Abstract
We previously reported that marchantin M (Mar) is an active agent to induce apoptosis in human prostate cancer (PCa), but the molecular mechanisms of action remain largely unknown. Here, we demonstrate that Mar potently inhibited chymotrypsin-like and peptidyl-glutamyl peptide-hydrolyzing activities of 20S proteasome both in in vitro and intracellular systems and significantly induced the accumulation of polyubiquitinated proteins in PCa cells. The computational modeling analysis suggested that Mar non-covalently bound to active sites of proteasome β5 and β1 subunits, resulting in a non-competitive inhibition. Proteasome inhibition by Mar subsequently resulted in endoplasmic reticulum (ER) stress, as evidenced by elevated glucose-regulated protein 78 and CHOP, increased phospho-eukaryotic translation initiation factor 2α (eIF2α), splicing of X-box-binding protein-1 and dilation of the ER. However, Mar-mediated cell death was not completely impaired by a pan inhibitor of caspases. Further studies revealed that the Mar-induced cell death was greatly associated with the activation of autophagy, as indicated by the significant induction of microtubule-associated protein-1 light chain-3 beta (LC3B) expression and conversion. Electron microscopic and green fluorescent protein-tagged LC3B analyses further demonstrated the ability of autophagy induction by Mar. Time kinetic studies revealed that Mar induced a rapid and highly sustained processing of LC3B in treated cells and simultaneously decreased the expression of p62/SQSTM1. Pharmacological blockade or knockdown of LC3B and Atg5 attenuated Mar-mediated cell death. The autophagic response triggered by Mar required the activation of RNA-dependent protein kinase-like ER kinase/eIF2α and suppression of the phosphatidylinositol-3 kinase/Akt/mammalian target of rapamycin axis via preventing activation and expression of Akt. Our results identified a novel mechanism for the cytotoxic effect of Mar, which strengthens it as a potential agent in cancer chemotherapy.
Keywords: marchantin M, proteasome, ER stress, autophagic cell death, prostate cancer
Androgen deprivation therapy is the canonical treatment for patients with prostate cancer (PCa). However, the development of hormone-refractory prostate cancer (HRPC) has been observed to occur shortly after hormonal deprivation therapy, and few efficacious treatment options are available for curing or even improving the quality of life in patients with HRPC.
Docetaxel, a natural product derivative, is approved to offer some extent of symptomatic and survival benefits for HRPC patients. However, some of the patients are insensitive or even experience significant toxicity to docetaxel chemotherapy. Thus, identification of less toxic but effective novel agents for the treatment of HRPC is highly desirable.
Natural products have gained much attention in their unique structures and diverse bioactivities.1, 2 Marchantin M (Mar), like other naturally occurring macrocyclic bisbibenzyls with various biological activities,3, 4, 5 was recently found to exert an inhibitory effect on PCa cell proliferation via the activation of the caspase pathway.6 However, Mar-mediated inhibition of cell proliferation was partially rescued by z-VAD-fmk, a pan inhibitor of caspases, suggesting that caspase-independent mechanisms may also contribute to its cytotoxic effect on PCa cells. Besides caspase-mediated apoptosis, autophagy is recently implicated to be important in cell survival and death.
Autophagy is a catabolic pathway that maintains cellular homeostasis by eliminating damaged or aging organelles and unwanted proteins and mediating turnover of long-lived proteins. When cells suffer cellular stresses, the level of autophagy could be dramatically stimulated as a cytoprotective response to cope with stresses, resulting in adaptation and survival.7, 8, 9 However, dysregulated or excessive autophagy could cause autophagic cell death. Autophagic cell death appears to be responsible for the death of some cancer cells (especially when they defect essential apoptotic modulators) with increased autophagic flux.10 Recent studies have demonstrated that autophagy, including autophagic cell death, is often activated in tumor cells in response to multiple chemotherapeutic agents or radiation.11, 12 For example, proteasome inhibitor activates cellular protective autophagy via a phospho-eukaryotic translation initiation factor 2α (eIF2α)-dependent mechanism required for endoplasmic reticulum (ER) stress signaling.13 ER is essential for cell survival through modulating both the folding and trafficking of newly synthesized proteins as well as the balance of intracellular calcium concentration. Upon disruption of ER functions by excess loading of unfolded/misfolded proteins in the ER, cells trigger the unfolded protein response (UPR) to avert ER stress. It is the cell's inherent nature that prolonged or excessive ER stress naturally leads to cell death through apoptosis. Compelling evidence has demonstrated that ER stress inducers could also activate autophagy involved in either cell survival or cell death, depending on the cell type and stimulus.14, 15, 16 Therefore, considering the functional duality of autophagy in the stress-related regulation of cell survival and death, induced autophagic cell death in malignant cell types can be regarded as a potential strategy in cancer therapy.17, 18, 19
In this study, we report the novel finding that Mar induced autophagy-dependent cell death, which was accompanied with the induction of ER stress and the inhibition of proteasome activity.
Results
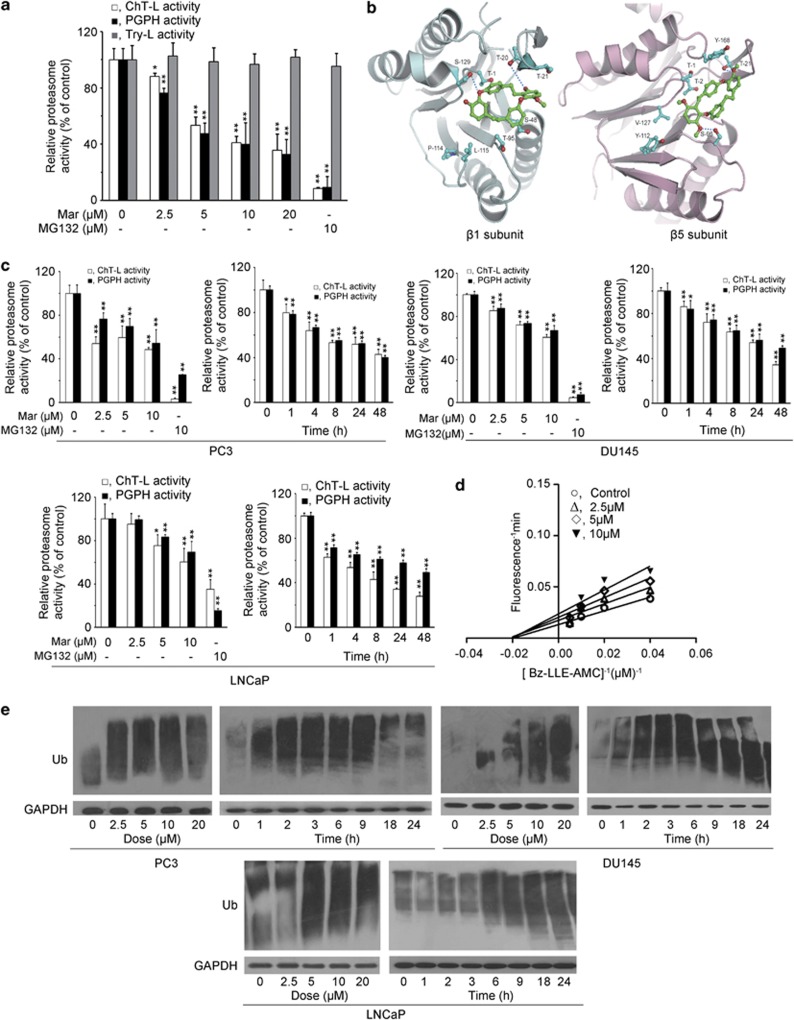
Mar inhibits the proteasome activity in PCa cells
Our recent studies demonstrated that Mar was able to induce cell cycle arrest and apoptosis in PC3 cells.6 Based on the nature of its structure, Mar was somewhat similar to tea polyphenols, bearing with phenol groups, which may act as tyrosine mimic binding to β5 subunit of proteasome.20, 21 As to whether Mar is also an active chemical to inhibit proteasome activity, we initiated our studies to examine the effects of Mar on the purified 20S proteasome by monitoring chymotrypsin-like (ChT-L), trypsin-like (Try-L) and peptidyl-glutamyl peptide-hydrolyzing (PGPH) activities in vitro. As shown in Figure 1a, both ChT-L and PGPH activities were significantly decreased with increasing concentrations of Mar; the IC50 values for ChT-L and PGPH activities were 6.99 μM and 5.33 μM, respectively. However, Mar hardly inhibited Try-L activity of the 20S proteasome. MG132, a known proteasome inhibitor for a positive control, showed more potent inhibition on the proteasome ChT-L and PGPH activities (Figure 1a). As the ChT-L and PGPH activities were mediated by the β5 and β1 subunits of proteasome, we docked the Mar molecule into β5 and β1 subunits to investigate their interactions. The docking results revealed that Mar bound to β5 and β1 subunits at the active sites with a conformation suitable for proteasome inhibition (Figure 1b). The docked free energy was −7.4 kcal/mol and −6.5 kcal/mol, respectively. The docking images also indicated that Mar interacted with β1 and β5 subunits through hydrogen and hydrophobic interactions, suggesting that the inhibitory effect of Mar could be reversible (Figure 1b). We next investigated the alteration in proteasome activity in PCa cells exposed to Mar. Treatment of PC3 cells with Mar led to marked suppression of both ChT-L and PGPH activities in a dose- and time-dependent manner (Figure 1c). Similar results were observed in DU145 and LNCaP cells (Figure 1c). Dynamic changes in the PGPH activity of 20S proteasome were also examined in response to Mar. As shown in Figure 1d, the plot for the PGPH activity displayed characteristics of non-competitive inhibition, and the Km value was determined to be 48.52 μM. Furthermore, in accordance with proteasome inhibition, Mar caused dose-dependent accumulation of ubiquitinated proteins in PCa cells (Figure 1e). The ubiquitinated proteins were noticeable as early as 1 h after treatment and sustained at high levels up to 9 h and then dropped down following 18 h of exposure in PC3 and DU145 cells. In LNCaP cells, Mar-triggered polyubiquitinated proteins were observed at 6 h and sustained up to 24 h. Thus, the data demonstrated that Mar is a potent proteasome inhibitor in PCa cells.
Figure 1.
Mar inhibits ChT-L and PGPH activities of proteasome. (a) Purified human 20S proteasome was incubated with Mar. ChT-L, Try-L and PGPH activities were monitored with specific fluorescent substrates. Relative proteasome activity represented the percentage of fluorescence compared with the control. *P<0.05, **P<0.01, compared with the control (neither Mar nor MG132 addition). Data shown are means±S.D., n=3. (b) The stereo view of interactions between Mar molecule and the β1 (left panel) and β5 subunits (right panel) of proteasome. Green, Mar; cyan-blue, side residues of amino acids in β1 and β5 subunits. The residues interacting between Mar and β1 and β5 of 20S proteasome are labeled in red. The sequence alignment of β1 between the model and human proteasome is shown below, and the interacting residues are shown in bold: model 1TTLAFR7 16DSRTTTGAYIANRVTDKLTQHDTIWCCRSGSAA53; human 35TTLAFK41 51DSRTTTGSYIANRVTDKLTPIHDRIFCCRSGSAA84; model 94LTAGLIIAGWDERHGGQVYSIPL115 124YAIGGSGS131; human 128LMAGIIIAGWDPQEGGQVYSVPM150 159 FAIGGSGS166. The sequence alignment of β5 between the model and human proteasome is shown below, and the interacting residues are shown in bold: model 1TTTLAFRFQGGIIVAVDSRATAGNW25 44TMAGGAA50 93AGLSMGT99; human 1TTTLAFKFRHGVIVAADSRATAGAY25 44TMAGGAA50 93MGLSMGT99; model 108PTIYYVDSDGTRLKGDIFCVGSG130 163AHRDAYS169; human 108PGLYYVDSEGNRISGATFSVGSG130 163TYRDAYS169. (c) PC3, DU145 and LNCaP cells were treated with Mar (2.5, 5 and 10 μM) for 24 h or 10 μM Mar for the indicated time, and proteasome activities in whole cell lysates were measured using fluorescent substrates. Relative proteasome activity represented the percentage of fluorescence compared with the control. MG132 (10 μM) served as a positive control. **P<0.01, compared with the control (neither Mar nor MG132 addition or 0 h). Data shown are means±S.D., n=3. (d) Lineweaver–Burk plot for the PGPH activity of 20S proteasome in the presence or absence of Mar. (e) Analysis of polyubiquitinated proteins in PCa cells exposed to Mar (0, 2.5, 5 and 10 μM) for 24 h or 10 μM Mar for the indicated time
Mar induces ER stress in PCa cells
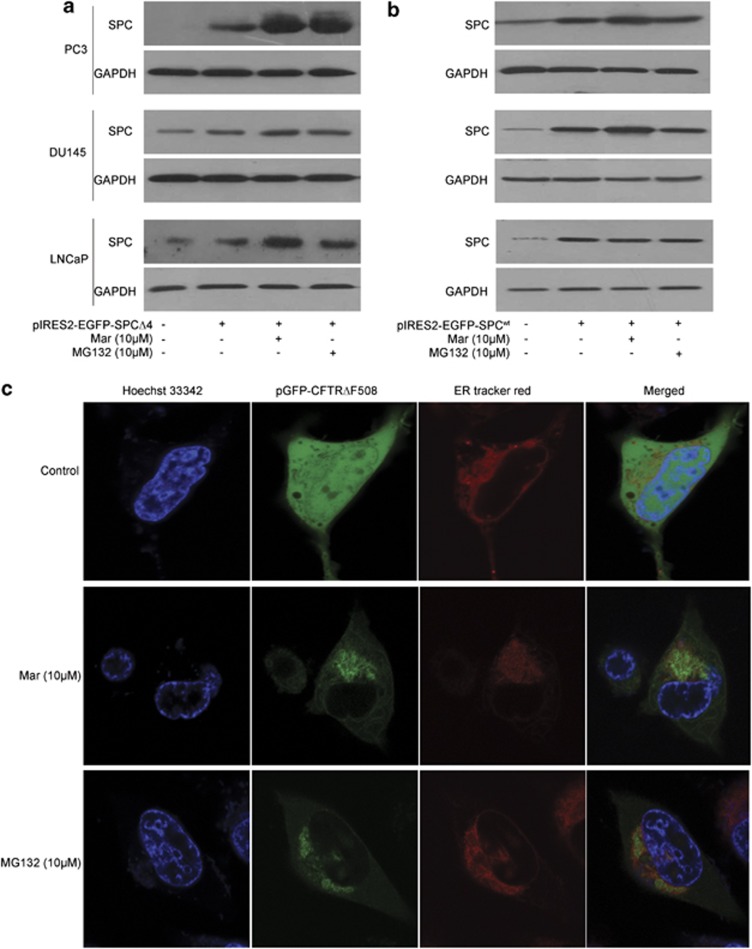
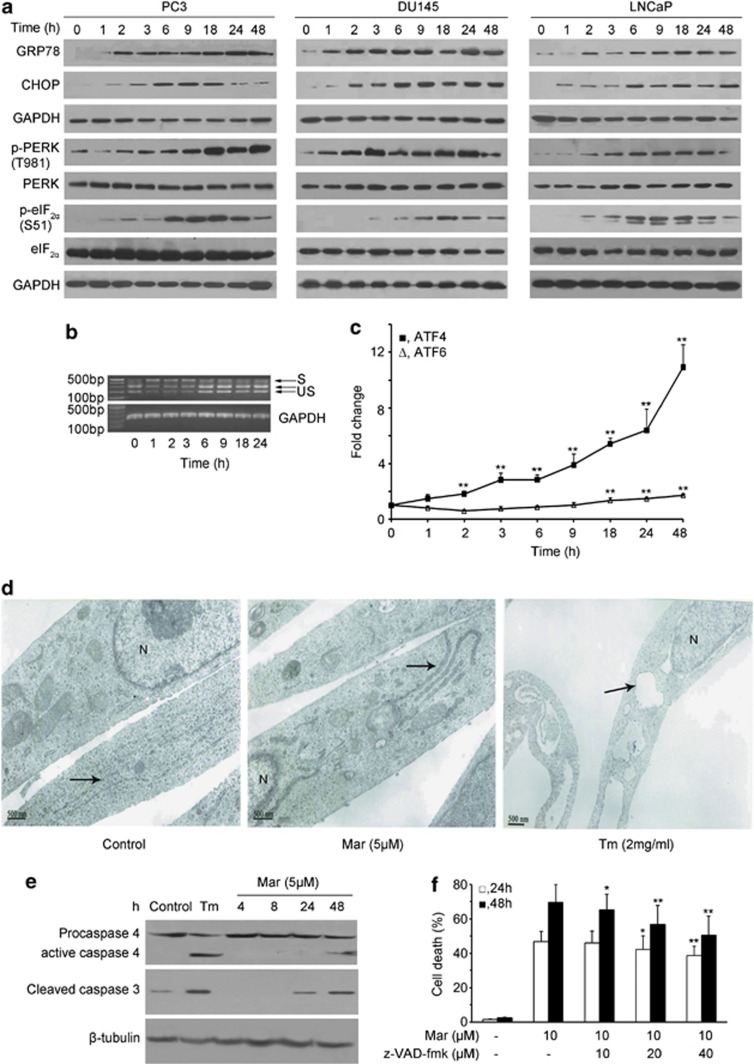
As proteasome activity is essential for eliminating excess misfolded/unfolded proteins exported from ER lumen to cytosol through the ERAD pathway, we next examined whether Mar disrupted ERAD due to the inhibition of proteasome activity. The results in Figures 2a and b show that, after transfection of SPCΔ4, a specific substrate of ERAD, Mar dramatically increased SPCΔ4 levels in all three transfected cells, similar to the observation in MG132 treatment, whereas neither Mar nor MG132 affected SPCwt levels, which served as a control. Effect of Mar on ERAD was also monitored by the localization of a green fluorescent protein (GFP) in PC3 cells transfected with pGFP-CFTRΔF508, another substrate of ERAD. As shown in Figure 2c, in the absence of Mar, GFP in transfected cells was detected in both cytoplasm and ER, whereas Mar-treated cells displayed primarily green fluorescence in the ER. Similar observations were shown in cells treated with MG132 (Figure 2c). These results indicated that Mar exerted antiproteasome activity and prevented ERAD and may retrogradely activate ER stress. We next examined whether Mar triggers ER stress by detecting its several well-documented molecular markers.22 As shown in Figure 3a, the expression of the glucose-regulated protein 78 (GRP78), a sensor of ER stress, increased markedly following a short exposure to Mar and was sustained at high levels throughout the duration of the treatment in three PCa cell lines. Induction of the ER stress-associated proapoptotic marker CHOP by Mar was predominantly observed at 6 h time point and dropped down to the basal level by 24 h in treated cells (Figure 3a), but it was maintained at elevated levels up to 48 h in LNCaP and DU145 cells (Figure 3a). Similar to the observation of CHOP, eIF2α phosphorylation was upregulated in response to Mar for 6 h and gradually decreased after treatment in three PCa cell lines; however, the total protein level of eIF2α was not affected by Mar. The above-mentioned data indicated that the Mar-induced prolonged ER stress was involved in the event of cell death in PCa cells. To further investigate the effects of Mar on the ER stress, three important ER stress response transducers X-box-binding protein-1 (XBP1), activating transcription factor 6 (ATF6) and activating transcription factor 4 (ATF4) were also examined in Mar-treated cells. As shown in Figure 3b, the spliced form of XBP1 mRNA, a transcription factor that induces expression of genes related with protein folding or degrading unfolded proteins, increased in PC3 cells exposed to Mar as early as 1 h and then decreased with longer treatment, suggesting that the IRE1/XBP1 pathway was activated following a short exposure to Mar. Real-time PCR analysis revealed that the ATF4 mRNA levels were largely increased by Mar and sustained up to 48 h during treatment, and the levels of ATF6 were slightly increased in Mar-treated cells (Figure 3c), suggesting the induction of expression of genes involved in restoring ER homeostasis. Additionally, transmission electron microscopy revealed that the ER was moderately dilated in cells exposed to Mar, less than that of tunicamycin (Tm), which is a well-demonstrated inducer of ER stress (Figure 3d). The above-mentioned data indicated that the inhibition of proteasome by Mar resulted in prolonged ER stress and loss of translational control in PCa cells.
Figure 2.
Mar disrupts ERAD. Analysis of the degradation of SPCΔ4 (a) and SPCwt (b) in PCa cells transfected with pIRES2-EGFP-SPCΔ4 and pIRES2-EGFP-SPCwt for 48 h and then treated with Mar (10 μM) for 24 h. (c) Analysis of CFTRΔF508 accumulation in ER by fluorescent staining. After transfection of PC3 cells with pGFP-CFTRΔF508 for 48 h and exposure to Mar (10 μM) for additional 24 h, cells were stained with ER-Tracker Red and Hoechst 33342. MG132 was used as a positive control
Figure 3.
Mar triggers ER stress in PCa cells. (a) Analysis of the effect of Mar on proteins associated with ER stress by western blotting. (b) Analysis of XBP1 splicing after Mar (10 μM) treatment in PC3 cells by RT-PCR. S, spliced XBP1; US, unspliced XBP1. (c) Measurement of ATF4 and ATF6 mRNA expressions in PC3 cells exposed to Mar by real-time RT-PCR. **P<0.01, compared with the control (0 h treatment). Data shown are means±S.D., n=3. (d) Changes of ER in cells treated with Mar (5 μM) using a transmission electron microscope. Tm (2 mg/ml) was taken as a positive control. The ER is indicated by black arrows. N, nucleus. Bar, 500 nm. (e) Analysis of caspase-4 and caspase-3 activation in Mar-treated cells by western blotting. (f) Effects of the apoptosis inhibitor z-VAD-fmk on Mar-induced cell death in PC3 cells. PC3 cells were incubated with z-VAD-fmk for 2 h prior to Mar treatment. Cell death was assayed by PI exclusion; *P<0.05, **P<0.01. Comparison between groups treated with Mar plus z-VAD-fmk or Mar alone. Data shown are means±S.D., n=3
Based on the importance of caspase-4 in ER-mediated apoptosis, we next examined its expression and caspase-3 activation in cells exposed to Mar. As shown in Figure 3e, activation of procaspase-4 was observed after 48 h treatment with Mar, leading to the induction of procaspase-4 cleavage to proteolytic fragments in treated cells, which in turn cleaved caspase-3 (Figure 3e). However, Tm caused more apparent cleavage of procaspase-4 at 24 h than Mar treatment (Figure 3e). To understand the role of caspase-dependent apoptosis in Mar-induced cell death, we tested Mar-induced cell death in the presence of z-VAD-fmk, a broad-spectrum inhibitor of caspases. The results in Figure 3f show that, after exposure of cells to Mar with the increasing concentrations of z-VAD-fmk, cell death was moderately reduced, but not completely blocked, suggesting that both caspase-dependent and caspase-independent pathways are required for Mar-induced cell death in PC3 cells.
Mar stimulates autophagy in PCa cells
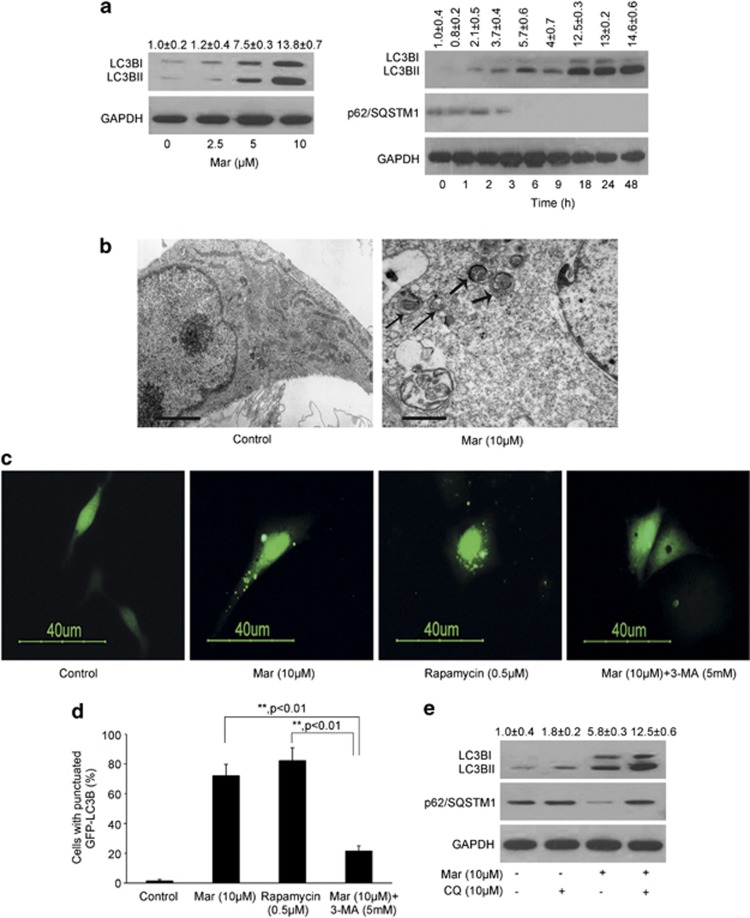
Autophagy could be increased as a compensatory means of protein degradation when the proteasome pathway is blocked.13 We next determined whether caspase-independent cell death processing by Mar is due to proteasome inhibition with subsequent autophagic cell death. As shown in Figure 4a and Supplementary Figures 1a and e, Mar rapidly upregulated autophagy levels in treated cells, as judged by the accumulation of the autophagy marker protein microtubule-associated protein-1 light chain-3 beta (LC3B) and the accumulation of lipidated LC3BII in a concentration-dependent manner. Kinetic study revealed that Mar-induced autophagic flux was observed at 2 h, which became more evident at 18 h, and then was sustained at high levels during 48 h treatment indicated by increased processing of LC3BI to LC3BII (Figure 4a and Supplementary Figure 1a). However, Mar-induced autophagy was observed at 6 h in LNCaP cells. Sequestome-1 protein (p62/SQSTM1), which was implicated to directly degrade by autophagy, was reduced after 3 h in PC3 and DU145 and 6 h in LNCaP in response to Mar accordingly (Figure 4a and Supplementary Figures 1a and e). Electron microscopy of Mar-treated PC3 cells showed the formation of double- or multi-membranes engulfing high electron-density substances, evidenced as autophagosomes or autolysosomes (Figure 4b).
Figure 4.
Mar treatment induces autophagy in PC3 cells. (a) Analysis of LC3B expression and conversion in PC3 cells treated with Mar by western blotting. OD of LC3BII relative to the control (0 h) was quantified as mean±S.D., n=3. (b) Electron micrographs showed autophagy induced by Mar. The thick arrows, autolysosome; the thin arrows, autophagosome. Bar, 1 μm. (c) Analysis of LC3B punctate in PCa cells transfected with GFP-LC3B in the presence or absence of Mar and 3-MA. Rapamycin (0.5 μM) was used as a positive control. PCa cells were pretreated with 5 mM 3-MA for 2 h prior to Mar treatment for 24 h. (d) Percentage of cells with punctuate GFP-LC3B to all GFP-positive cells. The cells with over five GFP-LC3B dots were chosen as positive cells. The values represented are means±S.D. for 200 cells each. **P<0.01. Data shown are means±S.D., n=3. (e) The accumulation of p62 and LC3BII was assessed in PC3 cells incubated with CQ (10 μM) for 2 h prior to Mar treatment for 24 h by western blot analysis. OD of LC3BII relative to the control was quantified as mean±S.D., n=3
Mar-induced autophagy was also judged by punctate dots of a GFP-tagged form of LC3B (GFP-LC3B). The cells transfected with the GFP-LC3B expression plasmid showed diffuse green fluorescence, whereas Mar treatment caused significantly punctated green fluorescence in the cytosol, similar to the observation in rapamycin-treated cells (Figures 4c and d and Supplementary Figures 1b, c, f and g). Notably, the induction of autophagy by Mar was blocked by 3-methyladenine (3-MA), an inhibitor of the class III phosphatidylinositol-3 kinase (PI3K) Vps34, as indicated by much lower levels of GFP-LC3B puncta when compared with cells treated with Mar alone (Figures 4c and d and Supplementary Figures 1b, c, f and g). Increased LC3BI to LC3BII conversion could occur when autophagy was either stimulated or blocked.23 To further explore whether autophagic flux can be induced by Mar, we chose chloroquine (CQ), an agent that prevents the formation of autolysosomes by inhibiting autophagosome–lysosome fusion and LC3BII degradation, to determine whether the increased LC3BII level by Mar was a result of increased autophagosomal formation or a defect in the fusion process. The result showed that Mar-stimulated processing of LC3B was enhanced accompanying with p62 increased in the presence of CQ, whereas no detectable change was observed in cells exposed to CQ alone (Figure 4e and Supplementary Figures 1d and h), suggesting that Mar has the potential to promote autophagosome formation. Thus, Mar was effective in inducing a prolonged autophagy that might be required for its effect in cell death promotion.
Blockade of autophagy attenuates Mar-induced cell death
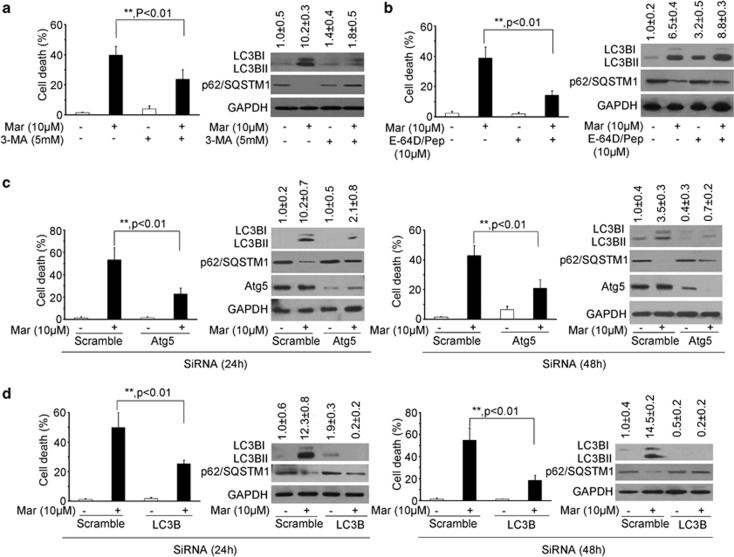
To assess the impact of autophagic flux in Mar-induced cell death, we analyzed cell viability and cell death by pretreating PC3 cells with autophagy inhibitors. As shown in Figure 5a and Supplementary Figure 2a, Mar treatment led to the accumulation of LC3BII and decrease of p62, whereas this effect was significantly blocked in the presence of 3-MA and accompanied with an increase in cell viability and reduction in cell death. Similarly, Mar-mediated induction of LC3BII was enhanced in combination with E-64D/Pepstatin A, another autophagy inhibitor that prevents lysosomal enzyme activation, and cell proliferation was significantly restored and cell death was markedly decreased in cotreatment (Figure 5b and Supplementary Figure 2b). Driven by these observations, we further confirmed the role of autophagic flux in the action of Mar by knockdown of specific autophagy-related genes. Atg5 is a crucial component involved in autophagic vesicle nucleation.7 Knockdown of endogenous levels of Atg5 by Atg5-targeting siRNA dramatically increased cell viability and blocked cell death after 24 and 48 h transfection followed by Mar treatment (Figure 5c and Supplementary Figure 2c). Mar-induced LC3BII levels were markedly suppressed when Atg5 was downregulated by siRNA. Similar to the observations of Atg5, reduction of LC3B or Atg7 by siRNA significantly alleviated cytotoxic activity of Mar (Figure 5d and Supplementary Figures 3 and 4). Taken together, our data clearly demonstrated that Mar activated an autophagic flux and promoted autophagy-dependent cell death.
Figure 5.
Inhibition of autophagy rescues cell death in Mar-treated PC3 cells. The cell death of PC3 cells exposed to Mar alone or combined treatment. Left panel, cell death by PI exclusion. Right panel, immunoblot analysis of LC3BII, Atg5 and p62 levels in PC3 cells after treatments. OD of LC3BII relative to the control was quantified as mean±S.D., n=3. (a) PC3 cells treated with 10 μM Mar or 10 μM Mar plus 5 mM 3-MA for 24 h. (b) PC3 cells treated with 10 μM Mar or 10 μM Mar plus 10 μM E-64D/10 μM Pepstatin A (Pep) for 24 h. (c) Knockdown of Atg5 by siRNA for 24 or 48 h in PC3 cells and then treated with 10 μM Mar. (d) Knockdown of LC3B by siRNA for 24 or 48 h in PC3 cells prior to Mar treatment. **P<0.01. Data shown are means±S.D., n=3
ER stress and mammalian target of rapamycin signaling involved in the activation of autophagy by Mar
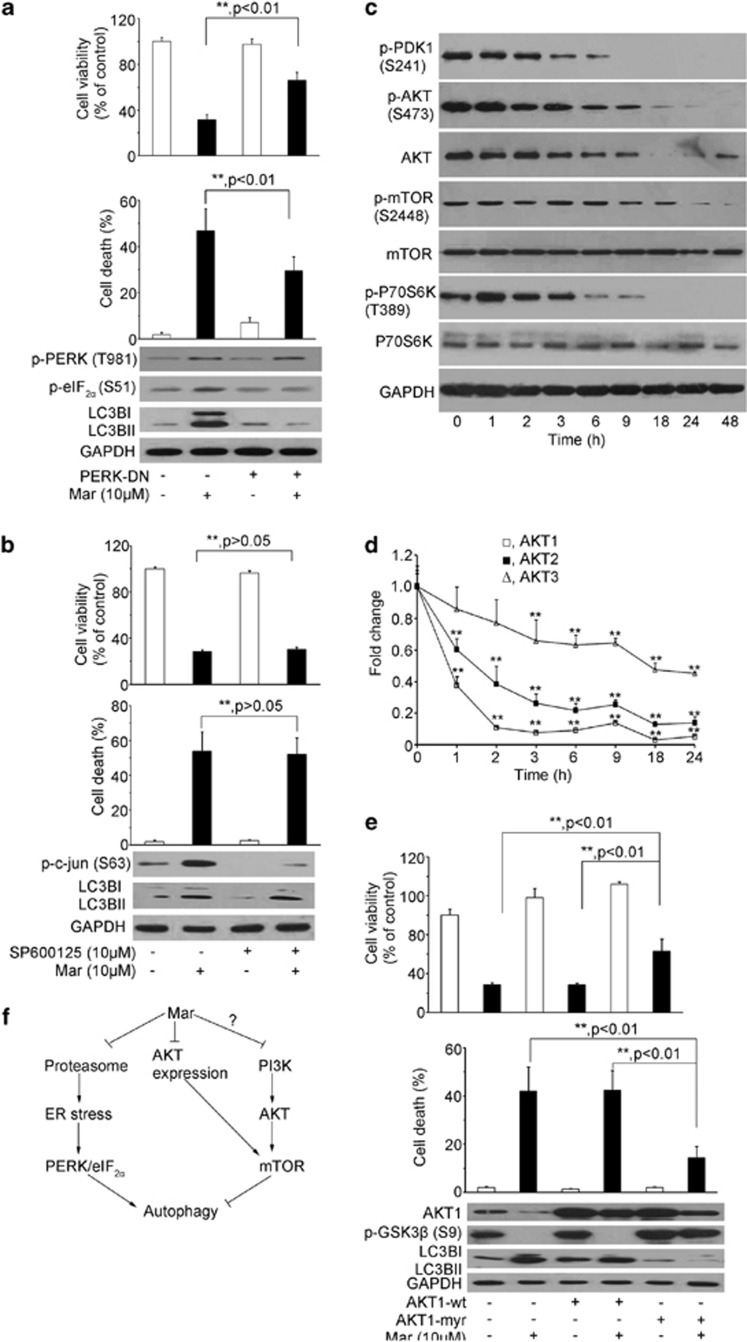
As Mar-mediated inhibition of proteasome activity led to the activation of ER stress and autophagy, we next examined certain modulator proteins that have been reported to establish the linkage between ER stress and autophagy. It has been reported that the PRK (RNA-dependent protein kinase)-like ER kinase (PERK)/eIF2α pathway in response to ER stress may be involved in autophagy activation.7 To explore a link between PERK/eIF2α signaling and autophagic activation in response to proteasome inhibition by Mar, we performed transfection with dominant-negative PERK (PERK-DN) expression plasmid to impair the function of PERK and examined whether autophagy was activated in the presence of Mar. The results in Figure 6a show that, inactivation of PERK by PERK-DN attenuated eIF2α phosphorylation and had little effect on cell proliferation, whereas Mar-induced eIF2α phosphorylation was blunted by PERK-DN, leading to the blocking of LC3BII accumulation and partial restoration of viable cells as well as decreased cell death. The similar results were observed by knockdown of PERK with siRNA (Supplementary Figures 5a–c). Additionally, IRE1/JNK signaling is also implicated to link ER stress and autophagy activation.7 The results in Figure 6b revealed that activation of c-Jun was evidenced in response to Mar, and SP600125, an inhibitor of JNK, profoundly abrogated Mar-triggered phospho-c-Jun in PC3 cells. However, either LC3B processing or cell proliferation by Mar hardly changed in the presence of SP600125. These results indicated the importance of the PERK/eIF2α pathway, but not IRE1/JNK signaling, in the linking of Mar-induced ER stress and autophagy when proteasome was inhibited.
Figure 6.
Signaling pathways involved in Mar-induced autophagy in PC3 cells. (a) Effect of PERK/eIF2α on Mar-mediated autophagy activation and cell death induction. After transfection of PERK-DN for 24 h, cells were treated with Mar and subjected for analysis. Upper panel, cell viability assay by MTT. Middle panel, cell death analysis by PI exclusion. Lower panel, determination of protein changes by western blotting assay. **P<0.01. Data shown are means±S.D., n=3. (b) Effect of JNK on Mar-activated autophagy and cell death. PC3 cells were pretreated with 10 μM SP600125 for 2 h prior to Mar treatment for 24 h. Cell viability (upper panel) and cell death (middle panel) of PC3 cells were measured by MTT and PI exclusion. Changes of p-c-Jun and LC3B were analyzed by western blotting. Data shown are means±S.D., n=3. (c) Analysis of changes of regulatory proteins in the PI3K/mTOR pathway in PC3 cells treated with Mar by western blotting. (d) Analysis of Akt mRNA levels in Mar-treated cells by qPCR assays. (e) PC3 cells transfected with Akt1-wt or Akt1-myr were treated with 10 μM Mar for 24 h. Cell viability was investigated by MTT (upper panel), and cell death was determined by PI exclusion (middle panel). **P<0.01. Data shown are means±S.D., n=3. (f) Schematic diagram represents mechanisms of action of Mar to trigger autophagy in PCa cells
Compelling evidence demonstrates that the PI3K/Akt/mammalian target of rapamycin (mTOR) pathway is the major regulatory signal of autophagy. Inactivation of mTOR is considered as a key step in autophagy activation. We therefore investigated whether Mar triggered autophagy also through the inhibition of PI3K/Akt/mTOR signaling. As shown in Figure 6c, phospho-Akt as well as phospho-3-phosphoinositide-dependent protein kinase-1 (PDK1) was pronouncedly reduced by Mar and sharply fell to much lower levels after 6 h treatment, in agreement with the result that a corresponding accumulation in LC3BII was evident following 6 h treatment and became more robust thereafter as shown in Figure 4a. Of interest, the expression of total Akt protein was noticeably impaired following 3 h treatment and decreased up to 48 h by Mar (Figure 6c). The attenuation of Akt by Mar was further confirmed at mRNA levels, and the expression of Akt3 decreased much lower than that of Akt1and Akt2 (Figure 6d). Therefore, we made efforts to fine-tuning the activity of Akt in Mar-mediated autophagy by overexpression of the wild-type Akt1 (Akt1-wt) and the constitutively activated form of Akt1 (Akt1-myr). Forced expression of Akt1-wt was unable to increase phospho-GSK3β, a downstream target of Akt, as well as the cell viability in the presence of Mar (Figure 6e). However, PC3 cells transfected with Akt1-myr showed significant increases in phospho-GSK3β and cell viability, accompanied with a decrease in LC3BII levels. These results indicated that Mar was able to inhibit PI3K activity, leading to the inactivation of Akt. Meanwhile, upon treatment with Mar, decreases in the phospho-mTOR and its downstream effector phospho-P70S6K were detected after 6–9 h treatments, yet total mTOR and P70S6K remained unchanged (Figure 6c). Based on these findings, we proposed a model by which Mar inhibited proteasome and blocked ERAD, resulting in ER stress, which promoted caspase-dependent apoptosis as well as induced autophagy by the PERK/eIF2α pathway. Also, Mar suppressed PI3K/Akt/mTOR signaling, contributing to the induction of autophagy (Figure 6f).
Discussion
The 26S proteasome and autophagy are considered as two major routes responsible for the quality control of protein. Proteasome degrades polyubiquitinated proteins that are involved in many cellular processes including cell proliferation and survival. Usually, cancer cells are more sensitive to proteasome inhibition than normal cells. Therefore, selective inhibitors of proteasome are considered to be a promising future for anticancer drug development.
In this study, we first identified Mar as a novel non-competitive inhibitor of proteasome by non-covalent binding to the active site Thr1Oγ of the β subunits because Mar does not bear electrophilic groups in its structure. This is similar to TMC-95A, a natural product that is linked non-covalently to the active site of β subunits,24 resulting in competitive inhibition. As irreversible inhibitors of proteasome usually cause normal cell apoptosis and death, reversible inhibitors are more valuable in drug discovery, suggesting that Mar may represent a promising candidate in PCa treatment.
Proteasome inhibition-mediated disruption of ERAD results in the accumulation of misfolded or unfolded proteins, which in turn induces ER stress and activates the UPR. Generally, ER stress promotes cell survival through the attenuation of global protein synthesis and the acceleration of protein folding. However, prolonged or severe ER stress induced apoptosis and activated the autophagy.25, 26 ER stress is one of the autophagy inducers and has attracted increasing attention recently. PERK/eIF2α signal is a canonical pathway that mediates autophagy during ER stress. PERK phosphorylates eIF2α, resulting in general protein translation inhibition, as well as selectively promotes some genes including ATF4 translation. ATF4 upregulated LC3 and ATG5 transcription in severe hypoxia or proteasome inhibition to enhance autophagic protection.27, 28 Although ATF4 dramatically induced in Mar-treated PC3 cells (Figure 3c), ATG genes except LC3B showed little change (data not shown). Moreover, inhibition of ATF4 expression by siRNA scarcely affects the turnover of LC3BI (Supplementary Figures 5d–f). Interestingly, the cell death significantly decreased, suggesting that Mar-induced ATF4 mediated cell death, which was associated with the upregulation of ATF3, a downstream target of ATF4 (our unpublished data). Although ER stress-activated autophagy is usually considered to be a cytoprotective mechanism, ER stress enhances autophagy and triggers cell death through the inhibition of the Akt/mTOR pathway in certain circumstances.8, 29 Here, we report that Mar-induced cell death was impaired by pharmaceutical and genetic autophagy inhibition, indicating that Mar-triggered autophagy is important for its cytotoxicity in PCa cells. PI3K/Akt/mTOR signaling cascade plays a key role in promoting cell proliferation and survival as well as autophagy blocking. Constitutive activation of PI3K/Akt/mTOR signaling might make cancer cells more sensitive to mTOR blockade because it would render cells more dependent on the pathway for growth.30, 31 Our studies revealed that Mar inhibited PI3K activity and downregulated Akt expression and activity, leading to repression of the PI3K/Akt/mTOR pathway, which is constitutively activated in PC3 cells due to the PTEN-null. Inactivation of PI3K/Akt/mTOR signaling is implicated to be important for Mar-promoted cell death because blocking of autophagy or overexpression of constitutively activated Akt resulted in impairment of cytotoxicity of Mar. Further investigations will be necessary to clarify the mechanisms by which Mar regulates the functions and/or expression of Akt and PI3K in PC3 cells.
Of note, autophagy inhibition could not completely reverse cell death induced by Mar, similar to the results using the apoptosis inhibitor alone. Combined inhibition of caspases and autophagy almost completely rescued Mar-induced cell death and improved cell viability in PCa cells (Supplementary Figure 6), indicating that both caspase and autophagic cell death signaling are required to Mar-mediated cytotoxicity.
Most conventional chemotherapeutic agents induce apoptosis; however, emerging evidence indicates that many cancer cells are chemoresistant and defective in apoptosis induction. Genistein, a natural compound, can induce both apoptotic and autophagic cell death, exerting the potential to overcome chemoresistance via alterations in apoptotic signaling.32 Compared with MG132, Mar exerted lower inhibition on proteasome activity, indicating that many efforts should be made to increase the efficiency by modification of Mar structure or preparation of its derivatives; however, Mar showed lower cytotoxicity to RWPE-1, an immortalized prostate epithelial cell line (Supplementary Figure 7). Our studies identified novel capability of Mar, which triggers apoptotic and autophagic cell death in PCa cells, indicating the potential application of Mar in cancer therapy.
Materials and Methods
Chemicals and reagents
Mar was purified from Asterella angusta as described previously.33 Mar was dissolved in DMSO and stored as small aliquots at −20 °C. SP600125, 3-MA, CQ and Tm were purchased from Sigma-Aldrich (St. Louis, MO, USA). The purified 20S proteasome, z-VAD-fmk, E-64D, Pepstatin A, N-succinyl-Leu-Leu-Val-Tyr-7-amino-4-methylcoumarin (Suc-LLVY-AMC), benzyloxycarbonyl-L-leucyl-leucyl-glutamyl-methylcoumarylamide (Bz-LLE-AMC) and Bz-Val-Gly-Arg-7-amino-4-methylcoumarin (Bz-VGR-AMC) were purchased from Enzo Life Sciences (Plymouth Meeting, PA, USA). Rapamycin and MG132 were from Calbiochem (Darmstadt, Germany). ER-Tracker Red was obtained from Beyotime Institute of Biotechnology (Haimen, China).
Cell line
Human PC3 and DU145 cells were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China). LNCaP cell line was purchased from the American Type Culture Collection (Manassas, VA, USA). PC3 cells were grown in Dulbecco's modified Eagle's medium (Hyclone, Beijing, China) containing 10% fetal bovine serum (FBS) and antibiotics. LNCaP cell lines were grown in RPMI-1640 medium with 10% FBS and antibiotics. The cells were maintained in a humidified incubator with 5% CO2 at 37 °C.
Proteasome activity assay
Proteasome substrates Suc-LLVY-AMC, Bz-VGR-AMC and Bz-LLE-AMC were used to measure ChT-L, Try-L and PGPH activities of proteasome, respectively. In the in vitro assay, the substrates were incubated at 37 °C for 40 min with 20S proteasome in the presence or absence of Mar in assay buffer, and then enzyme activities were evaluated by measuring fluorescent activities of substrates using Mithras LB-940 (Berthold Technologies, Bad Wildbad, Germany). To measure activities of proteasome in cells, whole cell lysates were prepared from Mar-treated cells with lysis buffer (50 mM Tris–HCl (pH 8.0), 150 mM NaCl, 5 mM EDTA, 0.5% NP40, 2 mM DTT). Proteasome activities were determined by incubation of cell lysates with substrates in assay buffer. To assay effect of Mar on the catalytic kinetics of proteasome, Bz-LLE-AMC (25, 50, 100 and 200 μM) was incubated with Mar for measuring PGPH activity, and the data were analyzed by a double-reciprocal Lineweaver–Burk plot.
Transmission electron microscopy
PC3 cells were treated with vehicle or Mar and fixed with 2.5% glutaraldehyde in 0.l M phosphate buffer (pH 7.4). After being rinsed by phosphate buffer, cells were incubated with 1% osmium tetroxide. Ultrathin sections were cut and stained with 2% uranyl acetate. Cellular ultrastructure was analyzed using a JEM-1400 transmission electron microscope (JEOL, Tokyo, Japan).
Quantitative RT-PCR
Total RNA was extracted from Mar- or vehicle-treated cells using TRIZOL RNAiso Plus (Takara, Dalian, China) and reversely transcribed using the ReverTraAce qPCR RT kit (Toyobo, Shanghai, China). Quantitative PCR (qPCR) analysis was performed with SYBR Green reaction master mix (Toyobo) and by a real-time PCR system (Eppendorf International, Hamburg, Germany). The primers used for amplification were listed in Supplementary Information. Gene expression levels were normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Changes in the transcript levels were calculated using the ΔΔCt method.34
XBP1 splicing analysis
XBP1 transcripts were synthesized using XBP1-specific primers by RT-PCR. The PCR products were subjected to analyze spliced and unspliced XBP1 fragments with digestion by the PstI restriction enzyme as described previously.35
Cell viability and cell death assay
PC3 cells were transfected with plasmids and siRNAs and/or treated with Mar. Cell viability was detected using colorimetric 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT, Sigma) assay on a plate reader (Bio-Rad, Hercules, CA, USA). Cell death was measured by propidium iodide (PI) exclusion with flow cytometry. In some experiments, cells were exposed to the JNK inhibitor SP600125 (10 μM) for 2 h, autophagy inhibitor 3-MA (5 mM) for 2 h or lysosomal protease inhibitor E-64D/Pepstatin A (10 μM) for 2 h before treatment with Mar.
Transient transfection of plasmids and siRNAs
PC3 cells were transiently transfected with Akt1-wt, Akt1-myr, dominant-negative pcDNA3.1-hPEK-K621M (PERK-DN), pIRES2-EGFP-SPCΔ4, pIRES2-EGFP-SPCwt (wild type), pGFP-CFTRΔF508 or siRNA duplex oligonucleotides targeting Atg5 and LC3B (GenePharmcon, Shanghai, China) using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA). Empty vectors PCMV6 and pcDNA3.1 served as controls. After 24 or 48 h transfection, cells were treated with Mar or vehicle for an additional 24 h, and cell lysates were subjected for western blot assay. Cell viability and cell death following treatment were determined by MTT and PI exclusion assays as described above. The siRNA sequences were listed in Supplementary Information.
Microscopy for GFP-LC3B puncta
PC3 cells were transfected with a GFP-tagged LC3B-expressing vector (GFP-LC3B) prior to Mar or rapamycin (0.5 μM) treatment for 24 h. The GFP-LC3B puncta in response to chemicals were observed under a fluorescence microscope (Olympus IX71; Olympus Co., Tokyo, Japan).
Western blot assay
After transfection and/or treatment with chemicals, cell lysates were prepared using RIPA buffer containing fresh protease inhibitor mixture (50 μg/ml aprotinin, 0.5 mM phenylmethanesulfonyl fluoride, 1 mM sodium orthovanadate, 10 mM sodium fluoride and 10 mM β-glycerophosphate). Equal amounts of proteins were subjected to western blot assays as described previously.6 The following antibodies were used: anti-LC3B (Novus Biologicals, Littleton, CO, USA); anti-GRP78, anti-CHOP, anti-eIF2α, anti-phospho-eIF2α (Ser51), anti-Atg5, anti-mTOR, anti-phospho-mTOR (Ser2448), anti-Akt, anti-phospho-Akt (Ser473), anti-phospho-GSK3β (Ser9), anti-phospho-PDK1 (Ser241), anti-P70S6K, anti-p-P70S6K (Thr389), anti-phospho-c-Jun (Ser63) and anti-p62/SQSTM1 (Cell Signaling Technology, Danvers, MA, USA); anti-β-tubulin, anti-Ub, anti-SP-C, PERK, p-PERK (Thr981), ATF4 (CREB-2) and anti-GAPDH (Santa Cruz Biotechnology, Dallas, TX, USA); anti-Akt1 (Shanghai Excell Biology, Shanghai, China); and cleaved caspase-3 (Epitomics, Burlingame, CA, USA).
CFTRΔF508 localization
PC3 cells were transfected with pGFP-CFTRΔF508 for 48 h. After being treated with Mar, the medium was removed and the cells were incubated with ER-Tracker Red and Hoechst 33342 for 30 min. Fluorescent image was monitored by laser confocal microscopy (LSM780, Zeiss, Oberkochen, Germany).
Docking
Computer prediction of ligand binding was carried out using AutoDock Vina.36 The searching parameter was set at twice as default value 16 to ensure the space coverage. Ligand file was first prepared by Chem Office and then edited by Autodock Tools (http://autodock.scripps.edu). All rotatable bonds were accepted during the docking. The subunits of β1 and β5 of yeast 20S proteasome crystal structure (PDB 2F16) were used as protein models, with the ligand deleted. The size of the docking grid was selected to cover the entire docking molecule.
Statistical analysis
The data were presented as the mean±S.D. of at least three independent experiments. The Student's t-test was used to determine statistical differences between treated and control groups, and P<0.05 was considered statistically significant.
Acknowledgments
We would like to thank Professor Jinquan Chen for kindly providing the plasmids expressing Akt1-wt and Akt1-myr, Professor Xuejun Jiang for the gift of GFP-LC3B vector, Professor Sokol V Todi for pGFP-CFTRΔF508 and Professor Timothy E Weaver for pIRES2-EGFP-SPCwt and SPCΔ4. This study was supported by the National Natural Science Foundation of China (30973551, 30925038), 973 Program (2013CB910900) and Shandong Scientific Technology Program (2012GSF11909).
Glossary
- ER
endoplasmic reticulum
- eIF2α
eukaryotic translation initiation factor 2α
- GRP78
glucose-regulated protein 78
- GADD153/CHOP
the growth arrest–DNA damage gene 153
- PERK
PRK (RNA-dependent protein kinase)-like ER kinase
- XBP1
X-box-binding protein-1
- LC3B
microtubule-associated protein-1 light chain-3 beta
- mTOR
mammalian target of rapamycin
- PDK1
3-phosphoinositide-dependent protein kinase-1
- PI3K
phosphatidylinositol-3 kinase
- 3-MA
3-methyladenine
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on the Cell Death and Disease website (http://www.nature.com/cddis)
Edited by T Brunner
Supplementary Material
References
- Barnes S. Role of phytochemicals in prevention and treatment of prostate cancer. Epidemiol Rev. 2001;23:102–105. doi: 10.1093/oxfordjournals.epirev.a000773. [DOI] [PubMed] [Google Scholar]
- Newman DJ, Cragg GM, Snader KM. Natural products as sources of new drugs over the period 1981–2002. J Nat Prod. 2003;66:1022–1037. doi: 10.1021/np030096l. [DOI] [PubMed] [Google Scholar]
- Cheng A, Sun L, Wu X, Lou H. The inhibitory effect of a macrocyclic bisbibenzyl riccardin D on the biofilms of Candida albicans. Biol Pharm Bull. 2009;32:1417–1421. doi: 10.1248/bpb.32.1417. [DOI] [PubMed] [Google Scholar]
- Huang WJ, Wu CL, Lin CW, Chi LL, Chen PY, Chiu CJ, et al. Marchantin A, a cyclic bis(bibenzyl ether), isolated from the liverwort Marchantia emarginata subsp. tosana induces apoptosis in human MCF-7 breast cancer cells. Cancer Lett. 2010;291:108–119. doi: 10.1016/j.canlet.2009.10.006. [DOI] [PubMed] [Google Scholar]
- Xi GM, Sun B, Jiang HH, Kong F, Yuan HQ, Lou HX. Bisbibenzyl derivatives sensitize vincristine-resistant KB/VCR cells to chemotherapeutic agents by retarding P-gp activity. Bioorg Med Chem. 2010;18:6725–6733. doi: 10.1016/j.bmc.2010.07.055. [DOI] [PubMed] [Google Scholar]
- Xu AH, Hu ZM, Qu JB, Liu SM, Syed AK, Yuan HQ, et al. Cyclic bisbibenzyls induce growth arrest and apoptosis of human prostate cancer PC3 cells. Acta Pharmacol Sin. 2010;31:609–615. doi: 10.1038/aps.2010.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–293. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin L, Wang Z, Tao L, Wang Y. ER stress negatively regulates AKT/TSC/mTOR pathway to enhance autophagy. Autophagy. 2010;6:239–247. doi: 10.4161/auto.6.2.11062. [DOI] [PubMed] [Google Scholar]
- Harhaji-Trajkovic L, Vilimanovich U, Kravic-Stevovic T, Bumbasirevic V, Trajkovic V. AMPK-mediated autophagy inhibits apoptosis in cisplatin-treated tumour cells. J Cell Mol Med. 2009;13:3644–3654. doi: 10.1111/j.1582-4934.2009.00663.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazi B, Bursch W, Fimia GM, Nardacci R, Piacentini M, Di Sano F, et al. Fenretinide induces autophagic cell death in caspase-defective breast cancer cells. Autophagy. 2008;4:435–441. doi: 10.4161/auto.5669. [DOI] [PubMed] [Google Scholar]
- Wang M, Tan W, Zhou J, Leow J, Go M, Lee HS, et al. A small molecule inhibitor of isoprenylcysteine carboxymethyltransferase induces autophagic cell death in PC3 prostate cancer cells. J Biol Chem. 2008;283:18678–18684. doi: 10.1074/jbc.M801855200. [DOI] [PubMed] [Google Scholar]
- Fujiwara K, Iwado E, Mills GB, Sawaya R, Kondo S, Kondo Y. Akt inhibitor shows anticancer and radiosensitizing effects in malignant glioma cells by inducing autophagy. Int J Oncol. 2007;31:753–760. [PubMed] [Google Scholar]
- Zhu K, Dunner K, Jr, McConkey DJ. Proteasome inhibitors activate autophagy as a cytoprotective response in human prostate cancer cells. Oncogene. 2010;29:451–462. doi: 10.1038/onc.2009.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WB, Feng LX, Yue QX, Wu WY, Guan SH, Jiang BH, et al. Paraptosis accompanied by autophagy and apoptosis was induced by celastrol, a natural compound with influence on proteasome, ER stress and Hsp90. J Cell Physiol. 2012;227:2196–2206. doi: 10.1002/jcp.22956. [DOI] [PubMed] [Google Scholar]
- Ullman E, Fan Y, Stawowczyk M, Chen HM, Yue Z, Zong WX. Autophagy promotes necrosis in apoptosis-deficient cells in response to ER stress. Cell Death Differ. 2008;15:422–425. doi: 10.1038/sj.cdd.4402234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong JT, Xu Y, Yi HW, Su J, Yu HM, Xiang XY, et al. The BH3 mimetic S1 induces autophagy through ER stress and disruption of Bcl-2/Beclin 1 interaction in human glioma U251 cells. Cancer Lett. 2012;323:180–187. doi: 10.1016/j.canlet.2012.04.009. [DOI] [PubMed] [Google Scholar]
- Aits S, Gustafsson L, Hallgren O, Brest P, Gustafsson M, Trulsson M, et al. HAMLET (human alpha-lactalbumin made lethal to tumor cells) triggers autophagic tumor cell death. Int J Cancer. 2009;124:1008–1019. doi: 10.1002/ijc.24076. [DOI] [PubMed] [Google Scholar]
- Ambrosini G, Musi E, Ho AL, de Stanchina E, Schwartz GK. Inhibition of mutant GNAQ signaling in uveal melanoma induces AMPK-dependent autophagic cell death. Mol Cancer Ther. 2013;12:768–776. doi: 10.1158/1535-7163.MCT-12-1020. [DOI] [PubMed] [Google Scholar]
- Hoyer-Hansen M, Bastholm L, Mathiasen IS, Elling F, Jaattela M. Vitamin D analog EB1089 triggers dramatic lysosomal changes and Beclin 1-mediated autophagic cell death. Cell Death Differ. 2005;12:1297–1309. doi: 10.1038/sj.cdd.4401651. [DOI] [PubMed] [Google Scholar]
- Nam S, Smith DM, Dou QP. Ester bond-containing tea polyphenols potently inhibit proteasome activity in vitro and in vivo. J Biol Chem. 2001;276:13322–13330. doi: 10.1074/jbc.M004209200. [DOI] [PubMed] [Google Scholar]
- Smith DM, Daniel KG, Wang Z, Guida WC, Chan TH, Dou QP. Docking studies and model development of tea polyphenol proteasome inhibitors: applications to rational drug design. Proteins. 2004;54:58–70. doi: 10.1002/prot.10504. [DOI] [PubMed] [Google Scholar]
- Egger L, Madden DT, Rheme C, Rao RV, Bredesen DE. Endoplasmic reticulum stress-induced cell death mediated by the proteasome. Cell Death Differ. 2007;14:1172–1180. doi: 10.1038/sj.cdd.4402125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010;140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M, Koguchi Y, Huber R, Kohno J. Crystal structure of the 20S proteasome:TMC-95A complex: a non-covalent proteasome inhibitor. J Mol Biol. 2001;311:543–548. doi: 10.1006/jmbi.2001.4869. [DOI] [PubMed] [Google Scholar]
- Faitova J, Krekac D, Hrstka R, Vojtesek B. Endoplasmic reticulum stress and apoptosis. Cell Mol Biol Lett. 2006;11:488–505. doi: 10.2478/s11658-006-0040-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Endoplasmic reticulum stress: a new pathway to induce autophagy. Autophagy. 2007;3:160–162. doi: 10.4161/auto.3653. [DOI] [PubMed] [Google Scholar]
- Rouschop KM, van den Beucken T, Dubois L, Niessen H, Bussink J, Savelkouls K, et al. The unfolded protein response protects human tumor cells during hypoxia through regulation of the autophagy genes MAP1LC3B and ATG5. J Clin Invest. 2010;120:127–141. doi: 10.1172/JCI40027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milani M, Rzymski T, Mellor HR, Pike L, Bottini A, Generali D, et al. The role of ATF4 stabilization and autophagy in resistance of breast cancer cells treated with Bortezomib. Cancer Res. 2009;69:4415–4423. doi: 10.1158/0008-5472.CAN-08-2839. [DOI] [PubMed] [Google Scholar]
- Salazar M, Carracedo A, Salanueva IJ, Hernandez-Tiedra S, Lorente M, Egia A, et al. Cannabinoid action induces autophagy-mediated cell death through stimulation of ER stress in human glioma cells. J Clin Invest. 2009;119:1359–1372. doi: 10.1172/JCI37948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neshat MS, Mellinghoff IK, Tran C, Stiles B, Thomas G, Petersen R, et al. Enhanced sensitivity of PTEN-deficient tumors to inhibition of FRAP/mTOR. Proc Natl Acad Sci USA. 2001;98:10314–10319. doi: 10.1073/pnas.171076798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downward J. PI 3-kinase, Akt and cell survival. Semin Cell Dev Biol. 2004;15:177–182. doi: 10.1016/j.semcdb.2004.01.002. [DOI] [PubMed] [Google Scholar]
- Gossner G, Choi M, Tan L, Fogoros S, Griffith KA, Kuenker M, et al. Genistein-induced apoptosis and autophagocytosis in ovarian cancer cells. Gynecol Oncol. 2007;105:23–30. doi: 10.1016/j.ygyno.2006.11.009. [DOI] [PubMed] [Google Scholar]
- Qu J, Xie C, Guo H, Yu W, Lou H. Antifungal dibenzofuran bis(bibenzyl)s from the liverwort Asterella angusta. Phytochemistry. 2007;68:1767–1774. doi: 10.1016/j.phytochem.2007.04.036. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Zu K, Bihani T, Lin A, Park YM, Mori K, Ip C. Enhanced selenium effect on growth arrest by BiP/GRP78 knockdown in p53-null human prostate cancer cells. Oncogene. 2006;25:546–554. doi: 10.1038/sj.onc.1209071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.