Abstract
Aberrant regulation of cholesterol homeostasis is associated with obesity as well as multiple types of cancer. However, the mechanism behind these is largely missing. Here, we show that microRNA (miRNA)-128-2 is not only a pro-apoptotic microRNA but it also alters the expression of genes involved in cellular cholesterol homeostasis. Cholesterol efflux via ATP-binding cassette transporters (ABCA1 and ABCG1) is a mechanism for cells to eliminate excess cholesterol and prevent cellular cholesterol accumulation. The regulation of these pathways is complex with transcriptional regulation by sterol-regulatory element-binding protein (SREBP) and liver X receptor/retinoid X receptor (RXR) transcription factors but poorly understood at the post-transcriptional levels. MiR-128-2 increases the expression of SREBP2 and decreases the expression of SREBP1 in HepG2, MCF7 and HEK293T cells independent of sirtuin 1 (SIRT1) status. MiR-128-2 inhibits the expression of ABCA1, ABCG1 and RXRα directly through a miR-128-2-binding site within their respective 3′untranslated regions. The administration of miR-128-2 leads to decline in the protein and mRNA levels of ABCA1, ABCG1 and RXRα. Conversely, anti-miRNA treatment leads to increased ABCA1, ABCG1 and RXRα expression. The inverse correlation between miR-128-2 and its targets viz. ABCA1 and ABCG1 was also established during high-fat diet in different mice tissues. Our data show that cholesterol efflux is attenuated by miR-128-2 overexpression and, conversely, stimulated by miR-128-2 silencing. Further, we also observed the induction of ER stress response by miR-128-2. In this study, we provide the first evidence of miR-128-2 to be a new regulator of cholesterol homeostasis. Our study shows dual role of miR-128-2, as a pro-apoptotic molecule as well as a regulator of cholesterol homeostasis.
Keywords: miRNA, cholesterol, SREBP2, ABCA1, ABCG1
Cholesterol is a lipid component of cell membranes that regulates membrane fluidity and is a part of membrane signaling systems. It also functions in intracellular transport, nerve conduction and cell proliferation.1 It is not only important in the pathogenesis of cardiac diseases but insufficient or excessive cellular cholesterol results in several pathological processes including diabetes, atherosclerosis, metabolic syndrome and neurodegenerative diseases like Alzheimer's.2, 3, 4, 5, 6 Recently, connection between aberrant regulation of cholesterol homeostasis and multiple types of cancer has also been established.7, 8, 9, 10, 11
Cholesterol levels are maintained through a tightly regulated and complex mechanism that includes the de novo biosynthesis, internalization of exogenous cholesterol and efflux of excess cholesterol. These mechanisms are regulated by transcription factors such as sterol-regulatory element-binding protein (SREBPs) and Liver X receptors (LXRs). The SREBP transcription factors regulate the expression of genes involved in cholesterol biosynthesis and cellular uptake.12 LXR/retinoid X receptor (RXR) functions as a sensor of cellular cholesterol concentration and mediates cholesterol efflux by inducing the transcription of key cholesterol shuffling vehicles namely, ATP-binding cassette transporters (ABCA1 and ABCG1) and apolipoprotein E and thus maintains cellular sterol homeostasis.13, 14
Recently, a class of noncoding RNAs called microRNAs (miRNAs) have emerged as critical regulators of gene expression, acting predominantly at the post-transcriptional levels.15, 16, 17 miRNAs hybridize to mRNA sequences that are frequently located in the 3′ UTR (untranslated region), and a single miRNA could target more than one hundred mRNAs. Although the role for miRNAs in regulating multiple physiological processes including apoptosis, cell differentiation and proliferation is well known,18, 19, 20, 21, 22 the importance of these tiny RNAs in regulating cholesterol and lipid metabolism is still in infancy. Till date, only miR-33,23 miR-122,24 miR-75825 and miR-37026 have been reported to regulate cholesterol homeostasis.
Our earlier studies showed that miR-128-2 induces apoptosis and is an endogenous regulator of Sirtuin 1 (SIRT1).27, 28 In the current study, we showed that miR-128-2 also directly targets ABC transporters (ABCA1 and ABCG1) and RXRα and regulates cellular cholesterol efflux. Furthermore, miR-128-2 significantly increased the expression of SREBP2 in HepG2, MCF7 and HEK293T cells. In conclusion, we identified that miR-128-2 is not only a pro-apoptotic molecule but is also a regulator of cholesterol metabolism, thus might be opening new avenues for the treatment of diseases besides cancer.
Results
MiR-128-2 induces apoptosis and modulates sterol regulatory element-binding proteins (SREBPs)
Previously we have shown that miR-128-2 induces apoptosis in a p53-dependent and -independent manner by downregulation of SIRT1.28 Literature reveals that SIRT1, a NAD-dependant deacetylase, is a key regulator of energy homeostasis and has a role in lipid and glucose metabolism. Hence, we next hypothesized that miR-128-2 might be regulating cholesterol and lipid metabolism also. Our hypothesis was further strengthened during the Ingenuity Pathway Analysis of our microarray data (described earlier, GEO accession number GSE3129728), where we observed cholesterol metabolism and fatty acid biosynthesis as one of the topmost toxicology functions and canonical pathways, respectively, (Supplementary Figures S1a and b).
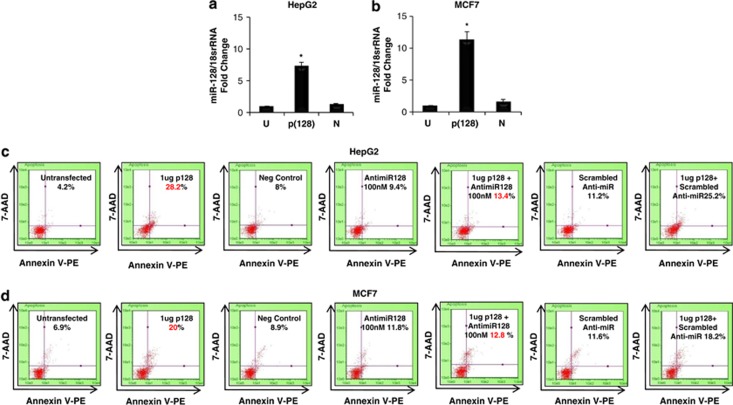
To test our hypothesis that miR-128-2 regulates cholesterol and fatty acid homeostasis, we herein transfected HepG2, MCF7 and HEK293T cells with p(128) (miR-128-2 expression vector) and confirmed the increase in the levels of mature form of miR-128-2 by 7.15-fold, 11.2-fold and 8.8-fold in HepG2, MCF7 and HEK293T cells, respectively, using TaqMan-based real-time PCR assay (Figures 1a and b, Supplementary Figure S1c). Consistent to our previous findings, the pro-apoptotic nature of miR-128-2 was also assessed using annexin asssay in all the three cell lines. The annexin assay showed that the annexin-positive cells increased to 28.2±0.8% (P<0.05) after overexpression of p(128) in HepG2 cells as compared with 4.2±0.2% (P<0.05) in untransfected HepG2 cells (Figure 1c). Similar increase in the number of annexin-positive cells was also observed after p(128) overexpression in MCF7 as well as HEK293T (Figure 1d, Supplementary Figure S1d).
Figure 1.
(a and b) Taqman assay for mature miR-128 in HepG2 (a) and MCF7 (b) cells showing overexpression of miR-128-2 in cells transfected with 4 μg p(128). n=3±S.E.M.; *P<0.05. U=untransfected, p(128)=cells transfected with 4 μg p(128), N=negative control. (c and d) Annexin V-staining shows that miR-128 induces apoptosis in HepG2 (c) and MCF7 (d) cells as described in ‘Materials and methods'. % here indicates the percentage of dead cells. Representative of three independent experiments is shown here (n=3; P<0.05)
Literature reveals that the uptake and synthesis of cholesterol is regulated by SREBPs, whereas cholesterol efflux is regulated by LXRs12, 29 (Supplementary Figure S1e). SREBP1 targets genes involved in fatty acid metabolism and SREBP2 targets genes specific to cholesterol synthesis.30 Hence, we next assessed the transcript levels of SREBP2 and SREBP1 in HepG2, MCF7 and HEK293T cells after overexpression of p(128). Transfection of p(128) significantly increased the mRNA levels of SREBP2 (by 1.75-fold in HepG2, by 1.67-fold in MCF7 and by1.73-fold in HEK293T) and decreased the mRNA levels of SREBP1 (by 1.81-fold in HepG2, by 1.85-fold in MCF7 and by 1.72-fold in HEK293T) as compared with negative control (Supplementary Figures S2a and b). Consistent to RT-PCR data, miR-128-2 significantly increased the levels of cleaved form (active) of SREBP2 and decreased the levels of SREBP1 in HepG2, MCF7 and HEK293T cells (Figures 2a–d and Supplementary Figures S2c and d) as compared with negative control. To determine the role of SIRT1 in modulating SREBP transcription factors, western blot analysis and real-time assay were performed for SREBP2 and SREBP1 in the presence or absence of SIRT1 siRNA/open reading frame (ORF) in all the three cell lines. Downregulation of SIRT1 by SIRT1 siRNA and upregulation of SIRT1 by SIRT1 ORF were confirmed by western blot analysis (Supplementary Figures S2g and h). Our western blot data showed increase in the levels of cleaved form (active) of SREBP2 in all the cell lines (Figures 2e, f, i and j, Supplementary Figures S2e and f) irrespective of SIRT1 expression. However, only in HepG2 cells SREBP1 expression was altered in the presence of SIRT1 siRNA/ORF in concordant to miR-128-2. In consistent to miR-128-2 modulation of SREBPs, significant changes were observed by real-time assay after the transfection of SIRT1 siRNA/ORF only in HEK293T cells for SREBP2 and only in MCF7 cells for SREBP1 (Supplementary Figures S2a and b). All these results suggest that SREBPs are regulated in a SIRT1-independent manner.
Figure 2.
MiR-128-2 regulates genes of cholesterol/lipid homeostasis. (a) Western blot analysis for cleaved SREBP2 (a and b, e and f, i and j) and SREBP1 (c and d, g and h, k and l) expression after overexpression of p(128) (a–d), after overexpression of SIRT1 siRNA (e–h) and after overexpression of SIRT1 ORF (i–l) in HepG2 and MCF7 cells. β-Tubulin was used as a loading control. U=untransfected, p(128)=cells transfected with 4 μg p(128), N=negative control, SIRT1 siRNA (at 100 nM dose)/scrambled siRNA/ SIRT1 ORF. n=3±S.E.M.; *P<0.05 for all experiments
MiR-128-2 regulates genes of cholesterol homeostasis
To confirm our hypothesis that miR-128-2 regulates cholesterol and fatty acid homeostasis, we checked the expression of HMG-CoA synthase (HMGCS), 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR) and low-density lipoprotein receptor (LDLR),31 LXRα LXRβ RXRα, ABCA1 and ABCG1 at the transcriptional level after the overexpression of p(128) in all the three cell lines. MiR-128-2 significantly increased the transcript levels of HMGCS1 and LDLR in all the cell lines as compared with negative control (Supplementary Figures S3a and b). Transcript levels of HMGCR significantly decreased in HepG2, insignificantly decreased in MCF7 and did not change in HEK293T cells as compared with their respective negative controls (Supplementary Figure S3c). We further observed slight to moderate but significant increase in LXRα levels in HepG2 and MCF7 cells, whereas no significant changes occurred in HEK293T after the overexpression of p(128) (Supplementary Figure S3d). miR-128-2 also significantly decreased the transcript levels of LXRβ RXRα ABCA1 and ABCG1 in all three cell lines as compared with negative control (Supplementary Figures 3e–h). In addition, miR-128-2 significantly decreased the mRNA levels of fatty acid metabolism genes such as acetyl CoA carboxylase alpha, fatty acid synthase, Stearoyl-CoA desaturase 132, 33, 34, 35, 36 in HEK293T cells, and this may be due to decrease in SREBP1 protein after the overexpression of p(128) (Supplementary Figures S4a–c). Significant changes were not observed in cholesterol and fatty acid homeostasis genes at the RNA levels in the presence and absence of SIRT1 (Supplementary Figures S3a–h), thereby confirming that SIRT1 is not having a role in their modulation.
MiR-128-2 directly targets ATP-binding cassette transporters—ABCA1 and ABCG1
It is well established that miRNAs regulate gene expression post-transcriptionally by binding to the 3′ UTR and repress protein production by destabilizing the mRNA or translational silencing.37 Hence, we next performed in-silico analyses to determine whether any of the cholesterol metabolism gene(s) is predicted target of miR-128-2. Using different target prediction algorithms, TargetScan, miRanda, RNAHybrid,38, 39, 40 we determined that the transcripts of ABCA1, ABCG1, RXRα, PPARα and LDLR harbored the putative binding sites for miR-128-2 in their respective 3′UTRs.
There was complete complementarity between the seed region of miR-128-2 and ABCA1 3′UTR (2592–2598 nucleotide) and ABCG1 3′UTR (34–40 nt) along with a few supplementary pairing sites at the 3′end of miR-128-2 (Figures 3a and g). This perfect complementarity binding site was highly conserved among different species of mammals, which further strengthened the likelihood of a physiological role of miR-128-2 in regulating cholesterol homeostasis (Supplementary Figures S5a and b). To determine whether ABCA1 and ABCG1 are direct targets of miR-128-2, HEK293T cells were transfected with ABCA1/ABCG1 luciferase reporter constructs containing their full 3′UTRs in the presence or absence of p(128) and/or anti-miR-128 in a dose-dependent manner. Co-transfection of p(128) resulted in a decrease in luciferase activity by 40 and 60% in ABCA1 and 30 and 66% in ABCG1 reporter constructs as compared with negative control, in HEK293T cells (Figures 3b and h). However, this suppression was relieved by transfection with anti-miR-128 in a dose-dependent manner as compared with scrambled anti-miR (SAM) (Figures 3b and h), thereby demonstrating that miR-128-2 directly targets ABCA1 and ABCG1 3′UTR.
Figure 3.
MiR-128-2 directly targets ATP-binding cassette transporters- ABCA1 (a–f) and ABCG1 (g–l). (a) Schematic representation of ABCA1 mRNA with the predicted binding site of miR-128-2 in its 3′UTR. Complete complementarity between first eight nucleotides of miR-128-2 (seed sequence from 5′ end) and its target ABCA1 3′UTR (from 2592 to 2598 nt) along with a few supplementary pairing sites at the 3′end of miR-128-2 is shown in red colored, bold letters. (b) Luciferase activity was measured in HEK293T cells after co-transfection with ABCA1 3′UTR (wild-type) with either 200 or 400 ng p(128) or negative control or in combination with 400 ng p(128)+anti-miR-128 at 30 or 60 nM or with SAM. Luminescence was measured at 24 h post transfection. The luciferase activity relative to pMIR-control vector (intact) was plotted. The bar diagram represents mean±S.E.M. for three independent experiments. *P<0.05. (c–d) Western blot analyses of ABCA1 protein expression in HepG2 after 24 h of transfection of p(128) (c) or anti-miR-128 (d) in a dose-dependent manner. β-Tubulin served as a loading control. Bar diagram represents the integrated densitometric values normalized to β-tubulin. U=untransfected, 2=cells transfected with 2 μg p(128), 4=cells transfected with 4 μg p(128), N=negative control, AM 100=cells transfected with 100 nM anti-miR-128, AM 200=200 nM anti-miR-128, SAM=Scrambled anti-miR. (e and f) RT-PCR analyses of ABCA1 mRNA in p(128) (e) or anti-miR-128 (f) transfected HepG2 cells. 18s rRNA was used for the normalization. n=3±S.E.M.; *P<0.05 for c–f panels. AM=anti-miR-128, SAM=scrambled anti-miR. (g–l) represents data for similar experiments for ABCG1 in HepG2 cells
Because of very low expression of ABCA1 and ABCG1 in HEK293T cells, the endogenous levels of ABCA1/ABCG1 were determined after overexpression of p(128) in a dose-dependent manner in HepG2 and MCF7 cells. MiR-128-2 significantly decreased the ABCA1 and ABCG1 protein levels (Figures 3c and i and Supplementary Figures S6a and e), and the suppressive effect was abolished by anti-miR-128 in a dose-dependent manner in both the cell lines (Figures 3d and j and Supplementary Figures S6b and f). Simultaneously, ABCA1/ABCG1 mRNA levels were also found to be significantly decreased after overexpression of p(128) in comparison with negative control (Figures 3e and k and Supplementary Figures S6c and g) and markedly increased after transfection with anti-miR-128 in comparison with SAM in HepG2, MCF7 and HEK293T cells (Figures 3f and l and Supplementary Figures S6d and h). Similar results were observed in HEK293T cells also (Supplementary Figures S5c and d).
MiR-128-2 negatively regulates nuclear hormone receptor RXRα
During the target search for miR-128-2, we observed another miR-128-2 response element in the 3′UTR of RXRα using the same algorithms as described above. Complete complementarity was found between the 2–7 nt (hexamer) of seed region of miR-128-2 and the 3′UTR of RXRα (from 3804 to 3810 nt) (Figure 4a). Multiple sequence alignment revealed the conservation of this hexamer among different species of mammals (Supplementary Figure S5e). To assess whether RXRα is a direct target of miR-128, a dual-luciferase reporter assay was performed. HEK293T cells were transfected with RXRα reporter construct in absence or in presence of miR-128-2 or in combination with anti-miR-128. MiR-128-2 decreased the luciferase activity by 25 and 60% in a dose-dependent manner, and anti-miR-128 increased the luciferase activity by 40 and 55% at 30nM and 60 nM concentration, respectively, in HEK293T cells (Figure 4b). We also assessed the changes in the endogenous levels of RXRα after the overexpression of p(128) and found that miR-128-2 significantly decreased the protein levels of RXRα as compared with negative control in HepG2 and MCF7 cells in a dose-dependent manner (Figures 4c i-ii). Furthermore, our results also show that RNA levels of RXRα were decreased by miR-128-2 and increased by anti-miR in HepG2 cells, thereby confirming that miR-128-2 directly targets RXRα (Figures 4d and e).
Figure 4.
MiR-128-2 negatively regulates nuclear hormone receptor RXRα. (a) Schematic representation of RXRα mRNA with miR-128-2-binding site (seed hexamer) in its 3′UTR along with a few supplementary binding sites, shown in red color and bold letters. (b) Luciferase activity was measured as described in Materials and methods. The luciferase activity relative to pMIR-control vector (intact) was plotted. The bar diagram represents mean±S.E.M. for three independent experiments. *P<0.05. (c) Western blot analyses of RXRα protein expression in HepG2 (c(i)) and MCF7 cells (c(ii)) after 24 h of transfection of p(128). β-Tubulin served as a loading control. Bar diagram represents the integrated densitometric values normalized to β-tubulin. (d and e) RT-PCR analyses of RXRα mRNA in p(128) (d) or anti-miR-128 (e) transfected HepG2 cells. 18s rRNA was used for the normalization. U=untransfected, p(128)=cells transfected with 4 μg p(128), N=negative control, AM=cells transfected with 100 nM anti-miR-128, SAM=scrambled anti-miR. n=3±S.E.M.; *P<0.05 for c–e panels
Taken together, all these results suggest that miR-128-2 inhibits the expression of human ABCA1, ABCG1 and RXRα by targeting their respective 3′UTRs. Whether PPARα and LDLR are direct targets of miR-128-2, needs validation.
miR-128-2 regulates ABCA1, ABCG1 and RXRα in a SIRT1-dependent and -independent manner
To elucidate the role of SIRT1 in post-transcriptional regulation of ABCA1, ABCG1 and RXRα, we next performed western blot analysis in HepG2 and MCF7 cells in the presence or absence of SIRT1 siRNA/ORF. We observed that SIRT1 siRNA significantly downregulates, whereas SIRT1 ORF upregulates ABCA1 in both HepG2 and MCF7 cells (Figures 5a–d). This suggests that miR-128-2 might be regulating ABCA1 in both HepG2 and MCF7 cells in a SIRT1-dependent manner. However, we observed that miR-128-2 regulates ABCG1 in a SIRT1-dependent manner in HepG2 cells and in a SIRT1-independent manner in MCF7 cells (Figures 5a–d). Our data further show that miR-128 downregulates RXRα in a SIRT1-independent manner in both HepG2 and MCF7 cells (Figures 5a–d).
Figure 5.
Role of SIRT1 in regulation of ABCA1, ABCG1 and RXRα. (a–b) Western blot analysis for ABCA1, ABCG1 and RXRα expression after overexpression of SIRT1 siRNA (a) and after overexpression of SIRT1 ORF (b) in HepG2 cells. (c–d) represents western blot data for ABCA1, ABCG1 and RXRα expression after overexpression of SIRT1 siRNA (c) and after overexpression of SIRT1 ORF (d) in MCF7 cells. β-Tubulin served as a loading control. Bar diagram represents the integrated densitometric values normalized to β tubulin. n=3±S.E.M.; *P<0.05 for all panels
Taken together, these results suggest that miR-128-2 inhibits the expression of ABCA1, ABCG1 and RXRα in a SIRT1-dependent and -independent manner.
Alteration of miR-128-2 expression modulates cholesterol efflux
Literature reveals that T0901317 is a LXR agonist, which increases the expression of LXR target genes including ABCA1 and ABCG1 and thus facilitates cholesterol efflux.41 To further confirm our findings, we examined the endogenous levels of ABCA1 and ABCG1 in the presence or absence of this ligand after the overexpression of p(128) or anti-miR-128. Transfection of HepG2 cells with p(128) repressed the stimulation of ABCA1 and ABCG1 protein levels by T0901317 as compared with negative control (Figures 6a and c). However, treatment with anti-miR-128 reversed the effect of miR-128-2 by increasing the stimulation of ABCA1 and ABCG1 protein levels by T0901317, in comparison with SAM (Figures 6b and d). ABCA1 and ABCG1 are associated with export of excess cellular cholesterol, hence, we next hypothesized that miR-128-2-dependent repression of ABCA1, ABCG1 and RXRα might affect cellular cholesterol efflux in vitro also. To test this, cells were treated with BODIPY-labeled cholesterol and transfected with p(128)/negative control/anti-miR-128/SAM along with LXR agonist T0901317 as a positive control as described in Materials and methods. Twenty-four hours post transfection, the BODIPY fluorescence was measured in the supernatants. Cholesterol efflux was significantly reduced by miR-128-2 in all the cell lines by the repression of ABCA1 and ABCG1 as compared with LXR agonist T0901317 (Figure 6e). However, antagonism of endogenous miR-128-2 by anti-miR-128 increased the cholesterol efflux from HepG2, MCF7 and HEK293T cells in comparison with SAM (Figure 6e). As the overexpression of p(128) caused reduction in cholesterol efflux and increased SREBP2 levels at both RNA and protein levels (Supplementary Figures S2a and c and Figures 2a and b), we speculated that it could cause cellular cholesterol accumulation. Therefore, cellular cholesterol levels were quantified after the overexpression of p(128) and anti-miR-128 in HEK293T and HepG2 cells. As shown in Figure 6f, transfection of both the cell lines with p(128) enhanced the cellular cholesterol levels by 1.59-fold in HepG2 cells and by 2.57-fold in HEK293T as compared with their respective negative controls. Conversely, silencing miR-128-2 using anti-miR-128 resulted in decreased cellular cholesterol levels by 1.96-fold in HepG2 and by 1.89-fold in HEK293T cells in comparison with p(128) (Figure 6f). Taken together, these data suggest that cellular miR-128-2 levels modulate cholesterol efflux.
Figure 6.
Alteration of miR-128-2 expression modulates cholesterol efflux. (a–d) Western blot analysis of ABCA1 (a and b), ABCG1 (c and d) and β-tubulin in HepG2 cells transfected with p(128) or negative control (a,c), anti-miR-128 or SAM (b,d) in the presence or absence of LXR agonist T0901317. Bar diagram represents the integrated densitometric values normalized to β-tubulin. Data are mean±S.E.M. from three independent experiments. *P<0.05. (e) BODIPY-cholesterol efflux assay in HepG2, MCF7 and HEK293T cells. Cells were incubated with 25 μM BODIPY-cholesterol for 2 h, rinsed and transfected with p(128) or anti-miR-128 as described in Materials and methods. LXR agonist T0901317 was also added to the cells wherever indicated. After 24 h, the BODIPY fluorescence intensity was measured. The data are expressed as the mean±S.E.M. of four independent experiments performed in triplicates. *P<0.05. (f) The total cholesterol levels were measured in HepG2 and HEK293T cells transfected with p(128) or anti-miR-128 and normalized to total protein. N=negative control, SAM=scrambled anti-miR. Data are mean±S.E.M. from three independent experiments. *P<0.05
Dietary regulation of miR-128 in mice tissues
The mature sequence of miR-128-2 is highly conserved among different species from Zebra fish to human (Supplementary Figure S7a), hence we next examined the in vivo distribution of miR-128 in different normal tissues of C57BL6 mice. miR-128 was highly expressed in the brain, heart and spleen as compared with the kidney and liver (Figure 7a). Highest expression of miR-128 in the brain was expected as miR-128 is a brain-enriched microRNA. To determine whether miR-128, ARPP21, SREBP2 and targets of miR-128 (ABCA1 and ABCG1) are regulated under different physiological conditions, henceforth, the expression of miR-128 and respective genes was measured in mice fed either normal or a high-fat diet for 14 weeks. C57BL6 mice treated with a high-fat diet showed a significant increase in body weight, serum triglyceride and cholesterol levels (Figures 7b–d). Levels of miR-128 were reduced upon high-fat diet as compared with normal diet in the brain, liver, kidney and heart and there was no change in the levels of miR-128 in the spleen (Figure 7e). Similar trend (as of miR-128-2) was observed for ARPP21, the gene that harbors miR-128-2 in its intron, in all the tissues except the spleen upon high-fat diet. Conversely, mRNA levels of ABCA1 and ABCG1 were increased in the brain, liver, kidney and heart, thus maintaining the inverse correlation with the expression of miR-128 (Figure 7e). Similar to miR-128 levels, SREBP2 mRNA levels were found to be decreased in corresponding tissues viz. the brain, liver and kidney but not in the heart and spleen upon high-fat diet as compared with normal diet (Figure 7e). This was expected as cells respond to high cholesterol levels by shutting down the cholesterol synthetic machinery (decreased SREBP2 expression).42 The reason behind the reduced expression of miR-128 during high-fat diet could be the decreased SREBP2 expression. Our search for the transcription factor binding sites in the ARPP21 gene by TRANSFAC software (TRANSFAC Professional 12.1, Biobase, Germany) revealed the presence of SREBP transcription factor binding site in the promoter region of ARPP21 gene (Supplementary Figure S7b).
Figure 7.
Dietary regulation of miR-128 in mice tissues. (a) Expression profile of miR-128 in different C57BL6 mice tissues by Taqman assay (n=4). (b–d) Body weight (b), serum triglycerides (c) and cholesterol levels (d) of C57BL6 mice on normal and high-fat diet were measured and plotted as graph. (e) Expression profile of miR-128, ARPP21, SREBP2, ABCA1 and ABCG1 in different tissues of C57BL6 mice fed a normal (ND) and high-fat diet (HFD). (n=4). Data are mean±S.E.M. from three independent experiments for all panels. *P<0.05
Several evidences in the literature suggested that cholesterol has a role in the regulation of programmed cell death under various stimuli.43, 44, 45 In the current study, we have also observed significant induction of ER stress inducible genes Bip, DDIT3 and ERO1α (Figures 8a–c) after overexpression of p(128) in HepG2 cells. Similar to our results, Seimon et al. and Feng et al.45, 46 recently demonstrated that cholesterol loading of ER membrane triggers unfolded protein response which eventually lead to the induction of apoptosis. We speculate that ER stress, cholesterol metabolism and pro-apoptotic activity of miR-128-2 are linked but this aspect needs further investigation.
Figure 8.
(a–c) Western blot analysis of ER stress-induced proteins Bip, DDIT3 and ERO1α and β-actin in HepG2 cells transfected with p(128) or negative control. U=untransfected, p(128)=cells transfected with 4 μg p(128), N=negative control. n=3±S.E.M.; *P<0.05. (d) A paradigm for cholesterol homeostasis by miR-128-2. Bioinformatic analysis by TRANSFAC search revealed SREBP as a transcription factor of ARPP21, the gene that encodes miR-128-2. In cells, low intracellular cholesterol conditions induce SREBP2, which might sit on the promoter of miR-128-2 and induce its expression. The activation of miR-128-2, in turn, decreases cholesterol efflux from the cell by targeting ABCA1, ABCG1 and RXRα and leads to the synthesis and accumulation of cellular cholesterol by further increasing the expression of SREBP2. Thus, the cells have managed to increase the cellular cholesterol during cholesterol depletion by activating the cholesterol synthesis and uptake machinery viz. SREBP2 and miR-128-2 and simultaneously also making sure to shut down the cholesterol export machinery via ABCA1, ABCG1 and RXRα. This feed forward loop might be employing in cells to ensure minimal loss of cholesterol during conditions of very low intracellular cholesterol that could otherwise compromise cell viability
Discussion
Cholesterol is an important component of cell membrane and also serves as a precursor for the synthesis of steroid hormones and bile acids. Here, we show that a miRNA that has been previously shown to be associated with cancer and apoptosis27, 47, 48 also influences the cholesterol metabolism. We found that miR-128-2 not only induces apoptosis but also post-transcriptionally regulates the cholesterol transporters ABCA1, ABCG1 and cholesterol sensor receptor RXRα by binding to their respective 3'UTRs and consequently ameliorates cholesterol efflux. We further observed that miR-128-2 regulates ABCA1 and ABCG1 in a SIRT1-dependent manner and regulates RXRα in a SIRT1-independent manner.
In contrary, it boosts intracellular cholesterol accumulation by increasing the expression of SREBP2 (at both transcriptional and translational levels) and blows down intracellular fatty acid levels by decreasing the expression of SREBP1 (at both transcriptional and translational levels). Similar to the findings of Puroshotham et al. and Walker et al.49, 50 we herein also observed that SREBPs are regulated in a SIRT1-independent manner. Our bioinformatic search by Transfac software further revealed the binding site of SREBP transcription factor in the promoter region of ARPP21, the gene that harbors miR-128-2 in its 18th intron (Supplementary Figure S7b).
We hereby propose a hypothetical model (feed forward loop) for the regulation of cholesterol homeostasis by miR-128-2 (Figure 8d). We speculate that low intracellular cholesterol (e.g. by Simvastatin) induces the SREBP2 expression (Supplementary Figure S7c) which might induce the expression of miR-128-2 from its host gene by sitting on its promoter. The activation of miR-128-2, in turn, prevents cholesterol from leaving the cell by targeting ABCA1, ABCG1 and RXRα and leads to the synthesis and accumulation of cellular cholesterol by further increasing the expression of SREBP2. Thus, SREBP2, besides having cholesterol synthesis genes (HMGCS1, HMGCR, LDLR) as its targets12, 31, 51 might also control a miR-128-2 that is not only pro-apoptotic but also regulates cholesterol metabolism in harmony with SREBP2. MiR-128-2 seems to be a new component joining the SREBP2 signaling pathway besides the miR-33a.23 It seems that the cells have a regulatory system to increase the cellular cholesterol during cholesterol depletion by activating the cholesterol synthesis and uptake machinery viz. SREBP2 and miR-128-2 and simultaneously also making sure to shut down the cholesterol export machinery via ABCA1, ABCG1 and RXRα. We anticipate that this feed forward loop might be employing in cells to ensure minimal loss of cholesterol during conditions of very low intracellular cholesterol that could otherwise compromise cell viability. Similar trend of SREBP2, ARPP21, miR-128 at RNA levels in mice tissues strengthens our bioinformatic predictions. However, our bioinformatic analyses merits further validation.
Although cholesterol is an important component of most cells, its inappropriate accumulation leads to obesity, atherosclerosis and diabetes.52, 53 Several epidemiological studies have revealed that obesity increases the risk of a number of cancers including breast, renal, immune (leukemia, lymphoma, myeloma), ovarian and hepatobiliary.54, 55, 56 However, the molecular mechanism linking the two is largely missing. Our results from mice experiments led us to further anticipate the mechanism underlying the molecular link between obesity and cancer. Hypercholesterolemia frequently occurs in patients with obesity and was also confirmed by us in mice fed a high-fat diet (Figure 7d). miR-128-2 was found to be significantly reduced in mice tissues (particularly in the brain, liver, kidney and heart) upon high-fat diet (Figure 7e). As miR-128-2 shows pro-apoptotic activity,27 its reduced expression in obese mice might confer resistance to apoptosis and therefore might increase the risk of cancer. Thus, our study provides the deep insight into the molecular link between obesity and cancer.
miRNAs have tremendous therapeutic potential for the treatment of various diseases. We believe miR-128-2 acts at an interface of cancer and obesity by being acting as a pro-apoptotic molecule as well as a regulator of cholesterol homeostasis. This seems to be interesting as same miRNA is having dual roles. By harboring a binding site for a key cholesterol transcription factor SREBP2 in promoter region of its host gene, miR-128-2, regulates cholesterol homeostasis in symphony with SREBP2 along with apoptosis. Our results suggest that miR-128-2 modulation by antagomiRs may be a useful therapeutic strategy for treating cardiovascular diseases. However, therapeutic miRNA manipulations warrant further investigations.
Materials and Methods
Animal experiments
Male C57BL6 mice (n=4 per group) were purchased from National Institute of Nutrition, Hyderabad and housed in cages in the animal house of the institute. Six- week-old mice were fed regular normal diet containing 10% of the calories or a high-fat diet containing 60% calories derived from fat for 14 weeks. The experiments were performed with the approval of Animal Ethics Committee of Institute of Genomics and Integrative Biology, New Delhi, India. Body weight of mice on normal and high-fat diet were taken. After euthanasia, the brain, liver, spleen, kidney and heart samples were collected, stored at −80 °C and total RNA was harvested and the expression of miR-128, ARPP21, SREBP2 and its targets (ABCA1 and ABCG1) were determined using real-time PCR chemistry.
Plasmid constructs
The sequences of the 3′UTR of ABCA1, ABCG1 and RXRα were retrieved from ENSEMBL (www.ensembl.org/). The miRNA sequence was retrieved from miRBase (http://microrna.sanger.ac.uk/sequences/). For primer design, Primer3 software was used (http://frodo.wi.mit.edu). Precursor of miR-128-2 was cloned in pSilencer 4.1 vector (Ambion, Austin, TX, USA) as described27 and this miR-128-2 expression vector is designated as p(128) throughout the manuscript. In brief, a 705-base pair genomic sequence spanning mature miR-128 was amplified by PCR from human genomic DNA using the following primers: forward primer 5′-CCGCCGGGATCCGCAGAAAGTCAACCATGTCC-3′ and reverse primer 5′-CGCCGAAGCTTATCC TTGGCAAGAACTGCAC-3′ having BamH1 and HindIII restriction sites. Amplified fragment was cloned in pSilencer4.1 vector (Ambion) and designated as p(128). Negative control is the pSilencer 4.1 vector that express a hairpin siRNA with limited homology to any known sequence in human (Ambion). 3′UTR reporter construct of ABCA1 (ENST00000374736, insert size=3340 nt) and ABCG1 (ENST00000398449, insert size=861 nt) were purchased from OriGene (OriGene, Rockville, MD, USA). To generate the 3′UTR reporter construct of RXRα, a full-length 3′UTR of RXRα mRNA (ENST00000481739, 405 nt) containing the predicted binding sites for miR-128-2 was cloned into pMIR-REPORT Luciferase Vector (Ambion) between SpeI and HindIII restriction sites and designated as RXRα 3'UTR. The plasmids were sequenced to ensure accuracy.
Cell culture and transfections
HEK293T (Human embryonic kidney), HepG2 (Liver hepatocellular carcinoma) and MCF7 (Breast cancer) cell lines were procured from National Centre for Cell Sciences (NCCS), Pune, India and maintained in DMEM containing 10% (v/v) fetal calf serum, 100 Units/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin at 37 °C in a humidified atmosphere at 5% CO2.
Approximately 2.5–3 × 105 cells of HEK293T/MCF7 were seeded in six-well plates (for western and real-time PCR) and 5 × 104 cells were seeded in 24-well plates (for luciferase and cholesterol assays) for overexpression studies. Transfections of these cells were done with pre-miRNA plasmid namely p(128) or negative control and/or anti-miR-128 and/or SAM (Dharmacon, CO, USA) using Lipofectamine 2000 (Invitrogen, CA, USA) reagent according to manufacturer's protocol. HepG2 (1 × 106) cells were transfected with pre-miRNA plasmid namely p(128) or negative control and/or anti-miR-128 and/or SAM via electroporation using the nucleofector kit V and Nucleofector II device (Amaxa Biosystem, Lonza, MD, USA) (program T028). The cells were harvested 24 h post transfection by trypsinization and used for all the experiments. ON-TARGETplus SMARTpool siRNAs for human SIRT1 and Non-targeting siRNA SMARTpool were purchased from Dharmacon and transfected into the cells for 48 h. SIRT1 ORF construct was purchased from Addgene (Addgene plasmid 13735, Addgene, Cambridge, MA, USA).57 The cells were also treated with 10 μM synthetic LXR ligand TO901317 (Cayman Chemical, Ann Arbor, MI, USA) and 25 μM BODIPY-cholesterol (Top Fluor-Cholesterol, Avanti Polar Lipids, Alabaster, AL, USA) wherever indicated.
Luciferase reporter assays
HEK293T cells were seeded into 24-well plates 1 day before transfection. Co-transfection of 200 ng of firefly luciferase containing 3′UTR reporter construct (ABCA1 or ABCG1 or RXRα) and/or 200 and 400 ng of pre-miRNA plasmid namely p(128) or negative control along with 30 ng pRL-CMV plasmid that expressed renilla luciferase (Promega, Madison, WI, USA) using Lipofectamine 2000 (Invitrogen). Anti-miR-128 (30 or 60 nM; Dharmacon) and SAM were also used wherever indicated. After 24 h of transfection, luciferase activity was measured using the Dual-luciferase assay kit as described by the manufacturer (Promega). Firefly luciferase activity was normalized to renilla luciferase activity and then compared with that transfected with the pMIR-control vector (without ABCA1/ABCG1/RXRα 3′UTR). These experiments were performed in triplicates.
Real-time PCR and Taqman microRNA assay
Total RNA from transfected/untransfected cells/mice tissues was extracted with the Trizol reagent (Invitrogen) and reverse transcription was carried out with RevertAid H Minus first strand cDNA synthesis kit as per the manufacturer's instructions (Fermentas, Glen Burnie, MD, USA). Real-time PCR was performed using cDNA with SYBR Green PCR master mix (Applied Biosystems, Foster City, CA, USA) in an ABI Prism 7500 Sequence Detection System (Applied Biosystems) and amplifications were performed in triplicate and repeated thrice as described.58 Results were normalized with 18S rRNA and analysis was done using the Pfaffl's method.59 Primers used for the detection of expression levels of ABCA1, ABCG1, HMGCR, HMGCS1, LXRα, LXRβ, LDLR, RXRα, SREBP1, SREBP2 and 18s rRNA are listed in Supplementary Table S1. Primers specific for mice genes viz. ARPP21, ABCA1, ABCG1, GAPDH and SREBP2 are listed in Supplementary Table S2.
TaqMan microRNA assays (Applied Biosystems) that include specific RT primers and TaqMan probes were used to quantify the expression of mature miR-128 (AB Assay ID 002216), as described by the manufacturer. 18S rRNA (AB Assay ID 4333760F) was used for normalization. The reaction was incubated in a 7500 Real-Time PCR System (Applied Biosystems) in 96-well plates at 95 °C for 10 min, followed by 40 cycles of the following steps: 95 °C for 15 s and 60 °C for 1 min. The real-time PCR data were analyzed using Pfaffl's method.59
Cholesterol quantitation
Cholesterol content was determined in the human cells as well as in serum obtained from mice using cholesterol quantitation kit (BioVision, Milpitas, CA, USA) according to the manufacturer's instructions. In brief, 106 untransfected and transfected cells were lysed and lipids were extracted by homogenization with 200 μl of chloroform: isopropanol: NP-40 (7:11:0.1). These lipid extracts were vacuum dried for 30 min and the residues were dissolved in 200 μl cholesterol assay buffer provided with the kit. Cholesterol was estimated by spectrophotometry at λ=570 nm in a 96-well plate according to the manufacturer's instructions. The cholesterol levels were normalized to amounts of total cellular protein.60
BODIPY-cholesterol efflux assay
BODIPY-cholesterol was purchased (TopFluor-Cholesterol, Avanti Polar Lipids) and stock solution was prepared at 5 mM in DMSO. Briefly, HepG2/ MCF7 and HEK293T cells were cultured in 24-well plates and loaded with 25 μM of BODIPY-cholesterol in culture medium for 2 h at 37 °C. Cells were rinsed twice with physiological buffer (140 mM NaCl, 5 mM KCl, 1 mM CaCl2, 1 mM MgSO4, 5 mM glucose, 20 mM HEPES, pH 7.4) and then transfected with pre-miRNA plasmid namely p(128) or negative control or anti-miR-128 or SAM for 24 h. Ten micrometers synthetic LXR ligand T0901317 was also added to the untransfected cells for 18 h, to act as positive control. The cell supernatant was centrifuged for 5 min at 6800 g, and the BODIPY fluorescence intensity in the supernatants was measured in a plate reader (Tecan, Seestrasse, Mannedorf, Switzerland) at excitation 490±10 nm; and emission 520±20 nm as described in.61
Protein preparation and western blotting
We performed western blotting as described62 using untransfected and transfected cells (with p(128) and/or negative control and/or anti-miR-128 and/or SAM and/or SIRT1 siRNA and/or SIRT1 ORF) after 24 h of transfection. In brief, cells were trypsinized and lysed with modified RIPA buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% NP40, 0.25% Na-deoxycholate, 1 mM EDTA) containing protease inhibitors (1 μg/ml aprotinin, 1 μgml leupeptin, 1 μg/ml pepstatin, 1 mM PMSF, 1 mM sodium orthovanadate and 1 mM sodium fluoride) for 30 min on ice. The lysates were centrifuged at 12000 r.p.m. for 30 min at 4 °C and the supernatant was collected. Protein concentration was determined by BCA (Sigma, St Louis, MO, USA) method. Equal amount of proteins (30–50 μg) were separated on 10–12% SDS polyacrylamide gel electrophoresis and transferred to PVDF membrane (Mdi; Advanced Microdevices, Ambala Cantt, India). Membrane was blocked using 5% skimmed milk for 1 h at RT and incubated with primary antibodies in 1% skimmed milk for 2 h followed by incubation with their respective secondary antibody for 1 h. Primary antibodies for SREBP2, SREBP1, ABCA1, RXRα, SIRT1 proteins were from Santa Cruz (Santa Cruz Biotechnology, Santa Cruz, CA, USA). The primary antibody for ABCG1 was purchased from Abcam (Cambridge, MA, USA) and β-tubulin was purchased from Sigma (Sigma). The secondary antibodies were HRP-linked and blots were developed using enhanced chemiluminiscence (Thermo Scientific, Waltham, MA, USA). Integrated density values were calculated using AlphaImager 3400 (Alpha InnoTech, San Leandro, CA, USA). These values were then normalized to β tubulin. All experiments were repeated at least three times; representative results are presented here. Western blot analysis for ABCA1 and ABCG1 was done under the influence of TO901317 wherever indicated.
Quantitation of triglyceride contents in serum sample
Serum was obtained from the mice fed normal or high-fat diet. Triglyceride levels were estimated using Triglyceride quantification kit (BioVision) according to manufacturer's instructions.
Statistical analysis
Results are given as mean of three independent experiments±S.E.M. An independent Student's two-tailed t-test was performed using replicate values. Values of P<0.05 were considered statistically significant.
Acknowledgments
We thank Professor Samir K Brahmachari for project conceptualization. We also acknowledge Dr. JM Denu (For SIRT1 ORF construct), Vikas Yadav and Shruti for their help. This work was supported by Grants NWP0036 from the Council of Scientific and industrial research (CSIR). YKA and RS were supported with fellowship from CSIR.
Glossary
- APOE
Apolipoprotein
- SIRT1
Sirtuin 1
- ABCA1
ATP-binding cassette transporter A1
- ABCG1
ATP-binding cassette transporter G1
- RXRα
Retinoid X receptor
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by E Candi
Supplementary Material
References
- Fernandez C, Martin M, Gomez-Coronado D, Lasuncion MA. Effects of distal cholesterol biosynthesis inhibitors on cell proliferation and cell cycle progression. J Lipid Res. 2005;46:920–929. doi: 10.1194/jlr.M400407-JLR200. [DOI] [PubMed] [Google Scholar]
- Goedeke L, Fernandez-Hernando C. Regulation of cholesterol homeostasis. Cell Mol Life Sci. 2012;69:915–930. doi: 10.1007/s00018-011-0857-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss AB, Voloshyna I. Regulation of cerebral cholesterol metabolism in Alzheimer disease. J Investig Med. 2012;60:576–582. doi: 10.231/JIM.0b013e318246d973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ledesma MD, Dotti CG. Peripheral cholesterol, metabolic disorders and Alzheimer's disease. Front Biosci (Elite Ed) 2012;4:181–194. doi: 10.2741/e368. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Li P, Ye J. Lipid homeostasis and the formation of macrophage-derived foam cells in atherosclerosis. Protein Cell. 2012;3:173–181. doi: 10.1007/s13238-012-2025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH. MicroRNAs in cardiometabolic disease. Curr Atheroscler Rep. 2011;13:202–207. doi: 10.1007/s11883-011-0179-y. [DOI] [PubMed] [Google Scholar]
- Tania M, Khan MA, Song Y. Association of lipid metabolism with ovarian cancer. Curr Oncol. 2010;17:6–11. doi: 10.3747/co.v17i5.668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hager MH, Solomon KR, Freeman MR. The role of cholesterol in prostate cancer. Curr Opin Clin Nutr Metab Care. 2006;9:379–385. doi: 10.1097/01.mco.0000232896.66791.62. [DOI] [PubMed] [Google Scholar]
- Jacobs R, Voorneveld P, Kodach L, Hardwick J. Cholesterol metabolism and colorectal cancers. Curr Opin Pharmacol. 2012;12:690–695. doi: 10.1016/j.coph.2012.07.010. [DOI] [PubMed] [Google Scholar]
- Broitman SA, Cerda S, Wilkinson JT. Cholesterol metabolism and colon cancer. Prog Food Nutr Sci. 1993;1993;17:1–40. [PubMed] [Google Scholar]
- Zhang F, Du G. Dysregulated lipid metabolism in cancer. World J Biol Chem. 2012;3:167–174. doi: 10.4331/wjbc.v3.i8.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazra S, Rasheed A, Bhatwadekar A, Wang X, Shaw LC, Patel M, et al. Liver X receptor modulates diabetic retinopathy outcome in a mouse model of streptozotocin-induced diabetes. Diabetes. 2012;61:3270–3279. doi: 10.2337/db11-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaven SW, Tontonoz P. Nuclear receptors in lipid metabolism: targeting the heart of dyslipidemia. Annu Rev Med. 2006;57:313–329. doi: 10.1146/annurev.med.57.121304.131428. [DOI] [PubMed] [Google Scholar]
- Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:2009. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bushati N, Cohen SM. MicroRNA functions. Annu Rev Cell Dev Biol. 2007;23:175–205. doi: 10.1146/annurev.cellbio.23.090506.123406. [DOI] [PubMed] [Google Scholar]
- Chang TC, Mendell JT. microRNAs in vertebrate physiology and human disease. Annu Rev Genomics Hum Genet. 2007;8:215–239. doi: 10.1146/annurev.genom.8.080706.092351. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Krutzfeldt J, Stoffel M. MicroRNAs: a new class of regulatory genes affecting metabolism. Cell Metab. 2006;4:9–12. doi: 10.1016/j.cmet.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Suarez Y, Sessa WC. MicroRNAs as novel regulators of angiogenesis. Circ Res. 2009;104:442–454. doi: 10.1161/CIRCRESAHA.108.191270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, et al. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Ramirez CM, Dávalos A, Goedeke L, Salerno AG, Warrier N, Cirera-Salinas D, et al. MicroRNA-758 regulates cholesterol efflux through posttranscriptional repression of ATP-binding cassette transporter A1. Arterioscler Thromb Vasc Biol. 2011;31:2707–2714. doi: 10.1161/ATVBAHA.111.232066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI. MicroRNA-370 controls the expression of microRNA-122 and Cpt1alpha and affects lipid metabolism. J Lipid Res. 2010;51:1513–1523. doi: 10.1194/jlr.M004812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlakha YK, Saini N. MicroRNA-128 downregulates Bax and induces apoptosis in human embryonic kidney cells. Cell Mol Life Sci. 2011;68:1415–1428. doi: 10.1007/s00018-010-0528-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adlakha YK, Saini N. miR-128 exerts pro-apoptotic effect in a p53 transcription-dependent and -independent manner via PUMA-Bak axis. Cell Death Dis. 2013;4:e542. doi: 10.1038/cddis.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repa JJ, Turley SD, Lobaccaro JA, Medina J, Li L, Lustig K, et al. Regulation of absorption and ABC1-mediated efflux of cholesterol by RXR heterodimers. Science. 2000;289:1524–1529. doi: 10.1126/science.289.5484.1524. [DOI] [PubMed] [Google Scholar]
- Shimano H. Sterol regulatory element-binding proteins (SREBPs): transcriptional regulators of lipid synthetic genes. Prog Lipid Res. 2001;40:439–452. doi: 10.1016/s0163-7827(01)00010-8. [DOI] [PubMed] [Google Scholar]
- Krycer JR, Brown AJ. Cross-talk between the androgen receptor and the liver X receptor: implications for cholesterol homeostasis. J Biol Chem. 2011;286:20637–20647. doi: 10.1074/jbc.M111.227082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Hammer RE, Shimomura I, Brown MS, Goldstein JL. Overproduction of cholesterol and fatty acids causes massive liver enlargement in transgenic mice expressing truncated SREBP-1a. J Clin Invest. 1996;98:1575–1584. doi: 10.1172/JCI118951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimano H, Horton JD, Shimomura I, Hammer RE, Brown MS, Goldstein JL. Isoform 1c of sterol regulatory element binding protein is less active than isoform 1a in livers of transgenic mice and in cultured cells. J Clin Invest. 1997;99:846–854. doi: 10.1172/JCI119248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shimomura I, Brown MS, Hammer RE, Goldstein JL, Shimano H. Activation of cholesterol synthesis in preference to fatty acid synthesis in liver and adipose tissue of transgenic mice overproducing sterol regulatory element-binding protein-2. J Clin Invest. 1998;101:2331–2339. doi: 10.1172/JCI2961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato R, Okamoto A, Inoue J, Miyamoto W, Sakai Y, Emoto N, et al. Transcriptional regulation of the ATP citrate-lyase gene by sterol regulatory element-binding proteins. J Biol Chem. 2000;275:12497–12502. doi: 10.1074/jbc.275.17.12497. [DOI] [PubMed] [Google Scholar]
- Damiano F, Alemanno S, Gnoni GV, Siculella L. Translational control of the sterol-regulatory transcription factor SREBP-1 mRNA in response to serum starvation or ER stress is mediated by an internal ribosome entry site. Biochem J. 2010;429:603–612. doi: 10.1042/BJ20091827. [DOI] [PubMed] [Google Scholar]
- Cannell IG, Kong YW, Bushell M. How do microRNAs regulate gene expression. Biochem Soc Trans. 2008;36:1224–1231. doi: 10.1042/BST0361224. [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehmsmeier M, Steffen P, Hochsmann M, Giegerich R. Fast and effective prediction of microRNA/target duplexes. RNA. 2004;10:1507–1517. doi: 10.1261/rna.5248604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- Murthy S, Born E, Mathur SN, Field FJ. LXR/RXR activation enhances basolateral efflux of cholesterol in CaCo-2 cells. J Lipid Res. 2002;43:1054–1064. doi: 10.1194/jlr.m100358-jlr200. [DOI] [PubMed] [Google Scholar]
- Korn BS, Shimomura I, Bashmakov Y, Hammer RE, Horton JD, Goldstein JL, et al. Blunted feedback suppression of SREBP processing by dietary cholesterol in transgenic mice expressing sterol-resistant SCAP(D443N) J Clin Invest. 1998;102:2050–2060. doi: 10.1172/JCI5341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George KS, Elyassaki W, Wu Q, Wu S. The role of cholesterol in UV light B-induced apoptosis. Photochem Photobiol. 2012;88:1191–1197. doi: 10.1111/j.1751-1097.2011.01038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou MH, Yang G, Jiao S, Hu CL, Mei YA. Cholesterol enhances neuron susceptibility to apoptotic stimuli via cAMP/PKA/CREB-dependent up-regulation of Kv2.1. J Neurochem. 2012;120:502–514. doi: 10.1111/j.1471-4159.2011.07593.x. [DOI] [PubMed] [Google Scholar]
- Devries-Seimon T, Li Y, Yao PM, Stone E, Wang Y, Davis RJ, et al. Cholesterol-induced macrophage apoptosis requires ER stress pathways and engagement of the type A scavenger receptor. J Cell Biol. 2005;171:61–73. doi: 10.1083/jcb.200502078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B, Yao PM, Li Y, Devlin CM, Zhang D, Harding HP, et al. The endoplasmic reticulum is the site of cholesterol-induced cytotoxicity in macrophages. Nat Cell Biol. 2003;5:781–792. doi: 10.1038/ncb1035. [DOI] [PubMed] [Google Scholar]
- Papagiannakopoulos T, Friedmann-Morvinski D, Neveu P, Dugas JC, Gill RM, Huillard E, et al. Pro-neural miR-128 is a glioma tumor suppressor that targets mitogenic kinases. Oncogene. 2012;31:1884–1895. doi: 10.1038/onc.2011.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chao T, Li R, Liu W, Chen Y, Yan X, et al. MicroRNA-128 inhibits glioma cells proliferation by targeting transcription factor E2F3a. J Mol Med (Berl) 2009;87:43–51. doi: 10.1007/s00109-008-0403-6. [DOI] [PubMed] [Google Scholar]
- Purushotham A, Schug TT, Xu Q, Surapureddi S, Guo X, Li X. Hepatocyte-specific deletion of SIRT1 alters fatty acid metabolism and results in hepatic steatosis and inflammation. Cell Metab. 2009;9:327–338. doi: 10.1016/j.cmet.2009.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AK, Yang F, Jiang K, Ji JY, Watts JL, Purushotham A, et al. Conserved role of SIRT1 orthologs in fasting-dependent inhibition of the lipid/cholesterol regulator SREBP. Genes Dev. 2010;24:1403–1417. doi: 10.1101/gad.1901210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. J Lipid Res. 2003;44:2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- Van Rooyen DM, Larter CZ, Haigh WG, Yeh MM, Ioannou G, Kuver R, et al. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology. 2011;141:1393–1403. doi: 10.1053/j.gastro.2011.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Witztum JL. Atherosclerosis. the road ahead. Cell. 2001;104:503–516. doi: 10.1016/s0092-8674(01)00238-0. [DOI] [PubMed] [Google Scholar]
- Reeves GK, Pirie K, Beral V, Green J, Spencer E, Bull D. Cancer incidence and mortality in relation to body mass index in the Million Women Study: cohort study. BMJ. 2007;335:1134. doi: 10.1136/bmj.39367.495995.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renehan AG, Tyson M, Egger M, Heller RF, Zwahlen M. Body-mass index and incidence of cancer: a systematic review and meta-analysis of prospective observational studies. Lancet. 2008;371:569–578. doi: 10.1016/S0140-6736(08)60269-X. [DOI] [PubMed] [Google Scholar]
- Whitlock G, Lewington S, Sherliker P, Clarke R, Emberson J, Halsey J, et al. Body-mass index and cause-specific mortality in 900 000 adults: collaborative analyses of 57 prospective studies. Lancet. 2009;373:1083–1096. doi: 10.1016/S0140-6736(09)60318-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA. 2006;103:10230–10235. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra R, Dubey R, Saini N. Gene expression profiling indicate role of ER stress in miR-23a∼27a∼24-2 cluster induced apoptosis in HEK293T cells. RNA Biol. 2011;8:648–664. doi: 10.4161/rna.8.4.15583. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey R, Chhabra R, Saini N. Small interfering RNA against transcription factor STAT6 leads to increased cholesterol synthesis in lung cancer cell lines. PLoS One. 2011;6:e28509. doi: 10.1371/journal.pone.0028509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bidet M, Joubert O, Lacombe B, Ciantar M, Nehmé R, Mollat P, et al. The hedgehog receptor patched is involved in cholesterol transport. PLoS One. 2011;6:e23834. doi: 10.1371/journal.pone.0023834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goel A, Chhabra R, Ahmad S, Prasad AK, Parmar VS, Ghosh B, et al. DAMTC regulates cytoskeletal reorganization and cell motility in human lung adenocarcinoma cell line: an integrated proteomics and transcriptomics approach. Cell Death Dis. 2012;3:e402. doi: 10.1038/cddis.2012.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.