Abstract
In our previous study, we demonstrated that the BRCC2 (breast cancer cell 2) gene is a proapoptotic molecule that interacts with Bcl-XL. BRCC2 downregulation is associated with poor disease-free and overall survival in breast cancer. In this study, we aimed to investigate the role of BRCC2 in tumor suppression in breast cancer. In clinical breast cancer samples, we found that BRCC2 expression was significantly downregulated in cancer lesions compared with paired normal breast tissues. By silencing or overexpressing BRCC2 in breast cancer cells, we found that BRCC2 could inhibit cell growth and metastasis in vitro. An in vivo assay showed that BRCC2 not only dramatically inhibited breast cancer cell xenograft formation and growth but also inhibited breast cancer cell metastasis in a lung metastasis model. Moreover, we demonstrated that BRCC2 inhibited breast cancer metastasis via regulation of the Akt pathway. Thus, our study provided evidence that BRCC2 functions as a novel tumor suppressor in breast cancer and may be a potential therapeutic target for breast cancer management.
Keywords: BRCC2, tumor suppressor, breast cancer
Breast cancer has become the second leading malignancy among women in both developing and developed countries. Cancer statistics indicate that ∼226 870 new breast cancer cases were reported in 2012 in the United States, accounting for 29% of all new female cancer cases. Nearly 39 510 women were estimated to die as a result of breast cancer, accounting for 14% of all cancer deaths.1 Despite the advances that have been made in the treatment of breast cancer, the major cause of breast cancer deaths continues to be metastasis.
Breast cancer is a heterogeneous disease, and breast cancer metastasis is a multistep process that results from genetic alterations, including the activation of oncogenes and the loss of function of tumor suppressors.2, 3 Numerous metastasis-related oncogenes have been characterized in breast cancers. HER-2 (human epithelial receptor 2) is an example of an oncogene that, when activated, contributes to more aggressive behavior in breast cancers.4, 5 Metadherin (MTDH) is another oncogene that has an important role in breast cancer metastasis. The results of our previous studies have demonstrated that MTDH promotes metastasis and chemoresistance and is associated with poor clinical outcomes in breast cancer patients.6, 7, 8 In addition, many tumor suppressors, such as p53 and BRCA1, have been reported to be related to breast cancer metastasis. Though many targeted therapies have improved the survival of breast cancer patients,9 a proportion of patients will eventually die as a result of metastasis. Therefore, there is an urgent need to identify novel genes and reveal the detailed mechanisms underlying breast cancer metastasis.
Breast cancer cell 2 (BRCC2, also known as BH3-like motif containing, cell death inducer) was originally identified as an ∼1.2-kb transcript in the MDA-MB-231 human breast cancer cell line. The longest predicted open reading frame of BRCC2 is 862 bp, and its mRNA encodes a protein that is 108 amino acids in length. BRCC2 is an intronless gene that has been mapped to human chromosome 11q24.1 using fluorescence in situ hybridization.10, 11 The 11q24.1 region is an area that is characterized by the extensive loss of heterozygosity in cancers, and the loss of heterozygosity is related to poor prognoses.12, 13, 14 Previously, we found that BRCC2 downregulation was an independent prognostic factor of distant metastasis-free survival in breast cancer.15 In the present study, we aimed to explore the tumor suppressor function of BRCC2 in breast cancer. We found that the expression of BRCC2 was significantly downregulated in human breast cancer lesions compared with paired normal breast tissues. We also demonstrated that BRCC2 could inhibit the growth and metastasis of breast cancer cell lines, both in vitro and in vivo. Our present manuscript suggests that BRCC2 acts as a potential tumor suppressor in breast cancer.
Results
Expression of BRCC2 was downregulated in breast cancer tissues
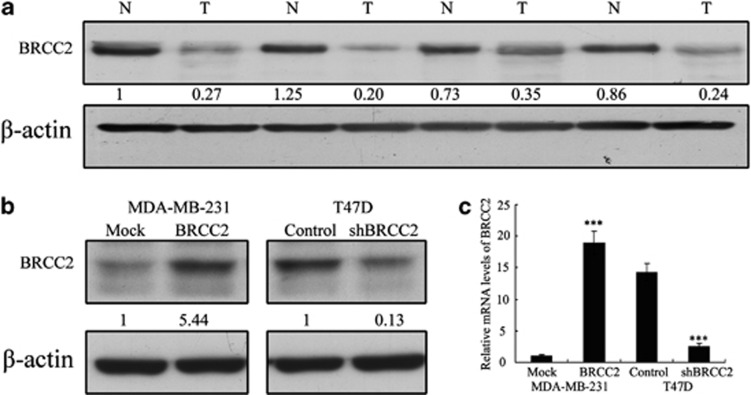
We previously demonstrated that BRCC2-negative tumors are associated with significantly lower metastasis-free and overall survival in breast cancer.16 These findings suggested that BRCC2 may function as a tumor suppressor in breast cancer. To test this hypothesis, we first compared the expression levels of BRCC2 in 39 breast cancer tissue samples to those in the adjacent normal tissues using western blotting. BRCC2 protein levels were found to be reduced in the tumor lesions compared with the matched normal tissue lesions in all of the samples (P=0.0012) (Figure 1a), suggesting a possible role for BRCC2 in the development or progression of breast cancer.
Figure 1.
Expression of BRCC2 in breast cancer tissues and the transfection efficiency of BRCC2 in breast cancer cell lines. (a) BRCC2 protein levels in tumor tissues and matched normal tissue lesions, as assessed using western blotting analyses. (b) The transfection efficiency of BRCC2 was analyzed by measuring protein levels by western blotting. (c) The transfection efficiency of BRCC2 was analyzed by measuring transcript levels using RT-PCR analyses. β-actin was used as a loading control. The expression of each protein was quantified as the densitometry value analyzed by SPSS 18.0 software. The data represent the means±S.D. of three independent experiments ***P<0.001
Establishment of stable BRCC2 transfectants in breast cancer cell lines
We used MDA-MB-231 cells to establish a stable cell line that constitutively overexpressed the BRCC2 protein with the aim of revealing the role that BRCC2 expression has in the development or progression of breast cancer. We also used shRNA to generate a stable BRCC2 knockdown in the T47D breast cancer cell line. The transfection efficiency was confirmed using western blotting and qRT-PCR analyses. As shown in Figures 1b and c, the MDA-MB-231 cells that had been transfected with the BRCC2 expression plasmid displayed significantly increased BRCC2 expression at both the mRNA and protein levels compared with the vector cell lines. In addition, the T47D cells that had been transfected with the BRCC2 shRNA plasmid displayed significantly decreased BRCC2 expression at both the mRNA and protein levels compared with the control cells.
BRCC2 inhibited breast cancer cell proliferation
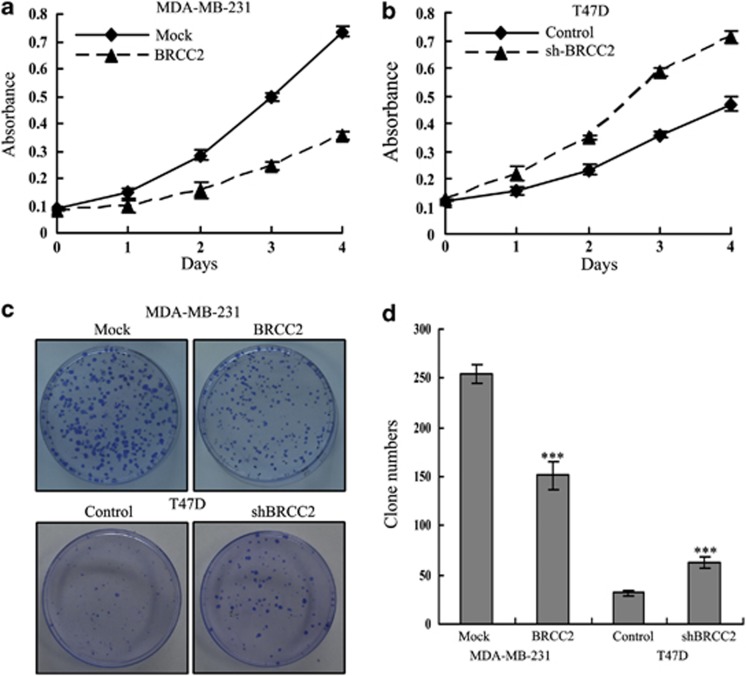
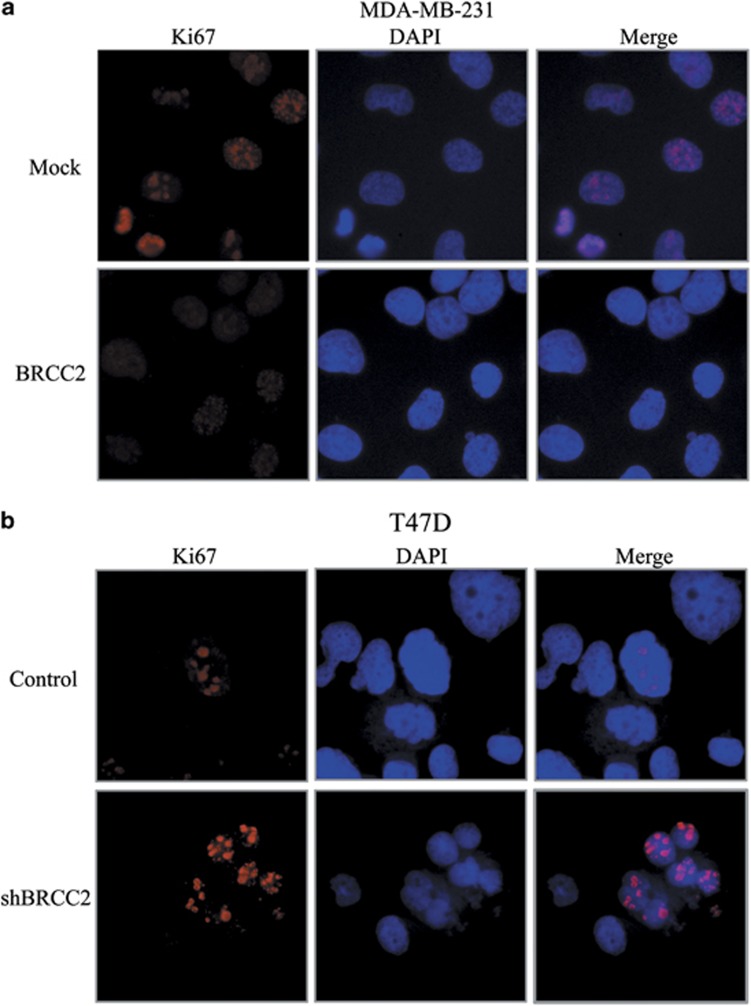
We first explored the effects of BRCC2 expression on cell growth using the MTT assay. As shown in Figure 2a, BRCC2 overexpression significantly inhibited the growth of MDA-MB-231 cells (P=0.034), whereas BRCC2 knockdown significantly enhanced the growth of T47D cells (P=0.017) (Figure 2b). Next, we performed a clonogenic assay to confirm the effects of BRCC2 on proliferation. We found that BRCC2 overexpression dramatically reduced the colony formation efficiency of MDA-MB-231 cells (P=0.0005), whereas the colony formation efficiency was dramatically increased in the BRCC2 shRNA cell lines (P=0.001) (Figures 2c and d). As Ki67 antigen is an important marker of cell proliferation, we next examined the Ki67 by immunofluorescence staining. As shown in Figure 3, we found that the overexpression of BRCC2 in MDA-MB-231 cells significantly downregulated Ki67 staining (P=0.008). Also knockdown of BRCC2 in T47D cells dramatically upregulated the staining of Ki 67 (P=0.0003). These results suggested that BRCC2 could significantly inhibit the proliferation of breast cancer cells.
Figure 2.
Effects of BRCC2 on proliferation in breast cancer cell lines. (a) Cell proliferation after BRCC2 overexpression in MDA-MB-231 cells was measured using MTT assays. (b) Cell proliferation after BRCC2 knockdown in T47D cells was measured using MTT assays. (c) The results of colony formation assays that were conducted in BRCC2-transfected breast cancer cell lines. (d) The summary graphs are presented for the colony formation assay that was outlined in c. The data represent the means±S.D. of three independent experiments ***P<0.001
Figure 3.
Effects of BRCC2 on Ki67 in breast cancer cell lines. (a) Immunofluorescence staining of Ki67 in MDA-MB-231-transfected cell lines. (b) Immunofluorescence staining of Ki67 in T47D-transfected cell lines. The percentage of Ki67-positive staining were counted and analyzed by a two-tailed Student's t-test
BRCC2 inhibited breast cancer cell mobility
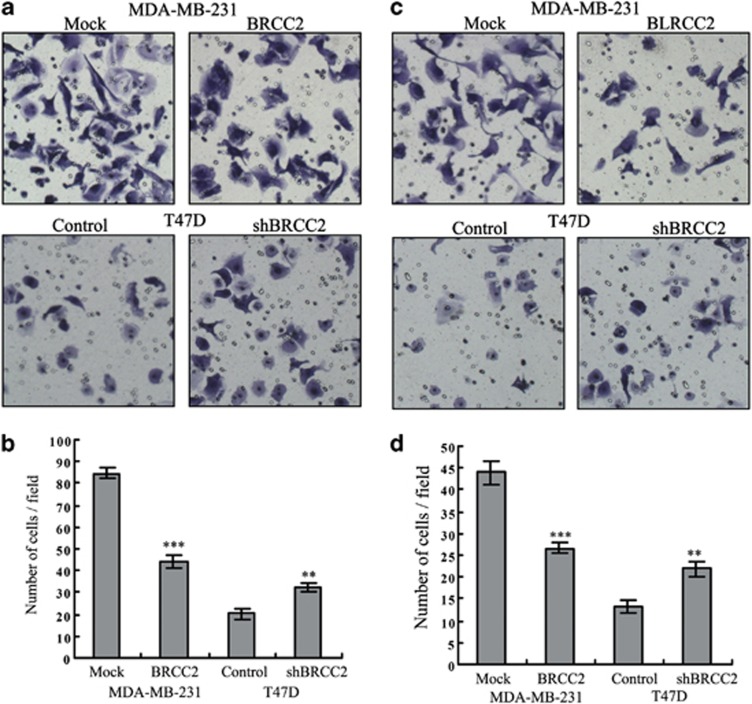
We next assessed whether BRCC2 could affect the ability of breast cancer cells to migrate and invade using a transwell assay. BRCC2 overexpression inhibited both migration (84.67±2.52 versus 44.33±3.21, P<0.0001) and invasion (44.00±2.65 versus 26.67±1.15, P=0.0004) in MDA-MB-231 cells. In addition, BRCC2 knockdown in T47D cells significantly promoted cell migration (20.33±2.52 versus 32.67±2.08, P=0.003) and invasion (13.33±1.53 versus 22.00±1.73, P=0.0003) (Figure 4). These results indicated that BRCC2 significantly inhibited the invasion and migration of breast cancer cells.
Figure 4.
Effects of BRCC2 on invasion and migration in breast cancer cells. (a) Migration ability was measured using Transwell assays in BRCC2-transfected MDA-MB-231 and T47D cells. (b) The summary graphs are presented for the experiment that was outlined in a. (c) Invasion ability was measured in BRCC2-transfected MDA-MB-231 and T47D cells. (d) The summary graphs are presented for the experiment that was outlined in c. The data represent the mean number of cells per field and are presented as the means±S.D. **P<0.01, ***P<0.001
BRCC2 inhibited tumorigenesis and metastasis in vivo
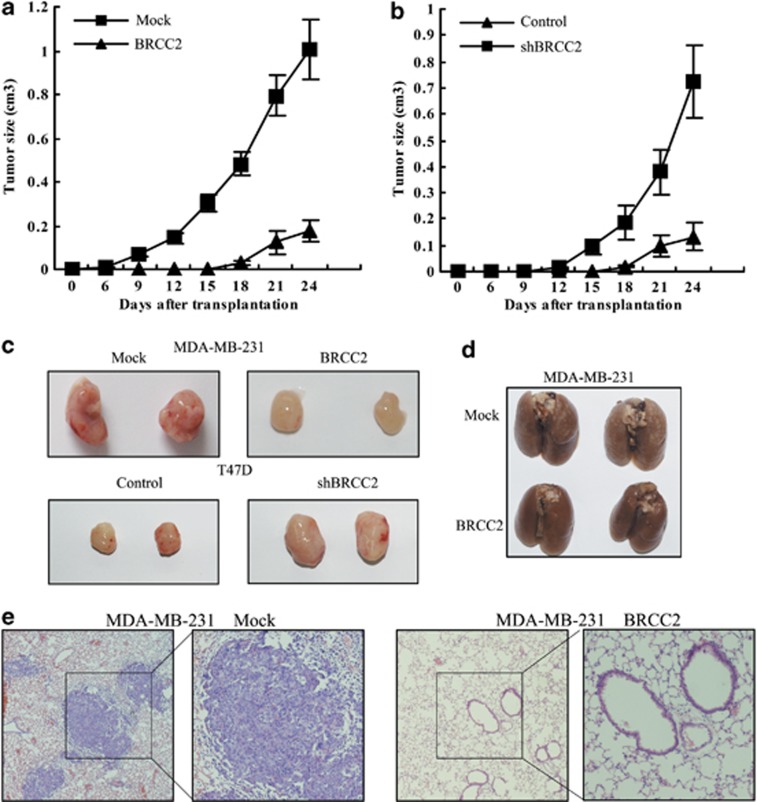
To explore the inhibitory effects of BRCC2 on tumorigenesis in vivo, the cells were injected subcutaneously into the flanks of nude mice. The diameters of the tumors were measured every 3 days. We found that the mice that had been injected with the control MDA-MB-231 cells formed tumors on the sixth day, while the mice that had been injected with BRCC2-overexpressing MDA-MB-231 cells did not form tumors until the fifteenth day. The tumors in the BRCC2 overexpression group grew very slowly compared with those in the mice that had been injected with the control cells during the subsequent days (Figure 5a). Similar results were observed in the T47D cells. We found that the BRCC2 knockdown cells formed tumors earlier and that the tumor volumes were much larger in those that were formed from the knockdown cells than in those that were formed from the control cells (Figure 5b). As shown in Figure 5c, the average tumor volume of MDA-MB-231 control was 1.01±0.135 cm3 while the BRCC2 group was only 0.175±0.08 cm3 (P=0.0004). Similar with these, the shBRCC2 group were dramatically larger than the control group in T47D cells (0.723±0.141 versus 0.134±0.05, P=0.002). These results suggested that BRCC2 inhibits breast cancer cell xenograft formation and growth in vivo.
Figure 5.
BRCC2 inhibited tumorigenesis and metastasis in vivo. (a) Growth curves of mammary tumors after the injection of BRCC2-overexpressing MDA-MB-231 and control cells into SCID mice. The error bars represent the means±S.D. (n=6). (b) Growth curves of mammary tumors after the injection of BRCC2-silenced T47D and control cells into SCID mice. The error bars represent the means±S.D. (n=6). (c) Representative tumors are presented from the experiments that were outlined in a and b. (d) Representative lungs that were harvested from the mice that had been injected in the lateral tail veins. (e) Representative hematoxylin and eosin staining of lung sections (magnification, × 40). The right panel is the high magnification (magnification, × 100) image for the picture that is marked in the box. Each group consisted of ten mice
We further investigated the effects of BRCC2 on breast cancer lung metastasis using MDA-MB-231 cells in vivo. An equal number of control and BRCC2-overexpressing breast cancer cells were injected into the lateral tail veins of 4- to 5-week-old nude mice. As shown in Figure 5d), all ten of the lungs that were isolated from the mice that had been injected with the control cells exhibited extensive metastases on the surfaces, while only one lung from the BRCC2-overexpressing mice displayed metastases (P<0.0001). The lungs were then fixed in 10% formalin and the lung sample sections were stained using hematoxylin and eosin. We found that the lungs that were isolated from the mice that had been injected with the control cells contained 28.52±6.38 metastatic breast cancer foci, whereas the mice that had been injected with cells in which BRCC2 had been upregulated displayed 5.31±3.87 metastatic foci under the microscope (P<0.0001). The metastatic foci in the control groups were also significantly larger than the BRCC2 group (Figure 5e). These results further confirmed that BRCC2 could significantly inhibit breast cancer metastasis in vivo.
BRCC2 inhibited tumor metastasis via activation of the Akt pathway
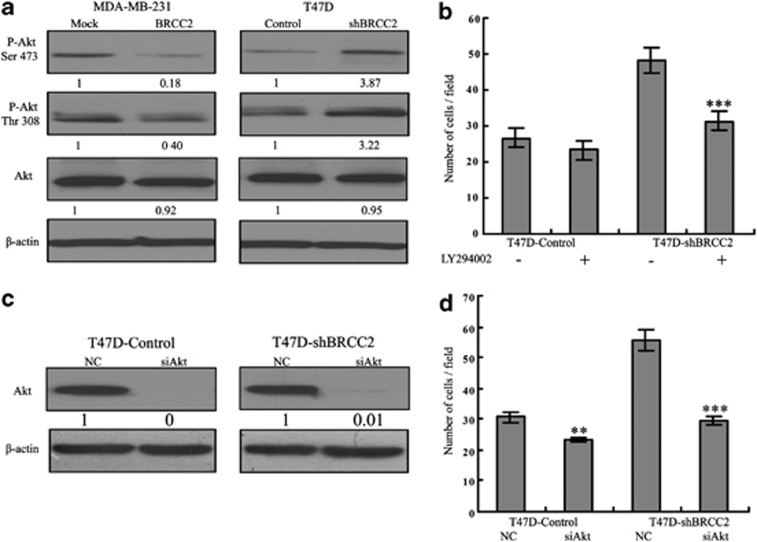
The PI3K/Akt pathway has important roles in the proliferation, migration and invasion of various cancer types, including breast cancer. Thus, we determined whether the Akt pathway was involved in BRCC2-mediated tumor metastasis. We evaluated the effects of BRCC2 on the Akt pathway in MDA-MB-231 and T47D cells by measuring the phosphorylation profile of Akt at Ser 473 and Thr 308. As shown in Figure 6a, upregulation of BRCC2 significantly inhibited the phosphorylation of Akt Ser 473 (P<0.0001), Akt Thr 308 (P<0.0001) and knockdown of BRCC2 dramatically activated the Akt Ser 473 and Thr 308 (both P<0.0001). To test whether the Akt pathway was involved in the anti-metastatic function of BRCC2, we pretreated the T47D cells with LY294002 for 1 h, after which point we detected the migration ability of the control and BRCC2-shRNA cells using a transwell assay. We found that the number of BRCC2-shRNA cells that had migrated was significantly decreased after LY294002 treatment, while there was only a slight decrease in the number of control T47D cells that had migrated (Figure 6b). We further tested the role of Akt in BRCC2-induced migration by knocking down Akt expression using siRNA. The Akt knockdown efficiency was detected using western blotting at 24 h after transfection (Figure 6c). As shown in Figure 6d, the migration ability that was induced by BRCC2 was obviously attenuated following Akt knockdown using siRNA (P=0.0003). The results confirmed that the Akt pathway was involved in BRCC2-mediated metastasis in breast cancer.
Figure 6.
The effects of BRCC2 on the Akt pathway. (a) The expression of Akt and the phosphorylation of Akt at Ser 473 and Thr 308 in BRCC2-transfected cells were examined using western blotting. β-actin was used as a loading control. The expression of each protein was quantified as the densitometry value analyzed by SPSS 18.0 software. (b) The summary graphs show the migration ability of BRCC2 knockdown T47D and its control cells after the cells had been pretreated with the Akt inhibitor LY294002 (100 nM). (c) The transfection efficiency of Akt siRNA 24 h after transfection was measured using western blot analyses. β-Actin was used as a loading control. The expression of each protein was quantified as the densitometry value analyzed by SPSS 18.0 software. (d) The summary graphs show the migration ability of BRCC2 knockdown T47D and its control cells after the cells had been transfected with Akt siRNA and its negative control. The data represent the mean number of cells per field and are presented as the means±S.D. **P<0.01, ***P<0.001
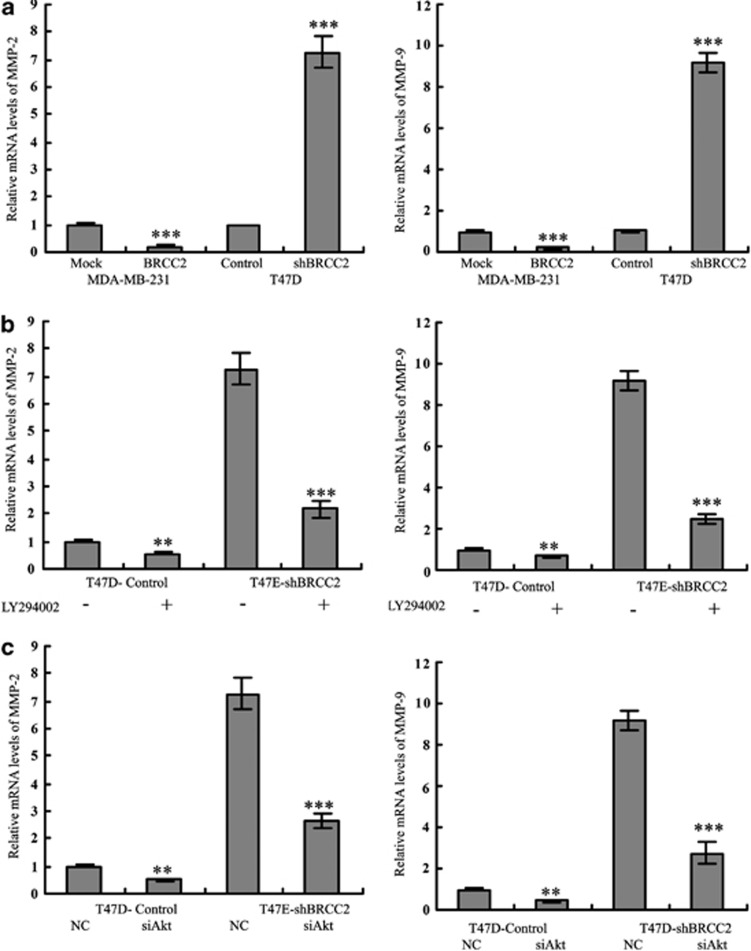
The matrix metalloproteinase (MMP) family has important roles in both tumor angiogenesis and metastasis.17, 18In addition, PI3K/Akt has been reported to enhance tumor progression by inducing MMP expression.19, 20 Therefore, we next examined whether BRCC2 inhibited tumor metastasis via activation of the Akt-mediated MMP pathway. As shown in Figure 7a, the mRNA levels of MMP-2 and MMP-9 were upregulated in the BRCC2 knockdown T47D cells and were decreased in the BRCC2-overexpressing MDA-MB-231 cells. Similar results were observed in the tumors that were isolated from the mice (data not shown). When we pretreated the BRCC2 knockdown cells with an Akt inhibitor or Akt siRNA, both MMP-2 and MMP-9 levels were dramatically downgegulated (Figures 7b and c, respectively). Our data demonstrated that BRCC2 inhibited cell mobility via Akt-mediated regulation of MMP-2 and MMP-9.
Figure 7.
BRCC2 inhibited migration and invasion via the Akt-mediated MMP-2 and MMP-9 pathways. (a) The levels of MMP-2 and MMP-9 in BRCC2-transfected breast cancer cell lines were detected by RT-PCR analyses. (b) The levels of MMP-2 and MMP-9 were examined using RT-PCT analyses after the cells had been pretreated with the Akt inhibitor LY294002 (100 nM). (c) The levels of MMP-2 and MMP-9 were examined using RT-PCR analyses after the cells had been transfected with Akt siRNA and its negative control. The data represent the means ±S.D. of at least three independent experiments **P<0.01, ***P<0.001
Discussion
Breast cancer is one of the most common cancers in the world. Despite the decline in breast cancer mortality, a number of breast cancer patients develop metastatic tumors even after surgical removal of the primary tumors. Therefore, metastasis continues to be the main obstacle to the effective treatment of breast cancer, and there is an urgent need to identify novel molecular factors that lead to the invasiveness and metastasis of breast cancer. In the present manuscript, we identified BRCC2 as a candidate target gene for the inhibition of breast cancer growth and metastasis.
BRCC2 is a proapoptotic molecule and is both a cytosolic and mitochondrial protein. BRCC2 has been reported to induce a caspase-dependent mitochondrial cell death pathway.11 Previously, we found that BRCC2 interacts with Bcl-XL and that the expression of BRCC2 is associated with Bax activation and increases in the cytosolic levels of cytochrome c. In addition, BRCC2 downregulation in breast cancer is correlated with poorer overall, cause-specific and local relapse-free survival and was determined to be an independent prognostic factor for distant metastasis-free survival.15 Cavalli et al.21 reported that loss of heterozygosity at the BRCC2 locus was more frequently detected in early-onset breast cancer patients. In addition, Nakanishi et al.22 reported that BRCC2 seems functionally relevant to the pathogenesis of pathological myopia and is potentially a candidate gene for high myopia in the Chinese Han population.23 All of these data strongly support a role for BRCC2 as a tumor suppressor gene.
Tumor suppressor genes can inhibit tumor growth and invasive and metastatic potential. Loss of tumor suppressor genes may lead to a malignant cancer phenotype. Previous studies have reported that the expression levels of tumor suppressor genes, such as p53, p21 and PTEN, were decreased in tumors compared with normal tissues.24, 25, 26 To confirm the tumor suppressor function of BRCC2, we first examined the levels of BRCC2 in breast cancer samples and matched normal breast tissue samples using western blotting. We found that BRCC2 was significantly reduced in cancers, which suggested that BRCC2 was a candidate tumor suppressor gene in breast cancer. To further explore the role of BRCC2 in breast cancer, we transfected breast cancer cells either to ectopically express BRCC2 or to inhibit its expression using RNA interference. Knockdown of BRCC2 in vitro significantly enhanced the proliferation, migration and invasion of breast cancer cells, while overexpression of BRCC2 inhibited cell growth and mobility. Our in vivo experiments also demonstrated that BRCC2 markedly inhibited tumorigenesis, growth and metastasis to the lungs. These data further supported the tumor suppressor role of BRCC2 in breast cancer.
The activated Akt pathway has been demonstrated to have an essential role in breast cancer proliferation, motility and invasion,27, 28, 29 and MMP-2 and MMP-9 have been shown to enhance cancer migration and invasion via regulation of the Akt pathway.30, 31 Higher expression of MMP-2 and MMP-9 were observed in breast cancer tissues compared with normal samples,32, 33 and higher levels of MMP-2 and MMP-9 were associated with both an increased risk of lymph node metastases and poor prognoses.34, 35 MMP-2 and MMP-9 degrade type I and IV collagen and breakdown the extracellular matrix, contributing to cancer metastasis. Therefore, we investigated whether BRCC2 suppressed tumorigenesis and metastasis in breast cancer via activation of the Akt-mediated MMP pathway. Our results indicated that the levels of p-Akt, MMP-2 and MMP-9 were significantly decreased in BRCC2-overexpressing cells, and p-Akt, MMP-2 and MMP-9 were upregulated in BRCC2 knockdown cells. When we pretreated the BRCC2 knockdown cells with an Akt inhibitor or Akt siRNA, the levels of MMP-2 and MMP-9 were dramatically decreased. In addition, the increased migration and invasion ability of BRCC2 knockdown cells was also inhibited. All of these data revealed that BRCC2 suppresses the migration and invasion activities of breast cancer cells via activation of the Akt-mediated MMP-2 and MMP-9 pathways. However, how BRCC2 might regulate the Akt pathway is still unknown. Previously studies have reported that Akt could be regulated by many genes, including PH domain leucine-rich repeats protein phosphatase,36 phosphoinositide-dependent kinase-1 (PDK1),37, 38 phosphatase and tensin homolog (PTEN),38 and miRNAs.39, 40 So in our following studies, we will revealed the detailed mechanism about how BRCC2 might regulate Akt pathway in breast cancers by using the global expression profile microarray and microRNA microarray.
In conclusion, we found that BRCC2 expression was generally lower in cancer lesions compared with matched non-tumor tissues. Our in vitro and in vivo data demonstrate that BRCC2 has a vital function in inhibiting cell mobility, which is at least partially controlled by the Akt-mediated MMP-2 and MMP-9 pathway. Thus, we propose that the candidate tumor suppressor gene BRCC2, together with the Akt signaling pathway, maybe an effective novel therapeutic target in the management of breast cancer.
Materials and Methods
Patients and tissue samples
A total of 39 breast cancer tissue samples, along with matched normal tissues, were used in this study. All of the samples were obtained from the Qilu Hospital of Shandong University between 2010 and 2011. For all of the patients who participated in this study, written informed consent was obtained, which was approved by the Ethical Committee of Shandong University.
Cell lines and reagents
The breast cancer cell lines (MCF-7 and T47D) were obtained from the American Type Culture Collection (ATCC, Rockville, MD, USA). Antibodies that had been raised against Akt and phosphorylated Akt (Ser 473 and Thr 308) were purchased from Cell Signaling Technology (Beverly, MA, USA), and a mouse anti-BRCC2 antibody was obtained from Abcam (Cambridge, MA, USA). The Akt inhibitor LY294002, signal silence Akt siRNA and its control siRNA were purchased from Cell Signaling Technology. All of the remaining reagents were obtained from Sigma-Aldrich (St. Louis, MO, USA), unless otherwise specified.
Plasmid construction and transfection
For overexpression, the cDNA representing the complete open reading frame of BRCC2 was cloned into the pcDNA3.1 vector (Invitrogen, Carlsbad, CA, USA) to generate the BRCC2 expression plasmid. The expression plasmid was verified by sequencing both strands and was used to transfect the MDA-MB-231 cells to establish the BRCC2 overexpression cell line. For BRCC2 RNA interference, the control and BRCC2 shRNA plasmids (sc-96672-SH) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). The transfection efficiency of BRCC2 was confirmed by western blotting and quantitative reverse-transcription PCR (qRT-PCR) analyses.
Western blotting analysis
Equal amounts of protein were separated using SDS polyacrylamide gels and were electrotransferred to polyvinylidene fluoride membranes (Millipore, Bedford, MA, USA). The membranes were immunoblotted overnight at 4 °C with primary antibodies, followed by their respective secondary antibodies. β-Actin was used as the loading control.
Quantitative reverse-transcription PCR
RNA was extracted using TRIzol reagent, according to the manufacturer's recommended protocol (Invitrogen). qRT-PCR was performed using Applied Biosystems (Foster City, CA, USA) StepOne and StepOne Plus Real-Time PCR Systems. GAPDH was used as a loading control. The experiments were repeated a minimum of three times to confirm the results.
Cell proliferation assay
A 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium (MTT) assay was used to assess cell proliferation. The cells were seeded and 20 μl of the MTT solution (5 mg/ml) was then added to each well at the indicated time. The absorbance at 490 nm was measured using a microplate reader (Bio-Rad, Hercules, CA, USA).
Clonogenic assay
The cells were seeded in 6-cm dishes at a density of 300 cells per dish. After incubation for 14 days, the colonies were fixed with methanol for 10 min and stained with crystal violet for 15 min, after which point the number of colonies containing more than 50 cells was scored.
Transwell assay
Migration and invasion assays were performed as previously described.8, 41 Invasion assays were performed in 24-well transwell chambers (BD Biosciences, Bedford, MA, USA) containing polycarbonate filters with 8-μm pores coated with Matrigel (BD Biosciences). First, the cells that were suspended in serum-free DMEM were added to the upper compartment of the chamber and medium containing 10% FBS was added to the lower compartment of the chamber. At the indicated timepoints, the number of cells that had migrated through the membrane and attached to the lower surface of the membrane was counted under a light microscope for a minimum of ten random visual fields. The migration assay was similar to the migration assay except that the upper side of the membranes was not coated with the Matrigel.
Immunofluorescence staining
The cells were grown on the sterile coverslips, and the cells were fixed with 4% paraformaldehyde and permeabilized using 0.1% Triton-X100. Cells were blocked with rabbit anti-Ki67 antibody followed by hodamine-conjugated anti-rabbit secondary antibody. Finally, the cells were further stained with 4, 6-diami-dino-2-pheny-lindole (DAPI).
In vivo tumorigenesis and metastasis assays
The in vivo tumorigenesis and metastasis assays were performed, as previously described.6, 42 Briefly, 1 × 106 cells were injected subcutaneously into the right flanks of severe combined immunodeficient (SCID) mice. For the T47D cells, a 17β-estradiol pellet (Innovative Research of America, Sarasota, FL) was implanted 2 days before cell injection. Tumor length (L) and width (W) were measured every 3 days, and tumor volume was calculated using the equation: volume=(W2 × L)/2. To produce the lung metastasis model, 5 × 105 cells were injected into the lateral tail veins of female SCID mice. After 6 weeks, the mice were killed and the lung tissues were harvested for use in further experiments. All of the animal experiments were performed with the approval of the Shandong University Animal Care and Use Committee.
Statistical analysis
The results were analyzed using SPSS 18.0 software (Chicago, IL, USA). Each experiment was repeated a minimum of three times. A two-tailed t-test was used to determine statistical significance. The results were presented as the means±S.D. P-values<0.05 were considered to be statistically significant.
Acknowledgments
This work was supported by National Natural Science Foundation of China (No. 81072150; No. 81172529; No. 81272903) and Shandong Science and Technology Development Plan (No. 2012GZC22115).
Glossary
- BRCC2
breast cancer cell 2
- MMP-2
matrix metalloproteinase-2
- MMP-9
matrix metalloproteinase-9
- MTT
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl tetrazolium
- shRNA
Short hairpin RNA
- siRNA
small interfering RNA
- qRT-PCR
quantitative reverse-transcription PCR
The authors declare no conflict of interest.
Footnotes
Edited by D Aberdam
References
- Siegel R, Naishadham D, Jemal A. Cancer statistics. CA Cancer J Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Chin L, Gray JW. Translating insights from the cancer genome into clinical practice. Nature. 2008;452:553–563. doi: 10.1038/nature06914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steeg PS. Tumor metastasis: mechanistic insights and clinical challenges. Nat Med. 2006;12:895–904. doi: 10.1038/nm1469. [DOI] [PubMed] [Google Scholar]
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987;235:177–182. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- Eichhorn PJ, Baselga J. HER2 signatures in breast cancer: ready to go to print. J Clin Oncol. 2010;28:1809–1810. doi: 10.1200/JCO.2009.26.7146. [DOI] [PubMed] [Google Scholar]
- Hu G, Chong RA, Yang Q, Wei Y, Blanco MA, Li F, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Wang X, Huo Q, Li X, Wang H, Schneider P, et al. The oncogene metadherin modulates the apoptotic pathway based on the tumor necrosis factor superfamily member TRAIL (Tumor Necrosis Factor-related Apoptosis-inducing Ligand) in Breast Cancer. J Biol Chem. 2013;288:9396–9407. doi: 10.1074/jbc.M112.395913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Kong X, Huo Q, Guo H, Yan S, Yuan C, et al. Metadherin enhances the invasiveness of breast cancer cells by inducing epithelial to mesenchymal transition. Cancer Sci. 2011;102:1151–1157. doi: 10.1111/j.1349-7006.2011.01919.x. [DOI] [PubMed] [Google Scholar]
- Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, et al. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719–726. doi: 10.1200/JCO.2002.20.3.719. [DOI] [PubMed] [Google Scholar]
- Cavalli LR, Santos SC, Broustas CG, Rone JD, Kasid UN, Haddad BR. Assignment of the BLID gene to 11q24.1 by fluorescence in situ hybridization. Cancer Genet Cytogenet. 2008;186:120–121. doi: 10.1016/j.cancergencyto.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broustas CG, Gokhale PC, Rahman A, Dritschilo A, Ahmad I, Kasid U. BRCC2 a novel BH3-like domain-containing protein, induces apoptosis in a caspase-dependent manner. J Biol Chem. 2004;279:26780–26788. doi: 10.1074/jbc.M400159200. [DOI] [PubMed] [Google Scholar]
- Gentile M, Olsen K, Dufmats M, Wingren S. Frequent allelic losses at 11q24.1-q25 in young women with breast cancer: association with poor survival. Br J Cancer. 1999;80:843–849. doi: 10.1038/sj.bjc.6690430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chunder N, Mandal S, Roy A, Roychoudhury S, Panda CK. Analysis of different deleted regions in chromosome 11 and their interrelations in early- and late-onset breast tumors: association with cyclin D1 amplification and survival. Diagn Mol Pathol. 2004;13:172–182. doi: 10.1097/01.pas.0000124337.49401.0b. [DOI] [PubMed] [Google Scholar]
- Climent J, Dimitrow P, Fridlyand J, Palacios J, Siebert R, Albertson DG, et al. Deletion of chromosome 11q predicts response to anthracycline-based chemotherapy in early breast cancer. Cancer Res. 2007;67:818–826. doi: 10.1158/0008-5472.CAN-06-3307. [DOI] [PubMed] [Google Scholar]
- Broustas CG, Ross JS, Yang Q, Sheehan CE, Riggins R, Noone AM, et al. The proapoptotic molecule BLID interacts with Bcl-XL and its downregulation in breast cancer correlates with poor disease-free and overall survival. Clin Cancer Res. 2010;16:2939–2948. doi: 10.1158/1078-0432.CCR-09-2351. [DOI] [PubMed] [Google Scholar]
- Bouwman P, Aly A, Escandell JM, Pieterse M, Bartkova J, van der Gulden H, et al. 53BP1 loss rescues BRCA1 deficiency and is associated with triple-negative and BRCA-mutated breast cancers. Nat Struct Mol Biol. 2010;17:688–695. doi: 10.1038/nsmb.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babykutty S, Priya SP, Nandini RJ, Kumar MA, Nair MS, Srinivas P, et al. Nimbolide retards tumor cell migration, invasion, and angiogenesis by downregulating MMP-2/9 expression via inhibiting ERK1/2 and reducing DNA-binding activity of NF-kappaB in colon cancer cells. Mol Carcinog. 2012;51:475–490. doi: 10.1002/mc.20812. [DOI] [PubMed] [Google Scholar]
- Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141:52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae IH, Park MJ, Yoon SH, Kang SW, Lee SS, Choi KM, et al. Bcl-w promotes gastric cancer cell invasion by inducing matrix metalloproteinase-2 expression via phosphoinositide 3-kinase, Akt, and Sp1. Cancer Res. 2006;66:4991–4995. doi: 10.1158/0008-5472.CAN-05-4254. [DOI] [PubMed] [Google Scholar]
- Hwang MK, Song NR, Kang NJ, Lee KW, Lee HJ. Activation of phosphatidylinositol 3-kinase is required for tumor necrosis factor-alpha-induced upregulation of matrix metalloproteinase-9: its direct inhibition by quercetin. Int J Biochem Cell Biol. 2009;41:1592–1600. doi: 10.1016/j.biocel.2009.01.014. [DOI] [PubMed] [Google Scholar]
- Cavalli LR, Noone AM, Makambi KH, Rone JD, Kasid UN, Haddad BR. Frequent loss of the BLID gene in early-onset breast cancer. Cytogenet Genome Res. 2011;135:19–24. doi: 10.1159/000330265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi H, Yamada R, Gotoh N, Hayashi H, Yamashiro K, Shimada N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5:e1000660. doi: 10.1371/journal.pgen.1000660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao F, Bai J, Chen W, Xue A, Li C, Yan Z, et al. Evaluation of BLID and LOC399959 as candidate genes for high myopia in the Chinese Han population. Mol Vis. 2010;16:1920–1927. [PMC free article] [PubMed] [Google Scholar]
- Fredersdorf S, Burns J, Milne AM, Packham G, Fallis L, Gillett CE, et al. High level expression of p27(kip1) and cyclin D1 in some human breast cancer cells: inverse correlation between the expression of p27(kip1) and degree of malignancy in human breast and colorectal cancers. Proc Natl Acad Sci USA. 1997;94:6380–6385. doi: 10.1073/pnas.94.12.6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, et al. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc Natl Acad Sci USA. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills GB, Lu Y, Fang X, Wang H, Eder A, Mao M, et al. The role of genetic abnormalities of PTEN and the phosphatidylinositol 3-kinase pathway in breast and ovarian tumorigenesis, prognosis, and therapy. Semin Oncol. 2001;28 (5 Suppl 16:125–141. doi: 10.1016/s0093-7754(01)90290-8. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Dillon RL, White DE, Muller WJ. The phosphatidyl inositol 3-kinase signaling network: implications for human breast cancer. Oncogene. 2007;26:1338–1345. doi: 10.1038/sj.onc.1210202. [DOI] [PubMed] [Google Scholar]
- Dillon RL, Marcotte R, Hennessy BT, Woodgett JR, Mills GB, Muller WJ. Akt1 and akt2 play distinct roles in the initiation and metastatic phases of mammary tumor progression. Cancer Res. 2009;69:5057–5064. doi: 10.1158/0008-5472.CAN-08-4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauvois B. New facets of matrix metalloproteinases MMP-2 and MMP-9 as cell surface transducers: outside-in signaling and relationship to tumor progression. Biochim Biophys Acta. 2012;1825:29–36. doi: 10.1016/j.bbcan.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Shi GM, Ke AW, Zhou J, Wang XY, Xu Y, Ding ZB, et al. CD151 modulates expression of matrix metalloproteinase 9 and promotes neoangiogenesis and progression of hepatocellular carcinoma. Hepatology. 2010;52:183–196. doi: 10.1002/hep.23661. [DOI] [PubMed] [Google Scholar]
- Stankovic S, Konjevic G, Gopcevic K, Jovic V, Inic M, Jurisic V. Activity of MMP-2 and MMP-9 in sera of breast cancer patients. Pathol Res Pract. 2010;206:241–247. doi: 10.1016/j.prp.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Shah FD, Shukla SN, Shah PM, Shukla HK, Patel PS. Clinical significance of matrix metalloproteinase 2 and 9 in breast cancer. Indian J Cancer. 2009;46:194–202. doi: 10.4103/0019-509X.52953. [DOI] [PubMed] [Google Scholar]
- McGowan PM, Duffy MJ. Matrix metalloproteinase expression and outcome in patients with breast cancer: analysis of a published database. Ann Oncol. 2008;19:1566–1572. doi: 10.1093/annonc/mdn180. [DOI] [PubMed] [Google Scholar]
- Wu ZS, Wu Q, Yang JH, Wang HQ, Ding XD, Yang F, et al. Prognostic significance of MMP-9 and TIMP-1 serum and tissue expression in breast cancer. Int J Cancer. 2008;122:2050–2056. doi: 10.1002/ijc.23337. [DOI] [PubMed] [Google Scholar]
- Liu J, Weiss HL, Rychahou P, Jackson LN, Evers BM, Gao T. Loss of PHLPP expression in colon cancer: role in proliferation and tumorigenesis. Oncogene. 2009;28:994–1004. doi: 10.1038/onc.2008.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A, Newton AC. Cellular signaling: pivoting around PDK-1. Cell. 2000;103:185–188. doi: 10.1016/s0092-8674(00)00110-0. [DOI] [PubMed] [Google Scholar]
- Williams MR, Arthur JS, Balendran A, van der Kaay J, Poli V, Cohen P, et al. The role of 3-phosphoinositide-dependent protein kinase 1 in activating AGC kinases defined in embryonic stem cells. Curr Biol. 2000;10:439–448. doi: 10.1016/s0960-9822(00)00441-3. [DOI] [PubMed] [Google Scholar]
- Bao L, Yan Y, Xu C, Ji W, Shen S, Xu G, et al. MicroRNA-21 suppresses PTEN and hSulf-1 expression and promotes hepatocellular carcinoma progression through AKT/ERK pathways. Cancer Lett. 2013;S0304-3835:376–385. doi: 10.1016/j.canlet.2013.05.007. [DOI] [PubMed] [Google Scholar]
- Zhang LY, Ho-Fun Lee V, Wong AM, Kwong DL, Zhu YH, Dong SS, et al. MicroRNA-144 promotes cell proliferation, migration and invasion in nasopharyngeal carcinoma through repression of PTEN. Carcinogenesis. 2013;34:454–463. doi: 10.1093/carcin/bgs346. [DOI] [PubMed] [Google Scholar]
- Li X, Xu B, Moran MS, Zhao Y, Su P, Haffty BG, et al. 53BP1 functions as a tumor suppressor in breast cancer via the inhibition of NF-kappaB through miR-146a. Carcinogenesis. 2012;33:2593–2600. doi: 10.1093/carcin/bgs298. [DOI] [PubMed] [Google Scholar]
- Zhu J, Li X, Kong X, Moran MS, Su P, Haffty BG, et al. Testin is a tumor suppressor and prognostic marker in breast cancer. Cancer Sci. 2012;103:2092–2101. doi: 10.1111/cas.12020. [DOI] [PMC free article] [PubMed] [Google Scholar]