Abstract
Oxidized bases in DNA have been implicated in cancer, aging and neurodegenerative disease. We have developed an approach combining single-cell gel electrophoresis (comet) with fluorescence in situ hybridization (FISH) that enables the comparative quantification of low, physiologically relevant levels of DNA lesions in the respective strands of defined nucleotide sequences and in the genome overall. We have synthesized single-stranded probes targeting the termini of DNA segments of interest using a polymerase chain reaction-based method. These probes facilitate detection of damage at the single-molecule level, as the lesions are converted to DNA strand breaks by lesion-specific endonucleases or glycosylases. To validate our method, we have documented transcription-coupled repair of cyclobutane pyrimidine dimers in the ataxia telangiectasia-mutated (ATM) gene in human fibroblasts irradiated with 254 nm ultraviolet at 0.1 J/m2, a dose ∼100-fold lower than those typically used. The high specificity and sensitivity of our approach revealed that 7,8-dihydro-8-oxoguanine (8-oxoG) at an incidence of approximately three lesions per megabase is preferentially repaired in the transcribed strand of the ATM gene. We have also demonstrated that the hOGG1, XPA, CSB and UVSSA proteins, as well as actively elongating RNA polymerase II, are required for this process, suggesting cross-talk between DNA repair pathways.
INTRODUCTION
Oxidative damage to DNA has been implicated in multiple human pathologies (1–3). It arises from attack by reactive oxygen species (ROS), generated either endogenously by cellular metabolic processes or environmentally from oxidizing agents and ionizing radiation. Oxidatively damaged bases in DNA are typically removed through base excision repair (BER). DNA glycosylases initiate this pathway by recognizing and releasing the altered bases, and the DNA is then incised at the resulting abasic sites; repair replication and ligation restore the integrity of the DNA. One of the major lesions induced by ROS is 7,8-dihydro-8-oxoguanine (8-oxoG) (4), which has the propensity to mispair with adenine, causing G·C to T·A transversions (5). Incorporation of adenosine opposite 8-oxoG during transcription generates mutant transcripts, which could direct the synthesis of faulty proteins (6).
Transcription is crucial to cell function and survival. Therefore, although DNA damage may be distributed randomly throughout the genome, repair of transcriptionally active sequences could be more urgent than that in silent domains of the genome. Transcription-coupled repair (TCR), a sub-pathway of nucleotide excision repair (NER) that targets the transcribed strands of active genes, has been well characterized for bulky DNA photoproducts and adducts (7–9). The methodologies used to study TCR of these lesions include the Southern blot approach (7,8) and the ligation-mediated polymerase chain reaction (LM-PCR) (10). These techniques require high doses of DNA damaging agents to induce detectable numbers of lesions. Recently developed methods use PCR amplification to detect oxidative base damage in DNA with high sensitivity. However, they cannot distinguish the specific type of DNA damage induced by ROS (11), and they do not permit examination of repair at the strand-specific level (12).
ROS attacks a wide variety of cellular components, including DNA, RNA, proteins (13) and lipids (14), rendering cells much less tolerant of ROS than of short wavelength ultraviolet (UV) irradiation inducing equivalent numbers of DNA lesions, as the latter targets mainly nucleic acids. Furthermore, >20 different oxidized DNA lesions can be induced by ROS (15). Thus, to study repair of low, physiologically relevant levels of specific oxidized DNA damage, new methodologies with high sensitivity and specificity are needed. Although it has been suggested that oxidation damage in DNA is repaired in a transcription-dependent manner (11,12,16), direct and reproducible evidence of preferential repair of specific oxidized DNA bases in the transcribed strands of active genes has been lacking.
We have developed an ultrasensitive approach combining single-cell gel electrophoresis (comet) with fluorescence in situ hybridization (FISH) using strand-specific probes, to facilitate the quantification of low, physiologically relevant levels of specific DNA lesions in each strand of defined DNA sequences, for comparison with that in the genome overall. In this approach, cells were exposed to UV irradiation or treated with potassium bromate to generate cyclobutane pyrimidine dimers (CPD) or 8-oxoG in DNA, respectively. After incubation in cell culture medium for various periods of time to allow repair, cells were mixed with agarose and layered on microscope slides. Incubation of UV-damaged cells with T4 endonuclease V (17) or oxidatively damaged cells with human 8-oxoG DNA glycosylase (hOGG1) (18) generated single-strand breaks specifically at the respective lesion sites. Upon electrophoresis under alkaline conditions, DNA containing single-strand breaks becomes unwound and migrates away from the nucleus, the head of the comet, to form the tail. The percentage of DNA in the comet tail reflects the frequency of strand breaks and is used to quantify global genomic repair (GGR). A schematic representation of the assay is shown in Figure 1a.
Figure 1.
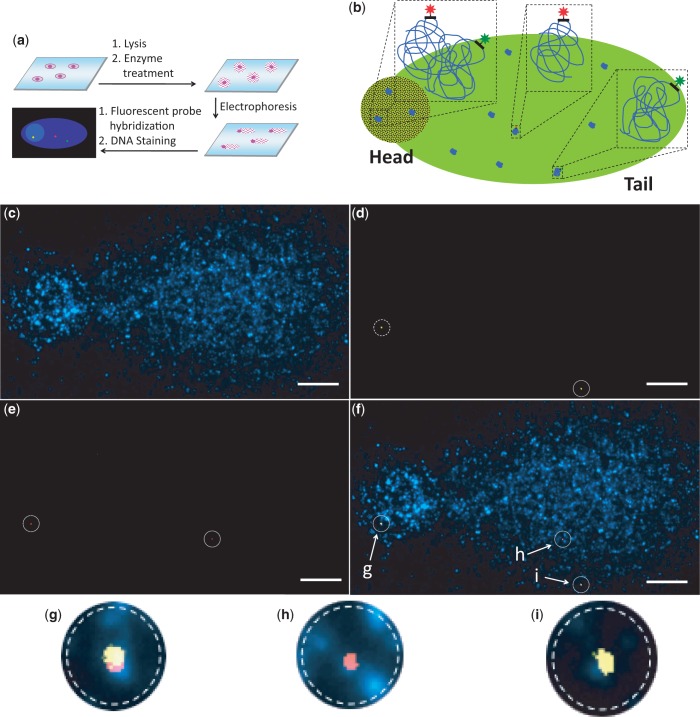
Comet-FISH with strand-specific probes. (a) After DNA-damaging treatment, cells are lysed, incubated with endonucleases or glycosylases and subjected to electrophoresis. Hybridization of strand-specific probes to the termini of the DNA segments of interest permits the quantification of TCR; staining the bulk of the DNA allows the analysis of GGR. (b) Schematic representation of comet-FISH. Adjacent green and red probe signals indicate an intact DNA strand with no lesions within the segment; separated green and red probe signals suggest a damaged DNA strand with a lesion within the segment. Representative comet-FISH images of a cell damaged with UV showing (c) the bulk DNA stained with DAPI, (d) Alexa 488-labeled probes targeting the 3′ regions of the transcribed ATM strands, (e) Alexa 594-labeled probes targeting the 5′ regions of the transcribed ATM strands and (f) an overlay of Figure 1c, d and e (scale bars, 5 µm); (g), (h) and (i) show enlargements of the probes signals from panel f.
We have previously used the comet-FISH approach with FISH signals localized in the comet head or tail to represent intact or damaged DNA segments, respectively (19). However, lesions induced in a region adjoining the undamaged DNA segment of interest could also result in FISH signals appearing in the comet tail. In addition, damaged DNA segments near nuclear matrix-associated regions have limited mobility during electrophoresis, which may lead to their FISH signals being localized in the comet head (20). To address these issues, we designed and synthesized single-stranded green and red fluorescent probes that hybridize specifically to the respective termini of the ataxia telangiectasia-mutated (ATM) gene. ATM is a protein kinase that phosphorylates several key proteins to activate DNA-damage checkpoints (21). The choice of the ATM gene to study TCR of low levels of lesions was based on the following criteria: this gene is highly expressed in human fibroblasts throughout the cell cycle (22), suggesting that the template strand of the ATM gene is actively transcribed; the ‘non-transcribed’ strand of the ATM gene is not used as the template for transcription of other genes; and because of the large size of the 146-kb ATM gene, a significant percentage of the ATM strands can be damaged with very low doses of DNA-damaging agents.
The conformation of the DNA in comet tails has been the subject of much speculation. A comprehensive study by Shaposhnikov et al. (23) postulates that in neutral comets, the DNA is double-stranded and arranged in linear arrays in loops that extend from anchorage points in the nuclear matrix, whereas in alkaline comets, the DNA is single-stranded and appears in globules that may be attached to the matrix or even to each other. These observations can be explained by the relative rigidity of double-stranded DNA, and the tendency of single-stranded DNA to acquire complex secondary structures on denaturation. Assuming that single-stranded DNA in the gel is a flexible polymer with 3D structures, the theoretical end-to-end distance of the ATM gene is <3 µm (24). This predicts that for intact DNA, the green and red FISH probes targeting the termini of the ATM gene should appear adjacent to each other when examined with ×100 magnification in a fluorescence microscope. However, when a lesion is induced within the gene and subsequently converted to a strand break, the resulting DNA fragments will migrate to different locations upon electrophoresis under denaturing conditions, as previously shown by Horváthová et al. (20). Consequently, the probes targeting the termini of the ATM gene should be separated, as shown in Figure 1b. Our experimental results confirmed this prediction. Thus, we concluded that the ATM strand contained a lesion when the distance between the green and red probes was >3 µm. In the representative images showing the bulk of the DNA (Figure 1c), Alexa 488- (Figure 1d) and Alexa 594- (Figure 1e) labeled probes hybridized to the respective ends of the ATM gene, and an overlay of the DNA and probes images (Figure 1f), the adjacent green and red signals in the comet head indicate an intact ATM strand, whereas the separated green and red signals in the comet tail document a damaged ATM strand.
We have validated this method by demonstrating TCR of CPD in the ATM gene in human fibroblasts irradiated with 0.1 J/m2 of UV, a dose ∼100-fold lower than those typically used in previous studies. We have documented that the CSB protein and actively transcribing RNA polymerase II (RNAP II) are required for TCR of low levels of CPD, and that the XPC protein is indispensable for repair of low levels of CPD in transcriptionally silent sequences, confirming previous results obtained with much higher levels of damage (9). Of major importance, we have documented preferential repair of correspondingly low levels of 8-oxoG in the transcribed strand of the ATM gene in wild-type human fibroblasts and have shown that this requires actively transcribing RNAP II, and the TCR-specific factors CSB and UVSSA. Moreover, we show that hOGG1 and XPA are essential for TCR of 8-oxoG, suggesting the participation of both BER and NER in this process.
MATERIALS AND METHODS
Chemicals were purchased from Sigma-Aldrich, and bioreagents were acquired from Invitrogen unless indicated otherwise. Biotinylated forward primers synthesized with 1-dimethoxytrityloxy-2-(N-biotinyl-4-aminobutyl)-propyl-3-O-(2-cyanoethyl)-(N,N-diisopropyl)-phosphoramidite and natural reverse primers were prepared by the Stanford University Protein and Nucleic Acid Facility. Absorbance spectra were obtained on a Nanodrop 2000 spectrophotometer (Thermo Scientific).
Synthesis of strand-specific probes for comet-FISH
To amplify the fragments of interest, bacterial artificial chromosome (BAC) clone RP11-455M10 (BACPAC) containing the ATM gene was used as the template in PCR (30 cycles at 94°C for 1 min, 55°C for 1 min, 72°C for 1 min, followed by the last extension at 72°C for 10 min). PCR was carried out on a GeneAmp PCR System 2400 (Perkin Elmer) with 320 ng of BAC clone RP11-455M10, 400 pmol primers, 20 nmol each dATP, dCTP and dGTP, 6.7 nmol dTTP, 13.3 nmol aminoallyl-dUTP, 2.5 U Taq polymerase and 10 µl 10× CoralLoad PCR buffer (Qiagen) in a total volume of 100 µl. Primers 1–44 and 45–88 with sequences as shown in Supplementary Table S1 were used for amplification of the 3′ region and the 5′ region of the ATM gene, respectively. Primers were selected using Gene Fisher, http://bibiserv.techfak.uni-bielefeld.de/genefisher2/). The PCR products were purified using a PCR purification kit (Qiagen). Subsequently, 8 µg of combined PCR products were incubated with 40 µg of streptavidin-coated beads at room temperature for 30 min. The strand without biotin was isolated from beads by incubation with 20 µl of 0.1 M NaOH at room temperature for 10 min. The strand with biotin was obtained by denaturing streptavidin with a solution containing 10 mM ethylenediaminetetraacetic acid (EDTA) and 95% formamide at 90°C for 3 min. Both strands were purified with a nucleotide removal kit (Qiagen). Equal amounts of the two complementary strands were mixed in hybridization buffer (10 mM Tris, 50 mM NaCl and 1 mM EDTA, pH 7.5), heated to 95°C for 5 min and allowed to slowly cool to room temperature to anneal. The PCR product, separated single-stranded DNA, and annealed double-stranded DNA were characterized by 1.5% standard agarose gel electrophoresis.
To fluorescently label the probes, single-stranded PCR products targeting the 3′ or 5′ region of the ATM TS or NTS were combined. The combined probes (1 µg) were mixed with 3 µmol NaHCO3, 20 µg Alexa fluorophore and 2 µl DMSO in a total volume of 10 µl and incubated at room temperature for 2 h. The fluorescently labeled probes were subsequently purified with a nucleotide removal kit and characterized by electrophoresis in 1.5% standard agarose gels. The labeling number of the generated fluorescent probes was determined by the equation:
NL is the number of fluorophore moieties per 100 bases. Abase is absorption of fluorescently labeled probes at 260 nm. Adye is absorption of fluorescently labeled probes at the maximum absorption wavelength of the fluorophore (Alexa 488: 492 nm, Alexa 594: 588 nm). εbase and εdye are the molar extinction coefficient of the base and the fluorophore, respectively (single-stranded DNA: 8910 cm−1M−1, Alexa 488: 62 000 cm−1M−1, Alexa 594: 80 400 cm−1M−1). α is defined as the ratio of absorption of the fluorophore at 260 nm and at the maximum absorption wavelength (Alexa 488: 0.30, Alexa 594: 0.43).
Cell culture and DNA-damaging treatment
GM00037F (wild-type) and NER-deficient AG06971 [xeroderma pigmentosum complementation group A (XP-A, defective in GGR and TCR)], GM00739 [Cockayne syndrome complementation group B (CS-B, defective in TCR)] and GM16684 [xeroderma pigmentosum complementation group C (XP-C, defective in GGR)] cell lines were obtained from NIGMS repository; TCR-defective Kps3 UV-sensitive syndrome (UVSS) primary skin fibroblasts, complementation group UVSS-A, were a gift from T. Itoh. hOGG1 knockdown cells (5504-001-01) were obtained from Trevigen. Cells were cultured in MEM supplemented with 10% fetal bovine serum, 2 mM glutamine and antibiotic/antimycotic solution. To generate UV-induced DNA damage, cells were washed twice with phosphate-buffered saline (PBS), irradiated with a Westinghouse IL782-30 germicidal lamp at 254 nm at a dose rate of 0.015 J/m2/s for 7 s; the cells were then incubated in culture medium for specified repair times. To induce oxidation damage in DNA, cells were washed twice with Hank’s buffered salt solution and incubated in Hanks’ buffered salt solution with 50 mM KBrO3 and 200 µM glutathione at room temperature for 1 h. Subsequently, cells were washed twice with PBS and incubated in culture medium for specified repair times. For experiments involving α-amanitin, cells were pre-incubated in culture medium with 10 µg/ml α-amanitin for 90 min. After inducing DNA damage, cells were further incubated in culture medium with 10 µg/ml α-amanitin for specified repair times.
Comet-FISH with strand-specific probes
Microscope slides (VWR) were coated with 1% agarose and stored at room temperature for at least 2 weeks. Cells were harvested, washed with PBS and suspended in PBS at 2 × 105 cells/ml. Eighty-five microliters of the mixture containing a 1:1 ratio of cell solution and 1.2% low-melting point agarose was added onto each agarose-coated slide. The slides were covered with cover slips and placed at 4°C for 30 min. After the cover slips were removed, the slides were incubated in lysis solution [2.5 M NaCl, 100 mM EDTA, 10 mM Tris–HCl, pH 10, with 10% dimethyl sulfoxide (DMSO) and 1% Triton X-100 added just before use] at 4°C overnight. For UV-treated cells, the slides were washed three times with 1× T4 endonuclease V reaction buffer (100 mM NaCl, 10 mM EDTA and 10 mM Tris, pH 8.0) at 4°C for 5 min and then incubated with a 1:500 dilution of T4 endonuclease V (a gift from S. Lloyd) in 1× reaction buffer at 37°C for 45 min in a humidified chamber. For KBrO3-treated cells, the slides were washed three times with 1× hOGG1 reaction buffer [1 mM EDTA, 1 mM dithiothreitol (DTT), 100 µg/ml bovine serum albumin and 20 mM Tris–HCl, pH 8.0] at 4°C for 5 min and subsequently incubated with hOGG1 (Trevigen) diluted 1:1000 in reaction buffer at 37°C for 45 min in a humidified chamber. After enzyme treatment, the slides were placed in a gel electrophoresis tank and incubated with cold electrophoresis buffer (300 mM NaOH and 1 mM Na2EDTA, pH > 13) for 40 min. Electrophoresis was conducted at 24 V and 300 mA for 30 min. After washing three times with neutralization buffer (0.4 M Tris, pH 7.5) for 5 min each at 4°C, the slides were incubated with 100% ethanol at 4°C for 30 min and with 0.5 M NaOH at room temperature for 25 min. The slides were dehydrated with 70, 85 and 95% ethanol in water for 5 min each at room temperature and left to air-dry.
To perform FISH, 10 µl of probe mixture [20 ng fluorescent probes, 10 µg Cot-I DNA, 10% dextran sulfate, 50% formamide and 2× saline-sodium citrate buffer (300 mM NaCl and 30 mM sodium citrate, pH 7.0)] was heated at 73°C for 5 min and incubated at 37°C for 20 min. Then, the prepared comet slides were incubated with the probe mixture in a humidified chamber at 37°C overnight. After washing twice with 50% formamide, 2× saline–sodium phosphate EDTA (SSPE) buffer (300 mM NaCl, 20 mM NaH2PO4 and 2 mM EDTA at pH 7.4) at 37°C for 10 min, once with 2× SSPE at 37°C for 10 min and once with 1× SSPE at room temperature for 5 min, the slides were incubated with 1 μg/ml 4′,6-diamino-2-phenylindole (DAPI) in 1× PBS at room temperature for 15 min. Subsequently, 20 µl of Prolong Gold antifade reagent was added to the air-dried slides, which were then covered with cover slips and stored at 4°C in the dark.
Comet-FISH analysis
The slides were imaged using an epifluorescence microscope (Nikon Eclipse 80i) through a ×100 objective. Images were captured with a Clara interline CCD camera (Andor) and NIS-Elements BR 3.10 imaging software. Chroma filters 49 000, 49 002 and 49 008 were used for DAPI, Alexa 488 and Alexa 594, respectively. GGR analysis was performed on 30 cells randomly selected at the center of each slide by using ImageJ software to acquire the density of DAPI stain and calculating the per cent of DNA in the comet tails, after subtracting the background fluorescence from a neighboring dark area of equivalent size. To study strand-specific repair of the ATM gene, 30 randomly selected cells were analyzed, and three independent experiments were conducted for each data point. Two sets of slides were processed in parallel and hybridized with probes for the TS or the NTS. Separated green and red spots with a distance of >3 µm were judged to indicate damaged DNA strands; adjacent green and red spots or those separated by <3 µm were considered intact DNA strands.
RESULTS
Design and synthesis of strand-specific probes for comet-FISH
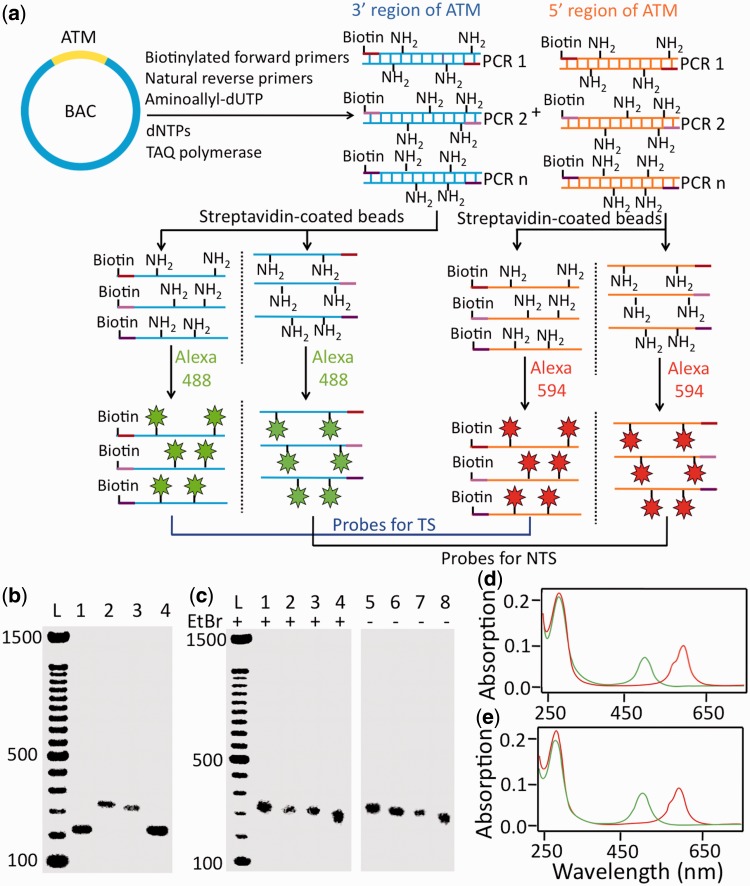
To study TCR of DNA lesions using the comet-FISH approach, strand-specific FISH probes are required to distinguish damage in the transcribed strand (TS) and the non-transcribed strand (NTS). Additionally, the probes should be ∼250 bases in length to achieve optimal gel penetration and ideal hybridization efficiency and specificity (25). To generate strand-specific probes with optimal length, we have developed a PCR-based approach to prepare fluorescent single-stranded DNA probes (Figure 2a) targeting the 10-kb regions at the 3′ and 5′ termini of the ATM gene. The probes were synthesized by using aminoallyl-dUTP, biotinylated forward primers and natural reverse primers to amplify the DNA segments of interest by PCR. To separate the two strands of PCR products, biotinylated PCR products were immobilized on streptavidin-coated beads. Under alkaline conditions, the strand without biotin was separated from the strand with biotin and released from the beads. Streptavidin was then denatured to release the strand with biotin. The resulting single-stranded probes, with or without biotin, exhibited reduced mobility in agarose gels compared with the double-stranded PCR products (Figure 2b), indicating that the two DNA strands have been successfully separated. This is further confirmed by the fact that the annealed product of the complementary single strands had the same mobility as the original double-stranded DNA (Figure 2b). Subsequently, we labeled the single-stranded probes targeting the 3′ and 5′ regions of the ATM gene with Alexa 488 and Alexa 594, respectively (Figure 2a). The amino groups on the PCR products generated by incorporation of aminoallyl-dUTP were coupled with N-hydroxysuccinimide (NHS) ester-modified fluorophores. The fluorescently labeled probes were purified and analyzed by gel electrophoresis. The reduced mobility of these probes (Figure 2c) compared with that of the double-stranded PCR products (Figure 2b) indicates that the probes are single-stranded. Co-localization of fluorophores and DNA on the agarose gel (Figure 2c) confirms that fluorophores have been successfully conjugated to the single-stranded probes. We also characterized the fluorescently labeled probes by absorption spectroscopy (Figure 2d and e). The relative peak height data further confirms that Alexa fluorophores are successfully coupled to single-stranded PCR products. From the absorption spectra, we calculated that about six Alexa fluorophores (Supplementary Table S2) were conjugated per 100 bases. This labeling number is in the optimal range to maximize the fluorescent signal from each probe molecule and to avoid self-quenching from fluorophores in close proximity (26). These single-stranded probes with optimal length and labeling number satisfy the key requirements necessary for applying the comet-FISH approach to study TCR of DNA lesions.
Figure 2.
Design and synthesis of strand-specific fluorescent probes for comet-FISH. (a) The 3′ and 5′ regions of the ATM gene are amplified by PCR using biotinylated forward primers, natural reverse primers and aminoallyl-dUTP. Subsequently, the biotinylated strands and the non-biotinylated strands of the PCR products are separated by streptavidin-coated beads. The probes targeting the 3′ and 5′ regions of the ATM gene are labeled with Alexa 488 and Alexa 594, respectively. Finally, all the biotinylated probes are combined as probes for the ATM TS, whereas all the non-biotinylated probes are mixed as probes for the ATM NTS. (b) Agarose gel containing the double-stranded PCR product (lane 1), single-stranded DNA with biotin (lane 2), single-stranded DNA without biotin (lane 3) and annealed double-stranded DNA (lane 4). (c) Agarose gel with fluorescently labeled strand-specific probes stained with ethidium bromide (left) or unstained (right). Probes for the 3′ region of the ATM TS (lanes 1 and 5), probes for the 3′ region of the ATM NTS (lanes 2 and 6), probes for the 5′ region of ATM TS (lanes 3 and 7), probes for the 5′ region of the ATM NTS (lanes 4 and 8). (d) Absorption spectra of probes targeting the 3′ region (green) and the 5′ region (red) of the ATM TS. (e) Absorption spectra of probes targeting the 3′ region (green) and the 5′ region (red) of the ATM NTS. The peaks centered at 260, 492 and 588 nm correspond to DNA, Alexa 488 and Alexa 594, respectively.
Requirement for CSB and actively transcribing RNAP II in TCR of low levels of CPD
TCR is a sub-pathway of NER that specifically removes transcription-blocking lesions in the template DNA strands of actively transcribing genes, but not in other regions of the genome, including those in the non-transcribed strands of active genes. It has been established that CSB and transcribing RNAP II are indispensable for TCR of CPD induced by UV irradiation at 10–20 J/m2, whereas XPC is required for repair of these high levels of CPD in transcriptionally silent sequences (9). Those earlier results were confirmed when we used our comet-FISH methodology to investigate TCR and GGR of CPD induced with 0.1 J/m2 of UV irradiation in wild-type, CSB and XPC cells. In most of the comets, we observed two sets of adjacent green and red signals (Supplementary Figure S1) or one set of adjacent signals and one set of separated signals (Figure 1c–f), indicating no damage or one damaged ATM strand, respectively. Two sets of well-separated green and red signals, resulting from damage in both ATM strands, were observed in 5–10% of the cells (Supplementary Figure S2).
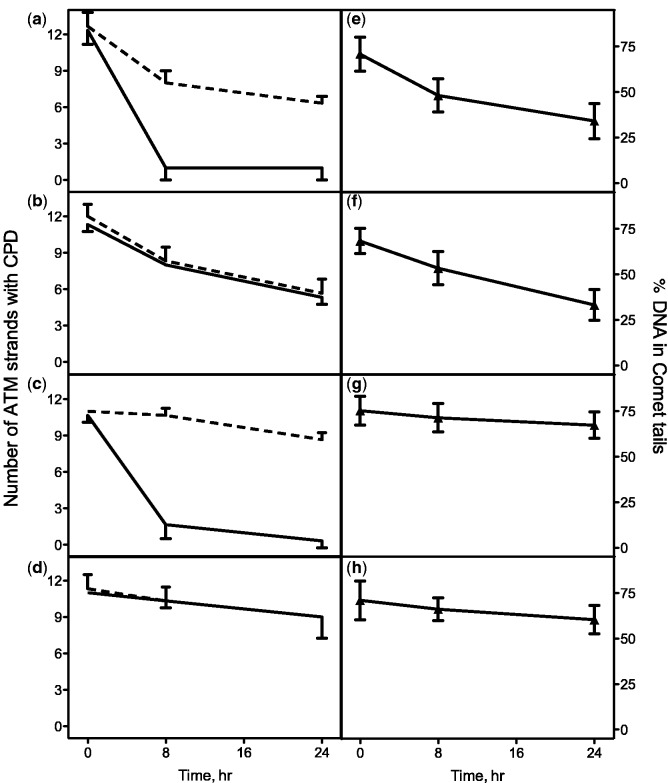
The number of strand breaks at CPD sites within the TS and the NTS of the ATM gene in 30 wild-type human fibroblasts was analyzed in triplicate experiments at 0, 8 and 24 h after UV irradiation (Figure 3a). In these experiments, ∼12 of the TS and NTS were damaged initially or 17% of ∼72 ATM strands (∼80% of cells in G0/G1 phase and ∼20% of cells in S and G2/M phases), indicating that the induction of lesions was similar in both strands, and that the likelihood of inducing more than one CPD within a single ATM strand is limited at this dose. Therefore, assuming that the majority of damaged strands contain only one CPD, we estimated that ∼1.3 CPD were induced in 106 bases with 0.1 J/m2 of UV irradiation. These results are consistent with those calculated by other methods (27,28). Within 8 h, most of the damaged TS were repaired, whereas only ∼4 NTS of 12 had been repaired. Even after 24 h, half of the damaged NTS remained unrepaired. These results indicate that TCR of low levels of CPD can be successfully quantified (P < 0.01) using the comet-FISH approach. On the other hand, the TS and NTS of CS-B cells were repaired at similar rates (Figure 3b), which closely resemble the repair rate of the NTS in wild-type cells. These results confirm that CSB is required for TCR of low levels of CPD. The repair kinetics of the TS in XPC cells (Figure 3c) closely resemble that in wild-type cells, confirming that XPC is not required for TCR of CPD. Repair of CPD in the NTS in XP-C cells was negligible, demonstrating that XPC is necessary for removal of CPD in transcriptionally silent sequences. When we used α-amanitin to inhibit RNAP II in XP-C cells, we observed that both the TS and NTS were unrepaired (Figure 3d), resembling the repair rate of the NTS in XP-C cells without α-amanitin treatment. These results indicate that actively transcribing RNAP II is required for TCR of CPD.
Figure 3.
Requirement for CSB and actively transcribing RNAP II for TCR of low levels of CPD. Left panels: 24 h time course of repair of CPD in the TS (solid lines) and NTS (dashed lines) of the ATM gene in (a) GM00037F (wild-type) cells, (b) GM00739 (CSB) cells, (c) GM16684 (XPC) cells and (d) α-amanitin-treated XPC cells. The results shown represent the numbers of ATM strands with CPD per 30 cells. The averages and standard deviations were obtained from three independent experiments. Right panels: 24 h time course of repair of CPD in the genome overall in (e) wild-type cells, (f) CSB cells, (g) XPC cells and (h) α-amanitin-treated XPC cells. The results shown represent the percentages of DNA in comet tails. The averages and standard deviations are calculated from 30 cells treated with T4 endonuclease V.
GGR of low levels of CPD was also quantified. In control experiments without T4 endonuclease V treatment, most of the DNA remained in comet heads (Supplementary Table S3), confirming that the specific lesion analyzed in this study is CPD. In wild-type (Figure 3e) and CSB cells (Figure 3f), the lesions in the genome overall were repaired at similar rates, showing that CSB is not required for repair of low levels of CPD in transcriptionally silent sequences. No GGR was detected in XPC cells (Figure 3g) or α-amanitin-treated XPC cells (Figure 3h), indicating that XPC is indispensable for the repair of low levels of CPD in transcriptionally silent sequences. In all experiments, the repair rates for the NTS closely resemble those in the genome overall, implying that low levels of CPD in the NTS of active genes are repaired in a similar manner to those in other transcriptionally silent sequences, as previously shown for much higher levels of CPD (9).
Requirement for CSB, UVSSA and actively transcribing RNAP II for preferential repair of 8-oxoG in the TS of ATM
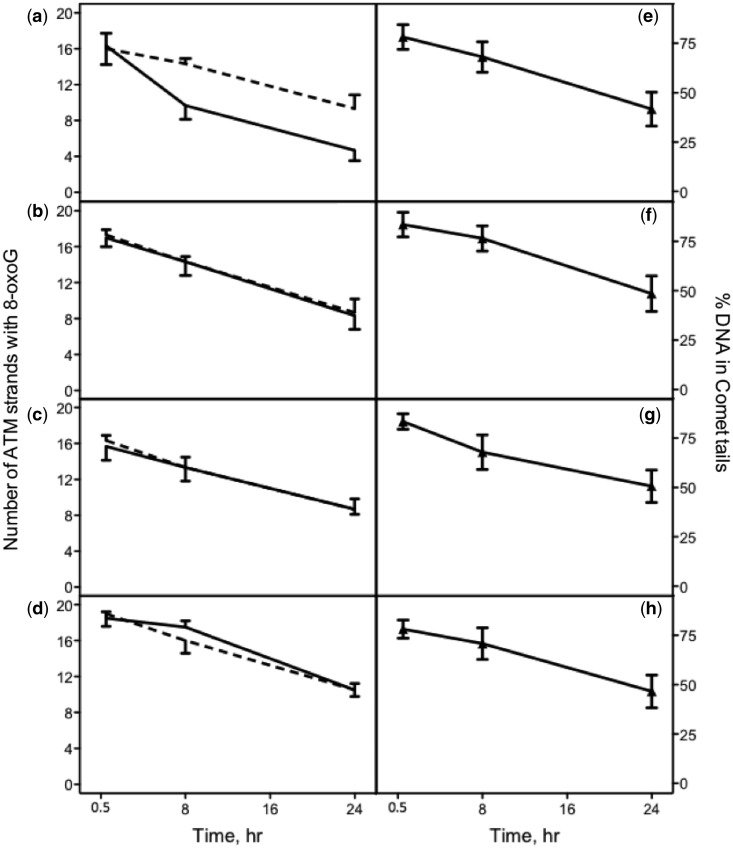
After validating the comet-FISH methodology by confirming TCR of low levels of CPD, we used the same approach to measure repair of 8-oxoG in the respective strands of the ATM gene and in the genome overall to determine whether 8-oxoG in the TS of actively transcribing genes is preferentially removed compared with those in transcriptionally silent sequences. To induce 8-oxoG and reduce endogenously generated ROS (29), we incubated wild-type cells with 50 mM potassium bromate and 200 µM glutathione at room temperature for 1 h. The number of DNA strands from the ATM gene with breaks induced by incision at 8-oxoG sites by hOGG1 in 30 wild-type cells was analyzed at 0.5, 8 and 24 h after removing the drugs (Figure 4a). In these experiments, ∼16 TS or NTS in ∼72 ATM strands were damaged 0.5 h after the KBrO3 treatment, indicating an even distribution of oxidized damage between the two strands. We calculated that ∼1.7 8-oxoG were detected in 106 bases under our conditions. After 8 and 24 h of repair, ∼7 and ∼12 damaged TS were repaired, whereas only ∼2 and ∼7 NTS were repaired, respectively. These results provide direct evidence that 8-oxoG in the TS of an actively transcribing gene are preferentially repaired compared with those in transcriptionally silent sequences.
Figure 4.
CSB, UVSSA and actively transcribing RNAP II are required for preferential repair of 8-oxoG in ATM. Left panels: 24 h time course of repair of 8-oxoG in the TS (solid lines) and NTS (dashed lines) of the ATM gene in (a) GM00037F (wild-type) cells, (b) GM00739 (CSB) cells, (c) Kps3 UVSS cells and (d) α-amanitin-treated wild-type cells. The results shown represent the average number of ATM strands with 8-oxoG per 30 cells. The averages and standard deviations were obtained from three independent experiments. Right panels: 24 h time course of repair of 8-oxoG in the genome overall in (e) wild-type cells, (f) CSB cells, (g) UVSS cells and (h) α-amanitin-treated wild-type cells. The results shown represent the percentages of DNA in comet tails. The averages and standard deviations were calculated from 30 cells treated with hOGG1.
To investigate whether CSB, UVSSA and actively transcribing RNAP II are required for preferential repair of 8-oxoG in the TS, we induced 8-oxoG in CS-B, UVSS-A and α-amanitin-treated wild-type cells, followed by quantification of repair in the respective strands of the ATM gene. The TS and NTS in CS-B cells (Figure 4b), UVSS-A cells (Figure 4c) and α-amanitin-treated wild-type cells (Figure 4d) were repaired at similar rates, which closely resemble the repair rate of the NTS in wild-type cells without the α-amanitin treatment (Figure 4a dashed line). These results indicate that CSB, UVSSA and actively transcribing RNAP II are required for preferential repair of 8-oxoG in the TS of active genes.
We also examined GGR of 8-oxoG. Without hOGG1 treatment, nearly 99% of the DNA remained in comet heads in cells after 30 min of repair (Supplementary Table S4), indicating that at that time, few strand breaks or alkali-labile sites were present in wild-type, CS-B and UVSS-A cells, and that the specific lesion analyzed in this study is 8-oxoG. Although hOGG1 also recognizes 2,6-diamino-4-hydroxy-5-formamidopyrimidine lesions, treatment with potassium bromate does not generate significant levels of these lesions (30). In wild-type cells (Figure 4e), CS-B cells (Figure 4f), UVSS-A cells (Figure 4g) and α-amanitin-treated wild-type cells (Figure 4h), the lesions in the genome overall were repaired at similar rates. In all the cells, the rates of removal of 8-oxoG from the NTS and from the genome overall were similar. Wild-type cells not treated with KBrO3 and incubated with hOGG1 had an average of 22.6 ± 10.9% DNA in comet tails, representing endogenously induced hOGG1-sensitive sites in addition to the small contribution of frank strand breaks and alkaline-sensitive sites stated earlier in the text. Similar levels of endogenous damage have been published by other groups (20).
DNA repair pathways required for TCR of 8-oxoG
Oxidized bases in DNA are repaired through the BER pathway, which is initiated by specialized glycosylases that recognize subsets of lesions, removing them to create abasic (AP) sites. AP sites in turn are cleaved by AP endonuclease activities within the same glycosylases, or by apurinic apyrimidinic endonuclease 1 (APE1) in humans. hOGG1 is the only human glycosylase dedicated to remove 8-oxoG from double-stranded DNA. We examined the role of hOGG1 in TCR of 8-oxoG by performing comet-FISH experiments with potassium bromate-treated cells that had been transfected with a vector expressing anti-hOGG1 shRNA. The treatment time was reduced from 60 to 40 min to obtain a lower effective dose, so that not all DNA would migrate to comet tails and to minimize the number of ATM strands containing more than one 8-oxoG. The data clearly show that hOGG1 is required for both GGR and TCR of 8-oxoG in human cells (Table 1A). According to the manufacturer, the knocked down hOGG1 cells (KD) retain 32% hOGG1 activity when compared with control cells; this remaining activity might explain the small amount of repair of 8oxoG observed. As expected, the overall incidence of 8-oxoG in the KD cells was increased (P < 0.0001) compared with that in the wild-type cells; control KD cells treated with KBrO3 but not incubated with hOGG1 also exhibited increased amounts of DNA in comet tails, 8.7 versus 1.5% in wild-type cells (Supplementary Table S4). The apparently similar numbers of 8-oxoG in ATM 30 min after treatment in wild-type cells (16.3 ± 2.1 and 16.0 ± 1.7 in the TS and NTS, respectively) and KD hOGG1 cells (Table 1) resulted from the different duration of the treatment, 60 min for wild-type and 40 min for KD cells; assuming a linear correlation between exposure time and damage, KD hOGG1 cells exposed to KBrO3 for 60 min would sustain lesion frequencies of 25.5 and 25.1 in the TS and NTS strands of ATM, respectively.
Table 1.
Repair of 8-oxoG in KD hOGG1 and in XPA cells
| Time, h | % DNA in tails (GGR) | % Lesions remaining | Breaks in ATM |
|||
|---|---|---|---|---|---|---|
| TS | % Lesions remaining | NTS | % Lesions remaining | |||
| A. KD hOGG1 cells | ||||||
| 0.5 | 93.5 ± 2.4 | 100 | 17.0 ± 1.0 | 100 | 16.7 ± 0.6 | 100 |
| 8 | 87.6 ± 7.5 | 93.7 | 15.7 ± 1.5 | 92.4 | 14.7 ± 0.6 | 88 |
| 24 | 77.8 ± 7.9 | 83.2 | 12.3 ± 1.2 | 72.4 | 13.0 ± 1.7 | 78 |
| Control | 8.7 ± 2.7 | |||||
| B. XPA cells | ||||||
| 0.5 | 92.9 ± 2.8 | 100 | 20.3 ± 2.5 | 100 | 19.7 ± 1.5 | 100 |
| 8 | 85.7 ± 4.3 | 92.2 | 16.3 ± 1.2 | 80.3 | 17.0 ± 1.0 | 86.3 |
| 24 | 67.3 ± 10.8 | 72.4 | 11.0 ± 1.7 | 54.2 | 11.0 ± 1.0 | 55.8 |
| Control | 9.5 ± 3.3 | |||||
The percent lesions remaining were calculated in relation to those observed at the 0.5 h time point. Control cells were treated with potassium bromate but not digested with hOGG1.
Our finding that CSB and UVSSA are needed for TCR of 8-oxoG suggests the involvement of NER. The XPA protein is an absolute requirement for both GGR and TCR of bulky DNA adducts and modifications such as CPD. Primary skin fibroblasts from an XP-A patient were treated with potassium bromate for 1 h, and repair of 8-oxoG was analyzed by comet-FISH. The results, shown in Table 1B, indicate that XP-A cells were deficient in TCR of 8-oxoG. We found a higher incidence of hOGG1-sensitive lesions in XP-A cells at 0.5 h, both in the genome and in ATM strands. Parlanti et al. (31) reported variable levels of 8-oxoG measured in different wild-type and XP-A cell lines; thus, we cannot attribute our observation to a defect in XP-A. Global repair of the lesions exhibited a slower rate when compared with that in wild-type cells, which had 87.1 and 53.3% of the lesions remaining 8 and 24 h after treatment, respectively. Background levels of DNA strand breaks were also elevated in KBrO3-treated XP-A cells (Supplementary Table S4).
Time course of repair of 8-oxoG
The data shown in Figure 4 indicate that repair of 8-oxoG takes place with relatively slow kinetics, whereas several reports state rapid repair with nearly 50% of the lesions removed rapidly after being induced (11,32). The design of our experiments included a 30-min incubation of the cells after removal of potassium bromate, to allow repair of frank single-strand breaks, and thus reduce the background of DNA in comet tails. Although this approach yielded clean results with very low damage in control cells, it may have caused an underestimation of the initial amount of damage. Indeed, harvest of wild-type cells immediately after 60-min incubation with KBrO3 resulted in 26.0 ± 1.7 and 26.3 ± 3.5 hOGG1-induced breaks in the TS and NTS of ATM, respectively. These results suggest that ∼40% of the 8-oxoG are repaired within the first 30 min of post-treatment incubation and are in agreement with the dual kinetics of repair of 8-oxoG previously reported, with a fast early phase and a slower later phase. KBrO3-treated cells not incubated with hOGG1 exhibited a small amount of damage, 10.9 ± 5.3% of DNA in comet tails on average, reflecting frank breaks or alkaline-sensitive sites that are not substrates for hOGG1. As aforementioned, after 30 min of repair, this background was reduced to 1.5% DNA in comet tails.
DISCUSSION
We have designed and synthesized strand-specific fluorescent probes and used them in an improved, ultrasensitive comet-FISH approach to quantify TCR and GGR of low, physiologically relevant amount of specific DNA lesions. This approach has been validated by confirming that CSB and actively elongating RNAP II are required for TCR of low levels of UV-induced CPD in the TS of the ATM gene, and that XPC is indispensable for GGR of low levels of CPD.
We have then documented the preferential repair of 8-oxoG in the TS of an active gene and have shown that actively transcribing RNAP II is required for this process. We have also demonstrated that CSB, UVSSA, hOGG1 and XPA are indispensable for TCR of 8-oxoG. We ascertained that the apparent slow course of repair of 8-oxoG was due to underestimation of the initial number of lesions, caused by our experimental design in which we allowed the cells a 30-min period of repair immediately after treatment with KBrO3 to eliminate frank strand breaks from the background. In our conditions, a fast phase of repair of 8-oxoG throughout the genome occurs during the first half hour, such that we found no significant differences in the number of lesions in the TS and NTS of ATM at the 30-min time point. Preferential repair of lesions in the TS becomes evident at later times, when repair in the genome overall and in the NTS slow down (Figure 4). Moreover, what we call ‘initial damage’ is actually the result of a steady-state of lesions induced plus lesions repaired within the entire hour of treatment, thus the kinetics of repair we show here might miss a portion of the fast phase.
Although the characteristic sun sensitivity in CS and UVSS patients is due to the defect in TCR of UV-induced lesions, the molecular basis for the severe developmental and neurological deficiencies in CS individuals remains unexplained. CS cells exhibit elevated levels of ROS and oxidative base damage in DNA (33–35), and defective mitochondrial function leading to increased amounts of ROS has been documented in these cells (36,37). These observations support the hypothesis that excessive endogenous DNA damage and/or defects in the processing of such damage leads to cell death, thus depleting essential tissues and organs.
Individuals with UV-sensitive syndrome are sun-sensitive and exhibit pigmentation anomalies. In contrast with most other DNA repair-deficient diseases, UVSS patients do not present with any neurological or developmental abnormalities. UVSS cells are UV-sensitive, defective in recovery of RNA synthesis after UV-irradiation and defective in TCR of UV-induced CPD; these characteristics are in common with those of CS cells (38). Experiments to test the sensitivity to treatment with oxidants, and host cell reactivation assays with plasmids containing 8-oxoG and thymine glycol lesions indicated that unlike CS cells, UVSS cells have wild-type capability to process oxidative base lesions (16). Given these observations, we anticipated that UVSS cells would exhibit preferential repair of DNA oxidation lesions in the transcribed strand of ATM, and we were, therefore, surprised to find that TCR of 8-oxoG is just as defective as in CS-B cells. This result warrants reconsideration of our model. The wild-type resistance to oxidation and the efficient recovery of expression of an oxidatively damaged reporter gene in UVSS cells could be explained by efficient transcriptional bypass of oxidized bases in the template DNA, implying that this process could be defective in CS cells. Indeed, there is evidence for a role of CSB in stimulation of transcription past 8-oxoG and thymine glycol lesions (39,40); similar roles for cyclosporin A or UVSSA have not been investigated as far as we know.
Most 8-oxoG lesions are repaired through the BER pathway. Although BER can be stimulated by certain NER factors such as CSB and XPG (the protein defective or absent in xeroderma pigmentosum complementation group G patients), the pathway does not require factors associated with NER (41). Our model for TCR implies that a paused RNAP II acts as an antenna to recruit repair factors. Various laboratories using in vitro assays with different nucleotide sequence contexts, promoters and RNA polymerases have detected either no transcription arrest by 8-oxoG (42,43) or varying degrees of arrest (39). Therefore, it is possible that only a subset of 8-oxoG blocks RNAP II directly. However, intermediates in the BER pathway including abasic sites and strand breaks can block transcription (9,44,45) and consequently initiate TCR of 8-oxoG. BER can also occur when a glycosylase such as Neil2 incises 8-oxoG within the single-stranded bubble generated by transcription, as suggested in human cells (12). TCR of oxidized lesions through any of these scenarios could require a crossover from BER to NER, which to date has not been demonstrated. Alternatively, BER factors could be recruited to stalled RNAP, as proposed by Banerjee et al. (12). Our results showing that hOGG1 and XPA are essential for TCR of 8-oxoG suggest a hypothetical scenario, depicted in Figure 5, involving lesion-specific glycosylases detecting and removing damaged or abnormal bases in actively transcribed DNA strands; the resulting AP sites or the single-strand breaks generated by AP-endonucleases arrest RNAP, leading to recruitment of TCR factors and downstream steps in NER. Further studies will be needed to elucidate the roles of various repair enzymes and pathways in TCR of oxidized DNA lesions.
Figure 5.
Model for TCR of oxidized base damage, initiated by BER and resolved by NER. A damaged base in a DNA segment (1) is recognized by a glycosylase and converted to an abasic site (2). This intermediate product, or the subsequent single-strand break generated by an AP endonuclease (3), is repaired via BER unless an elongating RNAP encounters either structure. Upon transcription arrest and recruitment of TCR factors, the lesion is repaired via NER and transcription may resume.
The comet-FISH assay with strand-specific probes that we have developed has the following advantages: (i) the homogenous size of the probes, 250 bases, leads to optimal gel penetration, hybridization efficiency and specificity; (ii) single-stranded probes permit the analysis of repair rates in the complementary DNA strands in defined sequences; (iii) using multiple probes labeled with the desired number of fluorophores to target the 3′ or 5′ of the DNA segments, our approach eliminates the signal amplification steps used in previous comet-FISH approaches (20); (iv) detecting damaged DNA strands at the single-molecule level rather than needing to amplify them by PCR (11,12), our method avoids the background generated by different amplification efficiencies; and (v) only specific lesions are recognized as substrates by endonucleases or glycosylases, leading to the high specificity of our method.
The comet-FISH approach with single-strand probes can quantify various DNA lesions and DNA modifications at physiological levels, allowing the study of damage induction and removal in each DNA strand in defined sequences at the single-molecule level, and in the genome overall in individual cells. These DNA lesions and modifications include different lesions corrected by NER or BER pathways, DNA single-strand breaks and double-strand breaks generated at chromosomal fragile sites (46) or induced by ionizing radiation, 5-formylcytosine and 5-carboxylcytosine involved in active DNA demethylation through BER (47) and ribonucleotides in DNA removed by ribonuclease H (48). By using multispectral fluorescent tags with distinguishable emission signatures (49) to label the strand-specific probes, the comet-FISH approach could enable the analysis of DNA lesions in multiple designated regions in the genome in single cells. Coupled with a high-throughput comet assay (50), our approach could simultaneously quantify TCR and GGR under a variety of conditions including different cell types, repair times, damaging agents and inhibitors of repair enzymes. This methodology could have wide applications, especially in drug screening to develop inhibitors for specific repair pathways to be used in conjunction with DNA damaging agents for synthetic lethality-based chemotherapy regimens.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–4 and Supplementary Figures 1–2.
FUNDING
Funding for open access charge: National Institute of Environmental Health Sciences [ES018834 to P.C.H.].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of our research group for helpful discussion.
REFERENCES
- 1.David SS, O’Shea VL, Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hasty P, Campisi J, Hoeijmakers J, van Steeg H, Vijg J. Aging and genome maintenance: lessons from the mouse? Science. 2003;299:1355–1359. doi: 10.1126/science.1079161. [DOI] [PubMed] [Google Scholar]
- 3.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J. 2003;17:1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 4.Klungland A, Rosewell I, Hollenbach S, Larsen E, Daly G, Epe B, Seeberg E, Lindahl T, Barnes DE. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl Acad. Sci. USA. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grollman AP, Moriya M. Mutagenesis by 8-oxoguanine: an enemy within. Trends Genet. 1993;9:246–249. doi: 10.1016/0168-9525(93)90089-z. [DOI] [PubMed] [Google Scholar]
- 6.Saxowsky TT, Meadows KL, Klungland A, Doetsch PW. 8-Oxoguanine-mediated transcriptional mutagenesis causes Ras activation in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:18877–18882. doi: 10.1073/pnas.0806464105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bohr VA, Smith CA, Okumoto DS, Hanawalt PC. DNA repair in an active gene: removal of pyrimidine dimers from the DHFR gene of CHO cells is much more efficient than in the genome overall. Cell. 1985;40:359–369. doi: 10.1016/0092-8674(85)90150-3. [DOI] [PubMed] [Google Scholar]
- 8.Mellon I, Spivak G, Hanawalt PC. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987;51:241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- 9.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nat. Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 10.Tornaletti S, Pfeifer GP. Slow repair of pyrimidine dimers at p53 mutation hotspots in skin cancer. Science. 1994;263:1436–1438. doi: 10.1126/science.8128225. [DOI] [PubMed] [Google Scholar]
- 11.Reis AM, Mills WK, Ramachandran I, Friedberg EC, Thompson D, Queimado L. Targeted detection of in vivo endogenous DNA base damage reveals preferential base excision repair in the transcribed strand. Nucleic Acids Res. 2012;40:206–219. doi: 10.1093/nar/gkr704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banerjee D, Mandal SM, Das A, Hegde ML, Das S, Bhakat KK, Boldogh I, Sarkar PS, Mitra S, Hazra TK. Preferential repair of oxidized base damage in the transcribed genes of mammalian cells. J. Biol. Chem. 2011;286:6006–6016. doi: 10.1074/jbc.M110.198796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cabiscol E, Piulats E, Echave P, Herrero E, Ros J. Oxidative stress promotes specific protein damage in Saccharomyces cerevisiae. J. Biol. Chem. 2000;275:27393–27398. doi: 10.1074/jbc.M003140200. [DOI] [PubMed] [Google Scholar]
- 14.Farmer EE, Mueller MJ. ROS-Mediated Lipid Peroxidation and RES-Activated Signaling. Annu. Rev. Plant. Biol. 2013;64:429–450. doi: 10.1146/annurev-arplant-050312-120132. [DOI] [PubMed] [Google Scholar]
- 15.Cadet J, Douki T, Gasparutto D, Ravanat JL. Oxidative damage to DNA: formation, measurement and biochemical features. Mutat. Res. 2003;531:5–23. doi: 10.1016/j.mrfmmm.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Spivak G, Hanawalt PC. Host cell reactivation of plasmids containing oxidative DNA lesions is defective in Cockayne syndrome but wild type in UV-sensitive syndrome fibroblasts. DNA Repair (Amst) 2006;5:13–22. doi: 10.1016/j.dnarep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 17.Sancar A, Rupp WD. A novel repair enzyme: UVRABC excision nuclease of Escherichia coli cuts a DNA strand on both sides of the damaged region. Cell. 1983;33:249–260. doi: 10.1016/0092-8674(83)90354-9. [DOI] [PubMed] [Google Scholar]
- 18.Radicella JP, Dherin C, Desmaze C, Fox MS, Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Spivak G, Cox RA, Hanawalt PC. New applications of the comet assay: comet-FISH and transcription-coupled DNA repair. Mutat. Res. 2009;681:44–50. doi: 10.1016/j.mrrev.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Horváthová E, Dusinská M, Shaposhnikov S, Collins AR. DNA damage and repair measured in different genomic regions using the comet assay with fluorescent in situ hybridization. Mutagenesis. 2004;19:269–276. doi: 10.1093/mutage/geh030. [DOI] [PubMed] [Google Scholar]
- 21.Canman CE, Lim DS, Cimprich KA, Taya Y, Tamai K, Sakaguchi K, Appella E, Kastan MB, Siliciano JD. Activation of the ATM kinase by ionizing radiation and phosphorylation of p53. Science. 1998;281:1677–1679. doi: 10.1126/science.281.5383.1677. [DOI] [PubMed] [Google Scholar]
- 22.Gately DP, Hittle JC, Chan GK, Yen TJ. Characterization of ATM expression, localization, and associated DNA-dependent protein kinase activity. Mol. Biol. Cell. 1998;9:2361–2374. doi: 10.1091/mbc.9.9.2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shaposhnikov SA, Salenko VB, Brunborg G, Nygren J, Collins AR. Single-cell gel electrophoresis (the comet assay): loops or fragments? Electrophoresis. 2008;29:3005–3012. doi: 10.1002/elps.200700921. [DOI] [PubMed] [Google Scholar]
- 24.Rechendorff K, Witz G, Adamcik J, Dietler G. Persistence length and scaling properties of single-stranded DNA adsorbed on modified graphite. J. Chem. Phys. 2009;131:095103. doi: 10.1063/1.3216111. [DOI] [PubMed] [Google Scholar]
- 25.Rapp A, Hausmann M, Greulich KO. The comet-FISH technique: a tool for detection of specific DNA damage and repair. Methods Mol. Biol. 2005;291:107–119. doi: 10.1385/1-59259-840-4:107. [DOI] [PubMed] [Google Scholar]
- 26.Levsky JM, Shenoy SM, Pezo RC, Singer RH. Single-cell gene expression profiling. Science. 2002;297:836–840. doi: 10.1126/science.1072241. [DOI] [PubMed] [Google Scholar]
- 27.Downes CS, Collins AR, Johnson RT. DNA damage in synchronized HeLa cells irradiated with ultraviolet. Biophys. J. 1979;25:129–150. doi: 10.1016/S0006-3495(79)85282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Douki T, Court M, Sauvaigo S, Odin F, Cadet J. Formation of the main UV-induced thymine dimeric lesions within isolated and cellular DNA as measured by high performance liquid chromatography-tandem mass spectrometry. J. Biol. Chem. 2000;275:11678–11685. doi: 10.1074/jbc.275.16.11678. [DOI] [PubMed] [Google Scholar]
- 29.Will O, Mahler HC, Arrigo AP, Epe B. Influence of glutathione levels and heat-shock on the steady-state levels of oxidative DNA base modifications in mammalian cells. Carcinogenesis. 1999;20:333–337. doi: 10.1093/carcin/20.2.333. [DOI] [PubMed] [Google Scholar]
- 30.Kasai H, Nishimura S, Kurokawa Y, Hayashi Y. Oral administration of the renal carcinogen, potassium bromate, specifically produces 8-hydroxydeoxyguanosine in rat target organ DNA. Carcinogenesis. 1987;8:1959–1961. doi: 10.1093/carcin/8.12.1959. [DOI] [PubMed] [Google Scholar]
- 31.Parlanti E, D'Errico M, Degan P, Calcagnile A, Zijno A, van der Pluijm I, van der Horst GT, Biard DS, Dogliotti E. The cross talk between pathways in the repair of 8-oxo-7,8-dihydroguanine in mouse and human cells. Free Radic. Biol. Med. 2012;53:2171–2177. doi: 10.1016/j.freeradbiomed.2012.08.593. [DOI] [PubMed] [Google Scholar]
- 32.Hollenbach S, Dhénaut A, Eckert I, Radicella JP, Epe B. Overexpression of Ogg1 in mammalian cells: effects on induced and spontaneous oxidative DNA damage and mutagenesis. Carcinogenesis. 1999;20:1863–1868. doi: 10.1093/carcin/20.9.1863. [DOI] [PubMed] [Google Scholar]
- 33.Tuo J, Müftüoglu M, Chen C, Jaruga P, Selzer RR, Jr, Brosh RM, Rodriguez H, Dizdaroglu M, Bohr VA. The Cockayne Syndrome group B gene product is involved in general genome base excision repair of 8-hydroxyguanine in DNA. J. Biol. Chem. 2001;276:45772–45779. doi: 10.1074/jbc.M107888200. [DOI] [PubMed] [Google Scholar]
- 34.Ropolo M, Degan P, Foresta M, D'Errico M, Lasigliè D, Dogliotti E, Casartelli G, Zupo S, Poggi A, Frosina G. Complementation of the oxidatively damaged DNA repair defect in Cockayne syndrome A and B cells by Escherichia coli formamidopyrimidine DNA glycosylase. Free Radic. Biol. Med. 2007;42:1807–1817. doi: 10.1016/j.freeradbiomed.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 35.D'Errico M, Parlanti E, Teson M, Degan P, Lemma T, Calcagnile A, Iavarone I, Jaruga P, Ropolo M, Pedrini AM, et al. The role of CSA in the response to oxidative DNA damage in human cells. Oncogene. 2007;26:4336–4343. doi: 10.1038/sj.onc.1210232. [DOI] [PubMed] [Google Scholar]
- 36.Scheibye-Knudsen M, Ramamoorthy M, Sykora P, Maynard S, Lin PC, Minor RK, III, Wilson DM, Cooper M, Spencer R, de Cabo R, et al. Cockayne syndrome group B protein prevents the accumulation of damaged mitochondria by promoting mitochondrial autophagy. J. Exp. Med. 2012;209:855–869. doi: 10.1084/jem.20111721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pascucci B, Lemma T, Iorio E, Giovannini S, Vaz B, Iavarone I, Calcagnile A, Narciso L, Degan P, Podo F, et al. An altered redox balance mediates the hypersensitivity of Cockayne syndrome primary fibroblasts to oxidative stress. Aging Cell. 2012;11:520–529. doi: 10.1111/j.1474-9726.2012.00815.x. [DOI] [PubMed] [Google Scholar]
- 38.Spivak G, Itoh T, Matsunaga T, Nikaido O, Hanawalt PC, Yamaizumi M. Ultraviolet-sensitive syndrome cells are defective in transcription-coupled repair of cyclobutane pyrimidine dimers. DNA Repair (Amst) 2002;1:629–643. doi: 10.1016/s1568-7864(02)00056-3. [DOI] [PubMed] [Google Scholar]
- 39.Pastoriza-Gallego M, Armier J, Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb-/Ogg1- DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 40.Charlet-Berguerand N, Feuerhahn S, Kong SE, Ziserman H, Conaway JW, Conaway R, Egly JM. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicholl ID, Nealon K, Kenny MK. Reconstitution of human base excision repair with purified proteins. Biochemistry. 1997;36:7557–7566. doi: 10.1021/bi962950w. [DOI] [PubMed] [Google Scholar]
- 42.Tornaletti S, Maeda LS, Kolodner RD, Hanawalt PC. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 43.Larsen E, Kwon K, Coin F, Egly JM, Klungland A. Transcription activities at 8-oxoG lesions in DNA. DNA Repair (Amst) 2004;3:1457–1468. doi: 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 44.Tornaletti S, Maeda LS, Hanawalt PC. Transcription arrest at an abasic site in the transcribed strand of template DNA. Chem. Res. Toxicol. 2006;19:1215–1220. doi: 10.1021/tx060103g. [DOI] [PubMed] [Google Scholar]
- 45.Kitsera N, Stathis D, Lühnsdorf B, Müller H, Carell T, Epe B, Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letessier A, Millot GA, Koundrioukoff S, Lachagès AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- 47.Nabel CS, Kohli RM. Demystifying DNA demethylation. Science. 2011;333:1229–12230. doi: 10.1126/science.1211917. [DOI] [PubMed] [Google Scholar]
- 48.Reijns MA, Rabe B, Rigby RE, Mill P, Astell KR, Lettice LA, Boyle S, Leitch A, Keighren M, Kilanowski F, et al. Enzymatic removal of ribonucleotides from DNA is essential for mammalian genome integrity and development. Cell. 2012;149:1008–1022. doi: 10.1016/j.cell.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Guo J, Wang S, Dai N, Teo YN, Kool ET. Multispectral labeling of antibodies with polyfluorophores on a DNA backbone and application in cellular imaging. Proc. Natl Acad. Sci. USA. 2011;108:3493–3498. doi: 10.1073/pnas.1017349108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wood DK, Weingeist DM, Bhatia SN, Engelward BP. Single cell trapping and DNA damage analysis using microwell arrays. Proc. Natl Acad. Sci. USA. 2010;107:10008–10013. doi: 10.1073/pnas.1004056107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.