Abstract
Glioblastomas (GBMs) are devastating tumors of the central nervous system, with a poor prognosis of 1-year survival. This results from a high resistance of GBM tumor cells to current therapeutic options, including etoposide (VP-16). Understanding resistance mechanisms may thus open new therapeutic avenues. VP-16 is a topoisomerase inhibitor that causes replication fork stalling and, ultimately, the formation of DNA double-strand breaks and apoptotic cell death. Autophagy has been identified as a VP-16 treatment resistance mechanism in tumor cells. Retinoblastoma protein (RB) is a classical tumor suppressor owing to its role in G1/S cell cycle checkpoint, but recent data have shown RB participation in many other cellular functions, including, counterintuitively, negative regulation of apoptosis. As GBMs usually display an amplification of the EGFR signaling involving the RB protein pathway, we questioned whether RB might be involved in mechanisms of resistance of GBM cells to VP-16. We observed that RB silencing increased VP-16-induced DNA double-strand breaks and p53 activation. Moreover, RB knockdown increased VP-16-induced apoptosis in GBM cell lines and cancer stem cells, the latter being now recognized essential to resistance to treatments and recurrence. We also showed that VP-16 treatment induced autophagy, and that RB silencing impaired this process by inhibiting the fusion of autophagosomes with lysosomes. Taken together, our data suggest that RB silencing causes a blockage on the VP-16-induced autophagic flux, which is followed by apoptosis in GBM cell lines and in cancer stem cells. Therefore, we show here, for the first time, that RB represents a molecular link between autophagy and apoptosis, and a resistance marker in GBM, a discovery with potential importance for anticancer treatment.
Keywords: cell death, cancer stem cells, tumor suppressor, VP-16, central nervous system tumor, macroautophagy
Glioblastoma (GBM) is the most prevalent form of primary central nervous system tumors with highly aggressive characteristics.1, 2 The average survival of GBM patients is 1 year and this prognosis has not been improved over the last decades,3 even with the adoption of antiangiogenic treatments, which only improves the condition of a subgroup of patients. Treatment for GBM patients consists of surgical resection, as well as radiotherapy and chemotherapy with temozolomide;4 nevertheless, the rate of recurrence is extremely high.5 The chemotherapeutic agent etoposide (VP-16) is generally reserved for treating recurrent tumors, in high doses, limiting its effectiveness because of acute and cumulative toxicity to normal tissues.6 Thus, a better understanding of the molecular regulation of VP-16 resistance mechanisms is important for the development of a more efficient therapy.
VP-16 is a topoisomerase type II inhibitor, also used for osteosarcoma, bladder and testicular tumor treatments.7 Biochemically, the inhibition of topoisomerases leads to replication fork stalling and, ultimately, formation of DNA double-strand breaks and apoptosis.8 Autophagy has been identified as a mechanism of tumor resistance to VP-16-induced apoptosis in tumor cells,9, 10, 11 including GBM cells. Katayama et al.12 showed that the pharmacological inhibition of the autophagic process, in its final steps, increased the apoptotic rate induced by the compound. This same cross talk between autophagy and apoptosis has also been shown for other chemotherapy agents, including temozolomide, arsenic trioxide, resveratrol, camptothecin, cisplatin and 5-fluoracil, in many types of tumor cells12, 13, 14, 15, 16 But still, the molecular mechanisms regulating the interplay between these two events are poorly understood.
Mutations in the retinoblastoma (RB) susceptibility gene (RB1) lead to childhood retinal cancer.17 This gene encodes a phosphoprotein that negatively regulates the cell cycle, and this activity is critical for the classic RB-mediated tumor suppression function.18 Upon mitogenic signaling, RB protein is inhibited by hyperphosphorylation, which disrupts RB transcriptional repression complexes to allow G1 to S phase transition.19 Knudsen et al.20 showed that RB also induces S-phase arrest, triggered by the intra S-phase checkpoint in the presence of DNA-damaging agents, such as cisplatin and VP-16. RB also participates in other cellular processes, such as terminal cell differentiation and maintenance of genetic stability.18, 21
Counterintuitively, evidences from the literature have also suggested that RB can exert an anti-apoptotic function. Haas-Kogan et al.22 showed that the expression of RB decreased radiation-induced apoptosis in the osteosarcoma RB-deficient cells Saos-2. It has also been shown that RB is cleaved by caspases during apoptosis induced by VP-16 and tumor necrosis factor-α (TNF-α).23, 24 The caspase-resistant RB, engineered in mice, inhibits inflammation-induced cell death in vivo,25, 26 and the TNF receptor 1 triggered apoptosis in mouse embryonic fibroblasts (MEFs).25 Moreover, the expression of caspase-resistant RB in cerebellar granular neurons reduces apoptosis caused by potassium deprivation.27 In addition, MEFs displaying a conditional knockout of RB are more susceptible to VP-16 and cisplatin-induced cell death.20 Chau et al.28 have further demonstrated, by using targeted mutations on the RB1 gene, that RB inhibition of VP-16-induced apoptosis is independent of RB growth suppression function.
The RB pathway is altered in 70% of human cancer types.29 In GBM, this pathway is altered in 78% of the cases, although mutations and homozygotic deletions of the RB1 gene itself appear in only 11% of them.30 Instead, the RB pathway is preferentially altered at components that lead to RB inactivation by hyperphosphorylation, which leads to suppression of its cell cycle blocker function.19
In this work, we tested whether RB, even inactivated by hyperphosphorylation, could promote resistance to VP-16 in GBM and in GBM cancer stem cells, which are more resistant to chemo- and radiotherapy2, 31, 32, 33, 34. We show here, for the first time, that RB is involved in the regulation of an interplay between autophagy and apoptosis, and promotes resistance of GBM cells to VP-16. Moreover, these results show that the hyperphosphorylated RB, found in GBM, is not only a determinant for the high levels of cell proliferation, but is also a determinant for chemotherapy resistance.
Results
RB knockdown using RNAi in GBM cell lines and in GSCs
To investigate the role of RB protein in the response of GBM cells to VP-16, we chose two GBM cell lines that display hyperphosphorylated RB (Supplementary Figure S1): U87 MG, an American Type Culture Collection (ATCC) cell line that does not express the CDK inhibitor p1635, and GBM95, isolated in our laboratory through biopsy of a recurrent GBM tumor36 and the GBM stem cell (GSC) line OB1 (refs 37, 38). By Immunoblotting analysis we were able to detect similar levels of total RB protein in GBM cell lines U87 and GBM95, and in GSC OB1 (Supplementary Figure S1A). Besides, RB phosphorylation at serine 807/811 was detected in the GBM cell lines and in GSCs (Supplementary Figure S1B). This confirms, as expected35, that the cell lines used in this work present hyperphosphorylated RB.
The efficiency of RB knockdown was of ∼70% in both GBM cell lines, and ∼80% in GSCs (Figures 1a–d). Twenty-four hours after transfection, 30% (Figures 1a and b) and 20% (Figures 1c and d) of residual RB was detected by western blotting in the silenced RB group (siRNA-RB) in comparison with the off-target non-silenced group (siRNA-Neg), in the GBM cell lines and in GSC OB1, respectively (Figures 1a–d). In all subsequent experiments, VP-16 treatment was initiated 24 h post-small interfering RNA (siRNA) transfection.
Figure 1.
RB knockdown in GBM cells and GSCs. (a) Representative western Blotting image of three independent experiments, comparing the levels of RB protein between non-silenced (siRNA-Neg) and silenced groups (siRNA-RB) of U87 and GBM95 cell lines. α-Tubulin was used as loading control. (b) Histogram comparing RB levels between silenced (siRNA-RB) and non-silenced groups (siRNA-Neg) of GBM cell lines U87 and GBM95. Data are represented as mean and S.E.M. of three independent experiments. (c) Representative western blotting image of three independent experiments, comparing the levels of RB protein between non-silenced (siRNA-Neg) and silenced groups (siRNA-RB) of GSCs OB1. α-Tubulin was used as loading control. (d) Histogram comparing RB levels in GSCs OB1 between silenced (siRNA-RB) and non- silenced groups (siRNA-Neg). Data are represented as mean and S.E.M. of three independent experiments. (e) VP-16 dose–response curves of GBM cell lines U87 and GBM95, and GSCs OB1. Graphic shows the % of survival as measured by MTT method by the indicated dose treatment of VP-16 for 24 h. Data are represented as mean and S.E.M. of three independent experiments
RB knockdown increased VP-16-induced apoptosis
To test whether RB knockdown could increase VP-16-induced apoptosis, we compared the percentage of terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL)-positive cells between silenced and non-silenced groups, 24 h after VP-16 treatment, in U87 and GBM95 cells. The concentration of 1 μM of VP-16 was chosen, as it decreased cell viability to ∼50% in both U87 and GBM95 cell lines, as seen in a VP-16 dose–response curve 24 h after treatment (Figure 1e). Twenty-four hours after 1 μM of VP-16 treatment, U87 cell cultures showed ∼2.5 times more TUNEL-positive cells in the RB-silenced group compared with the off-target non-silenced group (siRNA-Neg) (Figures 2a and c). As for GBM95 cells, the percentage of TUNEL-positive cells increased fourfold in the silenced RB group (Figures 2a and c). In both the GBM cell lines tested, RB knockdown also increased the cell death level measured by ethidium homodimer incorporation, which is thought to occur only in cells with disrupted plasma membrane39 (data not shown). Therefore, in both GBM cell lines tested, RB knockdown was capable of potentiating VP-16 treatment through increased apoptosis.
Figure 2.
RB knockdown increased apoptosis induced by VP-16. (a) Representative images of three independent experiments of TUNEL assay (red) in U87 and GBM95 cells after 24 h of 1 μM VP-16 treatment; experimental groups as indicated. Nuclei were stained with DAPI. Arrows indicate TUNEL-positive cells. Scale bar=10 μm. (b) Representative images of three independent experiments of TUNEL assay (red) in OB1 GSCs cells after 24 h of 30 μM VP-16 treatment; experimental groups as indicated. Nuclei were stained with DAPI. Arrows indicate TUNEL-positive cells. Scale bar=10 μm. (c) Percentage of TUNEL-positive U87 (black bars) and GBM95 (white bars) cells 24 h after 1 μM VP-16 treatment; experimental groups as indicated. **P<0.01 and ***P<0.005 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (d) Percentage of TUNEL-positive OB1 cells 24 h after VP-16 treatment in each experimental group indicated. ***P<0.005 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (e) Percentage of cleaved caspase-3 positive U87 (black bars) cells 24 h after 1 μM VP-16 treatment; experimental groups as indicated. *P<0,05 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (f) Representative phospho-p53 (ser 15) immunoblotting for U87 cells 24 h post-VP-16 treatment; experimental groups as indicated. α-Tubulin was used as loading control
GSCs have now been positioned in a centre stage for the development of cancer therapies because of its capacity for self-renewal and generation of varied differentiated cell types within a tumor mass.31 As we observed that RB knockdown increased VP-16-induced apoptosis in GBM cell lines, we wondered whether the same would be observed in GSCs.
To test this hypothesis, we used the GSC line OB1 refs37, 38, which has a high capacity to self-renew. This cell line expresses Nestin, a neural stem cell marker, SOX-2, an embryonic stem cell marker and CD133, a GSC marker (Supplementary Figure S2). We silenced RB in OB1 cells (Figures 1c and d) and compared the percentage of TUNEL-positive cells between silenced and non-silenced groups after VP-16 treatment. As seen in the VP-16 dose–response curves (Figure 1e), OB-1 GSCs are more resistant to VP-16 treatment than the GBM cell lines tested (U87 and GBM95); therefore, in this assay we used VP-16 at a concentration of 30 μM, as it resulted in 80% of OB1 survival (Figure 1e).
Our results showed that RB knockdown increased the percentage of TUNEL-positive OB1 cells by threefold after VP-16 treatment, resulting in over 85% of TUNEL-positive cells (Figures 2b and d). This result shows that RB knockdown increases VP-16-induced apoptosis in GSCs in a similar manner than that previously observed for GBM cells (Figure 2c). Cleaved caspase-3 immunostaining confirmed the effectiveness of RB silencing to increase VP-16-induced apoptotic cell death (Figure 2e and Supplementary Figure S3). Moreover, RB silencing increased p53 phosphorylation at serine 15 residue (Figure 2f), which leads to p53 accumulation and induction of apoptosis40 suggesting that RB knockdown induced an increase in p53-dependent apoptosis.
RB knockdown increased VP-16-induced apoptosis even in a p53-independent manner
To test whether RB knockdown could have a broader effect on GBM cells, we used three other GBM cell lines: A172, T98G, which is p53 mutated, and U373, which expresses low levels of RB. We performed RB knockdown in these cell lines (Figure 3c) and compared the percentage of TUNEL-positive cells between silenced (siRNA-RB plus VP-16) and non-silenced (siRNA-Neg plus VP-16) groups after VP-16 treatment. U373 cells, which naturally present lower levels of RB expression (Figure 3c), exhibited the highest sensitivity to VP-16-induced apoptosis (Figures 3a and b). Furthermore, RB knockdown in U373 cells increased even more the percentage of TUNEL-positive cells (Figures 3a and b). These results further confirm that lower levels of RB expression favors VP-16-induced apoptosis. RB knockdown also increased the levels of TUNEL-positive nuclei in A172 and even in T98G cells (Figures 3a and b). This later cell line express mutant p53, which is not activated, as measured by serine 15 phosphorylation, in the presence of VP-16 (Supplementary Figure S4). These results suggest that RB knockdown is able to increase VP-16-induced apoptosis independently of p53 activation, although the combinatory treatment is more efficient in cells that express wild-type p53. This data set shows that RB knockdown is able to increase VP-16-induced apoptosis in a wide range of genetic backgrounds of GBM cells.
Figure 3.
RB knockdown increased apoptosis induced by VP-16 in different GBM cell lines. (a) Representative images of three independent experiments of TUNEL assay (red) using A172, T98G and U373 cells after 24 h of 1 μM VP-16 treatment; experimental groups as indicated. Nuclei were stained with DAPI. Arrows indicate TUNEL-positive cells. Scale bar=10 μm. (b) Percentage of TUNEL-positive A172 (checkered bars), T98G (horizontal striped bars) and U373 (gray bars) cells 24 h after 1 μM VP-16 treatment; experimental groups as indicated. **P<0.01 and ***P<0.005 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (c) Representative western Blotting image of three independent experiments, comparing the levels of RB protein between non-silenced (siRNA-Neg) and silenced groups (siRNA-RB) of A172, T98G and U373 cell lines. α-Tubulin was used as loading control
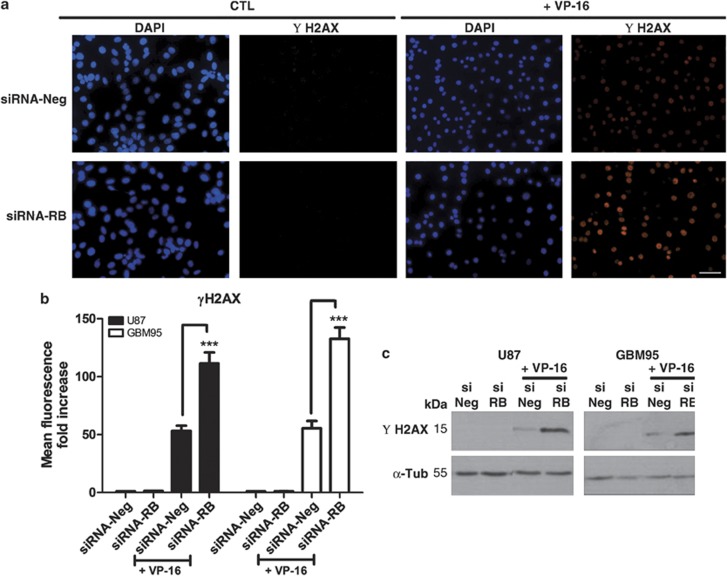
RB knockdown increased VP-16-induced DNA double-strand breaks
VP-16 generates DNA double-strand breaks that accumulate and ultimately trigger apoptosis.41 Therefore, we questioned whether the increase in VP-16-induced apoptosis, seen after RB knockdown, could be due to an increase in DNA double-strand breaks. To test that, we compared the levels of γ-H2AX staining between RB-silenced and non-silenced groups. γ-H2AX is a modified histone, which is γ-phosphorylated after DNA double-strand break.42 Our results showed that incubation with VP-16 for 1 h increased the fluorescence intensity of γ-H2AX immunostaining in both GBM cell lines tested (Figures 4a and b). However, the increase in γ-H2AX immunostaining induced by VP-16 was significantly higher in the RB-silenced group (siRNA-RB plus VP-16) when compared with the siRNA off-target control group (siRNA-Neg plus VP-16) (Figures 4a and b). Western blotting analysis confirmed that the amount of γ-H2AX was significantly higher in the RB-silenced group after VP-16 treatment (siRNA-RB plus VP-16) when compared with the siRNA off-target control group (siRNA-Neg plus VP-16), suggesting that VP-16 was more efficient to induce DNA damage in the RB-silenced group (Figure 4c).
Figure 4.
RB knockdown increased DNA double-strand breaks induced by VP-16. (a) Representative images of three independent experiments of immunofluorescence staining with anti-γH2AX antibody (red) after 1 h of VP-16 treatment in U87 cells; experimental groups as indicated. Nuclei were stained with DAPI. Scale bar=10 μm. (b) Histogram shows fold changes of red fluorescence intensity (mean red fluorescence/total cell number) over siRNA-Neg control group; after 1 h of VP-16 treatment of U87 (black bars) and GBM95 (white bars) cells; experimental groups as indicated. ***P<0.005 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (c) Representative γ-H2AX immunoblot of U87 and GBM95 cells 1 h post-VP-16 treatment; experimental groups as indicated. α-Tubulin was used as loading control
RB knockdown disrupted VP-16-induced autophagic process
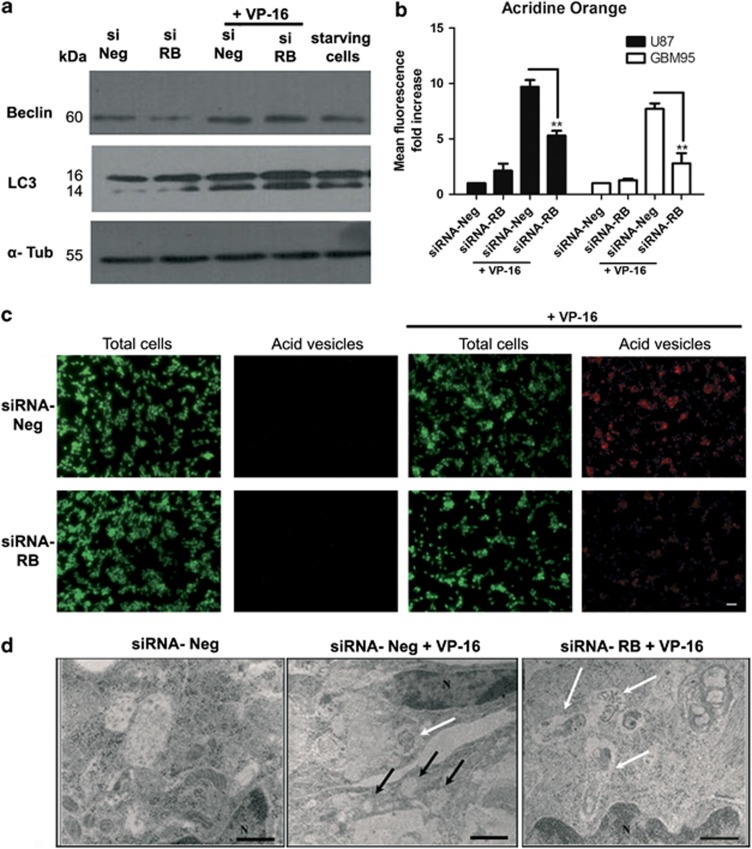
It has been postulated that autophagy may be a resistance mechanism of tumor cells in the presence of an apoptotic stimulus. For this reason, we decided to investigate whether VP-16 treatment could induce autophagy in GBM cells, and whether RB silencing could interfere with it. As autophagy is a multi-step process43 regulated at different points, we performed experiments to evaluate separate stages of this process.
First, we measured the expression of Beclin-1, a marker of autophagy induction, using starving cells as positive controls. As shown in Figure 5a, VP-16 treatment increased Beclin-1 expression in both non-silenced (siRNA-Neg plus VP-16) and silenced (siRNA-RB plus VP-16) groups, in the same manner, indicating that RB silencing does not alter VP-16-dependent autophagy induction. Next, we compared the levels of the lipidated form of microtubule-associated protein 1A/1B-light chain 3 (LC-3 II), among our experimental groups. In the autophagic process, LC-3 I is lipidated, becoming LC-3 II, which is incorporated into autophagosome membranes, where it is degraded when autophagosomes fuse with lysosomes to form autophagolysosomes.44 As seen in Figure 5a, VP-16 treatment increased, in all groups, the level of LC-3 II, which is detected at ∼14 kDa. Furthermore, the level of LC-3 II was higher in the silenced (siRNA-RB plus VP-16) than in the non-silenced (siRNA-Neg plus VP-16) group (Figure 5a). This could be due to a higher level of LC3-II formation, or to a lower level of LC3-II degradation in autophagolysosomes. As autophagolysosomes are acid compartments, we compared, through acridine orange labeling, the formation of acid vesicles, a final step in autophagic process.43 As noticed from Figures 5b and c, the increase in red fluorescence, depicting acid vesicles, is approximately two times lower in the silenced group (siRNA-RB plusVP-16) when compared with that of the non-silenced group (siRNA-Neg plus VP-16) in both cell lines, showing that VP-16-induced acid vesicle formation is lower in the RB-silenced group. This result, in conjunction with the accumulation of LC-3 II, suggests that RB silencing blocks VP-16-induced autophagic flux in its final steps. Electron microscopy data corroborates this hypothesis (Figure 5d), as it shows that the VP-16-induced autophagic vacuoles in the RB-silenced group (siRNA-RB plus VP-16) are frequently filled with electron dense material, similar to the cytosol (indicated by white arrows), possibly representing vacuoles containing non-degraded material due to autophagic flux blockage. The non-silenced group (siRNA-Neg plus VP-16) most frequently presents clear vacuoles (indicated by black arrows), which could represent vacuoles with total digested content.45
Figure 5.
RB knockdown impaired VP-16-induced autophagy. (a) Representative Beclin-1 and LC-3 immunoblots for U87 cells 24 h post-VP-16 treatment (1 μM); experimental groups as indicated. α-Tubulin was used as loading control. Starved cells were used as positive control of autophagy induction. Starvation was induced by incubating the cells for 3 h in Krebs-Ringer buffer. (b) Graphic shows changes in intensity of red fluorescence (indicating acid vesicles labeled with acridine orange) in U87 (black bars) and GBM95 (white bars) cells after 24 h of VP-16 treatment (1 μM). Changes in red fluorescence were represented as fold increase (mean red fluorescence/total cell number) over control (siRNA-Neg group); experimental groups as indicated. **P<0.01 by ANOVA and Tukey's post test. Data are represented as mean and S.EM. of three independent experiments. (c) Representative images of three independent experiments of acridine orange vital staining of U87 cells after 24 h of 1 μM VP-16 treatment; experimental groups as indicated. Acid vesicles are stained in red and total cells are represented by the nuclear–cytoplasmic green staining. Scale bar=10 μm. (d) Transmission electron microscopy of VP-16-treated cells (1 μM for 24 h) shows the presence of content filled autophagic vacuoles (white arrows) in both non-silenced and silenced groups (siRNA-Neg plus VP-16 and siRNA-RB plus VP-16 groups, respectively). Although non-silenced cells also show several clear vacuoles of which most of the content has been degraded (black arrows), RB-silenced cells show mainly content-filled vacuoles (white arrows), possibly indicating autophagic flux blockage. Control cells (siRNA-Neg) show vacuoles with hardly any content. N, nucleus. Scale bar=6 μm
RB knockdown blocked VP-16-induced autophagic flux
To test whether RB knockdown in fact blocked VP-16-induced autophagic flux in its final steps, we performed western blotting experiments to detect p62 levels. This protein is degraded in autophagolysosomes and, thus, its accumulation indicates a blockage in autophagic flux.43 Our results showed that VP-16 treatment induced a significant reduction of p62 levels in U87 cells (siRNA-Neg plus VP-16), which is expected when autophagy is induced. On the contrary, RB knockdown significantly increased p62 levels (siRNA-RB plus VP-16) (Figures 6c and e), thus confirming that RB knockdown blocked VP-16-induced autophagic flux.
Figure 6.
RB knockdown blocked VP-16-induced autophagic flux. (a) Representative confocal images of three independent experiments showing LAMP 1 – a lysosome marker (red) and LC3 – a autophagosome marker (green) colocalization in U87 cells after 24 h of 1 μM VP-16 treatment; experimental groups as indicated. Nuclei were stained with DAPI. Scale bar=20 μm. (a′) Higher magnifications of siRNA-Neg plus VP-16 and siRNA-RB plus VP-16 conditions images. Arrows indicate cells with LAMP 1/LC3 colocalization. Arrowheads indicate cells without LAMP 1/LC3 colocalization. Scale bar=20 μm. (b) Histogram showing the LAMP1/LC3 colocalization rate in U87 cells after 24 h of VP-16 treatment. Experimental groups as indicated. ***P<0.005 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (c) Histogram shows fold changes of p62 levels over siRNA-Neg control group; after 24 h of VP-16 treatment of U87 cells; experimental groups as indicated. ***P<0.005 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (d) Histogram shows fold changes VRK1 levels over siRNA-Neg control group; after 24 h of VP-16 treatment of U87 cells; experimental groups as indicated. *P<0.05 by ANOVA and Tukey's post test. Data are represented as mean and S.E.M. of three independent experiments. (e) Representative p62 and VRK1 immunoblots for U87 cells 24 h post-VP-16 treatment; experimental groups as indicated. α-Tubulin was used as loadisng control
To test whether the autophagic blockage induced by RB knockdown would be at the stage of autophagosomes fusion to lysosomes, we obtained confocal images of double staining to localize LC3, an autophagosome marker, and LAMP 1, a lysosome marker. Our results showed that VP-16 treatment induced the colocalization of LAMP 1 and LC3, and that RB knockdown decreased this colocalization (Figures 6a, a′ and b). Therefore, RB knockdown blocks VP-16-induced autophagic flux by somehow inhibiting the fusion of autophagosomes with lysosomes.
It has been shown that p53 induces the expression of damage-regulated autophagy modulator (DRAM).46 DRAM is a lysosomal protein that has been shown to regulate autophagy and is necessary for apoptosis.46 To test whether DRAM activity is affected by RB knockdown, we performed western blotting experiments to detect the levels of vaccinia-related kinase 1 (VRK1), which is downregulated by DRAM,47 as a way to indirectly analyze the activation of DRAM. Our results showed that VP-16 reduced the levels of VRK1 in U87 cells, and that RB knockdown significantly reduced it even more (Figures 6d and e). These results are consistent with RB knockdown increasing VP-16-induced DRAM activation.
Discussion
RB is a classical tumor suppressor owing to its role as a regulator of G1 to S phase checkpoint; however, novel functions exerted by this protein are being described in the literature, including the negative regulation of apoptosis.18, 22, 23, 24, 25, 26, 27 Here we showed that RB, even in its hyperphosphorylated state, when it is inactivated for its classical cell cycle blocker function, is able to block apoptosis in GBM cells. This result shows that RB anti-apoptotic function is independent from its cell cycle function, in accordance with what Chau et al.28 have shown. Furthermore, the presence of hyperphosphorylated RB in tumor cells, such as GBM, may not only allow high rates of cellular proliferation, but also resistance to therapy. The involvement of RB in VP-16 treatment response in GBM cells has not been explored so far. Our results showed that RB knockdown increased VP-16-induced apoptosis in GBMs, as well as in GSCs. It has already been described that conditional RB knockout increases susceptibility of MEFs to VP-16-induced cell death.20 Here we showed that the reduction of RB expression can also sensitize GBMs and GSCs to VP-16-induced apoptosis, demonstrating that RB anti-apoptotic function is still maintained in these tumor cells. The fact that we also observed this phenomenon in a recurrent GBM cell line (GBM95)36 increases the potential of clinical translation, as VP-16 treatment is mostly reserved for recurrent GBM patients.6
In this work we also showed that RB knockdown increased DNA double-strand breaks induced by VP-16 treatment in GBM cells, suggesting that RB normally blocks VP-16-induced DNA damage. Bosco et al.48 have shown that RB conditional knockout increases DNA double-strand breaks induced by cisplatin or camptothecin treatment due to ongoing replication following chemotherapeutic treatment in RB-deficient cells. Here we show that this property of RB is also valid for VP-16 treatment and is kept in GBM cells, indicating once again that the presence of RB in tumor cells can influence the response to chemotherapy. We believe that an increase in DNA double-strand breaks, followed by RB knockdown, leads to higher levels of apoptosis following VP-16 treatment in GBM cells, as we also observed an increase in p53 phosphorylation at serine 15 residue and in VP-16-induced apoptosis. However, our results showed that RB knockdown also potentiated VP-16 cytoxicity in the absence of p53 activation, showing that in different genetic backgrounds RB knockdown may act differently to increase VP-16-induced apoptosis.
The literature shows that autophagy may serve as a resistance mechanism in tumor cells, including GBM, as the pharmacological inhibition of its final steps increases chemotherapy-induced apoptosis.9, 10, 11, 13, 14, 15, 16 It has been postulated that upon stress stimuli, autophagy is able to remove mitochondria that had undergone outer membrane permeabilization, which would explain autophagy's role on apoptosis inhibition49 Here we showed that RB protein positively regulates the progression of VP-16-induced autophagic flux in GBM cells, and that at the same time it reduces VP-16-induced apoptosis. Huang et al.50 have shown that RB cleavage by caspase and bafilomycin 1, an inhibitor of the vacuolar type H(+)-ATPase, treatment had the same effect on TNF-α-induced cell death, which suggests that RB might have a role on acidification and/or vesicle trafficking. However, to our knowledge, this is the first report to show RB participation in the interplay between autophagy and apoptosis in the context of tumor resistance to therapy. Besides, we also show effectively that RB participates in the process of autophagosome/lysosome fusion.
Our work gives a valuable contribution not only to the understanding of the molecular mechanisms driving GBM resistance to VP-16, but also adds molecular regulation information to the body of knowledge constructed with numerous works, showing that in a universal manner pharmacological inhibition of autophagic flux improves chemotherapy-induced apoptosis.51, 52, 53
Finally, the study of the molecular regulation of GBM resistance to VP-16 is of utmost importance, as the effectiveness of VP-16 treatment is restricted because of its use in high doses, and consequent side effects, in recurrent GBM patients.6 Our work gives consistent evidences that the presence of hyperphosphorylated RB in GBM cells contributes to VP-16 resistance. This information creates the possibility of the adoption of a low-dosage VP-16 therapy for patients with non-RB-expressing GBM, providing a more effective therapy, with fewer side effects. Our results showing that the U373 cell line, which expresses low levels of RB, is more sensitive to VP-16 treatment compared with the other cell lines corroborates this possibility. For RB-expressing GBM patients, the combination of RB silencing with VP-16 would be one way of improving, in the future, the effectiveness of the therapy. Currently, there is a clinical trial involving the use of RNAi in GBM patients, which attests the viability of RNAi as therapeutic strategy for GBM, despite the need for improved delivery efficacy.54
Materials and Methods
Cell culture
Human GBM cell lines used were as follows: U87MG, which does not express the CDK inhibitor p1635 A172 (expresses RB and wild-type p53); T98G (mutant for p53) and U373MG (expresses low levels of RB) are originally from (ATCC, Manassas, VA, USA); GBM95 was isolated in our laboratory through the biopsy of a recurrent GBM tumor.36 All cell lines were maintained in Dulbecco's modified Eagle's medium-F12 (DMEM-F12) (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum. Culture flasks were kept at 37 °C and 5% CO2.
Non-adherent cultures of human GSCs (OB1),37, 38 which express RB and wild-type p53, were grown as floating spheres in 75 cm2 tissue culture flasks (2500–5000 cells/cm2), in DMEM-F12 supplemented with B27, N2 and G5 (supplements were purchased from Invitrogen, Carlsbad, CA, USA). Spheres were dissociated each week with a renewal of two third of the culture medium, as previously described.55 Cultures were maintained at 37 °C and 5% CO2.
Starvation was induced by incubating the cells for 3 h in Krebs-Ringer buffer (118 mM NaCl, 4.7 mM KCl, 1.2 mM KH2PO4, 1.2 mM MgSO4, 4.2 mM NaHCO3, 2 mM CaCl2, 10 mM glucose, 200 mM sulphinpyrazone and 10 mM Hepes, pH 7.4), as previously described.43
siRNA and transfection
The siRNA targeting RB (siRNA-RB) was synthesized by Ambion (Austin, TX, USA) (5′-GUUGAUAAUGCUAUGUCAA3′).50 Fluorescent non-targeting BLOCK-it siRNA (Invitrogen) was used as a negative control (siRNA-Neg) and to estimate transfection efficiency. Following normalization, we used the concentration of 30 nM for all siRNAs in further experiments, as it resulted in 80% transfection efficiency in both cell lines used. For transient gene silencing, cells displaying ∼70% confluence were transfected with siRNAs using RNAi Transfection Reagent (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions. After transfection, cells were cultured for 24 h, followed by treatment with 1 μM VP-16 (Instituto Biochimio, Rio de Janeiro, Brazil) for the indicated periods of time, or prepared for western blotting.
OB1 cells, at the 15th passage, were split as previously described,55 and were transfected with siRNA-RB or with non-targeting control. OB1 oncospheres were transiently transfected using the Amaxa Nucleofector Electroporator (Amaxa Biosystems, Gaithersburg, MD, USA). Briefly, cells were centrifuged (1000 rpm) and resuspended in 100 μl nucleofector solution (Amaxa Biosystems). siRNA (300 nM) were added to tubes and electroporation was performed using optimal Amaxa Nucleofector program X-005 (70% transfection efficiency). Twenty-four hours after transfection, cells were treated with 30 μM VP-16 (Biochimio) or prepared for western blotting.
Immunoblotting
Proteins from GBM cell lines were extracted in RIPA buffer (25 mM Tris-HCl pH 7.4, 1 mM EDTA, 0.1% SDS, 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 1 mM phenylmethylsulfonyl fluoride) and protease inhibitor cocktail (Sigma, St Louis, MO, USA). Protein concentration was measured using Bradford assay. Thirty micrograms of total proteins were loaded on SDS-PAGE gels, transferred into PVDF membrane and probed with primary antibodies anti-human RB protein (1 : 1000 – BD Pharmingen, San Diego, CA, USA), anti-RB 851 (1 : 5000),56 anti-H2AX (1 : 1000 – Millipore, Bedford, MA, USA), anti- phospho-p53 (ser 15) (1 : 1000 – Cell Signaling Technology, Beverly, MA, USA), anti-Beclin-1 (1 : 1000 – Cell Signaling), anti-LC-3 1A/1B (1 : 1000 – Cell Signaling), anti-P62/SQSTM1 (1 : 1000 – Cell Signaling), anti-VRK1 (1 : 1000 – Cell Signaling) and anti-α-Tubulin (1 : 10 000 – Sigma) overnight at 4 °C. HRP-conjugated secondary antibodies (anti-mouse and anti-rabbit, 1 : 3000 – Invitrogen) were incubated for 2 h at room temperature. Proteins were visualized by using ECL reagents as recommended by the manufacturer (Thermo, Waltham, MA, USA).
Transmission electron microscopy
Cells were fixed in fixative solution containing 2.5% glutaraldehyde, 4% paraformaldehyde and 0.1 M sodium cacodylate buffer pH 7.2 for 2 h at RT. The pellet was then washed twice with 0.1 M cacodylate buffer and post fixed in 1% OsO4 solution in 0.1 M cacodylate buffer for 40 min at RT. Following three times wash in PBS, the pellet was dehydrated with ethanol (35–100% 15 min for each step) and infiltrated overnight in a mix 2 : 1 ethanol: EPON. The next day, the mixture was changed to 1 : 1 ethanol: EPON for 24 h, and then the sample was embedded in EPON and placed at 60 °C for 48 h for polymerization. Sections were made using a Leica Ultramicrotome (Buffalo Grove, IL, USA) at 60 nm, collected using cupper grids and counterstained with uranyl acetate and lead citrate. Images were taken using a Zeiss EM900 transmission electron microscope (Zeiss, Oberkochen, Germany).
Immunofluorescence
Cells were fixed with 4% paraformaldehyde and permeabilized with 0.25% Triton X-100. Non-specific binding was blocked with 5% bovine serum albumin and the samples were then incubated with anti-LAMP1 conjugated to Cy3 (Sigma), anti-γH2AX (Millipore), anti-cleaved caspase-3 (Cell Signaling) or anti-LC-3 1A/1B (Cell Signaling) overnight at 4 °C. Cells were then incubated with anti-mouse ALEXA fluor-594 (Invitrogen), or anti-rabbit ALEXA fluor-488 (Invitrogen) for 2 h at room temperature. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole dihydrochloride (DAPI).
Fluorescence images were captured with the Nikon Eclipse TE300 microscope (Nikon do Brasil LTDA, São Paulo, Brazil) equipped with a CoolSNAP-Procf color, ROPER SCIENTIFICTM Photometrics digital camera (Nikon do Brasil LTDA), or using a confocal microscope (Leica TCS-SP5) equipped with a × 63 NA 1.40 oil-immersion objective. Cleaved caspase-3 immunostaining images were captured in Operetta High Content Imaging System (Perkin Elmer, Waltham, MA, USA) and quantification of the percentage of cleaved caspase-3-positive cells was done by the Harmony High Content Imaging and Analysis Software (Perkin Elmer).
To quantify H2AX staining, we used the ratio of fluorescence intensity (Image J software),57 over the total cell number (DAPI). The ratio obtained in siRNA-Neg control was used to normalize other experimental groups. Experiments were carried out in duplicates; for every experimental condition, at least 500 cells were counted.
Cell counting was done by using the Embryonic Stem Cell Counter (ESCC) software.58
The LAMP 1/LC3 colocalization rate was determined by confocal images using the software LAS AF (Leica Microsystems, Wetzlar, Germany).
TUNEL assay
TUNEL assay was performed according to manufacturer's instructions (In Situ Cell Death Detection Kit, Terminal Red – Roche Applied Science, Indianapolis, IN, USA). Nuclei were counterstained with DAPI. Fluorescence images were captured using the Nikon Eclipse TE300 microscope as above.
TUNEL quantification was done using the percentage of TUNEL-positive cells relative to total cells (DAPI). Experiments were carried out in duplicates, and for every experimental condition at least 500 cells were counted.
Cell counting was done by using the ESCC software.58
MTT cytotoxicity assay
For VP-16 dose–response curves, within the last 2 h of VP-16 treatment, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT – Sigma) was added to the media at a final concentration of 0.5 mg/ml. Cells were then resuspended and lysed in dimethyl sulfoxide. Absorbance was measured at 570 nm. Mean absorbance obtained for control condition was considered 100% of cell viability.
Supravital cell staining with acridine orange for acid vesicle formation
Cytoplasm and nuclei of acridine orange-stained cells normally fluoresce bright green and dim red, respectively, whereas acidic compartments fluoresce bright red. The intensity of red fluorescence is proportional to the degree of acidity.43 Acridine orange solution (1 mg/ml; Sigma) was used to stain cells for 20 min. Images of staining were obtained with the Nikon Eclipse TE300 microscope, as described above. To quantify acid vesicle formation, we used the ratio of red fluorescence intensity (Image J software)57 over total cell number (green staining). The ratio obtained for siRNA-Neg condition was used to normalize other experimental groups. Cell counting was done using the ESCC software58
Statistical analysis
GraphPad-Prism software was used to analyze data and plot curves. Data are represented as mean and S.E.M. ANOVA (analysis of variance) test and Tukey's post test were used to determine statistical significance of difference between data sets. P<0.05 was considered statistically significant.
Acknowledgments
We thank Rosenilde Carvalho de Holanda Afonso and Anna Carolina Carvalho da Fonseca for technical assistance. We thank Dr Jean Wang (UCSD) and Dr Leonardo Travassos (UFRJ) for donation of anti-RB 851 and anti-p62, respectively. We are grateful to Dr Silvya Stuchi Maria-Engler (USP) for donating the T98G cell line. This work was supported by the National Council for Scientific and Technological Development (CNPq), by the Brazilian Federal Agency for Support and Evaluation of Higher Education (CAPES), by the Carlos Chagas Filho Foundation for Research Support of the State of Rio de Janeiro (FAPERJ) and Cancer Foundation (Oncobiology Program UFRJ).
Glossary
- RB
retinoblastoma protein
- VP-16
etoposide
- GBM
glioblastoma
- siRNA
small interfering RNA
- RNAi
RNA interference
- GSC
GBM stem cell
- TNF-α
tumor necrosis factor-α
- MEF
mouse embryonic fibroblast
- TUNEL
terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling
- MTT
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- LC-3
microtubule-associated protein 1A/1B-light chain 3
- DMEM-F12
Dulbecco's modified Eagle's medium-F12
- DAPI
4',6-diamidino-2-phenylindole dihydrochloride
- DRAM
damage-regulated autophagy modulator
- VRK1
vaccinia-related kinase 1
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Raschellá
Supplementary Material
References
- Alves TR, Lima FR, Kahn SA, Lobo D, Dubois LG, Soletti R, et al. Glioblastoma cells: a heterogeneous and fatal tumor interacting with the parenchyma. Life Sci. 2011;89:532–539. doi: 10.1016/j.lfs.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Lima FR, Kahn SA, Soletti RC, Biasoli D, Alves T, da Fonseca AC, et al. Glioblastoma: therapeutic challenges, what lies ahead. Biochim Biophys Acta. 2012;1826:338–349. doi: 10.1016/j.bbcan.2012.05.004. [DOI] [PubMed] [Google Scholar]
- Maher EA, Furnari FB, Bachoo RM, Rowitch DH, Louis DN, Cavenee WK, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15:1311–1333. doi: 10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- Adamson C, Kanu OO, Mehta AI, Di C, Lin N, Mattox AK, et al. Glioblastoma multiforme: a review of where we have been and where we are going. Expert Opin Investig Drugs. 2009;18:1061–1083. doi: 10.1517/13543780903052764. [DOI] [PubMed] [Google Scholar]
- Grossman SA, Batara JF. Current management of glioblastoma multiforme. Semin Oncol. 2004;31:635–644. doi: 10.1053/j.seminoncol.2004.07.005. [DOI] [PubMed] [Google Scholar]
- Cheema TA, Kanai R, Kim GW, Wakimoto H, Passer B, Rabkin SD, et al. Enhanced antitumor efficacy of low-dose Etoposide with oncolytic herpes simplex virus in human glioblastoma stem cell xenografts. Clin Cancer Res. 2011;17:7383–7393. doi: 10.1158/1078-0432.CCR-11-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin EL, Osheroff N. Etoposide, topoisomerase II and cancer. Curr Med Chem Anti-Cancer Agents. 2005;5:363–372. doi: 10.2174/1568011054222364. [DOI] [PubMed] [Google Scholar]
- Karpinich NO, Tafani M, Rothman RJ, Russo MA, Farber JL. The course of etoposide-induced apoptosis from damage to DNA and p53 activation to mitochondrial release of cytochrome c. J Biol Chem. 2002;277:16547–16552. doi: 10.1074/jbc.M110629200. [DOI] [PubMed] [Google Scholar]
- Xie X, Le L, Fan Y, Lv L, Zhang J. Autophagy is induced through the ROS-TP53-DRAM1 pathway in response to mitochondrial protein synthesis inhibition. Autophagy. 2012;8:1071–1084. doi: 10.4161/auto.20250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abedin MJ, Wang D, McDonnell MA, Lehmann U, Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007;14:500–510. doi: 10.1038/sj.cdd.4402039. [DOI] [PubMed] [Google Scholar]
- Crowley LC, Elzinga BM, O'Sullivan GC, McKenna SL. Autophagy induction by Bcr-Abl-expressing cells facilitates their recovery from a targeted or nontargeted treatment. Am J Hematol. 2011;86:38–47. doi: 10.1002/ajh.21914. [DOI] [PubMed] [Google Scholar]
- Katayama M, Kawaguchi T, Berger MS, Pieper RO. DNA damaging agent-induced autophagy produces a cytoprotective adenosine triphosphate surge in malignant glioma cells. Cell Death Differ. 2007;14:548–558. doi: 10.1038/sj.cdd.4402030. [DOI] [PubMed] [Google Scholar]
- Guo XL, Li D, Hu F, Song JR, Zhang SS, Deng WJ, et al. Targeting autophagy potentiates chemotherapy-induced apoptosis and proliferation inhibition in hepatocarcinoma cells. Cancer Lett. 2012;320:171–179. doi: 10.1016/j.canlet.2012.03.002. [DOI] [PubMed] [Google Scholar]
- Li JL, Han SL, Fan X. Modulating autophagy: a strategy for cancer therapy. Chinese J Cancer. 2011;30:655–668. doi: 10.5732/cjc.011.10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippi-Chiela EC, Villodre ES, Zamin LL, Lenz G. Autophagy interplay with apoptosis and cell cycle regulation in the growth inhibiting effect of resveratrol in glioma cells. PLoS One. 2011;6:e20849. doi: 10.1371/journal.pone.0020849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pucer A, Castino R, Mirkovic B, Falnoga I, Slejkovec Z, Isidoro C, et al. Differential role of cathepsins B and L in autophagy-associated cell death induced by arsenic trioxide in U87 human glioblastoma cells. Biol Chem. 2010;391:519–531. doi: 10.1515/BC.2010.050. [DOI] [PubMed] [Google Scholar]
- Sparkes RS, Murphree AL, Lingua RW, Sparkes MC, Field LL, Funderburk SJ, et al. Gene for hereditary retinoblastoma assigned to human chromosome 13 by linkage to esterase D. Science. 1983;219:971–973. doi: 10.1126/science.6823558. [DOI] [PubMed] [Google Scholar]
- Burkhart DL, Sage J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat Rev Cancer. 2008;8:671–682. doi: 10.1038/nrc2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L. Tumour suppressor retinoblastoma protein Rb: a transcriptional regulator. Eur J Cancer. 2005;41:2415–2427. doi: 10.1016/j.ejca.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Knudsen KE, Booth D, Naderi S, Sever-Chroneos Z, Fribourg AF, Hunton IC, et al. RB-dependent S-phase response to DNA damage. Mol Cell Biol. 2000;20:7751–7763. doi: 10.1128/mcb.20.20.7751-7763.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viatour P, Sage J. Newly identified aspects of tumor suppression by RB. Dis Models Mechanisms. 2011;4:581–585. doi: 10.1242/dmm.008060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas-Kogan DA, Kogan SC, Levi D, Dazin P, T'Ang A, Fung YK, et al. Inhibition of apoptosis by the retinoblastoma gene product. EMBO J. 1995;14:461–472. doi: 10.1002/j.1460-2075.1995.tb07022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattman CL, Delach SM, Dou QP, Johnson DE. Sequential two-step cleavage of the retinoblastoma protein by caspase-3/-7 during etoposide-induced apoptosis. Oncogene. 2001;20:2918–2926. doi: 10.1038/sj.onc.1204414. [DOI] [PubMed] [Google Scholar]
- Tan X, Martin SJ, Green DR, Wang JY. Degradation of retinoblastoma protein in tumor necrosis factor- and CD95-induced cell death. J Biol Chem. 1997;272:9613–9616. doi: 10.1074/jbc.272.15.9613. [DOI] [PubMed] [Google Scholar]
- Chau BN, Borges HL, Chen TT, Masselli A, Hunton IC, Wang JY. Signal-dependent protection from apoptosis in mice expressing caspase-resistant Rb. Nat Cell Biol. 2002;4:757–765. doi: 10.1038/ncb853. [DOI] [PubMed] [Google Scholar]
- Borges HL, Bird J, Wasson K, Cardiff RD, Varki N, Eckmann L, et al. Tumor promotion by caspase-resistant retinoblastoma protein. Proc Natl Acad Sci USA. 2005;102:15587–15592. doi: 10.1073/pnas.0503925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutillier AL, Trinh E, Loeffler JP. Caspase-dependent cleavage of the retinoblastoma protein is an early step in neuronal apoptosis. Oncogene. 2000;19:2171–2178. doi: 10.1038/sj.onc.1203532. [DOI] [PubMed] [Google Scholar]
- Chau BN, Pan CW, Wang JY. Separation of anti-proliferation and anti-apoptotic functions of retinoblastoma protein through targeted mutations of its A/B domain. PLoS One. 2006;1:e82. doi: 10.1371/journal.pone.0000082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudsen ES, Knudsen KE. Tailoring to RB: tumour suppressor status and therapeutic response. Nat Rev Cancer. 2008;8:714–724. doi: 10.1038/nrc2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLendon R, Friedman A, Bigner D, Van Meir EG, Brat DJ, Mastrogianakis GM, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nduom EK, Hadjipanayis CG, Van Meir EG. Glioblastoma cancer stem-like cells: implications for pathogenesis and treatment. Cancer J. 2012;18:100–106. doi: 10.1097/PPO.0b013e3182452e0d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos B, Wan F, Farhadi M, Ernst A, Zeppernick F, Tagscherer KE, et al. Differentiation therapy exerts antitumor effects on stem-like glioma cells. Clin Cancer Res. 2010;16:2715–2728. doi: 10.1158/1078-0432.CCR-09-1800. [DOI] [PubMed] [Google Scholar]
- Zhuang W, Li B, Long L, Chen L, Huang Q, Liang Z. Induction of autophagy promotes differentiation of glioma-initiating cells and their radiosensitivity. Int J Cancer. 2011;129:2720–2731. doi: 10.1002/ijc.25975. [DOI] [PubMed] [Google Scholar]
- Chen J, Li Y, Yu TS, McKay RM, Burns DK, Kernie SG, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien WW, Domenech C, Catallo R, Salles G, Ffrench M. S-phase lengthening induced by p16(INK4a) overexpression in malignant cells with wild-type pRb and p53. Cell Cycle. 2010;9:3286–3296. doi: 10.4161/cc.9.16.12600. [DOI] [PubMed] [Google Scholar]
- Faria J, Romao L, Martins S, Alves T, Mendes FA, de Faria GP, et al. Interactive properties of human glioblastoma cells with brain neurons in culture and neuronal modulation of glial laminin organization. Diff Res Biol Diversity. 2006;74:562–572. doi: 10.1111/j.1432-0436.2006.00090.x. [DOI] [PubMed] [Google Scholar]
- Silvestre DC, Pineda JR, Hoffschir F, Studler JM, Mouthon MA, Pflumio F, et al. Alternative lengthening of telomeres in human glioma stem cells. Stem cells. 2011;29:440–451. doi: 10.1002/stem.600. [DOI] [PubMed] [Google Scholar]
- Galan-Moya EM, Le Guelte A, Lima Fernandes E, Thirant C, Dwyer J, Bidere N, et al. Secreted factors from brain endothelial cells maintain glioblastoma stem-like cell expansion through the mTOR pathway. EMBO Rep. 2011;12:470–476. doi: 10.1038/embor.2011.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aeschbacher M, Reinhardt CA, Zbinden G. A rapid cell membrane permeability test using fluorescent dyes and flow cytometry. Cell biology and toxicology. 1986;2:247–255. doi: 10.1007/BF00122693. [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Sonoda E, Sasaki MS, Buerstedde JM, Bezzubova O, Shinohara A, Ogawa H, et al. Rad51-deficient vertebrate cells accumulate chromosomal breaks prior to cell death. EMBO J. 1998;17:598–608. doi: 10.1093/emboj/17.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdalla FC, Abeliovich H, Abraham RT, Acevedo-Arozena A, Adeli K, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8:445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Minematsu-Ikeguchi N, Ueno T, Kominami E. Lysosomal turnover, but not a cellular level, of endogenous LC3 is a marker for autophagy. Autophagy. 2005;1:84–91. doi: 10.4161/auto.1.2.1697. [DOI] [PubMed] [Google Scholar]
- Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell. 2006;98:53–67. doi: 10.1042/BC20040516. [DOI] [PubMed] [Google Scholar]
- Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006;126:121–134. doi: 10.1016/j.cell.2006.05.034. [DOI] [PubMed] [Google Scholar]
- Valbuena A, Castro-Obregon S, Lazo PA. Downregulation of VRK1 by p53 in response to DNA damage is mediated by the autophagic pathway. PLoS One. 2011;6:e17320. doi: 10.1371/journal.pone.0017320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco EE, Mayhew CN, Hennigan RF, Sage J, Jacks T, Knudsen ES. RB signaling prevents replication-dependent DNA double-strand breaks following genotoxic insult. Nucleic Acids Res. 2004;32:25–34. doi: 10.1093/nar/gkg919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri MC, Zalckvar E, Kimchi A, Kroemer G. Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol. 2007;8:741–752. doi: 10.1038/nrm2239. [DOI] [PubMed] [Google Scholar]
- Huang X, Masselli A, Frisch SM, Hunton IC, Jiang Y, Wang JY. Blockade of tumor necrosis factor-induced Bid cleavage by caspase-resistant Rb. J Biol Chem. 2007;282:29401–29413. doi: 10.1074/jbc.M702261200. [DOI] [PubMed] [Google Scholar]
- Kanematsu S, Uehara N, Miki H, Yoshizawa K, Kawanaka A, Yuri T, et al. Autophagy inhibition enhances sulforaphane-induced apoptosis in human breast cancer cells. Anticancer Res. 2010;30:3381–3390. [PubMed] [Google Scholar]
- Amaravadi RK, Yu D, Lum JJ, Bui T, Christophorou MA, Evan GI, et al. Autophagy inhibition enhances therapy-induced apoptosis in a Myc-induced model of lymphoma. J Clin Invest. 2007;117:326–336. doi: 10.1172/JCI28833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu S, Takehara T, Hikita H, Kodama T, Tsunematsu H, Miyagi T, et al. Inhibition of autophagy potentiates the antitumor effect of the multikinase inhibitor sorafenib in hepatocellular carcinoma. Int J Cancer. 2012;131:548–557. doi: 10.1002/ijc.26374. [DOI] [PubMed] [Google Scholar]
- Rolle K, Nowak S, Wyszko E, Nowak M, Zukiel R, Piestrzeniewicz R, et al. Promising human brain tumors therapy with interference RNA intervention (iRNAi) Cancer Biol Ther. 2010;9:396–406. doi: 10.4161/cbt.9.5.10958. [DOI] [PubMed] [Google Scholar]
- Patru C, Romao L, Varlet P, Coulombel L, Raponi E, Cadusseau J, et al. CD133, CD15/SSEA-1, CD34 or side populations do not resume tumor-initiating properties of long-term cultured cancer stem cells from human malignant glio-neuronal tumors. BMC Cancer. 2010;10:66. doi: 10.1186/1471-2407-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JY, Knudsen ES, Welch PJ. The retinoblastoma tumor suppressor protein. Adv Cancer Res. 1994;64:25–85. doi: 10.1016/s0065-230x(08)60834-9. [DOI] [PubMed] [Google Scholar]
- Rasband WS.ImageJ US National Institutes of Health: Bethesda, MD, USA; 1997. Available from: http://imagej.nih.gov/ij/ . [Google Scholar]
- Geisa M, Faustino MG, Carlos J, de Lucena P, Stevens Rehen.Embryonic Stem Cell Counter 2009. Available from http://escc.inf.puc-rio.br/index.html .
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.