Abstract
Here we analyzed the function of the c-MYC-inducible basic helix–loop–helix leucine-zipper transcription factor AP4 in AP4-deficient mouse embryo fibroblasts (MEFs). Loss of AP4 resulted in premature senescence and resistance towards immortalization. Senescence was accompanied by induction of the cyclin-dependent kinase inhibitor-encoding genes p16, a known tumor suppressor, and p21, a previously described target for repression by AP4. Notably, AP4 directly repressed p16 expression via conserved E-box motifs in MEFs and human diploid fibroblasts. Senescence caused by AP4-deficiency was prevented by depletion of p16 and/or p21, demonstrating that these factors mediate senescence caused by AP4 loss. As senescence induced by the loss of AP4 was rescued by ectopic AP4, secondary lesions were not involved in causing premature senescence. Activation of c-MYC resulted in repression of p21 and p16 in AP4+/+, but not in AP4−/− MEFs. Furthermore, after combined expression of c-MYC and mutant RAS in MEFs, AP4 was required for colony formation, anchorage-independent growth and tumor formation in mice. In addition, combined ectopic expression of AP4 and mutant RAS in MEFs resulted in colony formation. However, additional loss of the p53 tumor suppressor was necessary for anchorage-independent growth and tumor formation of MEFs by combined AP4 and mutant RAS expression. In conclusion, this study identified AP4 as an oncogenic antagonist of cellular senescence. AP4 achieves this effect by direct repression of p16 and p21, and may thereby critically contribute to c-MYC function and tumor progression.
Keywords: AP4, c-MYC oncogene, p16 tumor suppressor, p21, senescence, transformation, cancer
The proto-oncogene c-MYC is commonly activated in human cancer by gene amplification, viral promoter insertion or chromosomal translocation but also due to mutations of upstream regulators (reviewed in Adhikary and Eilers1). c-MYC is highly expressed in proliferating cells and downregulated when cells cease to proliferate, for example, during differentiation. Deregulated c-MYC expression promotes cell proliferation and causes resistance to anti-mitogenic stimuli.2 Furthermore, constitutive expression of c-MYC sensitizes toward apoptosis (reviewed in Meyer et al.3). The c-MYC gene encodes a transcription factor of the basic/helix–loop–helix/leucine-zipper (bHLH-LZ) class, which binds to the E-box motif CACGTG (reviewed in Dang et al.4). However, the mechanisms that underlie the mitogenicity of c-MYC are only partially understood. It seems likely that the combined actions of multiple genes regulated by c-MYC contribute to the stimulatory effects of c-MYC on cell cycle re-entry and progression.5, 6 Several c-MYC-regulated genes encode components of the cell cycle machinery, which control G1/S-progression, such as cyclin D1/D2,7, 8 cyclin-dependent kinase 4 (CDK4)9 and CDC25A,10 or represent regulators of the G2/M progression, as MAD2.11 Accordingly, c-MYC also influences G2/M progression.11, 12 c-MYC activation was shown to activate p53, either by inducing DNA damage and/or by activating the expression of ARF.13, 14 c-MYC-induced DNA damage is presumably due to replicative stress caused by unscheduled DNA replication.15 The activation of p53 by c-MYC mediates apoptosis, representing a fail-safe mechanism against oncogene activation.16
In cells deficient for the c-MYC-target genes encoding CDK2, htert/telomerase or the WRN/Werner syndrome, protein activation of c-MYC was shown to result in cellular senescence, a permanent cell cycle arrest, which is accompanied by cellular enlargement and induction of senescence-associated β-galactosidase (SA-β-gal) activity.17, 18, 19, 20 Recently, SIRT1 was shown to mediate immortalization by c-MYC.21 SIRT1 encodes an NAD-dependent deacetylase, which inhibits p53 and other pro-apoptotic factors.
Similar to c-MYC, the AP4 protein belongs to the class of bHLH-LZ transcription factors. AP4 exclusively forms homodimers, which bind to the E-box motif CAGCTG, and thereby either represses or activates the expression of target genes.22, 23, 24, 25, 26 We previously identified the AP4 gene as a direct transcriptional target of c-MYC and showed that the gene encoding the CDK-inhibitor p21 is directly repressed by AP4 in human cells.27, 28 Notably, elevated expression of AP4 was observed in pancreatic cancer and colorectal cancer.27, 29 Furthermore, increased AP4 expression was shown to correlate with poor patient survival and metastasis in gastric, hepatocellular and colorectal carcinomas.30, 31, 32
In order to determine the function of AP4, we deleted the AP4 gene in mice using homologous recombination. Here we describe the analysis of AP4-deficient mouse embryo fibroblasts (MEFs) derived from these mice. Our results imply that AP4 suppresses senescence by direct repression of p16 and p21. This function of AP4 was required during spontaneous immortalization of MEFs, for c-MYC-mediated repression of p21 and p16, and for c-MYC/RAS-induced transformation and tumor formation. In addition, our analyses revealed that AP4 itself has oncogenic potential, as its co-expression with mutant RAS in p53-deficient MEFs enhanced tumor formation in mice. Taken together, these results show that AP4 is an important mediator of c-MYC functions and itself harbors oncogenic potential.
Results
Phenotypes of AP4-deficient MEFs
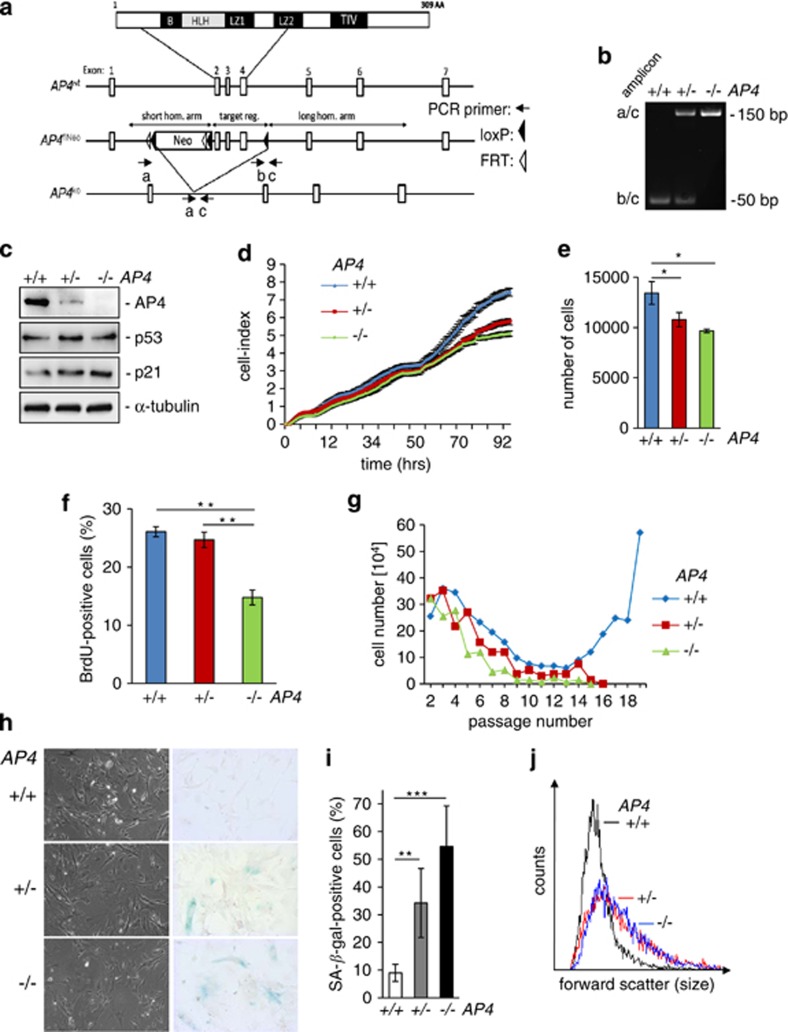
In order to determine the function of AP4, a conditional knock-out strategy was devised: the AP4 exons 2-4 were flanked by loxP sites and a neo cassette flanked by frt sites was introduced into the first intron of AP4 in murine ES cells. Subsequently, mice with germ-line deletion of AP4 were generated (Figure 1a; Supplementary Figures 1a and b), and MEFs were isolated and genotypes confirmed by genomic PCR (Figure 1b). AP4−/− mice were born at a normal Mendelian ratio upon intercrossing of heterozygous mice and were grossly normal and fertile (Hermeking et al., in preparation). MEFs were isolated from mice with AP4+/+, AP4+/− and AP4−/− genotypes in at least two different MEF isolations. In these AP4−/− MEFs, expression of the p21 protein was upregulate and AP4+/− displayed intermediate levels, whereas p53 expression was unchanged in AP4−/− MEFs (Figure 1c). These results further confirm our previous characterization of AP4 as a regulator of p21 in human cells.27 Furthermore, AP4-deficient MEFs showed decreased proliferation and DNA replication, as determined by 5-bromo-2′-deoxyuridine (BrdU) incorporation (Figures 1d, e and f and Supplementary Figure 1c). MEFs heterozygous for AP4 displayed an intermediate decrease in proliferation, but no significant change in BrdU incorporation, indicating that other processes downstream of AP4 besides DNA synthesis may be responsible for a decrease in proliferation. When subjected to a 3T3 protocol AP4+/+ MEFs entered a crisis with increasing passage number and immortalized cells emerged, which resumed exponential proliferation, as expected (Figure 1g). However, AP4+/− and AP4−/− MEFs were resistant to immortalization and ceased to proliferate. MEFs deficient for AP4 began to display an increase in size and expression of SA-β-gal (pH 6) at passage three, with cells heterozygous for AP4 showing an intermediate frequency of senescent cells (Figures 1h and i). The concomitant increase in size of senescent AP4−/− MEFs was confirmed by flow cytometric analysis of forward scatter (Figure 1j and Supplementary Figure 1d). These observations suggested that the immortalization defect caused by AP4 deficiency may have been due to the induction of premature senescence.
Figure 1.
AP4-deficient MEFs display decreased proliferation and premature senescence. (a) Schematic representation of the AP4/tcfAP4 knock-out strategy. On top, the domains of the AP4 protein are indicated. Below the organization of the AP4 gene before and after homologous recombination with a targeting construct containing loxP and frt sites for Cre- or Flp-mediated removal of exons 2–4 or the neomycin cassette, respectively, is provided. AA, amino acid; B, basic region; HLH, helix–loop–helix domain; LZ1 and LZ2, leucine zipper 1 and 2; Neo, neomycin resistance; TIV, an evolutionary conserved motif containing the amino acids TIV. (b) Genotyping of AP4-deficient MEFs using the indicated primer pairs. (c) The expression of the indicated proteins was determined by immunoblot analysis in asynchronously growing MEFs (passage 3) of the indicated genotypes. α-tubulin served as loading control. (d) Cell proliferation in medium with 10% serum was determined by impedance measurement. The cell index represents cell numbers. (e) The results in d were confirmed by cell counting using standard Neubauer-chambers at the end of the analysis (96 h). For d and e, 2 × 103 cells were seeded per well of 96-well format plate. (f) Determination of S-phase via BrdU incorporation of asynchronously proliferating MEFs with indicated genotypes. 8 × 104 cells were seeded into a 25-cm2 flask. (g) MEFs with indicated genotypes were subjected to a 3T3 protocol at 20% oxygen. (h) Representative detection of SA-β-galactosidase at pH 6 of MEFs (passage 3) with the indicated genotypes. (i) Quantification of SA-β-galactosidase-positive cells in MEFs with the indicated genotypes at passage 3. (j) Size determination of 5 × 103 MEFs (passage 3) by flow cytometry. Results in f, h and i were performed in two independent series with cells from independent MEF isolations. Results in e, f, i represent the mean±S.D. (n=6). Results in d, j represent the mean±S.D. (n=3). Significance level as indicated: *P<0.05, **P<0.01, ***P<0.001
Characterization of p16 as a direct AP4 target
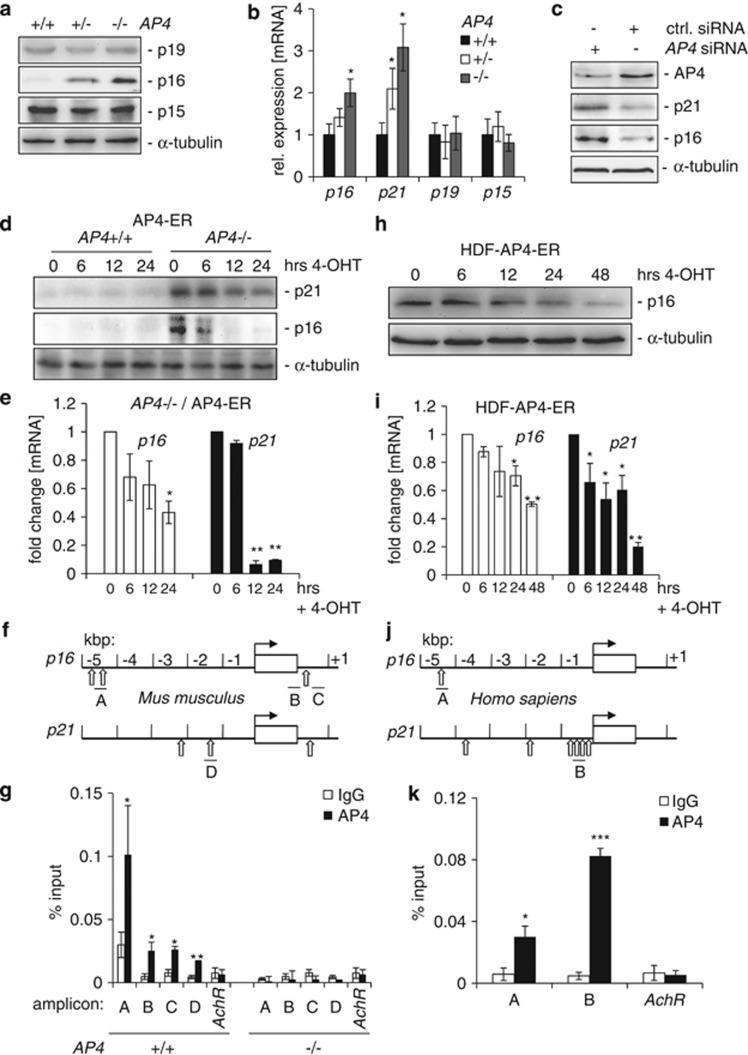
Next, we analyzed whether factors previously implicated in the regulation of senescence were deregulated in AP4-deficient MEFs. Indeed, the CDK inhibitors p16 and p21, but not p19/ARF and p15/INK4B/CDKN2B, were induced at protein and mRNA levels in AP4-deficient MEFs (Figures 1c, 2a and b). AP4+/− MEFs showed intermediate levels of p16 and p21 expression. When AP4+/+ MEFs were treated with small interfering RNAs (siRNAs) directed toward AP4 mRNA for 72 h, a similar increase in p21 and p16 expression was detected as in AP4-deficient MEFs (Figure 2c). Therefore, the increase in p21 and p16 expression caused by AP4 loss is presumably a direct consequence of decreased presence of AP4 at the respective promoters and not an indirect consequence of premature senescence. Conversely, activation of a conditional AP4-ER (estrogen receptor) fusion protein by addition of 4-hydroxytamoxifen (4-OHT) in AP4−/− MEFs resulted in a rapid repression of p16 and p21 at the protein and mRNA levels (Figures 2d and e). Chromatin immunoprecipitation (ChIP) and subsequent qPCR (qChIP) analyses showed that AP4 occupies E-boxes present ∼4.5 kbp upstream and 400 bp downstream of the p16 transcriptional start site (TSS) in MEFs (Figures 2f and g). The occupancy of the mouse p21 promoter by AP4, which was previously shown for human cells,27 was confirmed by qChIP (Figure 2g). Also in human diploid fibroblasts (HDFs) conditional AP4-ER activation resulted in the repression of p16 mRNA and protein, and the repression of p21 mRNA (Figures 2h and i). Analysis by qChIP revealed AP4 occupancy at an E-box present ∼4.5 kbp upstream of the p16 TSS and in the vicinity of the p21 TSS in HDFs (Figures 2j and k). Therefore, the transcriptional regulation of p16 and p21 by AP4 is conserved between species indicating functional importance.
Figure 2.
Direct repression of p16 and p21 by AP4. (a) The expression of the indicated proteins was determined by immunoblot analysis in asynchronously growing MEFs (passage 3). (b) The expression of the indicated mRNAs was determined by qPCR analysis in asynchronously growing MEFs (passage 3). (c) The expression of the indicated proteins was determined by immunoblot analysis in asynchronously growing AP4+/+ MEFs (passage 3) 72 h after treatment with AP4-specific siRNAs. (d) Immunoblot analysis of the indicated proteins in MEFs with indicated genotypes after AP4-ER activation by addition of 200 nM 4-OHT (4-hydroxytamoxifen) for the indicated periods. (e) qPCR analysis of p16 and p21 mRNAs after AP4-ER activation in AP4−/− MEFs by addition of 200 nM 4-OHT. (f) Schematic representation of the genomic organization of the indicated murine promoters. Vertical arrows indicate AP4-binding motifs (CAGCTG). Horizontal bars indicate qChIP amplicons. (g) qChIP analysis of genomic DNA co-precipitated with an AP4-specific or, as a reference, IgG antibody in AP4+/+ and AP4−/− MEFs. The mouse acetylcholine receptor (AchR) promoter, which lacks AP4-binding motifs, served as a negative control. (h) Immunoblot analysis of the indicated proteins in HDFs after AP4-ER activation by addition of 200 nM 4-OHT for the indicated periods. (i) qPCR analysis of p16 and p21 mRNAs after AP4-ER activation in HDFs by addition of 200 nM 4-OHT. (j) Schematic representation of the genomic organization of the indicated human promoters. Vertical arrows indicate AP4-binding motifs (CAGCTG). Horizontal bars indicate qChIP amplicons. (k) qChIP analysis of genomic DNA co-precipitated with an AP4-specific or, as a reference, IgG antibody in HDFs. The human acetylcholine receptor (AchR) promoter, which lacks AP4-binding motifs, served as a negative control. Primer pairs used for qPCR and qChIP are depicted in Supplementary Tables 2 and 3, respectively. Results in b represent the mean±S.E.M. (n=2) of cells from two different MEF isolations, in (e, g, i, k) the mean±S.D. (n=3) is depicted. Significance level as indicated: *P<0.05, **P<0.01. (a, c, d) Detection of α-tubulin served as loading control
p16 and p21 represent critical mediators of AP4 functions
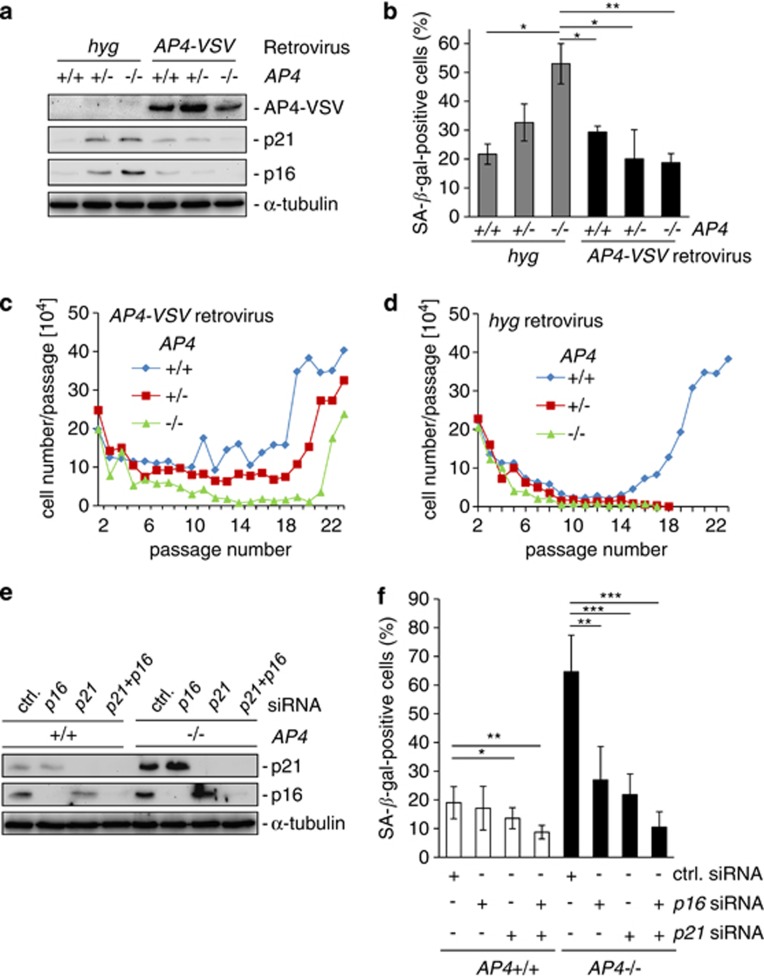
Re-introduction of AP4 by retroviral transfer suppressed the increased expression of p16 and p21 in AP4+/− and AP4−/− MEFs, whereas an infection with a virus not expressing AP4 had no effect (Figure 3a). Furthermore, ectopic expression of AP4 decreased the frequency of SA-β-gal-positive cells in AP4−/− and AP4+/− MEFs to that of AP4+/+ MEFs (Figure 3b). Moreover, retroviral reexpression of AP4 rescued the immortalization defect of MEFs deficient or heterozygous for AP4 in a 3T3 protocol, whereas a control retrovirus not expressing AP4 had no effect (Figures 3c and d). Therefore, AP4 is necessary for immortalization of MEFs and we can exclude that AP4−/− MEFs acquired a secondary lesion, which rendered them resistant to immortalization. To analyze whether the derepression of the AP4 targets p16 and p21 mediates the premature senescence of AP4-deficient MEFs, we downregulated p16 and p21 expression in AP4+/+ and AP4−/− MEFs using specific siRNAs, which were described previously in Lu et al.33 (Figures 3e and f): knock-down of either p16 or p21 significantly decreased the number of SA-β-gal-positive cells in AP4−/− MEFs, and a combined inactivation of p16 and p21 by siRNAs further decreased the number of senescent cells. These treatments also affected the initially lower frequency of senescent cells in AP4+/+ MEFs, albeit to a lesser extent. Therefore, the derepression of p21 and p16 caused by loss of AP4 is sufficient to mediate cellular senescence. Taken together, these results show that AP4 coordinates the expression of two central cell cycle regulators, to suppress senescence and thereby facilitate immortalization of MEFs. This property presumably contributes to the oncogenic functions of AP4 downstream of c-MYC.
Figure 3.
Derepression of p16and p21 mediates senescence in AP4-deficient MEFs. (a) MEFs with the indicated genotypes were infected with retrovirus expressing an AP4-VSV cassette or, as a control, only the hygromyin B resistance gene (hyg). The expression of the indicated proteins was determined by immunoblot analysis in asynchronously growing MEFs (passage 4). (b) Quantification of SA-β-galactosidase staining of MEFs at passage 4 with constitutive expression of AP4-VSV or hyg, as a control. (c and d) MEFs analyzed in (a) were subjected to a 3T3 protocol and counted at the indicate time points. (e) Western blot analysis of the indicated MEFs (passage 3) 96 h after transfection with the indicated siRNAs. (f) SA-β-galactosidase detection in the indicated MEFs (passage 3), 96 h after transfection of the indicated siRNAs. All transfections were performed with the same concentration of total siRNA (40 nM) by reconstitution with control siRNA. For every condition and genotype, biological triplicates were analyzed microscopically with five different fields for each replica. Results in e and f were obtained in two independent series with cells from independent MEF isolations (n=6). Results in b and f are expressed as mean±S.D. (n=6). Significance level as indicated: *P <0.05, **P<0.01, ***P <0.001. (a and e) Detection of α-tubulin served as loading control
AP4 as a mediator of c-MYC function
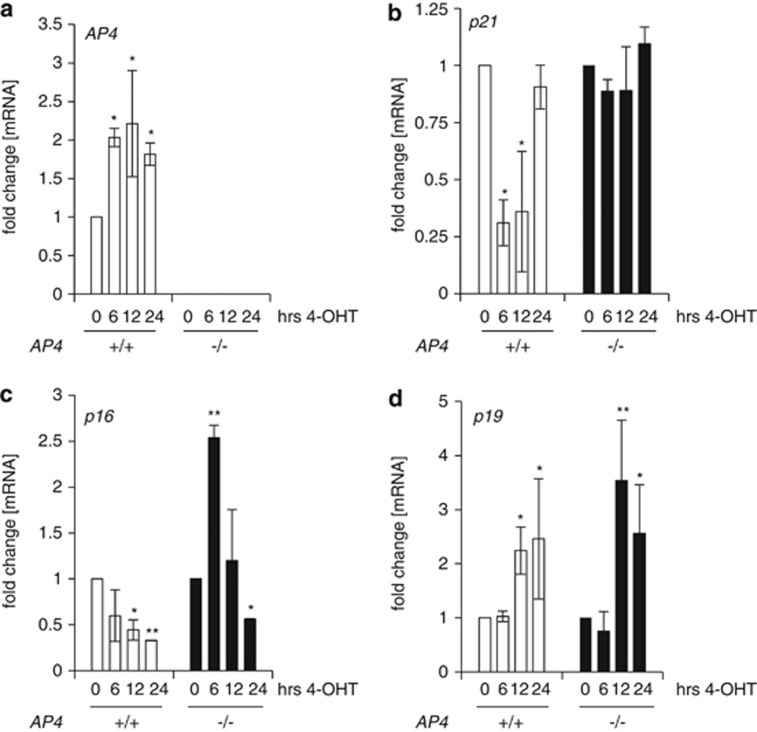
Next, we determined the role of AP4 for c-MYC-mediated gene regulation in MEFs. As in our previous analyses in human cell lines,27 AP4 expression was induced by c-MycER activation in serum-starved MEFs (Figure 4a). Therefore, the induction of AP4 by c-MYC is conserved between species. In AP4+/+ MEFs, both p21 and p16 were repressed within a few hours after activation of c-MycER (Figures 4b and c). However, in AP4-deficient MEFs, p21 levels remained unchanged within 24 h after c-MycER activation and p16 mRNA even showed a ∼2.5-fold induction after c-MYC activation. The expected induction of p19/ARF mRNA by c-MYC was not affected by deletion of AP4 (Figure 4d). Taken together, these results show that AP4 is a transcriptional target of c-MYC in MEFs and necessary for repression of p16 and p21 after c-MYC activation.
Figure 4.
c-MYC represses p16 and p21 via AP4. (a–d) MEFs with the indicated genotypes stably expressing retrovirally transduced c-MycER were cultured for 48 h at 0.5% serum before activation of c-MycER by addition of 200 nM 4-OHT for the indicated periods. qRT-PCR analyses of indicated mRNAs. Results in a–d represent the mean±S.E.M. (n=2). Significance level as indicated: *P<0.05, **P<0.01
Role of AP4 in cellular transformation
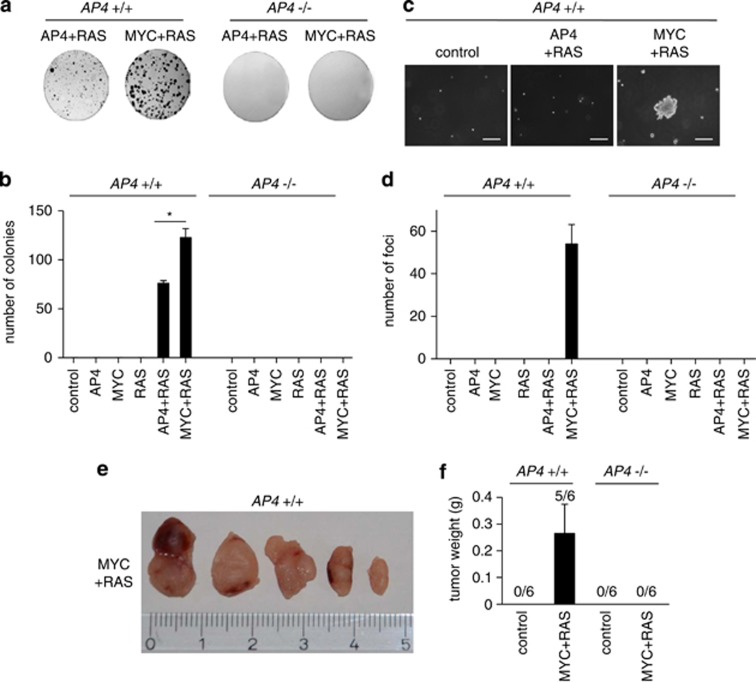
Next, we used a co-transformation assay to assess the putative oncogenic functions of AP4 and its requirement for c-MYC-induced transformation and tumorigenicity. Transformation of primary rodent cells requires at least two cooperating oncogenes.34, 35 Ectopic expression of the oncogenic H-RasG12V mutant (hereafter RAS) alone induces senescence in MEFs, which is accompanied by induction of p16.36 However, co-expression of RAS with an additional oncogene, such as c-MYC, which on its own can immortalize or induce apoptosis under growth-promoting or inhibitory conditions, respectively,37 results in transformed cells, which lose contact inhibition in monolayer culture, show anchorage-independent growth in soft agar and, after subcutaneous injection, form tumors in NOD/SCID mice. In a low density, colony formation assay, AP4+/+ MEFs transduced with AP4+RAS and MYC+RAS formed colonies as detected by crystal violet staining, whereas cells transduced with a retrovirus encoding either AP4, MYC or RAS, or a control virus did not form colonies (Figures 5a and b). In the soft agar assay, which monitors the ability of cells to proliferate in an anchorage-independent manner, only AP4+/+, but not AP4−/− MEFs transduced with a combination of MYC and RAS-expressing retroviruses formed detectable colonies (Figures 5c and d), indicating that AP4 is required to mediate this function of c-MYC. As the combination of AP4 and RAS did not yield colonies or foci in soft agar assays, c-MYC presumably regulates additional pathways/functions, which are not affected by AP4. When 1 × 106 viable AP4+/+ MEFs with combined expression of MYC and RAS were injected into the flanks of non-obese diabetic/severe combined immunodeficiency (NOD/SCID) mice, tumors consistently developed, whereas the same number of AP4−/− MEFs expressing MYC and RAS did not give rise to detectable tumors (Figures 5e and f). Therefore, AP4 is a required mediator of the co-transforming functions of c-MYC, which ultimately allows the formation of tumors in vivo.
Figure 5.
Requirement of AP4 for co-transformation by MYC and mutant RAS. (a) Colony formation analysis after retroviral transduction of AP4, c-MYC and mutant RAS. 1 × 103 AP4+/+ or AP4−/− MEFs expressing the indicated genes or, as a control, the retroviral vector only expressing hygromycin B or puromycin resistance genes were seeded onto a 10-cm diameter dish. All cells containing the indicated genes were additionally infected with viruses transducing the empty vector harboring the complementary resistance (see also Material and Methods). After 21 days, cells were stained with crystal violet and evaluated by counting with the Image J software. Representative results are shown. (b) Quantification of the colonies detected per plate. (c) Representative pictures of foci in soft agar. MEFs (passage 2) were infected with the indicated retroviruses. 1.6 × 104 cells were seeded per 35 mm dish and colonies were quantified 21 days later. × 100 magnification; scale bar, 100 μm. (d) Quantification of foci numbers in soft agar. (e) Tumor formation in xenograft assay: 6- to 8–week-old male NOD/SCID mice were sub-cutaneously injected with 106 AP4+/+ or AP4−/− MEFs (passage 6) expressing the indicated genes into each flank. After 3 weeks, tumors were resected and photographed. Scale in centimeters. (f) Weight of the resected tumors 3 weeks after injection. Results in (b, d, f) were obtained in two independent experimental series with MEFs from two different isolations in triplicates and represent the mean±S.D. (n=6). Significance level as indicated: *P<0.05, **P<0.01
p53 deficiency uncovers oncogenic functions of AP4
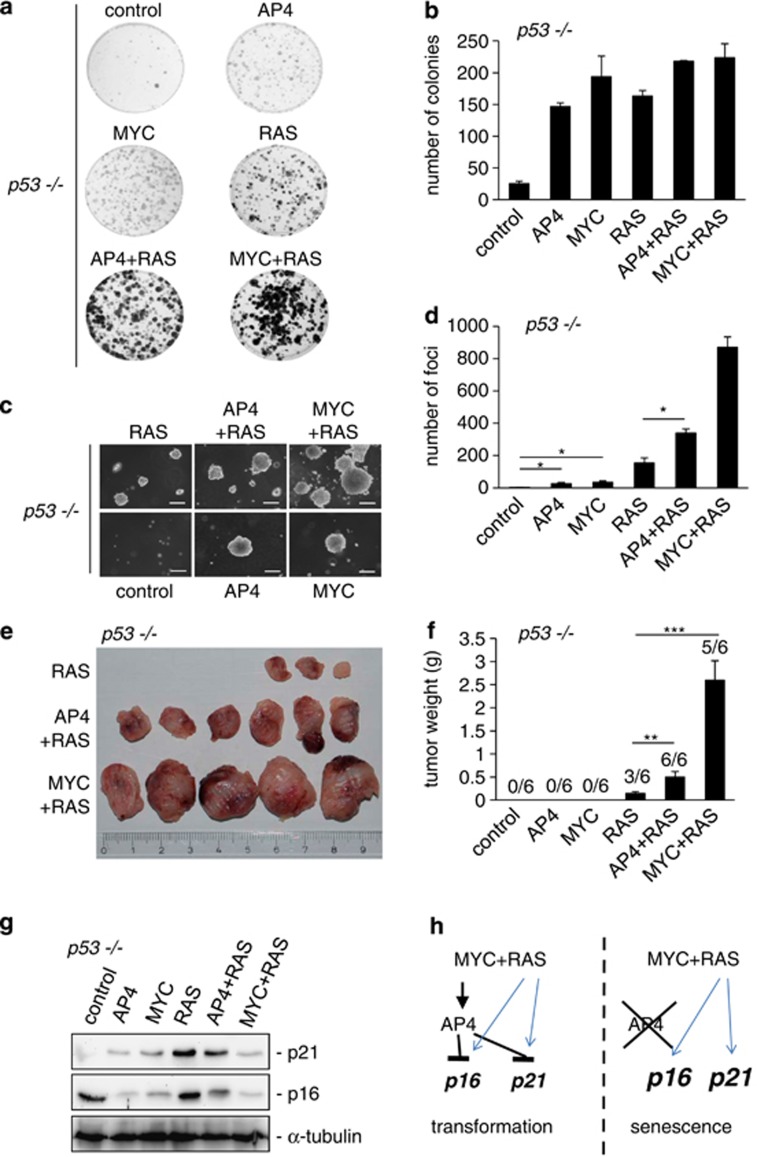
In the assays described above, ectopic expression of AP4 in combination with RAS allowed the outgrowth of MEFs in the colony formation assay, whereas this combination was not sufficient to mediate focus formation in soft-agar and tumor formation in mice. It has been reported that p53-deficient MEFs are transformed by RAS or MYC expression alone.38, 39 Therefore, we tested whether AP4 induces cellular transformation in a p53-deficient background. For this analysis, the same experimental conditions as used for AP4+/+ and AP4−/− MEFs shown in Figures 5a–f were employed. Indeed, p53−/− MEFs transduced with an AP4-encoding retrovirus alone gave rise to the formation of colonies with a frequency similar to that observed for MYC or RAS (Figures 6a and b). The number of colonies caused by RAS was similar to that of AP4 or MYC, but RAS-induced colonies displayed in increased cellular density as evidenced by a darker staining. Combined expression of AP4+RAS or MYC+RAS resulted in a further increase in colony number and cell density. In a soft-agar assay, expression of AP4 or MYC in p53−/− MEFs led to a significant increase in the colony number (Figures 6c and d). RAS expression alone was approximately fourfold more effective in forming colonies. Notably, the combined expression of AP4+RAS led to a further increase in the number of colonies, which was surpassed by the combination of MYC+RAS. Therefore, AP4 has a transforming capacity in the absence of p53, which is comparable to that of c-MYC. To further extend these findings, we injected p53-deficient MEFs transduced with the different vector combinations into immune-deficient mice (Figures 6e and f). Notably, AP4 co-expression increased the frequency of tumor formation of RAS-expressing MEFs to a similar extent as co-expression of c-MYC. Tumors derived from MEFs harboring AP4+RAS showed an increased size and weight when compared with those resulting from MEFs only expressing mutant RAS. Tumors derived from MYC+RAS-expressing MEFs showed an additional increase in tumor weight, again indicating that c-MYC performs additional tumorigenic functions. Western blot analyses of retrovirally transduced p53−/− MEFs before injection into NOD/SCID mice revealed that singular ectopic expression of AP4 or MYC represses p16 and slightly induces p21, whereas expression of RAS alone markedly induces p16 and p21 (Figure 5g). In contrast, cells expressing AP4+RAS or MYC+RAS showed decreased expression of p16 and p21 when compared with MEFs expressing RAS alone. Interestingly, the combined expression of MYC and mutant RAS led to the strongest repression of p16 and p21, which could explain the superior tumorigenicity of this oncogene combination. Taken together, these results show that AP4 activation alone is sufficient to transform cells in a p53-null background to a similar extent as c-MYC. Furthermore, AP4 effectively cooperates with RAS in the transformation of p53-deficient cells. This effect of AP4 may be explained by its ability to directly repress p16 and p21.
Figure 6.
p53-deficiency uncovers oncogenic functions of AP4. (a) Colony formation analysis as described under Figure 5a. 1 × 103 p53−/− MEFs expressing the indicated genes introduced by retroviral transduction and the empty vector control were plated in 10-cm diameter dishes. After 7 days, cells were stained with crystal violet. Representative results are depicted. (b) Quantification of the colonies detected per dish. (c) Focus formation assay in soft agar as in Figure 5c. Representative foci derived from p53−/− MEFs infected with the indicated retroviruses 7 days after seeding 1.6 × 104 cells are shown. × 100 Magnification; scale bar, 100 μm. (d) Quantification of foci in soft agar. (e) Xenograft tumor formation analysis: 6- to 8–week-old male NOD/SCID mice were subcutaneously injected with 106 p53−/− MEFs (passage 6) expressing the indicated genes into each flank. After 3 weeks, tumors were resected and photographed. Scale in centimeters. (f) Weight of the resected tumors 3 weeks after injection. (g) Western blot analysis of the indicated proteins in p53−/− MEFs (passage 6) was performed when cells were injected into mice. (h) Model summarizing the results of this study. Blue arrows indicate oncogenic signaling, which induces p16 and p21.45 Results in (b, d, f) were obtained in two independent experimental series with MEFs from two different isolations in triplicates and represent the mean±S.D. (n=6). Significance level as indicated: *P<0.05, **P<0.01, ***P<0.001
Discussion
By using a genetic approach, we were able to assign important cellular functions to the c-MYC-induced transcription factor AP4. We found that AP4 suppresses senescence via direct repression of p21 and p16. Thereby, AP4 presumably contributes to immortalization of MEFs during a 3T3 protocol. In addition, ectopic AP4 was sufficient for transformation of primary, p53-deficient MEFs and mediated tumor formation of these cells in mice in cooperation with activated RAS. Finally, AP4 was required for the oncogenic effects of c-MYC in MEFs, as AP4-deficient MEFs were resistant to focus and tumor formation induced by combined c-MYC/RAS expression.
Similar to the AP4-deficient MEFs analyzed here, CDK2 MEFs are not efficiently immortalized.40, 41 Compound CDK2/CDK4-deficient MEFs show premature senescence.42 Furthermore, silencing of CDK2 significantly decreases the proliferation rate of compound CDK4 and CDK6 knock-out MEFs.43 Therefore, at least one G1-CDK has to be expressed for successful cell cycle progression. In the light of these findings, it seems likely that the premature senescence observed in AP4-negative cells is due to the decreased activity of the CDK2/cyclin and other CDK/cyclin complexes, which results from the elevated levels of the CDK inhibitors p21 and p16 caused by the derepression of the corresponding genes in the absence of AP4. Recently, knockdown of AP4 by siRNAs in gastric cancer cell lines was shown to inhibit cell cycle progression and apoptosis by regulating the expression levels of p21, p53, caspase-3, cyclin D1, BCL-2 and BCL-xl.44 However, it remained unclear whether these regulations are directly mediated by AP4. Nonetheless, these results in combination with our previous results27 and the results presented here suggest that targeting AP4 may be a potential tumor therapeutic strategy.
As AP4 directly represses two effectors of senescence inducing pathways, p16 and p21, AP4 may participate in suppressing c-MYC-induced senescence. AP4 presumably suppresses expression of these genes by affecting the state of the chromatin at these promoters. Indeed, AP4 was shown to bind to HDAC1 and HDAC3 to mediate transcriptional repression of target genes.23, 24 Notably, other oncogenic factors, such as ID-1 and BMI-1, have previously been implicated in the transcriptional repression of p16,45, 46, 47 as well as other members of the polycomb group of transcription factors.48, 49 Therefore, the ability to repress p16 represents an important feature of oncogenic transcription factors. Similar to AP4-deficient MEFs, ID-1-deficient MEFs undergo premature senescence and show increased expression of p16 and decreased CDK2 and CDK4 kinase activity.50 Interestingly, the BMI-1 oncogene cooperates with c-MYC in murine lymphomagenesis.51, 52 Further analyses revealed that BMI-1-deficient MEFs show impaired cell cycle progression and undergo premature senescence.49 Interestingly, BMI-1 and other polycomb-group members occupy the same region of the p16 promoter as AP4, namely the first intron.53 c-MYC has been previously shown to repress p16 and p21 expression via several mechanisms.54, 55, 56 Furthermore, c-MYC has been reported to directly induce the expression of p19/ARF, which also belongs to the INK4 locus.13 This may explain why the deletion of AP4 did not affect the induction of p19/ARF by c-MYC activation here. However, the repression of p16 and p21 by c-MYC was strictly AP4 dependent in our analyses of MEFs described here. The requirement for AP4 does, however, not exclude the involvement of other regulatory mechanism, for example, via MIZ1, which may also be required for efficient repression of p16 and p21 by c-MYC.
Recently, we described AP4 as an inducer of epithelial–mesenchymal transition (EMT) and mediator of metastasis in human colorectal cancer cells.32 Other EMT-inducing transcription factors, such as TWIST and ZEB1, were shown to suppress senescence, which may represent a tumor suppressive fail-safe program against invasion and metastasis.57, 58, 59 Our findings indicate that AP4 also suppresses senescence and may thereby contribute to tumor progression in the context of EMT, invasion and metastasis.
As RAS activation was shown to induce p16 and premature senescence in a p53-independent manner in primary fibroblasts,36 a possible mechanism for the observed cooperation between AP4 and RAS lies in the ability of AP4 to repress p16. This may also extend to the cooperation between c-MYC and RAS, as c-MYC induces AP4, which would repress p16. Given the observed function of AP4 in transformation, AP4 may represent an important mediator of cellular transformation in human malignancies. This conclusion is also supported by the consistently elevated expression of AP4 in several types of human cancer (see also http://www.proteinatlas.org), which has been associated with poor patient survival in hepatocellular, gastric and colorectal cancer.30, 31, 32
The molecular and cellular defects identified here will guide the future studies of AP4 function in the context of pathological c-MYC activation and during physiological processes associated with c-MYC activation on the organismal level, as expansion of cell populations during development. Our results show that the AP4 gene is a critical hub, controlling cellular senescence and transformation. Therefore, AP4 may represent a useful target for tumor therapeutic interventions.
Materials and Methods
Generation of AP4-deficient mice
AP4-deficient mice and their phenotypes will be described elsewhere (Hermeking et al., in preparation). In short, the loxP-site-flanked exons 2–4 of AP4 were removed by crossing to CMV-Cre mice. All ES cells and mice used had a C57Bl/6 background. The Cre allele was removed by further crossing. The respective genotypes of the mice and MEFs were confirmed by specific PCR analyses (for primers see Supplementary Table 1). These mice were used to isolate MEFs (see below).
Isolation and cultivation of mouse embryonic fibroblasts and HDFs
MEFs were prepared from embryos at day E13.5. The uterus was removed and washed in phosphate-buffered saline (PBS). The yolk sacs were separated and the embryos were isolated. The viscera of each embryo were removed, and the embryo was washed twice in PBS, placed in trypsin-EDTA (Invitrogen, Karlsruhe, Germany), cut finely and incubated for 10 min at 37 °C. The cell suspension prepared from the embryo was washed with medium containing 10% fetal bovine serum (FBS) and plated in 100-mm culture dishes. MEFs and HDFs were routinely cultured in a humidified 5% CO2 and 20% O2 atmosphere at 37 °C. MEFs and HDFs were maintained in Dulbecco's modified Eagle medium (Invitrogen) containing 10% FBS. MEFs were cultivated in the presence of 100 U/ml penicillin and 0.1 mg/ml streptomycin. One passage corresponds to treatment with trypsin and subsequent dilution of the cells at a ratio of 1:3 every 3 days. 4-OHT (Sigma, St. Louis, MO, USA) was dissolved in ethanol (400 μM stock solution) and used at a final concentration of 200 nM. MEFs used in the same analysis were derived from littermates.
ChIP assay
MEFs were cultured as described above. Cross-linking was performed with formaldehyde (Merck, Darmstadt, Germany) at a final concentration of 1% and terminated after 5 min by addition of glycine at a final concentration of 0.125 M. Cells were harvested with SDS buffer (50 mM Tris pH 8.1, 0.5% SDS, 100 mM NaCl, 5 mM EDTA) and after pelleting resuspended in IP buffer (two parts of SDS buffer and one part of Triton dilution buffer (100 mM Tris-HCl pH 8.6, 100 mM NaCl, 5 mM EDTA, pH 8.0, 0.2% NaN3, 5.0% Triton X-100)). Chromatin was sheered by sonication (HTU SONI 130, G. Heinemann, Schwäbisch Gmünd, Germany) to generate DNA fragments with an average size of 500 bp. Preclearing and incubation with AP4 antibody (AbD Serotec, Puchheim, Germany) or the respective IgG control (M-7023, Sigma) for 16 h was performed as previously described.11 Washing and reversal of cross-linking was performed as described.60 Immunoprecipitated DNA was analyzed by qPCR and the enrichment was expressed as percentage of the input for each condition.60 The sequences of oligonucleotides used as qChIP primers are listed in supporting Supplementary Table 3.
Colony formation assay
For low-density, colony formation assays, 1 × 103 cells were seeded onto a 10-cm dish for 7–21 days. Subsequently, cells were stained with crystal violet as described above, photographed colonies were counted using Image J software (NIH, USA). MEFs were cultured as described above, plus 0.1 mM non-essential amino acids (Invitrogen) and 55 μM 2-mercaptoethanol (Invitrogen).
Detection of SA-β-gal
The β-galactosidase staining was performed as described previously.61 In short, 8 × 104 cells were plated per well of a 6-well plate. Twenty-four hours later the media was removed from the cells and washed twice with PBS. To fix the cells, cells were incubated in 0.5% glutaraldehyde containing PBS for 5 min at room temperature. Cells were washed three times with P6M (PBS pH 6, 1 mM MgCl2). Cells were stained by incubation in X-gal solution (PBS pH 6; 3 mM K3Fe(CN)6; 3 mM K4Fe(CN)6; 1 mg/ml X-gal (Sigma)) at 37 °C overnight. The frequency of blue β-galactosidase-positive senescent cells was determined on pictures obtained with a CCD camera.
Flow cytometric analysis of DNA synthesis and cell size
To monitor DNA synthesis, 50 μM BrdU (Roche, Mannheim, Germany) was added for 60 min at 37 °C. Next, cells were harvested by addition of trypsin and centrifuged at 300 × g for 5 min. After washing with PBS, cells were fixed by addition of ice-cold 70% ethanol and incubation for at least 30 min at −20 °C. Fixed cells were resuspended in 0.1 mg/ml pepsin, and DNA was denatured by incubation in 2 M HCl for 30 min at room temperature. After centrifugation at 500 × g, cells were resuspended in 0.1 M Na2B4O7. Cells were washed once with PBS and PTS buffer (PBS, 0.5% Tween-20, 2% FBS), respectively, and subsequently resuspended in 60 μl PTS+6 μl anti-BrdU-FITC antibody (BD Biosciences Pharmingen, Franklin Lakes, NY, USA) or an appropriate isotype control IgG and incubated for 30 min at room temperature in the dark. Next, cells were washed two times with PTS and resuspended in 500 μl PTS, 0.5 mg/ml RNase A (Sigma) and 50 μg/ml propidium iodide (Sigma). After incubation for 30 min at RT, cells were analyzed by flow cytometry (CFlow6, Accuri, Erembodegem, Belgium). For cell size analysis, cells were harvested by addition of trypsin and after washing subjected to flow cytometry using a CFlow6 device (Accuri) and forward scatter was determined.
Focus formation assay in soft agar
For the anchorage-independent growth assays in soft agar, 35-mm dishes were filled with base agar consisting of 0.5% agarose (Lonza, Basel, Switzerland) and full medium containing 10% FBS. Afterwards 1.6 × 104 cells dissolved in a 0.3% top agar mixture containing 10% FBS were placed on top of the base agar. 7–21 days later, cells were stained with a 0.01% crystal violet solution, photographed and foci counted using Image J software. MEFs were cultured as described above, plus 0.1 mM non-essential amino acids (Invitrogen) and 55 μM 2-mercaptoethanol (Invitrogen).
Plasmids and RNAi
For generation of the pBabe-puro-AP4-ER vector, the c-Myc ORF was removed by restriction with BamHI from pBabe-puro-c-MycER (kind gift from B Amati) and the AP4 ORF was inserted. pBabe-hyg-AP4-VSV was generated by isolation of the AP4-VSV fragment using BamHI restriction of pcDNA3-AP4-VSV27 and insertion into the pBabe-hyg vector. pBabe-hyg-c-Myc was generated by insertion of the c-Myc ORF, which was isolated from pBabe-puro-c-Myc (kind gift from M Eilers) by restriction with SalI and BamHI. pBabe-puro (Addgene, Cambridge, MA, USA; plasmid 1764) and pBabe-puro-HRasV12 (Addgene; plasmid 1768; both kindly provided by R Weinberg). siRNAs were transfected at 40 nM final concentration using HiPerFect reagent (Qiagen, Venlo, Netherlands). siRNA target sequences were as follows: AP4-specific siRNA: 5′-GUGAUAGGAGGGCUCUGUAG-3′ as described in Jung et al.27; p16-specific siRNA: 5′-GGAGUCCGCUGCAGACAGAT-3′ and p21-specific siRNA: 5′-AACGGUGGAACUUUGACUUCG-3′ were previously described in Lu et al.33 As a control, the Silencer negative control siRNA #1 (Ambion, Kaufungen, Germany) was used.
Phase-contrast microscopy
Images of cells in culture were captured using an Axiovert 25 microscope (Zeiss, Jena, Germany) with a Sony Digital Hyper HAD camera (Software: Kappa Image Base, Kappa Opto-electronics, Gleichen, Germany) or an Axiovert Observer Z.1 microscope connected to an AxioCam MRm camera with an Axiovision software (Zeiss).
Quantitative real-time PCR (qPCR)
Total RNA was isolated using the High Pure RNA Isolation Kit (Roche). cDNA was generated from 1 μg total RNA per sample using anchored oligo-dT primers (Reverse-iT First Strand Synthesis; ABgene, Waltham, MA, USA). qPCR was performed by using the LightCycler 480 (Roche) and the Fast SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA) as described previously.62 The sequences of oligonucleotides used as qPCR primers are listed in Supplementary Table 2.
Retroviral infections
For retrovirus production and ectopic expression in MEFs and HDFs, Phoenix-E and Phoenix-A packaging cells were transfected with pBabe vectors, respectively, using calcium phosphate precipitation. Twenty-four hours after transfection, retrovirus-containing supernatants were harvested, passed through 0.45-μm filters (Millipore, Billerica, MA, USA), and used to infect MEFs or HDFs in the presence of polybrene (8 μg/ml) four times in 4-h intervals. For transformation assays, a second round of infection with additional constructs was performed 48 h later. Selection was started 48 h later by addition of 2 μg/ml puromycin (Sigma) and 75 μg/ml hygromycin B (Sigma) for 5 days.
Senescence/immortalization assay
The 3T3 protocol was performed as previously described.17 Briefly, MEFs in passage 2 were seeded in a 10-cm cell culture dish at a density of 3 × 105 cells per dish. Three days later, cells were trypsinized, counted and seeded at a density of 3 × 105 cells per dish. Cells were stained with trypan blue and viable cells counted using a Neubauer chamber at each passage.
Statistical analysis
Statistical significance was determined using an unpaired two-tailed Student's t-test.
Tumor formation assays in NOD/SCID mice
For the analysis of in vivo tumorigenicity, 6- to 8-week-old age-matched male immunocompromised NOD/SCID mice were injected with 100 μl of living MEF cells (106 cells) subcutaneously into each flank. Three weeks after injection, mice were killed, the tumors were resected and their weight was determined. All animals were maintained in individually ventilated cages. All studies involving mice were conducted with approval by the local Animal Experimentation Committee.
Western blot analysis and antibodies
Cells were lysed in RIPA lysis buffer (50 mM Tris/HCl, pH 8.0, 250 mM NaCl, 1% NP40, 0.5% (w/v) sodium deoxycholate, 0.1% sodium dodecylsulfate, complete mini-protease inhibitors (Roche)). Lysates were sonicated and centrifuged at 16 060 × g for 15 min at 4 °C. Per lane, 30–80 μg of whole-cell lysate was separated using 7.5% or 12% SDS-acrylamide gels and transferred on Immobilon PVDF membranes (Millipore). For immunodetection, membranes were incubated with antibodies specific for p19 (Santa Cruz, Heidelberg, Germany), p15 (Santa Cruz), p16 (Santa Cruz), p21 (BD PharMingen, Franklin Lakes, NJ, USA), p53 (Novocastra, Nussloch, Germany), AP4 (AbD Serotec), VSV-G (Sigma) and α-tubulin (Sigma). Signals from HRP (horse-radish-peroxidase)-coupled secondary antibodies were generated by enhanced chemiluminesence (Perkin-Elmer Life Sciences, Boston, MA, USA) and recorded with a CCD camera (440CF imaging system, Eastman Kodak Co., Rochester, NY, USA).
Acknowledgments
We thank Bruno Amati (Milan, Italy), Martin Eilers (Würzburg, Germany) and Robert Weinberg (Cambridge, USA) for the plasmids. We are grateful to David Horst for advice and Angela Servatius for technical assistance. Work in the Hermeking lab is supported by the DFG/Deutsche Forschungsgemeinschaft, and in part by the Deutsche Krebshilfe, DKTK/German Cancer Consortium and the Rudolf-Bartling-Stiftung.
Author Contributions
RJ performed the analyses, planed experiments, analyzed data and wrote the paper, PJ contributed important material, HH conceived and supervised the study, analyzed results, planed experiments and wrote the paper.
Glossary
- bHLH-LZ
basic helix–loop–helix leucine-zipper
- BrdU
5-bromo-2′-deoxyuridine
- CDK
cyclin-dependent kinase
- ChIP
chromatin immunoprecipitation
- EMT
epithelial–mesenchymal transition
- ER
estrogen receptor
- FBS
fetal bovine serum
- HDF
human diploid fibroblast
- MEF
mouse embryonal fibroblast
- PBS
phosphate-buffered saline
- SA-β-gal
senescence associated-β-galactosidase
- siRNA
small interfering RNA
- TSS
transcription start site
- NOD/SCID
non-obese diabetic/severe combined immunodeficiency
- 4-OHT
4-hydroxytamoxifen
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellá
Supplementary Material
References
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Alexandrow MG, Kawabata M, Aakre M, Moses HL. Overexpression of the c-Myc oncoprotein blocks the growth-inhibitory response but is required for the mitogenic effects of transforming growth factor beta 1. Proc Natl Acad Sci USA. 1995;92:3239–3243. doi: 10.1073/pnas.92.8.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16:275–287. doi: 10.1016/j.semcancer.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Dang CV, O'Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Koh E, Daley GQ, Bernards R. A genetic screen to identify genes that rescue the slow growth phenotype of c-myc null fibroblasts. Oncogene. 2000;19:3330–3334. doi: 10.1038/sj.onc.1203639. [DOI] [PubMed] [Google Scholar]
- Nikiforov MA, Kotenko I, Petrenko O, Beavis A, Valenick L, Lemischka I, et al. Complementation of Myc-dependent cell proliferation by cDNA expression library screening. Oncogene. 2000;19:4828–4831. doi: 10.1038/sj.onc.1203880. [DOI] [PubMed] [Google Scholar]
- Philipp A, Schneider A, Vasrik I, Finke K, Xiong Y, Beach D, et al. Repression of cyclin D1: a novel function of MYC. Mol Cell Biol. 1994;14:4032–4043. doi: 10.1128/mcb.14.6.4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4 as a target of c-MYC. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galaktionov K, Chen X, Beach D. Cdc25 cell-cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- Menssen A, Epanchintsev A, Lodygin D, Rezaei N, Jung P, Verdoodt B, et al. c-MYC delays prometaphase by direct transactivation of MAD2 and BubR1: identification of mechanisms underlying c-MYC-induced DNA damage and chromosomal instability. Cell Cycle. 2007;6:339–352. doi: 10.4161/cc.6.3.3808. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Zetterberg A, Zhu J, Tlsty T, Bishop JM. Overexpression of MYC causes p53-dependent G2 arrest of normal fibroblasts. Proc Natl Acad Sci USA. 2000;97:10544–10548. doi: 10.1073/pnas.190327097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zindy F, Eischen CM, Randle DH, Kamijo T, Cleveland JL, Sherr CJ, et al. Myc signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12:2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vafa O, Wade M, Kern S, Beeche M, Pandita TK, Hampton GM, et al. c-Myc can induce DNA damage, increase reactive oxygen species, and mitigate p53 function: a mechanism for oncogene-induced genetic instability. Mol Cell. 2002;9:1031–1044. doi: 10.1016/s1097-2765(02)00520-8. [DOI] [PubMed] [Google Scholar]
- Dominguez-Sola D, Ying CY, Grandori C, Ruggiero L, Chen B, Li M, et al. Non-transcriptional control of DNA replication by c-Myc. Nature. 2007;448:445–451. doi: 10.1038/nature05953. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Eick D. Mediation of c-Myc-induced apoptosis by p53. Science. 1994;265:2091–2093. doi: 10.1126/science.8091232. [DOI] [PubMed] [Google Scholar]
- Campaner S, Doni M, Hydbring P, Verrecchia A, Bianchi L, Sardella D, et al. Cdk2 suppresses cellular senescence induced by the c-myc oncogene Nat Cell Biol 20101254–59.sup pp 1-14. [DOI] [PubMed] [Google Scholar]
- Wu KJ, Grandori C, Amacker M, Simon-Vermot N, Polack A, Lingner J, et al. Direct activation of TERT transcription by c-MYC. Nat Genet. 1999;21:220–224. doi: 10.1038/6010. [DOI] [PubMed] [Google Scholar]
- Wang J, Xie LY, Allan S, Beach D, Hannon GJ. Myc activates telomerase. Genes Dev. 1998;12:1769–1774. doi: 10.1101/gad.12.12.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hydbring P, Kapelle K, Vervoorts J, Diebold J, Luscher B, et al. The c-MYC oncoprotein, the NAMPT enzyme, the SIRT1-inhibitor DBC1, and the SIRT1 deacetylase form a positive feedback loop. Proc Natl Acad Sci USA. 2012;109:E187–E196. doi: 10.1073/pnas.1105304109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mermod N, Williams TJ, Tjian R. Enhancer binding factors AP-4 and AP-1 act in concert to activate SV40 late transcription in vitro. Nature. 1988;332:557–561. doi: 10.1038/332557a0. [DOI] [PubMed] [Google Scholar]
- Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J Biol Chem. 2006;281:12495–12505. doi: 10.1074/jbc.M511773200. [DOI] [PubMed] [Google Scholar]
- Kim MY, Jeong BC, Lee JH, Kee HJ, Kook H, Kim NS, et al. A repressor complex, AP4 transcription factor and geminin, negatively regulates expression of target genes in nonneuronal cells. Proc Natl Acad Sci USA. 2006;103:13074–13079. doi: 10.1073/pnas.0601915103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujimoto K, Ono T, Sato M, Nishida T, Oguma T, Tadakuma T. Regulation of the expression of caspase-9 by the transcription factor activator protein-4 in glucocorticoid-induced apoptosis. J Biol Chem. 2005;280:27638–27644. doi: 10.1074/jbc.M501304200. [DOI] [PubMed] [Google Scholar]
- Hu YF, Luscher B, Admon A, Mermod N, Tjian R. Transcription factor AP-4 contains multiple dimerization domains that regulate dimer specificity. Genes Dev. 1990;4:1741–1752. doi: 10.1101/gad.4.10.1741. [DOI] [PubMed] [Google Scholar]
- Jung P, Menssen A, Mayr D, Hermeking H. AP4 encodes a c-MYC-inducible repressor of p21. Proc Natl Acad Sci USA. 2008;105:15046–15051. doi: 10.1073/pnas.0801773105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P, Hermeking H. The c-MYC-AP4-p21 cascade. Cell Cycle. 2009;8:982–989. doi: 10.4161/cc.8.7.7949. [DOI] [PubMed] [Google Scholar]
- Friess H, Ding J, Kleeff J, Fenkell L, Rosinski JA, Guweidhi A, et al. Microarray-based identification of differentially expressed growth- and metastasis-associated genes in pancreatic cancer. Cell Mol Life Sci. 2003;60:1180–1199. doi: 10.1007/s00018-003-3036-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu BS, Zhao G, Yu HF, Chen K, Dong JH, Tan JW. High expression of AP-4 predicts poor prognosis for hepatocellular carcinoma after curative hepatectomy. Tumour Biol. 2012;34:271–276. doi: 10.1007/s13277-012-0547-4. [DOI] [PubMed] [Google Scholar]
- Xinghua L, Bo Z, Yan G, Lei W, Changyao W, Qi L, et al. The overexpression of AP-4 as a prognostic indicator for gastric carcinoma. Med Oncol. 2012;29:871–877. doi: 10.1007/s12032-011-9845-8. [DOI] [PubMed] [Google Scholar]
- Jackstadt R, Roeh S, Neumann S, Jung P, Hoffmann R, Horst D, et al. AP4 is a mediator of epithelial-mesenchymal transition and metastasis in colorectal cancer. J Exp Med. 2013;210:1331–1350. doi: 10.1084/jem.20120812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu ZH, Books JT, Ley TJ. YB-1 is important for late-stage embryonic development, optimal cellular stress responses, and the prevention of premature senescence. Mol Cell Biol. 2005;25:4625–4637. doi: 10.1128/MCB.25.11.4625-4637.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Land H, Chen AC, Morgenstern JP, Parada LF, Weinberg RA. Behavior of myc and ras oncogenes in transformation of rat embryo fibroblasts. Mol Cell Biol. 1986;6:1917–1925. doi: 10.1128/mcb.6.6.1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruley HE. Adenovirus early region 1A enables viral and cellular transforming genes to transform primary cells in culture. Nature. 1983;304:602–606. doi: 10.1038/304602a0. [DOI] [PubMed] [Google Scholar]
- Serrano M, Lin AW, McCurrach ME, Beach D, Lowe SW. Oncogenic ras provokes premature cell senescence associated with accumulation of p53 and p16INK4a. Cell. 1997;88:593–602. doi: 10.1016/s0092-8674(00)81902-9. [DOI] [PubMed] [Google Scholar]
- Evan GI, Wyllie AH, Gilbert CS, Littlewood TD, Land H, Brooks M, et al. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992;69:119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Sage J, Mulligan GJ, Attardi LD, Miller A, Chen S, Williams B, et al. Targeted disruption of the three Rb-related genes leads to loss of G(1) control and immortalization. Genes Dev. 2000;14:3037–3050. doi: 10.1101/gad.843200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo E, Bonetti P, Lazzerini Denchi E, Martinelli P, Zamponi R, Marine JC, et al. Nucleophosmin is required for DNA integrity and p19Arf protein stability. Mol Cell Biol. 2005;25:8874–8886. doi: 10.1128/MCB.25.20.8874-8886.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- Ortega S, Prieto I, Odajima J, Martin A, Dubus P, Sotillo R, et al. Cyclin-dependent kinase 2 is essential for meiosis but not for mitotic cell division in mice. Nat Genet. 2003;35:25–31. doi: 10.1038/ng1232. [DOI] [PubMed] [Google Scholar]
- Berthet C, Klarmann KD, Hilton MB, Suh HC, Keller JR, Kiyokawa H, et al. Combined loss of Cdk2 and Cdk4 results in embryonic lethality and Rb hypophosphorylation. Dev Cell. 2006;10:563–573. doi: 10.1016/j.devcel.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Malumbres M, Sotillo R, Santamaria D, Galan J, Cerezo A, Ortega S, et al. Mammalian cells cycle without the D-type cyclin-dependent kinases Cdk4 and Cdk6. Cell. 2004;118:493–504. doi: 10.1016/j.cell.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Liu X, Zhang B, Guo Y, Liang Q, Wu C, Wu L, et al. Down-regulation of AP-4 inhibits proliferation, induces cell cycle arrest and promotes apoptosis in human gastric cancer cells. PLoS One. 2012;7:e37096. doi: 10.1371/journal.pone.0037096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuilman T, Michaloglou C, Mooi WJ. Peeper DS. The essence of senescence. Genes Dev. 2010;24:2463–2479. doi: 10.1101/gad.1971610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006;127:265–275. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Lanigan F, Geraghty JG, Bracken AP. Transcriptional regulation of cellular senescence. Oncogene. 2011;30:2901–2911. doi: 10.1038/onc.2011.34. [DOI] [PubMed] [Google Scholar]
- Gil J, Bernard D, Martinez D, Beach D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat Cell Biol. 2004;6:67–72. doi: 10.1038/ncb1077. [DOI] [PubMed] [Google Scholar]
- Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature. 1999;397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- Alani RM, Young AZ, Shifflett CB. Id1 regulation of cellular senescence through transcriptional repression of p16/Ink4a. Proc Natl Acad Sci USA. 2001;98:7812–7816. doi: 10.1073/pnas.141235398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Lohuizen M, Verbeek S, Scheijen B, Wientjens E, van der Gulden H, Berns A. Identification of cooperating oncogenes in E mu-myc transgenic mice by provirus tagging. Cell. 1991;65:737–752. doi: 10.1016/0092-8674(91)90382-9. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Alexander WS, Barri G, Klinken SP, Adams JM. Novel zinc finger gene implicated as myc collaborator by retrovirally accelerated lymphomagenesis in E mu-myc transgenic mice. Cell. 1991;65:753–763. doi: 10.1016/0092-8674(91)90383-a. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guney I, Wu S, Sedivy JM. Reduced c-Myc signaling triggers telomere-independent senescence by regulating Bmi-1 and p16(INK4a) Proc Natl Acad Sci USA. 2006;103:3645–3650. doi: 10.1073/pnas.0600069103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herold S, Wanzel M, Beuger V, Frohme C, Beul D, Hillukkala T, et al. Negative regulation of the mammalian UV response by Myc through association with Miz-1. Mol Cell. 2002;10:509–521. doi: 10.1016/s1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- Wu S, Cetinkaya C, Munoz-Alonso MJ, von der Lehr N, Bahram F, Beuger V, et al. Myc represses differentiation-induced p21CIP1 expression via Miz-1-dependent interaction with the p21 core promoter. Oncogene. 2003;22:351–360. doi: 10.1038/sj.onc.1206145. [DOI] [PubMed] [Google Scholar]
- Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: an alliance against the epithelial phenotype. Nat Rev Cancer. 2007;7:415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- Ansieau S, Bastid J, Doreau A, Morel AP, Bouchet BP, Thomas C, et al. Induction of EMT by twist proteins as a collateral effect of tumor-promoting inactivation of premature senescence. Cancer Cell. 2008;14:79–89. doi: 10.1016/j.ccr.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Liu Y, El-Naggar S, Darling DS, Higashi Y, Dean DC. Zeb1 links epithelial-mesenchymal transition and cellular senescence. Development. 2008;135:579–588. doi: 10.1242/dev.007047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri GP, Lee X, Basile G, Acosta M, Scott G, Roskelley C, et al. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc Natl Acad Sci USA. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, et al. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle. 2011;10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.