Abstract
Myotonic dystrophy type-1 (DM1) is the most prevalent form of muscular dystrophy in adults. This disorder is an RNA-dominant disease, caused by expansion of a CTG repeat in the DMPK gene that leads to a misregulation in the alternative splicing of pre-mRNAs. The longer muscleblind-like-1 (MBNL1) transcripts containing exon 5 and the respective protein isoforms (MBNL142–43) were found to be overexpressed in DM1 muscle and localized exclusively in the nuclei. In vitro assays showed that MBNL142–43 bind the Src-homology 3 domain of Src family kinases (SFKs) via their proline-rich motifs, enhancing the SFK activity. Notably, this association was also confirmed in DM1 muscle and myotubes. The recovery, mediated by an siRNA target to Ex5-MBNL142–43, succeeded in reducing the nuclear localization of both Lyn and MBNL142–43 proteins and in decreasing the level of tyrosine phosphorylated proteins. Our results suggest an additional molecular mechanism in the DM1 pathogenesis, based on an altered phosphotyrosine signalling pathway.
Keywords: MBNL1, DM1, SFKs, muscle
Myotonic dystrophy type-1 (DM1, OMIM 160900) is an autosomal dominant multisystemic disorder with considerable phenotypic variation between individuals; clinical aspects include myotonia, progressive muscle weakness, cataracts, insulin resistance and cardiac conduction defects.1 The genetic mutation responsible for DM1 is an expanded (CTG)n, in the 3′-untranslated region (UTR) of the dystrophia myotonica protein kinase (DMPK) gene (OMIM 605377), on chromosome 19q13.2, 3, 4 DM1 pathogenesis is linked to the expression of expanded mutant RNA transcripts, which accumulate in the nuclei to form nuclear foci (ribonuclear inclusions).5 This clue prompted the theory that a toxic gain-of-function effect, resulting from these inclusions, alters the proportion-activity of two RNA-binding proteins: muscleblind-like-1 (MBNL1) and CUG-BP1. The splicing of several MBNL1 and CUG-BP1 pre-mRNA targets is subsequently severely altered in DM1-affected tissues. The disruption of the Mbnl1 gene in transgenic mouse models in fact recapitulates the splicing abnormalities of DM1 patients,6, 7, 8 as well as the myotonic-dystrophy-like phenotype, with skeletal muscle myotonia and cataracts. Moreover, the myotonic-dystrophy-like phenotype of mice expressing (CUG)n can be reversed by overproduction of MBNL1,9 and long-term MBNL1 overexpression prevents CUG-induced myotonia, myopathy and alternative splicing abnormalities in DM1 mice.10 MBNL1 is also sufficient to largely abolish DM1 foci in vivo.11 In humans, the MBNL1 gene contains 12 exons and the coding sequence is distributed over 10 exons (numbered 1 to 10 in this paper; Figure 1a). The inclusion/exclusion of exons 3, 5, 7 and 9 generates several mRNA isoforms12, 13 developmentally regulated and reportedly altered in DM tissues.7, 9, 14, 15 MBNL1 exons encode for protein domains with different functions: exons 1, 2 and 4 are essential for RNA binding,16 exons 5 and 6 for controlling the nuclear localization of MBNL1; exon 3 strongly enhances the affinity of MBNL1 for its pre-mRNA target sites; exons 3 and 6 encode for a splicing regulatory domain; and exon 7 enhances MBNL1 self-dimerization.17 Although MBNL1 isoforms have been partly characterized in human skeletal muscle, recent work on DM1-patient brain revealed the existence of a combination of foetal and adult isoforms, with the expression of additional, longer variants assuming different functional roles (still unexplored).9, 15 The MBNL1 protein contains three proline-rich motifs (PRMs), known to bind Src-homology 3 (SH3) domains present in many signalling proteins (Supplementary Figure SF1). This observation suggests that the different MBNL1 isoforms might regulate the activity and/or localization of the tyrosine (Tyr) kinases of the Src family (SFKs), triggering a signalling cascade mediated by Tyr phosphorylation. Interestingly, CUG-BP1 was also recently found hyperphosphorylated in DM1 tissues and in a DM1 mouse model.18
Figure 1.
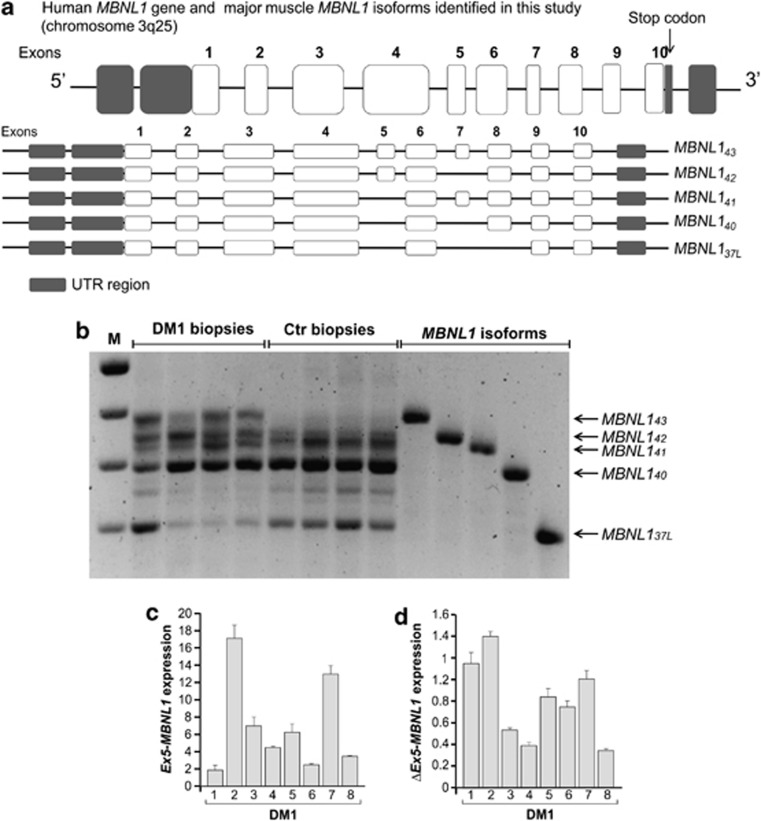
Qualitative and quantitative expression analysis of the major MBNL1 isoforms expressed in the muscle from DM1 patients and controls. (a) Representation of the MBNL1 gene and major muscle transcripts identified in this study. The human MBNL1 gene contains 10 coding exons, represented as white boxes; untranslated regions (UTRs) are shown in grey, and introns as lines. The names of each isoform and the exon order are indicated on the right following Pascual et al.19 (b) Representative gel electrophoresis of the RT-PCR products corresponding to the entire coding sequences of the MBNL1 gene. Five major muscular transcripts were identified (MBNL137L–40–41–42–43) that showed relative expression differences between four DM1 (lanes 2–5) and four control (lanes 6–9) muscle samples. (c and d) Results from Syber Green Real Time PCR analysis using primers allowing detection of MBNL1 isoforms (c) including (Ex5-MBNL142-43) and (d) not including (ΔEx5-MBNL140-41) exon 5 in muscle from DM1 (n=8, grey bars). DM1 samples express significantly higher level of Ex5-MBNL142–43 isoforms compared with controls (P<0.01 by Mann–Whitney test), whereas no differences were found in ΔEx5-MBNL137L–40–41 transcript expression. All experiments were in triplicate; the median value of controls was set as one, and the level of β-actin transcript was used for sample normalization
Intracellular localization, regulation of the alternative splicing of model pre-mRNA, ability to interact with SFKs through SH2 and SH3 domains and susceptibility to Tyr phosphorylation of MBNL1 isoforms in DM1 muscle and myotubes were the aspects specifically addressed by our investigation.
Results
MBNL1 42–43 isoforms are overexpressed in the muscle from DM1 patients compared with controls, and localize in the nuclei
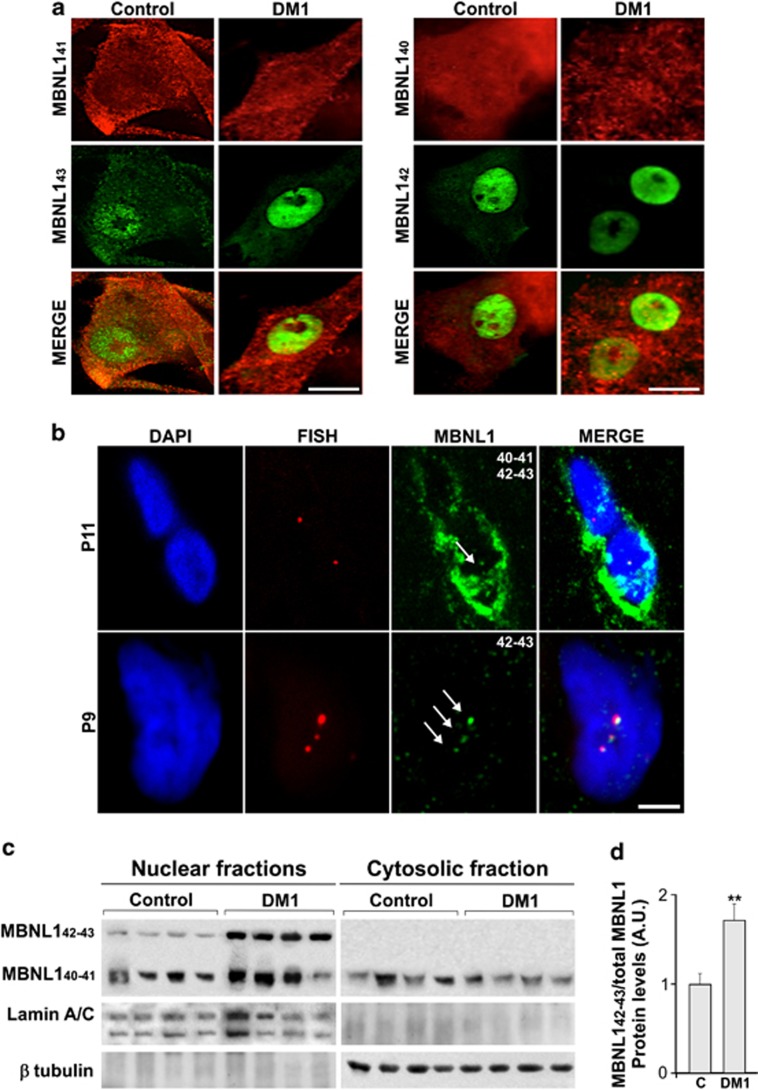
The complete splicing pattern of the MBNL1 gene was first determined in muscle tissues from DM1 (n=8) patients compared with controls (n=4), by RT-PCR analysis. The number assigned to each MBNL1 exon varies in the literature; here, we use the exon numbering of Pascual and co-workers19 and coding exons have been labelled from 1 to 10 (Figure 1a). Five major MBNL1 transcripts were identified; their relative expression in DM1 and control samples was found to be significantly different. The major transcripts expressed in the muscle from controls corresponded to MBNL140 (NM 207292.1) and MBNL141 (NM 021038.3) isoforms. We also identified and characterized a novel MBNL1 isoform lacking exons 5, 7 and 8 – named MBNL137L – in view of the predicted molecular weight (MW) of the encoded protein (Figure 1a). In the DM1 muscle, in addition to these MBNL1 transcripts, we observed higher levels of the MBNL142 (NM 207293.1) and MBNL143 isoforms (Figure 1b), whereas the expression of MBNL143 and MBNL142 was absent and greatly reduced, respectively, in the control muscle. The MBNL143 mRNA is expressed in human foetal brain and mouse skeletal muscle9, 15 and corresponds to the MBNL142 sequence of Pascual's classification with the addition of exon 7 (Figure 1a). RT-PCR analysis, using primers designed to discriminate between MBNL1 isoforms containing (Ex5-MBNL142–43) or not containing (ΔEx5-MBNL37L–40–41) exon 5, confirmed that DM1 tissues express higher levels of Ex5-MBNL142–43 mRNAs than controls. Ex5-MBNL142–43 isoforms were overexpressed in DM1 muscle 1.9- to 17.3-fold compared with controls, whereas ΔEx5-MBNL137L–40–41 transcripts vary over the range from 0.38 to 1.40 (Figures 1c and d). These results are in accordance with the previous observation that MBNL1 autoregulates the splicing of its own pre-mRNA inducing the exclusion of exon 5 by binding to a response element located in intron 4 of the gene.20 The functional depletion of the MBNL1 protein in DM1 tissues could therefore lead to the aberrant inclusion of exon 5 within the MBNL1 transcripts. We then subcloned the coding sequences of the MBNL140–41–42–43 major muscular MBNL1 isoforms into appropriate expression vectors to localize the encoded proteins. The exon 5-dependent nuclear localization of the MBNL142–43 isoforms has been reported in HeLa cells and control myoblasts.15, 17 Consistently, in DM1 myoblasts, the localization of MBNL140–41-V5 isoforms was diffuse in the cytosol and nucleus, whereas MBNL142–43-myc isoforms were exclusively nuclear (Figure 2a), with no differences between DM1 and control primary myoblasts (Figure 2a). This observation suggests that in this in vitro model, the MBNL1 cell localization is unaffected by the expression of the pathological CUG expansions. Two novel anti-MBNL1-specific antibodies (anti-P9 and -P11) were synthesized to distinguish the MBNL1 isoforms. Anti-P9, targeted to the 18 amino acids encoded by exon 5, was designed to recognize only the MBNL142–43 isoforms, whereas anti-P11, designed against the constitutive exon 6, binds to all isoforms (see Supplementary Figures S1 and S2). Immunofluorescence analysis showed that the antibody anti-P9 (MBNL142–43) colocalizes with the ribonuclear foci of DM1 muscle, as well as the anti-P11 antibody (MBNL140–41–42–43) (Figure 2b). However, the immunofluorescence signal of anti-P11 antibody revealed a nuclear and cytosolic expression, providing further evidence of a ubiquitous cell distribution of MBNL140–41 (Figure 2b). Consistent with the results of mRNA expression, western blotting (WB) analysis showed significant increase (P<0.01) of the signal recognized by the anti-P9 antibody (target to exon 5) in DM1 muscle nuclei, as compared with controls, but was absent in the cytoplasmic fraction of all tested samples (Figures 2c and d). Tests were performed to establish the extent to which the various MBNL1 isoforms differed in function, properties and interactions. Minigene assays on the cardiac troponin T (c-TNT) and insulin receptor (IR) pre-mRNAs splicing21, 22 revealed no differences in the in trans regulatory splicing properties of the characterized MBNL1 isoforms (Supplementary Figure S3).
Figure 2.
Nuclear localization and amount of MBNL142–43 isoforms in DM1 myoblasts and muscle tissue. (a) Representative confocal images of DM1 and control primary myoblasts, overexpressing MBNL140–41–42–43 isoforms. Immunofluorescence with specific anti-V5 (MBNL140–41) or -myc (MBNL142–43) tags was performed after 48 h. Scale bar, 10 μm. (b) Representative images of nuclear foci (red) from fixed DM1 muscle, obtained by fluorescence in situ hybridization with a Texas Red labelled (CAG)10 probe. The MBNL1 antibodies (green) anti-P9 and anti-P11 co-localized with foci. Anti-P9 was localized only in the nucleus, in contrast with anti-P11, which was also present in the cytosol. Scale bar, 2 μm. (c) Representative WB analysis of MBNL140–41–42–43 in nuclear and cytosolic fractions from four DM1 patients and four control muscle samples. Proteins, separated in 7.5–17.5% T30C4 SDS-PAGE gels, were blotted onto nitrocellulose and probed with anti-P9 (specific for MBNL142–43 isoforms) and anti-P11 antibodies. Lamin A/C and β-tubulin protein levels were used to normalize the samples from nuclear and cytosolic fractions, respectively. (d) The graph represents the mean±S.D. of at least three experiments. In each lane the ratio between the signal density of MBNL142–43 (upper band) and the sum of the signal density of the two MBNL1 bands (upper and lower) was calculated. The values were expressed as percentage of the value obtained in the control muscle, considered as reference unit. **Significant difference for P<0.01 by Mann–Whitney test
MBNL142–43 proteins interact with SFKs, enhance cellular Tyr phosphorylation and induce the nuclear translocation of Lyn Tyr kinase
The four above MBNL1 isoform sequences exhibit three PRMs, encoded by MBNL1 exon 2 (111PLQPVP116), exon 3 (161PGVPVP166) and exon 6 (295PPLPKRP301 of MBNL142–43 proteins, corresponding to amino acids 277–283 in MBNL140–41 proteins), respectively (Supplementary Material and Supplementary Figure S1); these sequences display the consensus motif for interaction with the SH3 domain within the SFKs. As the PRM binding to the SH3 domain of SFKs is described to bring about activation,23 turnover24 and subcellular localization25, 26 of SFKs themselves, we wondered whether the MBNL1 isoforms could affect these SFK properties, and/or whether Lyn could be implicated in the pathogenesis of DM1. SFKs are non-receptor Tyr kinases, classifiable into two groups, based on the phylogenetic tree,27 namely Src-related (Src, Yes, Fyn and Fgr) and Lyn-related SFKs (Lyn, Hck, Lck and Blk).28 Their common modular structure consist of (i) an N-terminal SH4 domain for acylation; (ii) the aforementioned SH3 domain; (iii) an SH2 domain, which recognizes phospho-Tyr (pTyr) motifs; and (iv) a kinase domain (SH1), followed by a C-terminal tail having a negative regulatory role when phosphorylated by the inhibitory kinases.23 SFKs are normally maintained in a closed, inactive conformation through two major intramolecular inhibitory interactions: the binding of the phosphorylated C-terminal tail to the SH2 domain, and the interaction of a polyproline type-II helical motif containing the core sequence P–X–X–P, where X is any amino acid, within the SH2 kinase linked with the SH3 domain.29 Conformational transition of SFKs from inactive to active form is induced by an array of factors causing dephosphorylation of the C-terminal tail by several Tyr phosphatases and/or displacement of the SH2 or SH3 domains by specific ligands, ultimately leading to disruption of the inhibitory intramolecular interactions.30, 31, 32, 33, 34
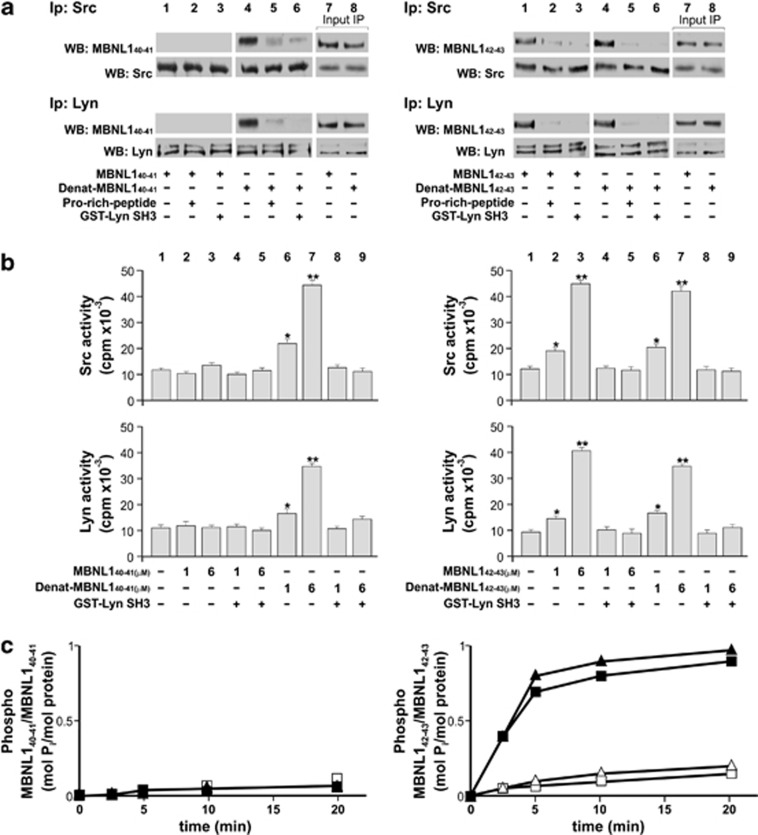
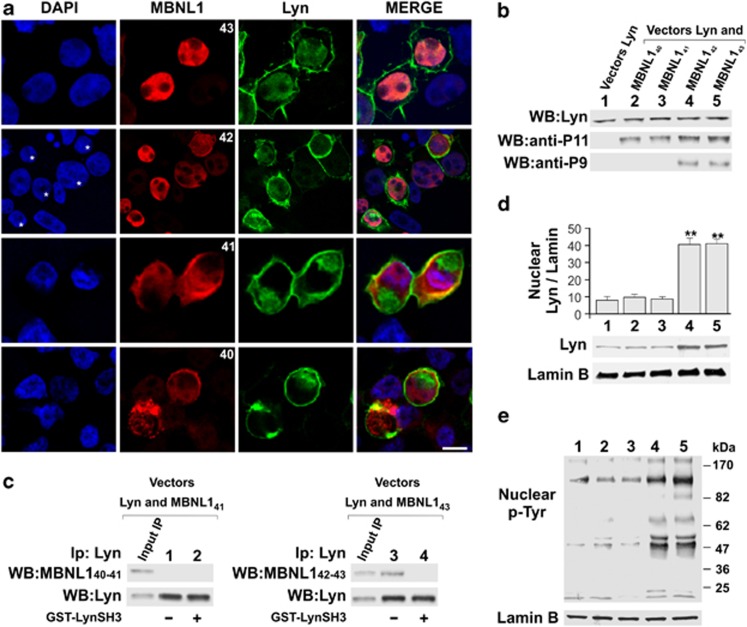
To assess whether MBNL140–41 and MBNL142–43 interacted with the SFK SH3 domain through their PRMs, we performed competition assays by incubating the MBNL1 isoforms with Lyn or Src (the prototypical members of the two SFK groups) in the absence or presence of a synthetic proline-rich peptide (KGGRSRLPLPPLPPPG), which is known to interact with Lyn SH3 domain, or the recombinant glutathione S-transferase (GST)-Lyn SH3 domain. The following immunoprecipitation with anti-Lyn and anti-Src antibodies showed that only the MBNL142–43 associated with Src and Lyn via interaction between the MBNL1 PRM and the SFK SH3 domain (Figure 3a, left panel). This interaction also determined stimulation of SFK activity (Figure 3b, right panel) and phosphorylation of MBNL142–43 itself (Figure 3c, right panel). Yet, denaturation by heating at 55 °C allowed MBNL140–41 to acquire the same ability to bind to and activate Src and Lyn as MBNL142–43 (Figures 3a and b, left panel), suggesting that the binding motif in the latter isoforms is accessible owing to a structural conformation conferred by the insert encoded by exon 5. As to phosphorylation, denaturation prevented recognition of MBNL142–43 as a substrate by Src and Lyn (data not shown), suggesting the significance of not only the interaction due to exposure of specific motifs but also of an appropriate three-dimensional structure. Surface plasmon resonance (SPR) studies confirmed the specific interaction of the Lyn kinase SH3 domain with only MBNL143 isoform (Supplementary Material and Supplementary Figure S4). Our in vitro data provide novel evidence of a strong functional discrimination between the long and short isoforms of MBNL1 in terms of interaction with SFKs and regulation of their activity. These novel functions of the MBNL1 proteins were also analyzed in 293T cells co-overexpressing Lyn and one MBNL140–41–42–43 isoform. As 293T cells express a low basal level of SFKs, the amount of Lyn-encoding plasmid was standardized to the expression of Lyn within HeLa cells (Supplementary Figure S5). Figure 4a shows colocalization of MBNL142–43 and Lyn in the nucleus, where Lyn translocated after the overexpression of MBNL142–43 proteins, and with Lyn being equally expressed under all the experimental conditions and regardless of the cotransfection with MBNL1 isoforms (Figure 4b). To assess the existence of a MBNL1/Lyn complex in vivo, immunoprecipitation assays with anti-Lyn antibody on 293T cells coexpressing Lyn and each of the MBNL1 isoforms were carried out. This experiment revealed that MBNL142–43 (but not MBNL140–41) co-immunoprecipitated with Lyn, this event occurring as a result of PRM/SH3 domain interaction, as confirmed by the competition assay with GST-Lyn-SH3 (Figure 4c). Moreover, the WB analysis of the nuclear fraction of these cells highlighted that the nuclear levels of Lyn rose up fourfold (Figure 4d, lanes 4 and 5 versus lane 1) when the MBNL142–43 was coexpressed, whereas the localization of Lyn remained unchanged in the presence of MBNL140–41 (Figure 4d, lanes 2 and 3 versus lane 1). To explore whether this interaction determined SFK activation in the nuclei, as a result of SH3 engagement by PRMs,35 WB analysis with anti-pTyr antibody revealed a dramatic increase in Tyr phosphorylation in the nuclear fraction of 293T cells when MBNL142–43, and not MBNL140–41, were coexpressed with Lyn (Figure 4e).
Figure 3.
Analysis of the interaction between the SH3 domain of the SFKs Src and Lyn and the proline-rich motif of recombinant MBNL140–41 and MBNL142–43 isoforms. (a) The interaction between SH3 domain of two SFK (Src and Lyn) and the proline-rich motifs of recombinant MBNL140–41 and MBNL142–43 isoforms. The 0.2 μg of recombinant MBNL140–41 or MBNL142–43 (as such or heated at 55 °C for 10 min, denat. MBNL1) was incubated with 0.1 μg of Src or Lyn in the absence or presence, respectively, of a proline-rich peptide or the GST-Lyn SH3 domain, for 10 min at 30 °C. Three-fourths of the sample underwent immunoprecipitation with anti-Src or anti-Lyn antibodies, respectively. Immunocomplexes were then probed with anti-P11 or anti-P9 antibodies, and subsequently with anti-Src or anti-Lyn antibodies. The presence of MBNL42 or MBNL43 (right panel), but not of MBNL40 or MBNL41 (left), indicates its association with the two SFKs and suggests that the stretch encoded by exon 5 might have a role in this interaction, possibly through a conformational change that renders the PRMs available to binding. Moreover, competition assays carried out in the absence or presence of a synthetic proline-rich peptide (5′-KGGRSRLPLPPLPPPG-3′) known to interact with the Lyn SH3 domain, or the GST-Lyn/SH3 domain, confirmed that this interaction was mediated by PRMs and the SH3 domain. The remainder of the sample underwent WB analysis (lanes 7 and 8) (b) As this interaction was also a prerequisite for enhancement of SFK activity relative to the basal state, we tested Tyr kinase activity of Src (top panel) and Lyn (bottom) on the Src-specific peptide substrate cdc2 in the absence (lane 1) or presence of increasing concentrations of MBNL141 (left panels, lanes 2–5) and denat. MBNL141 (left panels, lanes 6–9) or MBNL143 (right panels, lanes 2–5) and denat. MBNL143 (right panels, lanes 6–9), alternatively supplemented with the GST-Lyn SH3 domain (lanes 4 and 5, and 8–9) as described in the Materials and Methods section. (c) As this interaction was also a prerequisite for recognition of MBNL143 as a substrate by SFKs, we tested whether MBNL141 or MBNL143 (each 1 μM) were phosphorylated by Lyn and Src in the absence (solid triangles and solid squares, respectively) or presence (open triangles and open squares, respectively) of the GST-Lyn SH3 domain. At various time points, reactions were stopped and the phosphate incorporated was analyzed after SDS-PAGE on the Cyclone Plus. The amount is expressed as mol P incorporated/mol prot. (values represent the mean of three separate experiments±S.D.). As expected, only MBNL143 is phosphorylated by Src. The same set of experiments conducted with the MBNL140 and MBNL142 isoforms demonstrated that these two proteins behave similarly to MBNL141 and MBNL143, respectively (data not shown). Only MBNL142–43 isoforms were able to significantly increase Src and Lyn activity, suggesting a novel function of these long isoforms in kinase enzyme regulation. Data are expressed as mean±S.D. from three separate experiments. *P<0.05; ***P<0.001 by Kruskal–Wallis analysis
Figure 4.
Translocation of Lyn to the nuclei of 293T cells occurs only upon co-expression with MBNL142–43. (a) Representative confocal images of 293T cells co-transfected with plasmids carrying different MBNL1 isoforms (MBNL140–41–42–43 in pEF1-Myc/HisA vector) and Lyn (pCMV6-XL4/Lyn). Nuclei were labelled with DAPI (blue), MBNL1 with anti-P9 (MBNL142–43) or anti-P11 (MBNL140–41) antibodies (red) coupled to immunodetection of Lyn (green). Scale bar 10 μm. (b) Representative WB analysis of whole-cell lysates from 293T cells transfected with expression vectors for Lyn, MBNL140, MBNL141, MBNL142 and MBNL143 probed with anti-Lyn antibody (top strip), anti-P11 antibody (middle strip) and anti-P9 antibody (bottom strip). (c) Representative co-immunoprecipitation of four-fifths of whole-cell lysates from 293T cells, overexpressing MBNL141 (pEF1/MBNL141 (left panel), lanes 1 and 2), or MBNL143 (pEF1/MBNL143 (right panel), lanes 3 and 4) and Lyn (pCMV6-XL4/Lyn, lanes 1–4). Thirty-six hours after transfection, whole-cell lysates were immunoprecipitated with anti-Lyn in the absence (lanes 1 and 3) or presence (lanes 2 and 4) of the GST-Lyn SH3 domain. The immunocomplexes and one-fifth of the whole-cell lysates underwent WB analysis with anti-P11 or anti-P9 antibodies, respectively, and subsequently with anti-Lyn antibodies. The figure represents one of three independent experiments. The same set of experiments conducted with MBNL140 and MBNL142 isoforms demonstrated that these two proteins behave similarly to MBNL141 and MBNL143, respectively, again supporting the hypothesis that it is exon 5 that enables MBNL1s to interact with SFKs (data not shown). (d) Representative WB analysis of nuclear lysates from 293T cells transfected with expression vectors for Lyn (lanes 1–5), MBNL140 (lane 2), MBNL141 (lane 3), MBNL142 (lane 4) and MBNL143 (lane 5). The membranes were probed with anti-Lyn and with anti-Lamin B antibody as loading control. The resulting bands underwent densitometric analysis. The values (arbitrary units) represent the mean±S.D. of at least three experiments. Reference unit: lane 1 sample (only transfected with Lyn). **Significant difference for P<0.01 by Kruskal–Wallis analysis. (e) Representative WB analysis of nuclear lysates from 293T cells, transfected with expression vectors for Lyn (lanes 1–5), MBNL140 (lane 2), MBNL141 (lane 3), MBNL142 (lane 4) and MBNL143 (lane 5) and probed with anti-p-Tyr antibody. The membranes were reprobed with anti-Lamin B antibody as loading control
Muscle and myotubes from DM1 patients show increased nuclear localization of Lyn and Tyr hyperphosphorylation
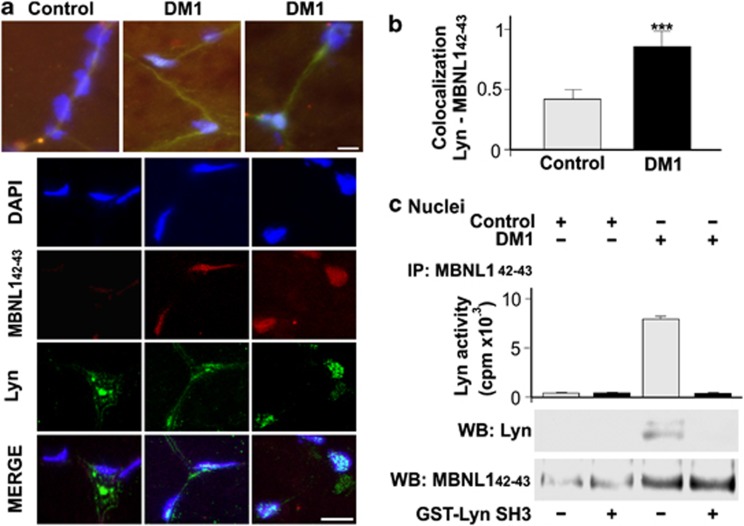
These data strongly suggest that MBNL142–43 protein isoforms, whose abundance is elevated in DM1 muscle, might induce an inappropriate activation of Lyn in DM1 muscle cell nuclei: as a direct test, we first examined muscle sections from DM1 patients (n=4) and controls (n=4). Confocal immunofluorescence images of the MBNL142–43 and Lyn proteins showed a nuclear localization of both proteins only in DM1 muscle (Figure 5a), confirmed by Manders overlap coefficient analysis (Figure 5b). Moreover, to assess whether the existence of an MBNL1/Lyn complex was also present in the nuclei of muscle tissue, co-immunoprecipitation analysis with anti-P9 (against MBNL142–43) antibody was performed on DM1 and control muscle samples. Consistently with the results obtained with 293T cells and in line with the increased amount of MBNL142–43 in DM1 muscle tissue (Figures 2c–d), we found that MBNL142–43 co-immunoprecipitated with Lyn only in the nuclear fraction of DM1 muscles, in parallel with an increased nuclear activity of Lyn (Figure 5c). This event occurred as a result of PRM/SH3 domain interaction, as confirmed by the competition assay with GST-Lyn-SH3 (Figure 5c).
Figure 5.
The nuclear colocalization of Lyn and MBNL142–43 was detected in DM1 muscle tissues. (a) Representative images at epifluorescent (low magnification) and confocal (high magnification) microscope of nuclei from muscle samples (four controls and four DM1 patients). The muscle sections were probed with anti-P9 (red) and anti-Lyn (green), nuclei were marked with DAPI. Scale bar, 10 μm. (b) Quantification of Lyn and MBNL142–43 signal colocalization in control and DM1 muscle samples. Values represent Manders' overlap coefficient (the fraction of total fluorescence signal of Lyn overlapping with MBNL142–43 signal), obtained from confocal images of nuclear and perinuclear regions as in (a) and as described in Materials and Methods section. Statistical significance was evaluated by two-tailed Student's t-test (n=50 nuclei, ***P<0.001). (c) MBNL142–43 immunoprecipitation from nuclear fractions, obtained from two controls and two DM1 muscle samples. Immunoprecipitate was used for Lyn Tyr kinase activity assay (upper panel) and for WB analysis (below), in the absence or presence of binding competitor domain GST-Lyn/SH3, as described in Materials and Methods section. Lyn co-immunoprecipitates with MBNL142–43 and displays Tyr kinase activity only in DM1 muscle samples. Both the interaction and the activity are abolished in the presence of competing GST-Lyn/SH3 domain
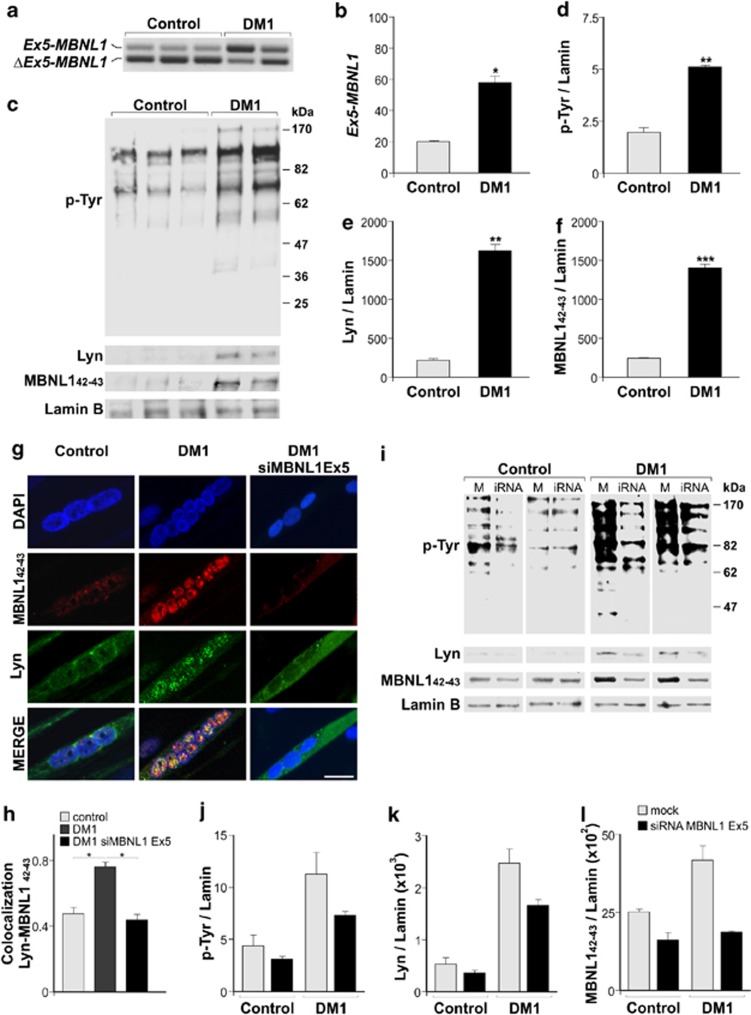
The same parameters were analyzed in 15-day-differentiated myotubes from two DM1 patients and three controls. As expected and reported elsewhere,36 RT-PCR-based analysis of MBNL1 splicing showed that DM1 myotubes had a statistically significant (2.8-fold) increase in the expression of Ex5-MBNL1 transcripts relative to controls (Figures 6a and b). Consistently, WB analysis demonstrated a significantly increased amount of Lyn (570%, P<0.02) and MBNL142–43 (680%, P<0.005) (Figures 6c, e and f) in the nuclei of the same DM1 myotubes, as compared with controls. The increased nuclear localization of Lyn in DM1 myotubes caused a Tyr hyperphosphorylation (250%, P<0.02), as revealed by WB analysis (Figures 6c and d). Immunofluorescence analysis with anti-Lyn and anti-P9 (against MBNL142–43) antibodies in 15-day-differentiated myotubes gave further confirmation of the MBNL142–43 and Lyn cellular distribution in DM1 and control cells (Figure 6g), validated by Manders' overlap coefficient analysis (Figure 6h).
Figure 6.
In 15-day-differentiated DM1 myotubes, nuclear MBNL142–43 isoforms are associated with increased presence and activity of Lyn in the nuclei. (a) RT-PCR analysis of MBNL1Ex5 and MBNL1ΔEx5 transcripts in 15-day-differentiated myotubes from three controls and two DM1 patients. (b) Quantification of MBNL1Ex5 transcript expressed as the percentage of the total MBNL1 transcripts. (c) Representative WB analysis with anti-p-Tyr, anti-Lyn, anti-P9 (against MBNL142–43) of nuclear lysates from 15-day-differentiated myotubes of three controls and two DM1 patients. Equal loading was monitored by anti-Lamin B antibody. (d–f) Quantification of at least three independent WB analyses as in (c). Values, normalized to the loading control, were expressed as mean±S.D. and compared with controls for statistical evaluation. *P<0.05; **P<0.02; ***P<0.005 by Mann–Whitney test. (g) Representative confocal images of 15-day-differentiated myotubes, from two controls and two DM1 patients, showing different localization of MBNL142–43 and Lyn (obtained using identical exposure time and acquisition setting). Myotubes from DM1 patients were also transfected with siMBNL1-Ex5, designed to target specifically the MBNL142–43 isoforms. Nuclei were labelled with DAPI (blue), MBNL1 with anti-P9 (red) coupled to immunodetection of Lyn (green). Scale bar 10 μm. (h) Quantification of Lyn and MBNL142–43 colocalization in control and DM1 myotubes. Values represent Manders' overlap coefficient (the fraction of total fluorescence signal of Lyn overlapping with MBNL142–43 signal), obtained from confocal images of nuclear and perinuclear regions as in (g) and as described in Materials and Methods section. Statistical significance was evaluated by two-tailed Student's t-test (n=30 nuclei, *P<0.05). (i) Representative WB analysis with anti-p-Tyr, anti-Lyn and anti-P9 (against MBNL142–43) antibodies of nuclear lysates from 15-day-differentiated myotubes of two controls and two DM1 patients, transfected with empty vector (M) and siMBNL1-Ex5 (iRNA). Equal loading was monitored by anti-Lamin B antibody. (j–l) Quantification of two WB analyses as in (i). Values, normalized to the loading control, were expressed as mean±S.D.
siRNA-mediated downregulation of MBNL142–43 isoforms in myotubes from DM1 patients restores cytoplasmic localization of Lyn
We used an siRNA-mediated approach to test whether both cytoplasmic localization of Lyn and increased nuclear Tyr phosphorylation could be restored in myotubes from DM1 patients. An siRNA specific to target MBNL1-exon 5 (siMBNL1-Ex5) was designed and tested on DM1 myotubes overexpressing Ex5-MBNL142–43 transcripts (Figure 6a). Splicing analysis, using primers designed to discriminate between Ex5-MBNL142–43 and ΔEx5-MBNL37L–40–41 isoforms, revealed a marked reduction of MBNL142–43 (from 70 to 85%), but not of the MBNL140–41 transcripts (from 97 to 115%) (data not shown). Immunofluorescence, on differentiated myotubes from DM1 patients 48 h after siRNA transfection, revealed that the signal corresponding to MBNL142–43 isoforms decreased in the nuclei along with that of Lyn, which instead increased almost throughout the myotube cytoplasm (Figure 6g). These data have been further validated by colocalization analysis, showing the significant decrease of Manders' overlap coefficient in DM1 siRNA-transfected myotubes (Figure 6h) to the control value. WB analysis confirmed the decrease in the nuclear amount of Lyn and MBNL142–43 to 67% and 44%, respectively, relative to the cells transfected with empty vector (mock) (Figures 6i, k and l), in parallel with a 35% decrease of Tyr phosphorylation (Figures 6i and j). These results strengthen the case for a functional link between the Ex5-containing MBNL1 isoforms and the Tyr kinase Lyn.
Discussion
MBNL1 expression is crucial in the DM pathophysiology, and the loss of MBNL1 protein leads to the spliceopathy in the muscle tissues of DM patients and mouse models.8, 37 The MBNL1 pre-mRNA itself undergoes tissue-specific alternative splicing.17 In this paper, we showed that muscle tissue from DM1 patients overexpresses MBNL142 and MBNL143 isoforms containing exon 5 (Ex5-MBNL142–43). In contrast, control muscle showed a prevalent expression of the MBNL140 and MBNL141 isoforms, without exon 5 (ΔEx5-MBNL37L–40–41). The altered splicing of the MBNL1 gene has also been reported in DM1 brain, whereas ΔEx5-MBNL40–41 isoforms are expressed in adult tissues.15 These data support the hypothesis that the DM1 phenotype is in part due to a change in the splicing pattern linked to the trans-dominant effect of the CUG and CCUG repeats, resulting in the mixed expression of adult and foetal isoforms of various proteins, including MBNL1 itself. The overexpression of Ex5-MBNL142–43 isoforms in the muscle from DM1 patients, ensuring their restrictive nuclear localization, indicates an important feature for the sequestration within the foci. To address this issue, we developed novel anti-MBNL1 antibodies that allow us to discriminate different MBNL1 isoforms on the basis of the inclusion/exclusion of sequences encoded by exon 5. We showed that all four MBNL1 isoforms analyzed can bind to the nuclear foci of CUG-expanded repeats in DM1 muscle tissue; however, MBNL142–43 protein levels are increased in the nuclei of DM1 muscle compared with controls.
Our data demonstrate for the first time that MBNL142–43 isoforms are able to interact with SFKs, in particular with Lyn, which is one of the most ubiquitous members of this class of Tyr kinases. As MBNL140–41 isoforms cannot form this complex, it is possible that the amino-acid stretch encoded by exon 5 enables MBNL1 protein to interact with SFKs, possibly through a conformational change whereby the PRMs become available for binding. The MBNL142–43–SFK interaction is also a prerequisite for enhancement of the SFK activity relative to the basal state, evidencing a strong functional discrimination between the long and short isoforms of MBNL1 in terms of interaction with SFKs and regulation of their activity. We also show that MBNL142–43 overexpression induces the nuclear translocation of Lyn and enhancement of its catalytic activity, resulting in increased levels of Tyr phosphorylation of nuclear proteins, which also occurs in DM1-patient myotubes. These findings are consistent with the accumulating evidence that SFKs, Lyn in particular, localize to a variety of intracellular organelles,38, 39, 40, 41 in addition to the plasma membrane42, 43, 44 Importantly, the acylation state—especially the combination of myristate and palmitoylate—of the N-terminal SH4 domain of SFKs, has been shown to be critical in directing SFKs to their final location.45, 46 However, recent studies have noted that an additional mechanism involved in targeting SFKs to cell compartments is the engagement of the SH3 domain by PRM-bearing proteins. Examples include p13, a human T-cell leukaemia virus-1 accessory protein with both a C-terminal PRM and a mitochondrial localization signal; this has been shown to bind to and carry SFKs into the mitochondria in a cell culture model.47 Another striking effect is the increase in SFK activity, with an elevation of Tyr phosphorylation within the specific compartment in which SFKs operate. We therefore hypothesize a mechanism by which the transfer of SFKs to various cell districts might exert a more general action on cell fate by modifying the phosphorylation state and the function of important substrates in the targeted cell compartments. The nucleus is another organelle to which Lyn has been shown to localize, with a role in the DNA damage response and cell cycle control by associating with and/or phosphorylating critical signalling molecules involved in cell survival and apoptosis.48, 49, 50, 51 Although far from fully elucidated, nuclear translocation of Lyn seems to depend on the level of its catalytic activity.52 In our study, MBNL142–43 seem to act as carriers for Lyn into the nucleus and they boost its catalytic activity; the effects of phosphorylation on the target proteins possibly representing an additional pathogenetic pathway in DM1, as proposed in Figure 7. Of the Tyr-phosphorylated proteins localized in the nuclei, the ‘RNA-processing' group consists predominantly of players involved in transcription and post-transcriptional events.53 Of the proteins found to be ‘RNA-related', six were hnRNP (heterogeneous nuclear ribonucleoparticles), known to complex with hnRNA and to influence pre-mRNA transcription, processing and export, ultimately regulating gene expression and RNA processing, in particular splicing and alternative splicing. In this scenario, phosphorylation of pre-mRNA splicing factors may either cause their removal from pre-mRNA processing events or result in changing the selection of a splice site.54, 55 The relationship between members of the Src family and splicing regulators has already been demonstrated and may coordinate and regulate apoptosis, thus promoting cell proliferation and formation of metastasis.56 Possibly, the phosphorylation of splicing regulators, mediated by MBNL142–43 overexpression and SFK activation, allows the cells to lower temporarily their active concentration, further aggravating the DM spliceopathy, resulting from the MBNL1 sequestration into the nuclear foci. Therefore, the altered pattern of MBNL1 expression may further contribute to the multisystemic manifestation of DM not only through the mere loss-of-function due to the titration of the RNA-binding proteins by the CUG expansions. The importance of these data is underscored by the ongoing research for functional differences between the MBNL1 isoforms, which, when better delineated, would open up the possibility that the MBNL1 proteins may have different, still unexplored functional roles. A recent paper by Wang et al.57 suggests that MBNL proteins, besides regulating the splicing, also contribute to mRNA translation, protein secretion and localization of alternative 3′-UTR isoforms.57 Our results broaden this scenario showing for the first time that MBNL142–43 proteins, overexpressed in the DM muscle, are Lyn-interacting partners that have an important role in regulating the nuclear-cytoplasmic shuttling and catalytic activity of Lyn itself, and suggest Lyn as a novel factor implicated in the onset and progression of DM1. In this respect, additional functional studies should help clarifying how the altered Lyn-dependent signalling pathways are affected by the dysregulation of Lyn itself, thereby paving the way for discovery of potential novel targets of therapeutic interest in DM1 muscle.
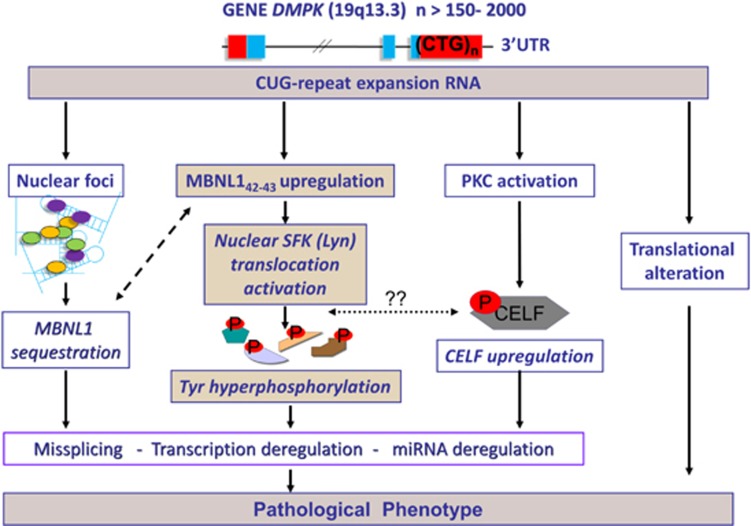
Figure 7.
Additional molecular pathogenetic mechanisms in DM1. Results suggest an additional altered mechanism in DM1 nuclei, triggered by CUG-containing DMPK transcripts and mediated by the longer Ex5-MBNL142–43 protein isoforms: Tyr hyperphosphorylation of nuclear proteins may influence and mediate changes in gene expression and RNA processing, expanding the known mechanisms of RNA toxicity in DM1 cells
Materials and Methods
Skeletal muscle biopsies
After written consent, DM1 (n=8) muscle samples were obtained from vastus lateralis by diagnostic open biopsies. Control samples (also from vastus lateralis, n=4) were obtained from subjects deemed free of neuromuscular disorders. Molecular diagnosis of DM1, using a combination of long PCR and Southern blot analysis in the peripheral blood, showed that DM1 patients considered in this study had a CTG expansion between 800 and 1500 repetitions.58
RNA extraction, RT-PCR and QRT-PCR analysis
Total RNA was extracted from frozen muscle biopsies and 1 μg of total RNA was reverse-transcribed according to the protocol of the High Capacity cDNA Archive kit (Applied Biosystem, Foster City, CA, USA). To assess the complete MBNL1 splicing pattern of muscle samples, PCR was performed using primers corresponding to sequences in exons 2 and 10 of the gene (numbered according to Pascual et al.;19 for primer sequences see Supplementary Table 1). The MBNL1 isoform expression levels were measured by SYBR Green semiquantitative QRT-PCR (Applied Biosystems) using primer pairs designed to amplify MBNL1 exons 4, 5 and 7 to distinguish MBNL140––41 from MBNL142–43. (Supplementary Table 1). The splicing pattern of the MBNL1 gene across exon 5 in DM1 myoblasts and myotubes has been determined as described previously.14 Transcripts from the exogenous IR and c-TNT minigenes have been measured using primers representing unique sequences in the cloning vectors (Supplementary Table 1).
Cloning of MBNL1 isoforms, molecular constructs and MBNL1 recombinant proteins
To clone the sequences of the MBNL140–41–42–43 isoforms, the entire coding sequence of the MBNL1 gene was amplified using the primers MBNL1-F and MBNL1-R listed in Supplementary Table 1. After sequencing, PCR products were cloned into pEF1-Myc/HisA and pEF4-V5/HisA (Invitrogen Carlsbad, CA, USA). Minigene B and minigene RTB300 have been described previously.21, 22 MBNL1 recombinant proteins have been produced in Escherichia coli and purified as described in the Supplementary Experimental Procedures.
Cell culture and plasmid transfection
Primary myoblasts were obtained and differentiated as described previously.36 Cells, plated 24 h before transfection at 80–90% confluence, were transiently transfected with 4 μg of plasmid DNA and Lipofectamine 2000 (Invitrogen). For siRNA experiments, myotubes from DM1 patients were transfected with 50 nM of siMBNL1-Ex5 and analyzed after 48 h. Rhabdomyosarcoma clones with permanent overexpression of MBNL141 and MBNL143 isoforms, named RD41 and RD43, respectively, were selected with G418.
Immunofluorescence analysis and RNA fluorescent in situ hybridization
Lyn and different MBNL1 isoforms were localized as detailed in the Supplementary Experimental Procedures. CUG-containing foci were detected in muscle sections and myoblasts as described.36 Colocalization was evaluated with JACoP plugin59 for the ImageJ software by calculating Manders' overlap coefficients M1 (fraction of A overlapping B) and M2 (fraction of B overlapping A). Threshold was arbitrarily set to non-saturating levels for Lyn and MBNL, whereas it was left saturated for 4′,6-diamidino-2-phenylindole (DAPI) images. Sets of seven M1 and M2 coefficients per pairwise analysis were collected by increasing the threshold. At least three different images were used for each condition. Data are shown as mean±S.E.M.
Anti-P9 and anti-P11 antibodies: generation, immunoblotting and immunoprecipitation
MBNL1 antibodies were raised in rabbits against amino acids 270–288 (TQSAVKSLKRPLEATFDL; this epitope is encoded by exon 5 of MBNL1 gene), hereafter defined as P9, and 294–308 (LPPLPKRPALEKTNG, encoded by exon 6 of MBNL1 gene), hereafter defined as P11, of the human MBNL143 protein (Supplementary Material and Supplementary Figure S1A) with an extra cysteine at the C terminus. Peptide synthesis, rabbit immunization and purification of anti-MBNL1 antisera and use of antibodies in immunoblotting and immunoprecipitation experiments are described in detail in the Supplementary Experimental Procedures.
Surface plasmon resonance
The interaction between MBNL1 isoforms and Lyn was performed as described below. Recombinant MBNL1 isoforms were immobilized on channels 1 and 3 of a His Cap research-grade sensor chip (Nomadics, ICx Nomadics Bioinstrumentation, Group, Oklahoma City, OK, USA) by nickel-affinity interaction using the manufacturer's protocols. Channel 2 was used as reference for all experiments. The Lyn kinase SH3 domain was the analyte and it was injected at 30 μl/min. SPR measurements were carried out in HBS buffer with 0.005% P20 surfactant at 20 °C using a SensiQ Pioneer Instrument (ICx Technologies, Stillwater, OK, USA). The data were analyzed with the Qdat evaluation analysis software (ICx Nomadics Bioinstrumentation, Group).
Phosphorylation and interaction assays
Tyr kinase assays were performed in 40 μl reaction mixture containing 50 mM Tris/HCl, pH 7.5, 10 mM MnCl2, 30 μM ATP/[γ-33P]ATP (specific activity 1000 c.p.m./pmol) (Perkin-Elmer, Waltham, MA, USA), 100 μM sodium orthovanadate, 200 μM Src-specific peptide substrate cdc2(6–20) as substrate and 20 ng of Src, or Lyn. Following incubation for 5 min at 30 °C, the reaction was blocked by adding 5 × SDS buffer and the samples were subjected to SDS-PAGE. Peptide phosphorylation was evaluated using the Cyclone Plus Storage Phosphor System (Perkin-Elmer). In time-course experiments, MBNL140–41 and MBNL142–43 (each 1 μM) were phosphorylated in the reaction mixture and the reactions were stopped at different time points as indicated above. The degree of protein phosphorylation was evaluated by analysis on the Cyclone Plus Storage Phosphor System (Perkin-Elmer). Interaction assay (between MBNL1 isoforms and Lyn) followed by immunoprecipitation were performed as described in the Supplementary Experimental Procedures.
Statistical analysis
Statistical analysis was performed only where three or more experimental values were available. Quantitative data were presented as mean±S.D. The statistical comparisons were performed using Mann–Whitney and Kruskal–Wallis tests, for comparison between two or more groups, respectively. In every analysis, values of P<0.05 were considered significant.
Acknowledgments
We thank T Cooper and N Webster at the College of Medicine, Houston, TX, USA for their gift of the c-TNT and IR minigenes, respectively; Dr. E Perissinotto at the University of Padua, Italy for assistance with the statistics; G Miotto at the University of Padua, Italy for the MS technical assistance; and I Holt at the RJAH Orthopaedic Hospital, Oswestry, UK for providing monoclonal antibodies to MBNL1. This work was supported by grants from AFM Grant No. 14631 to L Vergani; from Fondazione CARIPARO (Padua) and from Telethon Grant No. GGP10145 to L Vergani.
Glossary
- c-TNT
cardiac troponin T
- DAPI
4′,6-diamidino-2-phenylindole
- DM1
myotonic dystrophy type-1
- GST
glutathione S-transferase
- IR
insulin receptor
- MBNL1
muscleblind-like-1
- PRMs
proline-rich motifs
- pTyr
phosphotyrosine
- SFKs
Src family kinases
- SH
Src homology domain
- SPR
surface plasmon resonance
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Harper PS.Myotonic dystrophy2nd edn. Sautnders: London2001
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;69:385. doi: 10.1016/0092-8674(92)90418-c. [DOI] [PubMed] [Google Scholar]
- Fu YH, Pizzuti A, Fenwick RG, Jr, King J, Rajnarayan S, Dunne PW, et al. An unstable triplet repeat in a gene related to myotonic muscular dystrophy. Science. 1992;255:1256–1258. doi: 10.1126/science.1546326. [DOI] [PubMed] [Google Scholar]
- Mahadevan M, Tsilfidis C, Sabourin L, Shutler G, Amemiya C, Jansen G, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255:1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- Cho DH, Tapscott SJ. Myotonic dystrophy: emerging mechanisms for DM1 and DM2. Biochim Biophys Acta. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- Lin X, Miller JW, Mankodi A, Kanadia RN, Yuan Y, Moxley RT, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanadia RN, Shin J, Yuan Y, Beattie SG, Wheeler TM, Thornton CA, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci USA. 2006;103:11748–11753. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamberlain CM, Ranum LP. Mouse model of muscleblind-like 1 overexpression: skeletal muscle effects and therapeutic promise. Hum Mol Genet. 2012;21:4645–4654. doi: 10.1093/hmg/dds306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J Biol Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
- Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum Mol Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- Botta A, Rinaldi F, Catalli C, Vergani L, Bonifazi E, Romeo V, et al. The CTG repeat expansion size correlates with the splicing defects observed in muscles from myotonic dystrophy type 1 patients. J Med Genet. 2008;45:639–646. doi: 10.1136/jmg.2008.058909. [DOI] [PubMed] [Google Scholar]
- Dhaenens CM, Schraen-Maschke S, Tran H, Vingtdeux V, Ghanem D, Leroy O, et al. Overexpression of MBNL1 fetal isoforms and modified splicing of tau in the DM1 brain: two individual consequences of CUG trinucleotide repeats. Exp Neurol. 2008;210:467–478. doi: 10.1016/j.expneurol.2007.11.020. [DOI] [PubMed] [Google Scholar]
- Teplova M, Song J, Gaw HY, Teplov A, Patel DJ. Structural insights into RNA recognition by the alternate-splicing regulator CUG-binding protein 1. Structure. 2010;18:1364–1377. doi: 10.1016/j.str.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran H, Gourrier N, Lemercier-Neuillet C, Dhaenens CM, Vautrin A, Fernandez-Gomez FJ, et al. Analysis of exonic regions involved in nuclear localization, splicing activity, and dimerization of muscleblind-like-1 isoforms. J Biol Chem. 2011;286:16435–16446. doi: 10.1074/jbc.M110.194928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual M, Vicente M, Monferrer L, Artero R. The muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- Gates DP, Coonrod LA, Berglund JA. Autoregulated splicing of muscleblind-like 1 (MBNL1) pre-mRNA. J Biol Chem. 2011;286:34224–34233. doi: 10.1074/jbc.M111.236547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaki A, Nelson J, Webster NJ. Identification of intron and exon sequences involved in alternative splicing of insulin receptor pre-mRNA. J Biol Chem. 1998;273:10331–10337. doi: 10.1074/jbc.273.17.10331. [DOI] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Chong YP, Ia KK, Mulhern TD, Cheng HC. Endogenous and synthetic inhibitors of the src-family protein tyrosine kinases. Biochim Biophys Acta. 2005;1754:210–220. doi: 10.1016/j.bbapap.2005.07.027. [DOI] [PubMed] [Google Scholar]
- Trentin L, Frasson M, Donella-Deana A, Frezzato F, Pagano MA, Tibaldi E, et al. Geldanamycin-induced lyn dissociation from aberrant Hsp90-stabilized cytosolic complex is an early event in apoptotic mechanisms in B-chronic lymphocytic leukemia. Blood. 2008;112:4665–4674. doi: 10.1182/blood-2008-02-139139. [DOI] [PubMed] [Google Scholar]
- Samuels AL, Klinken SP, Ingley E. Liar, a novel lyn-binding nuclear/cytoplasmic shuttling protein that influences erythropoietin-induced differentiation. Blood. 2009;113:3845–3856. doi: 10.1182/blood-2008-04-153452. [DOI] [PubMed] [Google Scholar]
- Hassan R, Suzu S, Hiyoshi M, Takahashi-Makise N, Ueno T, Agatsuma T, et al. Dys-regulated activation of a src tyroine kinase hck at the golgi disturbs N-glycosylation of a cytokine receptor fms. J Cell Physiol. 2009;221:458–468. doi: 10.1002/jcp.21878. [DOI] [PubMed] [Google Scholar]
- Ingley E. Src family kinases: regulation of their activities, levels and identification of new pathways. Biochim Biophys Acta. 2008;1784:56–65. doi: 10.1016/j.bbapap.2007.08.012. [DOI] [PubMed] [Google Scholar]
- Engen JR, Wales TE, Hochrein JM, Meyn MA, 3rd, Banu Ozkan S, Bahar I, et al. Structure and dynamic regulation of src-family kinases. Cell Mol Life Sci. 2008;65:3058–3073. doi: 10.1007/s00018-008-8122-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicheri F, Moarefi I, Kuriyan J. Crystal structure of the src family tyrosine kinase hck. Nature. 1997;385:602–609. doi: 10.1038/385602a0. [DOI] [PubMed] [Google Scholar]
- Boonyaratanakornkit V, Scott MP, Ribon V, Sherman L, Anderson SM, Maller JL, et al. Progesterone receptor contains a proline-rich motif that directly interacts with SH3 domains and activates c-src family tyrosine kinases. Mol Cell. 2001;8:269–280. doi: 10.1016/s1097-2765(01)00304-5. [DOI] [PubMed] [Google Scholar]
- Moarefi I, LaFevre-Bernt M, Sicheri F, Huse M, Lee CH, Kuriyan J, et al. Activation of the src-family tyrosine kinase hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- Trible RP, Emert-Sedlak L, Smithgall TE. HIV-1 nef selectively activates src family kinases hck, lyn, and c-src through direct SH3 domain interaction. J Biol Chem. 2006;281:27029–27038. doi: 10.1074/jbc.M601128200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner EC, Smithgall TE. SH3-dependent stimulation of src-family kinase autophosphorylation without tail release from the SH2 domain in vivo. Nat Struct Biol. 2002;9:365–369. doi: 10.1038/nsb782. [DOI] [PubMed] [Google Scholar]
- Porter M, Schindler T, Kuriyan J, Miller WT. Reciprocal regulation of hck activity by phosphorylation of tyr(527) and tyr(416). effect of introducing a high affinity intramolecular SH2 ligand. J Biol Chem. 2000;275:2721–2726. doi: 10.1074/jbc.275.4.2721. [DOI] [PubMed] [Google Scholar]
- Weng Z, Thomas SM, Rickles RJ, Taylor JA, Brauer AW, Seidel-Dugan C, et al. Identification of src, fyn, and lyn SH3-binding proteins: Implications for a function of SH3 domains. Mol Cell Biol. 1994;14:4509–4521. doi: 10.1128/mcb.14.7.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro E, Rinaldi F, Malena A, Masiero E, Novelli G, Angelini C, et al. Normal myogenesis and increased apoptosis in myotonic dystrophy type-1 muscle cells. Cell Death Differ. 2010;17:1315–1324. doi: 10.1038/cdd.2010.33. [DOI] [PubMed] [Google Scholar]
- Osborne RJ, Lin X, Welle S, Sobczak K, O'Rourke JR, Swanson MS, et al. Transcriptional and post-transcriptional impact of toxic RNA in myotonic dystrophy. Hum Mol Genet. 2009;18:1471–1481. doi: 10.1093/hmg/ddp058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougoule C, Carreno S, Castandet J, Labrousse A, Astarie-Dequeker C, Poincloux R, et al. Activation of the lysosome-associated p61Hck isoform triggers the biogenesis of podosomes. Traffic. 2005;6:682–694. doi: 10.1111/j.1600-0854.2005.00307.x. [DOI] [PubMed] [Google Scholar]
- Kaplan KB, Swedlow JR, Varmus HE, Morgan DO. Association of p60c-src with endosomal membranes in mammalian fibroblasts. J Cell Biol. 1992;118:321–333. doi: 10.1083/jcb.118.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohn H, Le Cabec V, Fischer S, Maridonneau-Parini I. The src-family protein-tyrosine kinase p59hck is located on the secretory granules in human neutrophils and translocates towards the phagosome during cell activation. Biochem J. 1995;309 (Pt 2:657–665. doi: 10.1042/bj3090657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Nakayama Y, Horimoto S, Kuga T, Ikeda K, Kasahara K, et al. Involvement of golgi-associated lyn tyrosine kinase in the translocation of annexin II to the endoplasmic reticulum under oxidative stress. Exp Cell Res. 2006;312:1205–1217. doi: 10.1016/j.yexcr.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Kovarova M, Tolar P, Arudchandran R, Draberova L, Rivera J, Draber P. Structure–function analysis of lyn kinase association with lipid rafts and initiation of early signaling events after fcepsilon receptor I aggregation. Mol Cell Biol. 2001;21:8318–8328. doi: 10.1128/MCB.21.24.8318-8328.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee T, Pirinen N, Adler J, Pagakis SN, Parmryd I. Lipid rafts: Cell surface platforms for T cell signaling. Biol Res. 2002;35:127–131. doi: 10.4067/s0716-97602002000200003. [DOI] [PubMed] [Google Scholar]
- Gulle H, Samstag A, Eibl MM, Wolf HM. Physical and functional association of fc alpha R with protein tyrosine kinase lyn. Blood. 1998;91:383–391. [PubMed] [Google Scholar]
- McCabe JB, Berthiaume LG. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol Biol Cell. 1999;10:3771–3786. doi: 10.1091/mbc.10.11.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato I, Obata Y, Kasahara K, Nakayama Y, Fukumoto Y, Yamasaki T, et al. Differential trafficking of src, lyn, yes and fyn is specified by the state of palmitoylation in the SH4 domain. J Cell Sci. 2009;122:965–975. doi: 10.1242/jcs.034843. [DOI] [PubMed] [Google Scholar]
- Tibaldi E, Venerando A, Zonta F, Bidoia C, Magrin E, Marin O, et al. Interaction between the SH3 domain of src family kinases and the proline-rich motif of HTLV-1 p13: A novel mechanism underlying delivery of src family kinases to mitochondria. Biochem J. 2011;439:505–516. doi: 10.1042/BJ20101650. [DOI] [PubMed] [Google Scholar]
- Grishin AV, Azhipa O, Semenov I, Corey SJ. Interaction between growth arrest-DNA damage protein 34 and src kinase lyn negatively regulates genotoxic apoptosis. Proc Natl Acad Sci USA. 2001;98:10172–10177. doi: 10.1073/pnas.191130798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharbanda S, Yuan ZM, Rubin E, Weichselbaum R, Kufe D. Activation of src-like p56/p53lyn tyrosine kinase by ionizing radiation. J Biol Chem. 1994;269:20739–20743. [PubMed] [Google Scholar]
- Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, et al. p27 phosphorylation by src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, et al. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128:269–280. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Nakayama Y, Togashi Y, Obata Y, Kuga T, Kasahara K, et al. Nuclear localization of lyn tyrosine kinase mediated by inhibition of its kinase activity. Exp Cell Res. 2008;314:3392–3404. doi: 10.1016/j.yexcr.2008.08.019. [DOI] [PubMed] [Google Scholar]
- Bergstrom Lind S, Artemenko KA, Elfineh L, Mayrhofer C, Zubarev RA, Bergquist J, et al. Toward a comprehensive characterization of the phosphotyrosine proteome. Cell Signal. 2011;23:1387–1395. doi: 10.1016/j.cellsig.2011.03.021. [DOI] [PubMed] [Google Scholar]
- Misteli T. RNA splicing: What has phosphorylation got to do with it. Curr Biol. 1999;9:R198–R200. doi: 10.1016/s0960-9822(99)80128-6. [DOI] [PubMed] [Google Scholar]
- Stamm S. Regulation of alternative splicing by reversible protein phosphorylation. J Biol Chem. 2008;283:1223–1227. doi: 10.1074/jbc.R700034200. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Cai L, Zhu J, Chen M, Chen J, Li ZH, et al. Fyn requires HnRNPA2B1 and Sam68 to synergistically regulate apoptosis in pancreatic cancer. Carcinogenesis. 2011;32:1419–1426. doi: 10.1093/carcin/bgr088. [DOI] [PubMed] [Google Scholar]
- Wang ET, Cody NA, Jog S, Biancolella M, Wang TT, Treacy DJ, et al. Transcriptome-wide regulation of pre-mRNA splicing and mRNA localization by muscleblind proteins. Cell. 2012;150:710–724. doi: 10.1016/j.cell.2012.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifazi E, Vallo L, Giardina E, Botta A, Novelli G. A long PCR-based molecular protocol for detecting normal and expanded ZNF9 alleles in myotonic dystrophy type 2. Diagn Mol Pathol. 2004;13:164–166. [PubMed] [Google Scholar]
- Bolte S, Cordelières FP. A guided tour into subcellular colocalization analysis in light microscopy. J Microsc. 2006;224:213–232. doi: 10.1111/j.1365-2818.2006.01706.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.