Abstract
Elongation factor G (EF-G) is a GTPase that plays a crucial role in the translocation of tRNAs and mRNA during translation by the ribosome. We report a crystal structure refined to 3.6 Å resolution of the ribosome trapped with EF-G in the post-translocational state using the antibiotic fusidic acid. Fusidic acid traps EF-G in a conformation intermediate between the GTP and GDP forms. The interaction of EF-G with ribosomal elements implicated in stimulating catalysis, such as the L10-L12 stalk and the L11 region, and of domain IV of EF-G with P-site tRNA and mRNA shed light on various aspects of EF-G function in catalysis and translocation. The stabilization of the mobile stalks of the ribosome also results in a more complete description of its structure.
Introduction
The ribosome is the large molecular machine that uses an mRNA template to synthesize proteins, in a process referred to as translation. Ribosomes from all species consist of a large and a small subunit, which are termed 50S and 30S subunits in bacteria and consist of roughly 2/3rd RNA and 1/3rd protein. The last decade has seen a revolutionary change in our understanding of translation, largely as a result of the determination of high-resolution structures of the ribosomal subunits as well as, more recently, the entire ribosome (reviewed in ref. 1). Protein factors play crucial roles in all stages of translation, which can be divided roughly into initiation, elongation and termination. The hydrolysis of GTP by factors is required in all three stages of translation.
At the heart of translation is the elongation cycle, which consists of sequential addition of amino acids to the growing polypeptide chain directed by the mRNA codons. This process is catalyzed by two GTPase factors, elongation factors Tu (EF-Tu) and G (EF-G). At the beginning of the cycle, a nascent peptide chain linked to a tRNA is bound in the P site of the ribosome, while the A site remains empty. The incoming aminoacyl-tRNA is delivered to the A site of the ribosome as a ternary complex with EF-Tu and GTP. Following GTP hydrolysis and peptidyl transfer, the nascent peptide chain is linked to the new amino acid on the A-site tRNA through the formation of a peptide bond, leaving the P-site tRNA deacylated.
This assembly of tRNAs and mRNA must then move with respect to the ribosome by one codon in a process known as translocation, which is catalyzed by EF-G. In the first step of translocation, the aminoacyl ends of the tRNAs move with respect to the 50S subunit, so that the P- and A-site tRNAs move to the E (exit) and P sites respectively, creating hybrid P/E and A/P states of the tRNAs (2). The formation of the intermediate states of tRNA is coupled to a rotation or ratcheting of about 6 degrees of the 30S and 50S subunits relative to each other (3, 4). The binding of EF-G to this ratcheted state catalyzes the second step of translocation, which involves movement of the mRNA and the anticodon stem-loops of the tRNAs with respect to the 30S subunit, accompanied by a resetting of the ratchet. The hydrolysis of GTP by EF-G is known to precede and accelerate this second step (5). Translocation completes the elongation cycle and leaves the ribosome with tRNAs in the E and P sites, and an empty A site ready for the next amino-acyl tRNA. In addition to its role in translocation, EF-G is also required for ribosome recycling, where it acts in conjunction with ribosome recycling factor (reviewed in ref. 1).
Although biochemical studies have elucidated the overall role of EF-G in translocation, understanding the detailed mechanism of the process requires knowledge of the precise interactions EF-G makes with the ribosome in various states of translocation. Crystal structures of EF-G with and without bound GDP have been determined (6, 7). The structure of the ternary complex of EF-Tu with tRNA and the GTP analog GDPNP showed a surprising similarity in shape to that of EF-G (8). The GTPase domains of the two factors are similar, and Domain IV of EF-G protrudes from the body of the molecule in a similar manner to the tRNA anticodon stem-loop, suggesting that EF-G and the ternary complex bind the ribosome in a similar way.
To date, the main structural data on the binding of any GTPase factors to the ribosome come from cryoelectron microscopy (cryoEM). The complex of EF-G with the ribosome has been solved in both the GTP and GDP states of the factor (9-13). These structures show that the conformation of EF-G on the ribosome is significantly altered from that of the isolated crystal structures. They also reveal the orientation of the factor on the ribosome in the pre- and post-GTP hydrolysis states and the conformational changes in the ribosome itself that accompany binding of EF-G. Further details of the mechanism of translocation require higher-resolution structures, but crystallization of the ribosome in complex with any GTPase factor has been elusive.
In this study, we report a new crystal form of the 70S ribosome made possible by use of a mutant ribosome that eliminates the steric clash that excludes GTPase factors in all previously published crystal forms. We have used this form to determine the structure of the post-translocation state of the ribosome in complex with EF-G and fusidic acid, an antibiotic that allows GTP hydrolysis and translocation by EF-G but prevents its release from the ribosome. The structure, which is refined to 3.6 Å (I/σ=2 at 3.7 Å), reveals conformational changes in the ribosome involved in GTPase activation; details of the interaction of domain IV of EF-G with the ribosome, mRNA and tRNA; and insights into the mechanism of fusidic acid, which functions only in complex with EF-G bound to the ribosome.
Crystallization and structure determination of the ribosome in the post-translocation state with EF-G and fusidic acid
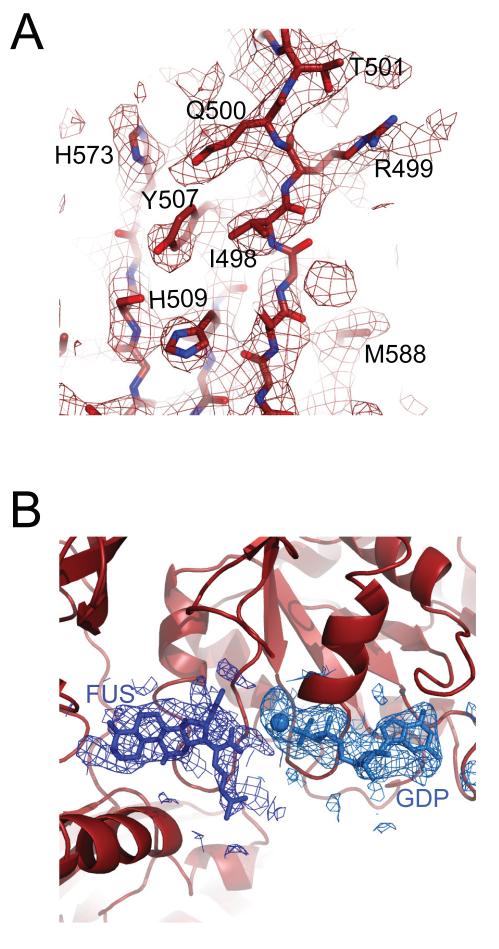
In all previously reported crystal forms of the ribosome from both Escherichia coli and Thermus thermophilus (14-16) a lattice contact with ribosomal protein L9 from a neighboring molecule would result in a steric clash with a bound GTPase factor. This interaction of L9 is so strong that our previous attempts to crystallize EF-G in complex with the ribosome resulted in a high-resolution crystal form in which the L9 contact had displaced the factor from the ribosome (16). This prompted us to truncate the gene for L9 in T. thermophilus. Previously established methods for genetic manipulation in this species (17) were used to generate the strain T. thermophilus HB8-MRC-MSAW1. Ribosomes from this strain were used to crystallize the post-translocational state with mRNA, deacylated tRNA in the P and E-sites and EF-G with GDP and fusidic acid. This complex crystallized in the space group P21 with cell dimensions of 291.8× 270.4× 402.4Å and β = 91.7 ° and diffracted to beyond 3.5 Å resolution. The structure was solved by molecular replacment using the empty ribosome (from ref. 16) as a starting model. Clear difference Fourier density was seen for the tRNAs, mRNA, GDP and EF-G, in which well-ordered side chains of the factor were clearly distinguishable (Fig. 1a). Difference Fourier maps also clearly showed the locations of GDP and fusidic acid (Fig.1b). The final structure after inclusion of all ligands was refined using data to 3.6 Å resolution with a final R/Rfree of 0.227/0.260 (Table S1; details in Supporting Online Material).
Figure 1.
Unbiased difference Fourier electron density maps for (A) EF-G and (B) the ligands fusidic acid (FUS) and GDP with its coordinated Mg2+ ion in the ribosome. All figures were made using PyMol (45)
Overall structure of EF-G in the ribosome
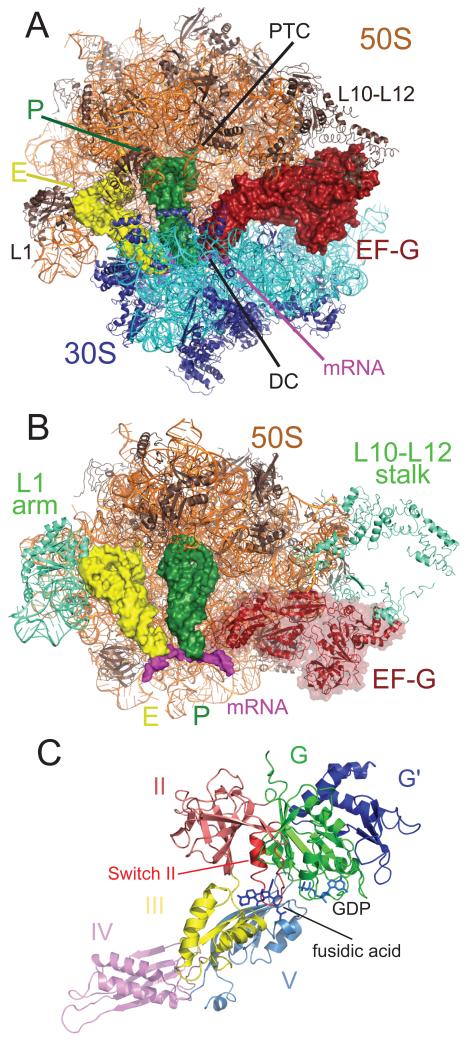
The overall conformation of EF-G in the ribosome (Fig. 2a) agrees with previous cryoEM studies either on an EF-G fusidic acid complex (9, 11, 12) or EF-G with a non-hydrolyzable GTP analogue bound to the ratcheted ribosome (12, 13). As expected, in the crystal structure here, the ribosomal subunits are in the canonical rather than ratcheted state. The binding of EF-G has made mobile elements of the ribosome better ordered, so that the L1 stalk and the N-terminal domain (NTD) of L10 are visible even in high-resolution maps allowing us to build them in entirety (Fig. 2b). Maps calculated at lower resolution (5.0 Å) allowed us to place the helical extension of L10, four copies of the NTD of L12 as well as a copy of the C-terminal domain (CTD) of L12 that makes contact with EF-G and L11. The ordering of the L1 stalk, which makes contact with E-site tRNA on the other side of P-site tRNA, must be indirect since EF-G does not directly contact it.
Figure 2.
EF-G in the post-translocational state of the ribosome. A. Overall view of EF-G in the ribosome. EF-G is shown in reddish-brown, the 50S subunit on top is shown in orange, the 30S subunit below is shown in cyan, P-site tRNA in green, and E-site tRNA in yellow and the mRNA in purple. The decoding center (DC) in the 30S subunit and the peptidyl transferase center (PTC), and the L1 and L10-L12 stalks are indicated as shown.B. Global changes in the 50S subunit as a result of EF-G binding. The mobile regions of the 50S subunit are indicated in teal, and include the L1 stalk (both RNA and protein), the L11 region, and the the L10-L12 stalk. C.The structure of EF-G bound to fusidic acid and GDP in the ribosome. The various domains of EF-G are colored as shown.
The conformation of EF-G on the ribosome (Fig. 2c) is different from that of the isolated EF-G with or without GDP (6, 7, 18) and the structure of EF-G-2 bound to the GTP analog GDPNP (13). An alignment using the G domain, which contains the nucleotide binding site, shows that the most striking conformational change from the isolated structures is a re-orientation of domain IV, although domains III and V have also moved significantly (Fig. S1). A sequence alignment with E. coli EF-G is shown in Fig. S2. Throughout this paper, RNA helices are numbered using the standard Brimacombe nomenclature, prefixed by H for 23S RNA and h for 16S RNA, and RNA residues are numbered using the E. coli sequence.
EF-G is held in position by contacts with both the 50S and 30S subunits (Figs. 2a and S3a). The G domain, domain III and domain V all surround the sarcin-ricin loop (SRL; 2660 loop or H95), the ribosomal element closest to the nucleotide-binding site, as inferred from previous cryoEM studies (Fig. S3b). The binding of EF-G also appears to have ordered the C-terminus of L6 near the SRL and domain V. The β-barrel domain II contacts h5 and h15 (360-380) in the shoulder of the 30S subunit (Fig. S3c). Domain III, which is required for GTP hydrolysis (19), makes contact with both the 30S subunit through interactions with h5 and protein S12, and the SRL in the 50S subunit (Fig. S3d). Domain V makes several contacts with the 50S subunit including the L11-binding region (H43/44; 1070/1095 loops) and the adjacent H89 (2475 loop) (Fig. S3e). Finally, domain IV is inserted into the 30S decoding center where it contacts P-site tRNA and mRNA, and the EF-G-specific G′ domain makes a contact with a copy of the L12 CTD, which bridges it to the NTD of L11. These two interactions are discussed in detail below.
Interaction of domain IV with mRNA and tRNA in the decoding center
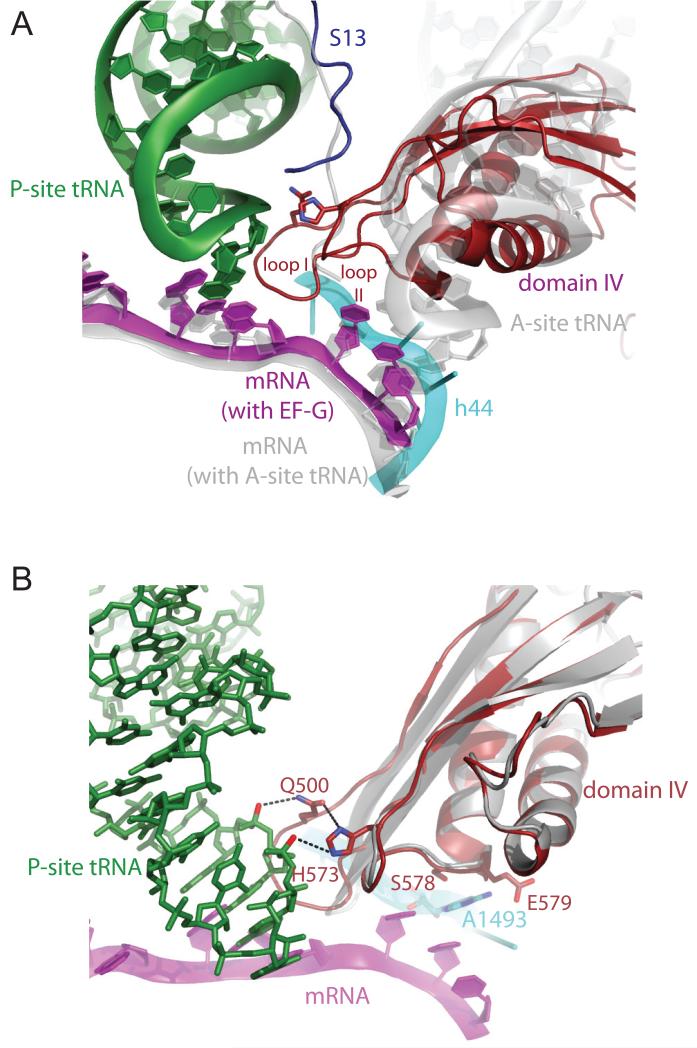
Domain IV, which occupies the A site of the 30S subunit, makes extensive contacts with mRNA, P-site tRNA and the decoding center of the 30S subunit (Fig. 3). Unlike A-site tRNA (20), the contacts of domain IV are made with the P-site tRNA and codon rather than with the A-site codon (Fig. 3a.).
Figure 3.
Interactions of domain IV of EF-G with tRNA and mRNA. A. Interactions of domain IV of EF-G with P-site tRNA and mRNA. Loop 1 is inserted into the minor groove between P-site tRNA and its codon. In gray are shown elements from a superposition of the 70S structure with A-site tRNA (20), showing conformational changes in the mRNA and protein S13 on EF-G binding. Unlike A-site tRNA, domain IV of EF-G does not make extensive interactions with the A-site codon. B. Details of interactions with domain IV in the decoding site. The residues Q500 from loop I and H573 from loop II form a network of interactions with P-site tRNA and each other; Loop 1 has changed conformation relative to its structure in domain IV of the isolated protein (gray) (2BM0 from ref. 18), to insert into the minor groove between P-site tRNA and mRNA. Residues S578 and E579 make interactions with the universally conserved A1493 of 16S RNA.
Domain IV contains two loops at its tip, loop I (496-509) and loop II (567-579). The two conserved glycines (502-503) facilitate a tight turn of loop I that allows it to be inserted into the minor groove between P-site tRNA and codon (Fig. 3a). When bound to the ribosome, the highly conserved loop I is in a different conformation from that in isolated structures (Fig. 3b). Two conserved residues, Q500 in loop I and H573 in loop 2 participate in a network of interactions (Fig. 3b). H573 can form hydrogen bonds with a phosphate oxygen of U37 of the P-site tRNA, as well as with the carbonyl oxygen of Q500. The amide of Q500 can in turn form a hydrogen bond with a phosphate oxygen of A38 of P-site tRNA. Finally, S578 and E579 are both within hydrogen bonding distance of the universally conserved A1493 of 16S RNA (Fig. 3b).
The structure clearly rationalizes the previous finding that mutation of H573 or insertions in loop II between 576 and 579 significantly reduce translocation without affecting EF-G binding or GTP hydrolysis (21, 22). In addition to these interactions with tRNA and mRNA, domain IV also makes contacts with both h35 in the head of the 30S subunit and with h44 at the decoding center. Thus although domain IV and A-site tRNA both bind in the same location in the 30S subunit, their interactions are quite different; In particular, there appear to be no direct contacts of domain IV with the A-site codon, at least in the post-translocational state. This is not unexpected, since the codon that has been translocated into the A site would not be accessible to EF-G prior to translocation. It is possible that the contacts of the tip of domain IV with the P-site tRNA and codon as seen in the current structure initially occur when they are in the A site and are maintained throughout their translocation into the P site, thus helping to avoid frameshifting during the process. If so, one of the early events in translocation by EF-G must be the displacement by loop I of the ribosomal bases G530, A1492 and A1493 that line the minor groove of the A-site codon-anticodon helix during decoding (23), and may explain why antibiotics that stabilize those bases in the minor groove also inhibit translocation (24).
Catalytic activity of EF-G
Translocation is a highly complex process involving large-scale movements of the ribosomal subunits and mRNA and tRNA. The structure presented here represents the post-translocational state after GTP hydrolysis trapped with fusidic acid. Interestingly, it retains some features of the GTP state of EF-G and thereby sheds light on the catalysis of GTP hydrolysis.
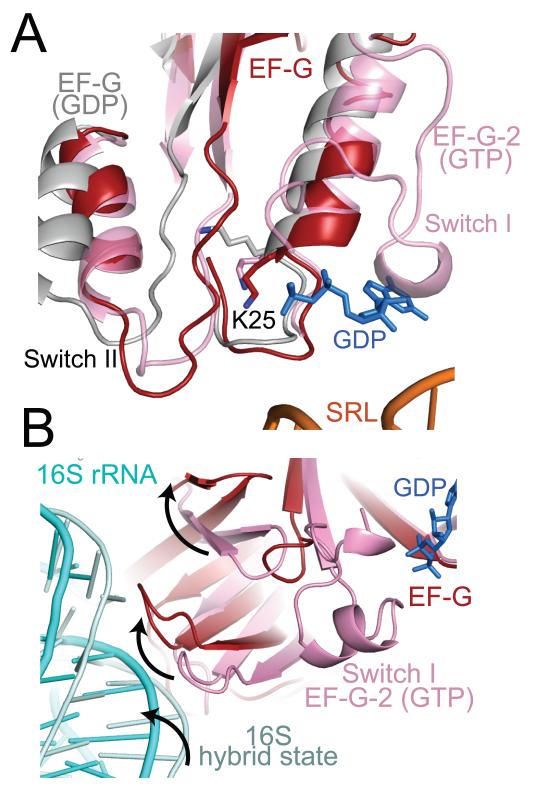
The structure reveals atomic details of the environment of the catalytic site. The GDP molecule is clearly visible along with a coordinated Mg2+ ion (Fig.1b). The closest ribosomal element to the catalytic site is the sarcin-ricin (2660) loop of 23S RNA, as expected from cryoEM studies (12, 13). In the GTP state, as represented by the structure of EF-G-2 with GDPNP(13), the nucleotide is protected from hydrolysis by two switch regions (40-66 and 80-102) which are ordered. In the structure here, the switch I loop has changed conformation, becoming disordered in the process, thus allowing access by water to the gamma phosphate. However, the switch II loop (80-102) is in the same conformation as the GTP state of EF-G-2 rather than of other GDP structures of EF-G where this loop has moved away from the nucleotide (Fig. 4a)(7, 18, 25). This suggests that the factor has been frozen in an intermediate state between GTP hydrolysis and the GDP-bound conformation after release from the ribosome.
Figure 4.
The environment of the nucleotide binding site in EF-G bound to the ribosome in the presence of fusidic acid. A. A superposition shows that the switch I region of EF-G-2 in the GTP-bound form (pink)(13) has become disordered in the fusidic-acid complex structure. However switch II (red) in the structure is close to the GTP form of EF-G-2, and different from the altered conformation of the GDP form of EF-G in isolation (gray)(2BM0 from ref. 18). Similarly, K25, which interacts with the gamma phosphate in the GTP form, remains in that form in this structure rather than adopt the altered conformation (gray) of the GDP form of EF-G. B. Conformational changes of the β–hairpins and 16S movement in domain II relative to the GTP form of EF-G-2 (13). This movement is in the same direction as the movement of 16S RNA (cyan) relative to that of the ratcheted ribosome (light blue) that was aligned using the 50S subunit(46).
EF-G lacking domain III has reduced GTPase activity although it binds GTP normally (19). Domain III interacts with both S12 and h5 (360 stem) of 16S RNA in the 30S subunit as well as the SRL in the 50S subunit (Fig. S3d). Thus by contacting both subunits, domain III can sense the conformation of the ribosome, in particular the difference between ratcheted and non-ratcheted subunits. These contacts may play a role in triggering GTP hydrolysis, in a manner similar to what has been proposed for EF-Tu (Schmeing et al., accompanying manuscript).
The β-hairpins (314-328 and 354-366) in domain II have moved relative to the GTP form of EF-G-2, and are similar to the GDP form of EF-G (Fig. 4b). In the GTP form, these hairpins would make contact with the switch I helix, and their movement may be correlated with the destabilization of the switch I region during GTP hydrolysis. It is interesting to note that at least one of the hairpins interacts with the shoulder of 16S RNA, so their movement away from the switch I helix could track a rotation of the 30S relative to the 50S subunit. This may thus hint at how GTP hydrolysis is coupled to the movement of the ribosomal subunits from the ratcheted form back to the canonical form during the second step of translocation.
A detailed understanding of the mechanism of GTPase activation and GTP hydrolysis by EF-G will require high-resolution structures of the pre-translocational ribosome with EF-G before and just after GTP hydrolysis. Remarkably, to date there is no such structure by any method; reported cryoEM structures of the ribosome complexed with EF-G and a GTP analogue do indeed show a ratcheted state of the ribosome but do not contain an A-site tRNA (e.g. refs. 12, 13). Moreover domain IV of EF-G in these structures is in almost the same conformation as the structure here, and would be incompatible with the A-site tRNA that would be present in a pre-translocated state.
Fusidic acid
Fusidic acid prevents the release of EF-G after GTP hydrolysis and translocation. It was originally thought to allow only a single round of GTP hydrolysis by EF-G (26) but recent studies suggest that multiple rounds of GTP hydrolysis can occur prior to trapping the ribosome in the post-translocational state (27). However, fusidic acid has low affinity for isolated EF-G, so it has not previously been possible to visualize it directly when bound to the factor alone.
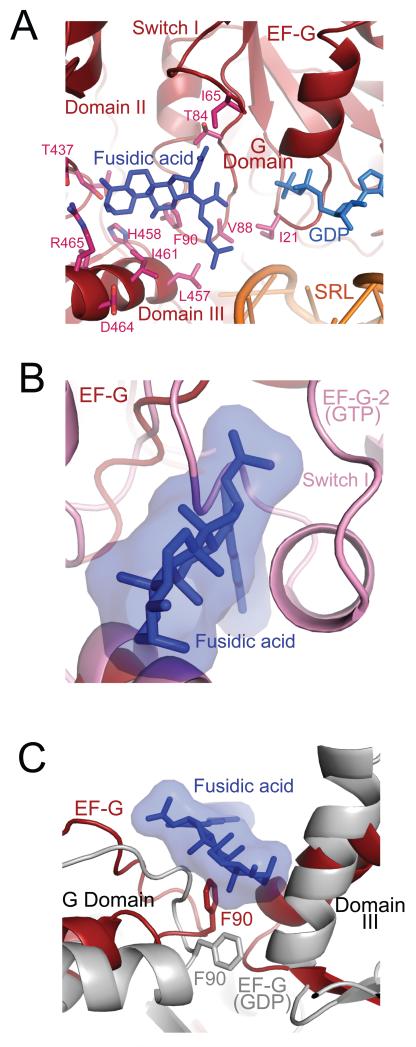
Clear density can be seen for fusidic acid (Fig. 1b) in a pocket surrounded by the switch II region of the G-domain, and domains II and III (Fig. 5a). We have placed the molecule in the most plausible orientation taking into account the shape of the density and the nature of potential interactions; however alternative orientations that are rotated or flipped by 180 degrees are also possible. The location is consistent with predictions from analysis of mutational data on fusidic acid (18, 28). Fusidic acid would clash with the closed conformation of the switch I loop as observed in the GTP-bound form of EF-G-2 (Fig. 5b) (13), and binding must therefore require an open, disordered form of the switch I loop. Since the opening of the switch I loop would expose the gamma phosphate to water, this suggests that the binding of fusidic acid must occur after GTP hydrolysis.
Figure 5.
Fusidic acid bound to EF-G in the ribosome. A. Interactions of fusidic acid (FA) in a pocket lined by domains of EF-G, showing amino acids whose mutation gives rise to fusidic acid resistance. B. A superposition of switch I from the GTP form of EF-G-2 (13) shows it would clash with fusidic acid (FA). C. Conformational changes between the fusidic-acid-bound structure compared to the GDP-bound form of EF-G in isolation (2BM0 from ref. 18) (gray). F90, switch II of the G domain and domain III are all in a conformation that is different from the GDP form of EF-G.
Two EF-G mutations, T84A and G16V give rise to fusidic acid resistance and hypersensitivity respectively (29). T84 is involved in a direct contact with fusidic acid. While the mutation of G16 to valine stabilizes the loop that is part of switch II in a conformation (18) similar to the fusidic acid bound structure described here, leading to increased fusidic acid affinity and thus sensitivity. Mutation of F90 greatly reduces fusidic acid sensitivity (30, 31). This residue interacts directly with fusidic acid in a manner similar to that observed in other fusidic-acid binding proteins (32, 33).
The conformation of F90 is similar to that in the GTP form of EF-G-2, but different from that in the GDP bound EF-G structures (Fig. 5c) (7, 18, 25) where it interacts with a helix of domain III. The direct interaction of F90 with fusidic acid would thus prevent the conformational change of switch II that would normally occur after GTP hydrolysis and the transmission of this change to domain III. In addition, K25, which interacts with the γ-phosphate in the GTP form of EF-G-2 (13), remains in a similar conformation rather than in the altered form observed in the GDP structures (Fig 4a) (7, 18, 25), where it makes a hydrogen bond with T84 of the switch II loop. Finally, fusidic acid also makes direct interactions with domain III, stabilizing a conformation that is different from that of the normal GDP form (Fig. 5a and c). The altered conformations of the switch II region and domain III relative to domain II would result in stabilization of domain IV in the 30S subunit binding pocket. Without fusidic acid, the conformation of domain IV that is preferred in the GDP form would result in the disruption of its contacts with the 30S subunit and destabilize the factor on the ribosome, leading to its release.
Thus the main effect of fusidic acid appears to be to lock switch II in a conformation similar to the GTP form even after GTP hydrolysis, and as a result prevent the transmission of conformational changes to domains III and IV. Although the location of fusidic acid is different from that of sordarin, which blocks the release of EF-2, the eukaryotic counterpart of EF-G (34), or of aurodox, which like kirromycin blocks the release of EF-Tu after GTP hydrolysis (35), all three antibiotics may act by preventing large scale conformational changes in the elongation factors that are required for their release from the ribosome after GTP hydrolysis.
The L10-L12 stalk and the L11 region
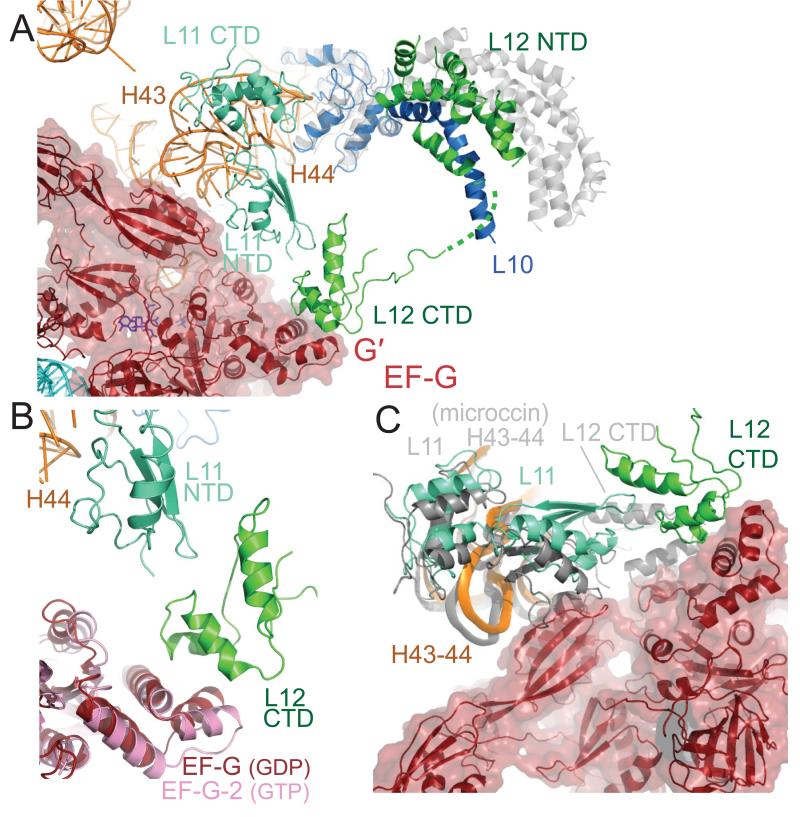
Although at some distance from each other on the ribosome, both the L10-L12 stalk and the L11 region have been implicated in EF-G function. Both EF-Tu and EF-G have greatly diminished GTPase activity on ribosomes lacking the CTD of L12 (36, 37). The L11 region of the 50S subunit (including protein L11 and the 1065/1096 stem loops of 23S RNA) is the binding site of the antibiotic thiostrepton, which inhibits steps in translocation after GTP hydrolysis, such as phosphate release (27, 38). Kinetic measurements suggest that thiostrepton does not prevent binding of EF-G or GTP hydrolysis, but inhibits subsequent steps such as the release of inorganic phosphate and translocation (38, 39).
The structure shows that EF-G interacts directly with both the L11 region and the CTD of L12 (Fig. 6a). In the presence of bound EF-G, the long alpha helix at the C-terminus of L10 is bent relative to its structure in the isolated L10-(L12)6 complex (37) (Fig.6a). This allows the L10-L12 stalk to bend towards the L11 region and as a result, a copy of the CTD of L12 interacts with both the NTD of L11 and the G′ domain of EF-G (Fig. 6b), in agreement with previous cryoEM data (40). The conformation of the G′ domain is similar to that in the GDP structures of EF-G and brings it into closer contact with the CTD of L12 as compared to the GTP form of EF-G-2, consistent with previous mutagenesis data (41). It is possible that this movement of the G′ domain may transmit conformational changes required for phosphate release, and as seen in Fig. 6b, there is also a shift in the G domain adjacent to the G′ domain which provides a path to the nucleotide binding site.
Figure 6.
Interaction of EF-G with the L11 region and the L10-L12 stalk. A. Overview showing that EF-G interacts directly with the L11 RNA (helices 43 and 44) through domain V. The L10-L12 stalk has bent towards the L11 region relative to the structure of the isolated stalk (gray) (37) as judged by superimposing the globular part of L10. As a result, a copy of the CTD of L12 bridges the G′ domain of EF-G and the NTD of L11 by interacting with both of them. B. Conformational changes in the G′ domain and domain V of EF-G in the GDP-fusidic acid form in the ribosome (reddish-brown) as compared to the GTP form (pink) (13). C. Comparison of the structure of EF-G trapped in the post-translocational state of the ribosome with the structure of the 50S subunit bound to micrococcin (43). Domain V of EF-G interacts with helices 43 and 44 of 23S RNA that bind L11. A superposition of the micrococcin-bound 50S structure (gray) using all of 23S RNA shows that helices 43 and 44 (gray) superimpose reasonably well on the EF-G ribosome complex, but the NTD of L11 would clash with domain V of EF-G. The CTD of L12 is in the micrococcin-bound 50S structure is also in a very different location and orientation compared to the current structure where it interacts with the G′ domain.
Domain V of EF-G interacts with the tip of the 1067 and 1095 loops of the L11 region of 23S RNA. The conformation of this region of 23S RNA appears to have only minimal changes compared to the isolated L11-RNA complex(42) or a native 50S structure (43). The binding of micrococcin, which binds to the L11 region and enhances GTPase activity by EF-G also stabilizes a CTD of L12 near L11 (43). Superposition of the micrococcin-bound 50S structure shows that the L11 CTD and RNA superimpose reasonably well, but the L11 NTD is in a different orientation that would clash with domain V of EF-G in the current structure (Fig. 6c). Most interestingly, the location of the CTD of L12 in the micrococcin structure is quite different from its location here in the presence of EF-G (Fig. 6c), and is closer to the SRL and the nucleotide binding site.
In the presence of thiostrepton, which also binds to the L11 region but inhibits EF-G function, the L11 NTD would also be brought closer to EF-G rather than away from it, resulting in a similar clash with EF-G as micrococcin(43). Thus, as suggested previously (43), the thiostrepton-bound form of the L11 region may allow EF-G binding and GTP hydrolysis, but in the absence of a compensating stabilization of the CTD of L12, may be sub-optimal for other functions such as phosphate release and may reduce the overall affinity of EF-G, thus explaining its loss from ribosomes on centrifugation (44).
The role of the CTD of L12 appears to be complex. It appears to be required for initial binding and GTPase activation for both EF-Tu and EF-G, even in single-turnover experiments (36, 37). On the other hand, mutations that would disrupt its interactions with the G′ domain inhibit phosphate release without affecting GTPase activation (41). Given the two quite different locations for the CTD of L12 in the micrococcin bound 50S structure (43) and the EF-G complex of the ribosome here (Fig. 6c), it is tempting to speculate that the former location, which could be common to EF-Tu and EF-G, may be involved in GTPase activation, whereas the latter location, which specifically involves the G′ domain is involved in the release of inorganic phosphate. Clearly, studies with the 70S ribosome in other states, as well as with micrococcin, will help to clarify this issue.
Conclusion
This work reports a crystal form obtained from ribosomes lacking protein L9, that allows crystallization of the ribosome in the presence of GTPase factors. The structure reported here reveals details of the ribosome complexed with EF-G trapped in the post-translocational state by the antibiotic fusidic acid. It reveals many aspects of EF-G function, in addition to providing a more complete model of the ribosome that includes the entire L1 stalk and much of the L10-L12 stalk. The close interactions of domain IV of EF-G with the 30S subunit suggest how it could facilitate translocation without slippage of the reading frame. The structure shows changes in the crucial switch regions around the nucleotide-binding pocket that accompany GTP hydrolysis and may couple it to movement of the subunits. It reveals the location of fusidic acid and shows how the antibiotic may trap EF-G on the ribosome by preventing conformational changes required for its release after translocation. The structure also sheds light on the interactions of EF-G with the L12 stalk and the L11 binding region. Further understanding of EF-G function in translation will require its structure in complex with the in other states, e.g. prior to GTP hydrolysis and translocation as well as in post-termination complexes with ribosome recycling factor. Finally, this work paves the way for the study of other GTPase factors bound to the ribosome.
Supplementary Material
Acknowledgments
We thank Clemens Schulze-Briese, Martin Fuchs and Meitian Wang for help with data collection at the Swiss Light Source, Joachim Frank for coordinates of the ratcheted ribosome obtained from modeling into cryoEM maps, Jendrik Hentschel for help with crystallization trials, Martin Schmeing and Rebecca Voorhees for help with data collection, making figures and critical feedback, and Cajetan Neubauer and Leong Ng for useful discussions. VR was supported by the Medical Research Council (UK), a Wellcome Trust programme grant, and awards from the Agouron Institute and the Louis-Jeantet Foundation. Coordinates for the structure have been deposited as four split entries with codes 2wri, 2wrj, 2wrk and 2wrl.
Footnotes
References
- 1.Ramakrishnan V. Cell. 2002;108:557. doi: 10.1016/s0092-8674(02)00619-0. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Noller HF. Nature. 1989;342:142. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 3.Frank J, Agrawal RK. Nature. 2000;406:319. doi: 10.1038/35018597. [DOI] [PubMed] [Google Scholar]
- 4.Ermolenko DN, et al. Nat Struct Mol Biol. 2007;14:493. doi: 10.1038/nsmb1243. [DOI] [PubMed] [Google Scholar]
- 5.Rodnina MV, Savelsbergh A, Katunin VI, Wintermeyer W. Nature. 1997;385:37. doi: 10.1038/385037a0. [DOI] [PubMed] [Google Scholar]
- 6.Aevarsson A, et al. Embo J. 1994;13:3669. doi: 10.1002/j.1460-2075.1994.tb06676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Czworkowski J, Wang J, Steitz TA, Moore PB. Embo J. 1994;13:3661. doi: 10.1002/j.1460-2075.1994.tb06675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nissen P, et al. Science. 1995;270:1464. doi: 10.1126/science.270.5241.1464. [DOI] [PubMed] [Google Scholar]
- 9.Agrawal RK, Penczek P, Grassucci RA, Frank J. Proc Natl Acad Sci. 1998;95:6134. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. Nat Struct Biol. 1999;6:643. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 11.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Cell. 2000;100:301. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 12.Valle M, et al. Cell. 2003;114:123. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 13.Connell SR, et al. Mol Cell. 2007;25:751. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 14.Schuwirth BS, et al. Science. 2005;310:827. [Google Scholar]
- 15.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Cell. 2006;126:1065. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 16.Selmer M, et al. Science. 2006;313:1935. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 17.Cameron DM, et al. J Bacteriol. 2004;186:5819. doi: 10.1128/JB.186.17.5819-5825.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansson S, Singh R, Gudkov AT, Liljas A, Logan DT. J Mol Biol. 2005;348:939. doi: 10.1016/j.jmb.2005.02.066. [DOI] [PubMed] [Google Scholar]
- 19.Martemyanov KA, Gudkov AT. J Biol Chem. 2000;275:35820. doi: 10.1074/jbc.M002656200. [DOI] [PubMed] [Google Scholar]
- 20.Voorhees RM, Weixlbaumer A, Loakes D, Kelley AC, Ramakrishnan V. Nat Struct Mol Biol. 2009;16:528. doi: 10.1038/nsmb.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savelsbergh A, Matassova NB, Rodnina MV, Wintermeyer W. J Mol Biol. 2000;300:951. doi: 10.1006/jmbi.2000.3886. [DOI] [PubMed] [Google Scholar]
- 22.Martemyanov KA, Yarunin AS, Liljas A, Gudkov AT. FEBS Lett. 1998;434:205. doi: 10.1016/s0014-5793(98)00982-x. [DOI] [PubMed] [Google Scholar]
- 23.Ogle JM, et al. Science. 2001;292:897. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 24.Peske F, Savelsbergh A, Katunin VI, Rodnina MV, Wintermeyer W. J Mol Biol. 2004;343:1183. doi: 10.1016/j.jmb.2004.08.097. [DOI] [PubMed] [Google Scholar]
- 25.al-Karadaghi S, Aevarsson A, Garber M, Zheltonosova J, Liljas A. Structure. 1996;4:555. doi: 10.1016/s0969-2126(96)00061-5. [DOI] [PubMed] [Google Scholar]
- 26.Bodley JW, Zieve FJ, Lin L. J Biol Chem. 1970;245:5662. [PubMed] [Google Scholar]
- 27.Seo HS, et al. Biochemistry. 2006;45:2504. doi: 10.1021/bi0516677. [DOI] [PubMed] [Google Scholar]
- 28.Laurberg M, et al. J Mol Biol. 2000;303:593. doi: 10.1006/jmbi.2000.4168. [DOI] [PubMed] [Google Scholar]
- 29.Martemyanov KA, Liljas A, Yarunin AS, Gudkov AT. J Biol Chem. 2001;276:28774. doi: 10.1074/jbc.M102023200. [DOI] [PubMed] [Google Scholar]
- 30.Johanson U, Hughes D. Gene. 1994;143:55. doi: 10.1016/0378-1119(94)90604-1. [DOI] [PubMed] [Google Scholar]
- 31.Nagaev I, Bjorkman J, Andersson DI, Hughes D. Mol Microbiol. 2001;40:433. doi: 10.1046/j.1365-2958.2001.02389.x. [DOI] [PubMed] [Google Scholar]
- 32.Murray IA, et al. J Mol Biol. 1995;254:993. doi: 10.1006/jmbi.1995.0671. [DOI] [PubMed] [Google Scholar]
- 33.Zunszain PA, Ghuman J, McDonagh AF, Curry S. J Mol Biol. 2008;381:394. doi: 10.1016/j.jmb.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jorgensen R, et al. Nat Struct Biol. 2003;10:379. doi: 10.1038/nsb923. [DOI] [PubMed] [Google Scholar]
- 35.Vogeley L, Palm GJ, Mesters JR, Hilgenfeld R. J Biol Chem. 2001;276:17149. doi: 10.1074/jbc.M100017200. [DOI] [PubMed] [Google Scholar]
- 36.Mohr D, Wintermeyer W, Rodnina MV. Biochemistry. 2002;41:12520. doi: 10.1021/bi026301y. [DOI] [PubMed] [Google Scholar]
- 37.Diaconu M, et al. Cell. 2005;121:991. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 38.Rodnina MV, et al. Proc Natl Acad Sci. 1999;96:9586. doi: 10.1073/pnas.96.17.9586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pan D, Kirillov SV, Cooperman BS. Mol Cell. 2007;25:519. doi: 10.1016/j.molcel.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Mol Cell. 2005;20:723. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 41.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. EMBO J. 2005;24:4316. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wimberly BT, Guymon R, McCutcheon JP, White SW, Ramakrishnan V. Cell. 1999;97:491. doi: 10.1016/s0092-8674(00)80759-x. [DOI] [PubMed] [Google Scholar]
- 43.Harms JM, et al. Mol Cell. 2008;30:26. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 44.Cameron DM, Thompson J, March PE, Dahlberg AE. J Mol Biol. 2002;319:27. doi: 10.1016/S0022-2836(02)00235-8. [DOI] [PubMed] [Google Scholar]
- 45.W. L. DeLano, http://www.pymol.org (2006).
- 46.Agirrezabala X, et al. Mol Cell. 2008;32:190. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.