1, DKK3 and WIF1 in cases with high HOXC6 expression suggesting that transcriptional silencing of these HOXC6 target genes might be elicited by the factor itself. This idea seems unlikely at first glance, since classical HOX factors act predominantly as transcriptional activators. There are however exceptions to the rule. For instance, HOX target genes may become repressed upon interactions with SMAD proteins that are activated by BMP signaling [33]. In order to explain our findings, we would like to propose the idea that the changed composition of transcription factors (and cofactors) in the nuclear milieu of prostate cancer cells may lead to a switch of HOXC6 function at some of its target genes (Figure 5B). An analogous case in prostate cancer is the altered target gene spectrum of the androgen receptor caused by changes in the expression of interacting transcription factors like HOXB13 and FOXA1 [34,35]. Accordingly, our data call for further detailed research on the HOXC6 paradox in prostate cancer which should yield important results for clinical application as well as insights into basic mechanisms of transcription.
Acknowledgements
We thank our lab members Christiane Hader and Christian Arsov for help and support as well as Rainer Engers for histopathological evaluation of the tissue samples used. We gratefully acknowledge financial support by the Deutsche Krebshilfe and Deutsche Forschungsgemeinschaft.
References and Notes
- 1.Nelson W.G., de Marzo A.M., Yegnasubramanian S. Epigenetic alterations in human prostate cancers. Endocrinology. 2009;150:3991–4002. doi: 10.1210/en.2009-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schulz W.A., Hoffmann M.J. Epigenetic mechanisms in the biology of prostate cancer. Semin. Cancer Biol. 2009;19:172–180. doi: 10.1016/j.semcancer.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Hoque M.O. DNA methylation changes in prostate cancer: Current developments and future clinical implementation. Expert Rev. Mol. Diagn. 2009;9:243–257. doi: 10.1586/erm.09.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perry A.S., Watson R.W., Lawler M., Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat. Rev. Urol. 2010;7:668–680. doi: 10.1038/nrurol.2010.185. [DOI] [PubMed] [Google Scholar]
- 5.Pfeifer G.P., Rauch T.A. DNA methylation patterns in lung carcinomas. Semin. Cancer Biol. 2009;19:181–187. doi: 10.1016/j.semcancer.2009.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shah N., Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10:361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 7.Waltregny D., Alami Y., Clausse N., de Leval J., Castronovo V. Overexpression of the homeobox gene HOXC8 in human prostate cancer correlates with loss of tumor differentiation. Prostate. 2002;50:162–169. doi: 10.1002/pros.10045. [DOI] [PubMed] [Google Scholar]
- 8.Economides K.D., Capecchi M.R. Hoxb13 is required for normal differentiation and secretory function of the ventral prostate. Development. 2003;130:2061–2069. doi: 10.1242/dev.00432. [DOI] [PubMed] [Google Scholar]
- 9.Hombría J.C., Lovegrove B. Beyond homeosis—HOX function in morphogenesis and organogenesis. Differentiation. 2003;71:461–476. doi: 10.1046/j.1432-0436.2003.7108004.x. [DOI] [PubMed] [Google Scholar]
- 10.Miller G.J., Miller H.L., van Bokhoven A., Lambert J.R., Werahera P.N., Schirripa O., Lucia M.S., Nordeen S.K. Aberrant HOXC expression accompanies the malignant phenotype in human prostate. Cancer Res. 2003;63:5879–5888. [PubMed] [Google Scholar]
- 11.Huang L., Pu Y., Hepps D., Danielpour D., Prins G.S. Posterior Hox gene expression and differential androgen regulation in the developing and adult rat prostate lobes. Endocrinology. 2007;148:1235–1245. doi: 10.1210/en.2006-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ke X.S., Qu Y., Rostad K., Li W.C., Lin B., Halvorsen O.J., Haukaas S.A., Jonassen I., Petersen K., Goldfinger N., et al. Genome-wide profiling of histone h3 lysine 4 and lysine 27 trimethylation reveals an epigenetic signature in prostate carcinogenesis. PLoS One. 2009;4:e4687. doi: 10.1371/journal.pone.0004687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen M.M., Abate-Shen C. Molecular genetics of prostate cancer: New prospects for old challenges. Genes Dev. 2010;24:1967–2000. doi: 10.1101/gad.1965810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ramachandran S., Liu P., Young A.N., Yin-Goen Q., Lim S.D., Laycock N., Amin M.B., Carney J.K., Marshall F.F., Petros J.A., Moreno C.S. Loss of HOXC6 expression induces apoptosis in prostate cancer cells. Oncogene. 2005;24:188–198. doi: 10.1038/sj.onc.1207906. [DOI] [PubMed] [Google Scholar]
- 15.McCabe C.D., Spyropoulos D.D., Martin D., Moreno C.S. Genome-wide analysis of the homeobox C6 transcriptional network in prostate cancer. Cancer Res. 2008;68:1988–1996. doi: 10.1158/0008-5472.CAN-07-5843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bibikova M., Chudin E., Arsanjani A., Zhou L., Garcia E.W., Modder J., Kostelec M., Barker D., Downs T., Fan J.B., et al. Expression signatures that correlated with Gleason score and relapse in prostate cancer. Genomics. 2007;89:666–672. doi: 10.1016/j.ygeno.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 17.Moreno C.S. The Sex-determining region Y-box 4 and homeobox C6 transcriptional networks in prostate cancer progression: Crosstalk with the Wnt, Notch, and PI3K pathways. Am. J. Pathol. 2010;176:518–527. doi: 10.2353/ajpath.2010.090657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wissmann C., Wild P.J., Kaiser S., Roepcke S., Stoehr R., Woenckhaus M., Kristiansen G., Hsieh J.C., Hofstaedter F., Hartmann A., et al. WIF1, a component of the Wnt pathway, is down-regulated in prostate, breast, lung, and bladder cancer. J. Pathol. 2003;201:204–212. doi: 10.1002/path.1449. [DOI] [PubMed] [Google Scholar]
- 19.Florl A.R., Steinhoff C., Müller M., Seifert H.H., Hader C., Engers R., Ackermann R., Schulz W.A. Coordinate hypermethylation at specific genes in prostate carcinoma precedes LINE-1 hypomethylation. Br. J. Cancer. 2004;91:985–994. doi: 10.1038/sj.bjc.6602030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Joesting M.S., Perrin S., Elenbaas B., Fawell S.E., Rubin J.S., Franco O.E., Hayward S.W., Cunha G.R., Marker P.C. Identification of SFRP1 as a candidate mediator of stromal-to-epithelial signaling in prostate cancer. Cancer Res. 2005;65:10423–10430. doi: 10.1158/0008-5472.CAN-05-0824. [DOI] [PubMed] [Google Scholar]
- 21.Lodygin D., Epanchintsev A., Menssen A., Diebold J., Hermeking H. Functional epigenomics identifies genes frequently silenced in prostate cancer. Cancer Res. 2005;65:4218–4227. doi: 10.1158/0008-5472.CAN-04-4407. [DOI] [PubMed] [Google Scholar]
- 22.Kawano Y., Kitaoka M., Hamada Y., Walker M.M., Waxman J., Kypta R.M. Regulation of prostate cell growth and morphogenesis by Dickkopf-3. Oncogene. 2006;25:6528–6537. doi: 10.1038/sj.onc.1209661. [DOI] [PubMed] [Google Scholar]
- 23.Zenzmaier C., Untergasser G., Hermann M., Dirnhofer S., Sampson N., Berger P. Dysregulation of Dkk-3 expression in benign and malignant prostatic tissue. Prostate. 2008;68:540–547. doi: 10.1002/pros.20711. [DOI] [PubMed] [Google Scholar]
- 24.Kawano Y., Diez S., Uysal-Onganer P., Darrington R.S., Waxman J., Kypta R.M. Secreted Frizzled-related protein-1 is a negative regulator of androgen receptor activity in prostate cancer. Br. J. Cancer. 2009;100:1165–1174. doi: 10.1038/sj.bjc.6604976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa V.L., Henrique R., Ribeiro F.R., Carvalho J.R., Oliveira J., Lobo F., Teixeira M.R., Jerónimo C. Epigenetic regulation of Wnt signaling pathway in urological cancer. Epigenetics. 2010;5:343–351. doi: 10.4161/epi.5.4.11749. [DOI] [PubMed] [Google Scholar]
- 26.Yee D.S., Tang Y., Li X., Liu Z., Guo Y., Ghaffar S., McQueen P., Atreya D., Xie J., Simoneau A.R., et al. The Wnt inhibitory factor 1 restoration in prostate cancer cells was associated with reduced tumor growth, decreased capacity of cell migration and invasion and a reversal of epithelial to mesenchymal transition. Mol. Cancer. 2010;9:162. doi: 10.1186/1476-4598-9-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eckerich C., Zapf S., Ulbricht U., Müller S., Fillbrandt R., Westphal M., Lamszus K. Contactin is expressed in human astrocytic gliomas and mediates repulsive effects. GLIA. 2006;53:1–12. doi: 10.1002/glia.20254. [DOI] [PubMed] [Google Scholar]
- 28.Su J.L., Yang C.Y., Shih J.Y., Wei L.H., Hsieh C.Y., Jeng Y.M., Wang M.Y., Yang P.C., Kuo M.L. Knockdown of contactin-1 expression suppresses invasion and metastasis of lung adenocarcinoma. Cancer Res. 2006;66:2553–2561. doi: 10.1158/0008-5472.CAN-05-2645. [DOI] [PubMed] [Google Scholar]
- 29.Veeck J., Wild P.J., Fuchs T., Schuffler P.J., Hartmann A., Knüchel R., Dahl E. Prognostic relevance of Wnt-inhibitory factor-1 (WIF1) and Dickkopf-3 (DKK3) promoter methylation in human breast cancer. BMC Cancer. 2009;9:217. doi: 10.1186/1471-2407-9-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz W.A., Alexa A., Jung V., Hader C., Hoffmann M.J., Yamanaka M., Fritzsche S., Wlazlinski A., Müller M., Lengauer T., et al. Factor interaction analysis for chromosome 8 and DNA methylation alterations highlights innate immune response suppression and cytoskeletal changes in prostate cancer. Mol. Cancer. 2007;6:14. doi: 10.1186/1476-4598-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chan S.L., Cui Y., van Hasselt A., Li H., Srivastava G., Jin H., Ng K.M., Wang Y., Lee K.Y., Tsao G.S., et al. The tumor suppressor Wnt inhibitory factor 1 is frequently methylated in nasopharyngeal and esophageal carcinomas. Lab. Invest. 2007;87:644–650. doi: 10.1038/labinvest.3700547. [DOI] [PubMed] [Google Scholar]
- 32.Marques R.B., Dits N.F., Erkens-Schulze S., van Weerden W.M., Jenster G. Bypass mechanisms of the androgen receptor pathway in therapy-resistant prostate cancer cell models. PLoS One. 2010;5:e13500. doi: 10.1371/journal.pone.0013500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X., Nie S., Chang C., Qiu T., Cao X. Smads oppose Hox transcriptional activities. Exp. Cell Res. 2006;312:854–864. doi: 10.1016/j.yexcr.2005.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Norris J.D., Chang C.Y., Wittmann B.M., Kunder R.S., Cui H., Fan D., Joseph J.D., McDonnell D.P. The homeodomain protein HOXB13 regulates the cellular response to androgens. Mol. Cell. 2009;35:405–416. doi: 10.1016/j.molcel.2009.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang D., Garcia-Bassets I., Benner C., Li W., Su X., Zhou Y., Qiu J., Liu W., Kaikkonen M.U., Ohgi K.A., et al. Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature. 2011;474:390–394. doi: 10.1038/nature10006. [DOI] [PMC free article] [PubMed] [Google Scholar]