Abstract
In metazoans, replication-dependent histone mRNAs end in a stem-loop structure instead of the poly(A) tail characteristic of all other mature mRNAs. This specialized 3′ end is bound by stem-loop binding protein (SLBP), a protein that participates in the nuclear export and translation of histone mRNAs. The translational activity of SLBP is mediated by interaction with SLIP1, a middle domain of initiation factor 4G (MIF4G)-like protein that connects to translation initiation. We determined the 2.5 Å resolution crystal structure of zebrafish SLIP1 bound to the translation–activation domain of SLBP and identified the determinants of the recognition. We discovered a SLIP1-binding motif (SBM) in two additional proteins: the translation initiation factor eIF3g and the mRNA-export factor DBP5. We confirmed the binding of SLIP1 to DBP5 and eIF3g by pull-down assays and determined the 3.25 Å resolution structure of SLIP1 bound to the DBP5 SBM. The SBM-binding and homodimerization residues of SLIP1 are conserved in the MIF4G domain of CBP80/20-dependent translation initiation factor (CTIF). The results suggest how the SLIP1 homodimer or a SLIP1–CTIF heterodimer can function as platforms to bridge SLBP with SBM-containing proteins involved in different steps of mRNA metabolism.
INTRODUCTION
The vast majority of eukaryotic mRNAs contain a string of adenosines at the 3′ end of the transcript, the so-called poly(A) tail. The poly(A) tail is added in the nucleus after transcription and is the binding site for the poly(A)-binding protein (PABP) [reviewed in (1)]. PABP is an integral part of the mature messenger ribonucleoprotein particle (mRNP) that is exported to the cytoplasm. In the cytoplasm, PABP interacts with the eukaryotic initiation factor 4G (eIF4G), which in turn is recruited to the 5′ end of the transcript (2,3). The network of interactions connecting the 3′ and 5′ ends of an mRNA promotes translation initiation and hampers degradation [reviewed in (4)]. Mammalian mRNAs transcribed from replication-dependent histone genes are the exception to the norm. Histones are massively produced during S-phase to assemble the newly synthesized genomic DNA into chromatin and their expression bypasses many of the conventional mechanisms used by other cellular mRNAs [reviewed in (5,6)]. One of the idiosyncratic features of these histone mRNAs is that they lack a poly(A) tail. Instead, they end with a stem-loop structure in the 3′ UTR that is the binding site for the stem-loop binding protein (SLBP, also known as hairpin-binding protein or HBP) (7,8). SLBP plays pivotal roles in the metabolism of histone mRNA, much in the same way as PABP for other cellular mRNAs [reviewed in (5,6)].
SLBP is synthesized just before the entry into S-phase (9) and is imported into the nucleus by the importin α-importin β transport factors (10). SLBP is believed to bind the stem-loop co-transcriptionally and to stabilize the interactions leading to 3′ end processing (11). The mature histone mRNP with bound SLBP is rapidly exported to the cytoplasm via the canonical mRNA transport factor TAP (10,12,13). In mammalian cells, SLBP appears to have an active role in the export process, as depletion of SLBP by RNA interference results in a substantial nuclear accumulation of fully processed histone mRNAs and consequently leads to cell-cycle arrest (14–16). In the cytoplasm, SLBP is required for efficient translation of histone mRNAs and protects them from degradation (17,18). SLBP is eventually phosphorylated and degraded by the proteasome at the end of the S-phase (19).
SLBP has a modular domain organization characterized by large regions of intrinsic disorder. The central region of SLBP folds on binding the histone RNA stem-loop, resulting in a tight interaction with nanomolar affinity (20,21). The central and the C-terminal regions of SLBP are involved in 3′ end processing (22). The N-terminal region contains a short segment of ∼15 residues that is essential for the activation of histone mRNA translation (18). The translation–activation segment of SLBP is located near the phosphorylation sites and is the binding site for a middle domain of initiation factor 4G (MIF4G)-like protein known as SLBP-binding protein 1 (SLIP1) or MIF4GD (23). SLIP1 cooperates with SLBP to activate histone mRNA translation (23) and interacts, either directly or indirectly, with the eIF3 translation initiation complex (17,24). In contrast to SLBP, SLIP1 is an essential protein in cultured mammalian cells (23). How the small compact fold of SLIP1 might harbor multiple functional sites to recruit different proteins and enhance translation is unclear. Also unknown is whether SLIP1 might have additional functions. In this work, we elucidated the determinants of the SLIP1–SLBP interaction and shed light on how SLIP1 can directly and concomitantly recruit SLBP together with proteins involved in translation initiation and nuclear export.
MATERIALS AND METHODS
Protein purification
Human and zebrafish full-length (f.l.) SLIP1 were subcloned as TEV-cleavable GST-tagged proteins and expressed at 18°C in Escherichia coli BL21(DE3) cells using auto-inducing medium (25). They were purified by affinity chromatography (GSH-Sepharose 4B, GE Healthcare) followed by cleavage of the GST tag by TEV protease and size-exclusion chromatography (Superdex 200, GE Healthcare). For pull-down assays, the GST tag was left uncleaved. A similar protocol was used to express and purify zebrafish SLBP residues 89–105 (SLBP89–105), zebrafish DBP51–32 and human DBP5 f.l. (residues 1–479), DBP5 ΔN (residues 68–479), DBP5 ΔNΔC (residues 68–302) and DBP51–51. Mutations were introduced into f.l. human DBP5 by the Quik-Change site-directed mutagenesis system (Stratagene) and expressed and purified as described above. Human and zebrafish SLIP1–SLBP and SLIP1–DBP5 complexes were reconstituted by mixing equimolar amounts of the purified proteins for 2 h at 4°C and further purified by size exclusion chromatography (in 20 mM Hepes, pH 7.5, 100 mM NaCl and 1 mM DTT). All samples were tested for monodispersity by coupling a Superdex 200 column to a static light scattering device (TDA302, Viscotech).
Crystallization and structure determination
The complex of Danio rerio (D.r.) SLIP1 and SLBP89–105 was concentrated to 16 mg ml−1 and subjected to crystallization experiments at 18°C using sitting-drop vapor diffusion. Crystals were obtained using a reservoir solution consisting of 0.17 M K/Na-tartrate, 0.085 M Na-citrate, pH 5.6, 1.7 M ammonium sulfate and 15% glycerol. The complex of D.r. SLIP1 and DBP51–32 crystallized at 18°C at a concentration of 20 mg ml−1 using a reservoir solution consisting of 0.05 M MES, pH 6.0, 18% (w/v) PEG 8000, 0.2 M Ca-Acetate. The crystals were cryoprotected with the addition of 15% (w/v) PEG 400 to the reservoir solution before data collection. Diffraction data were collected at 100 K at the Swiss Light Source synchrotron (SLS Villigen, Switzerland) and were processed with XDS (26). The crystals of D.r. SLIP1–SLBP89–105 belong to spacegroup P6522, contain one molecule per asymmetric unit and diffracted to 2.5 Å resolution (statistics in Table 1). The crystals of D.r. SLIP1–DBP51–32 belong to spacegroup P6122, contain two molecules per asymmetric unit and diffracted to 3.25 Å resolution (statistics in Table 1). Both structures were solved by molecular replacement with the program Phaser (27) using D.r. SLIP1 (PDB entry 2I2O) as a search model. The models were manually built using Coot (28) and refined using Refmac5 (29) and Phenix (30). The SLIP1–SLBP89–105 structure was refined to Rfree of 24.3%, Rwork of 19.2% and good stereochemistry (statistics in Table 1). In the case of the SLIP1–DBP51–32 structure, the two independent molecules in the asymmetric unit were refined using tight non-crystallographic symmetry restraints, which were released in the final stages of refinement. The final model of DBP51–32 has an Rfree of 27.2%, Rwork of 24.5% and more 98% of the residues in the most favored regions of the Ramachandran plot (statistics in Table 1).
Table 1.
Data collection and refinement statistics
| SLIP1–SLBP | SLIP1–DBP5 | |
|---|---|---|
| Data collection | ||
| Space group | P6522 | P6122 |
| Cell dimensions | ||
| a, b, c (Å) | 70.65, 70.65, 242.55 | 85.22, 85.22, 258.4 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Resolution (Å) | 43.07-2.50 (2.60-2.50) | 86.13-3.25 (3.45-3.25) |
| Rmeas | 11.4 (75.9) | 8.3 (68.2) |
| I/σI | 13.18 (2.01) | 16.34 (2.30) |
| Completeness (%) | 99.9 (99.9) | 99.8 (100) |
| Redundancy | 6.14 (6.1) | 4.5 (4.6) |
| Refinement | ||
| Resolution (Å) | 43.07 - 2.50 | 73.77 - 3.25 |
| No. reflections | 81081 | 75822 |
| Rwork/Rfree | 19.2/24.3 | 24.5/27.2 |
| No. atoms | ||
| Protein | 1955 | 3618 |
| Water | 62 | 15 |
| B-factors | ||
| Protein | 32 | 100 |
| Water | 27 | 81 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.0066 |
| Bond angles (°) | 1.11 | 0.93 |
| Ramachandran (%) | ||
| Outliers | 0 | 0 |
| Favored | 98.0 | 98.3 |
In vitro binding assays
RNA-binding experiments were performed as previously described (31) using 5′ end-biotinylated U20 ssRNA (Dharmacon). Briefly, 2 µg of DBP5 proteins was incubated in binding buffer in the presence or absence of 1 mM AMPPNP and 170 nM U20 RNA. Samples were incubated overnight at 4°C. Magnetic streptavidin beads (50 µg; Dynal) were added for 1 h. Beads were washed three times with 0.5 ml binding buffer. Samples were eluted in sodium dodecyl sulphate (SDS) loading buffer and were analyzed by SDS polyacrylamide gel electrophoresis (PAGE).
GST-pull-down experiments with purified proteins were carried out as described (31). About 2 µg of GST-tagged recombinant DBP5 proteins were immobilized on 20 µl of glutathione sepharose beads (GE Healthcare) per binding reaction. After a 2-h incubation, beads were washed three times with 1 ml binding buffer (20 mM Hepes, pH 7.5, 100 mM NaCl, 1 mM DTT, 10 mM MgCl2 supplemented with 0.1% (v/v) Nonidet P40). A 2-fold excess of SLIP1 proteins was added to a final volume of 100 µl binding buffer per reaction. After a 2-h incubation, beads were washed three times with 1 ml binding buffer. Bound proteins were eluted with SDS sample buffer and analyzed with SDS PAGE.
GST pull-down experiments with in vitro translated proteins were carried out as previously described (23) by labeling proteins with 35S-methionine in rabbit reticulocyte lysate and carrying out the pull-down reaction with different GST-SLIP1 fusion proteins.
Isothermal titration calorimetry (ITC) experiments were performed on a VP-ITC microcalorimeter from Microcal at 25°C equipped with a 300 ml syringe (Microcal, GE healthcare). SLIP1 and the different DBP5 constructs tested were dialyzed in the same gel filtration buffer overnight. We titrated 10 µM of SLIP1 in a cell volume of 1.44 ml with 100 µM of different DBP5 constructs in 28 injections of 10 µl volumes at 5 min intervals. The released heat was obtained by integrating the calorimetric output curves. The Kd values and binding ratios were calculated with the Origin5 software supplied with the instrument.
In vivo protein interactions
HeLa cells were stably transformed with an N-terminally tagged HA-DBP5 cloned in pcDNA3 and stable cells expressing the tagged DBP5 in similar amounts to the endogenous protein were selected. Immunoprecipitations were carried out as previously described (23), after precipitation with either anti-HA antibody to precipitate the tagged DBP5 or with anti-SLIP1 antibody to precipitated endogenous SLIP1. We were able to detect both the tagged DBP5 (with the anti-HA antibody) and the endogenous DBP5 when the immunoprecipitation was carried out with the anti-SLIP1 from untransfected cells.
RESULTS AND DISCUSSION
Structure determination of SLIP1 bound to the N-terminus of SLBP
Previous studies identified residues 68–83 of Xenopus SLBP1 as the translation activation segment that is essential for binding to SLIP1 (18,23). This region is intrinsically disordered with a propensity to form a transient helical structure (21,32). To obtain diffracting crystals of a SLIP1–SLBP complex, we took advantage of the evolutionary conservation of these proteins. SLIP1 is overall highly conserved, with the Xenopus, human and zebrafish orthologues sharing >70% sequence identity. The SLIP1-binding region of SLBP is also highly conserved in vertebrates.
The complex between zebrafish SLIP1 and a fragment of zebrafish SLBP encompassing residues 89–105 (SLBP89–105, corresponding to Xenopus 68–84) yielded crystals that diffracted to 2.5 Å resolution and contain one complex in the asymmetric unit. We determined the structure by molecular replacement using the SLIP1 coordinates deposited in the PDB (code 2I2O) from the Center for Eukaryotic Structural Genomics Consortium (Madison, Wisconsin). The final atomic model of the SLIP1–SLBP complex is refined to Rfree of 24.3%, Rwork of 19.2% and good stereochemistry (Table 1). The structure consists of residues 7–218 of SLIP1 and residues 89–105 of SLBP (with an additional N-terminal GPLGSM sequence from the expression vector that mediates crystal contacts with a neighboring SLIP1 molecule in the lattice). Comparison with the apo structure of SLIP1 indicates that binding of SLBP does not cause major conformational changes. More than 90% of the amino-acid residues superpose over all atom positions with a root mean square deviation (r.m.s.d.) of <0.6 Å.
SLIP1 is a non-canonical MIF4G-like protein
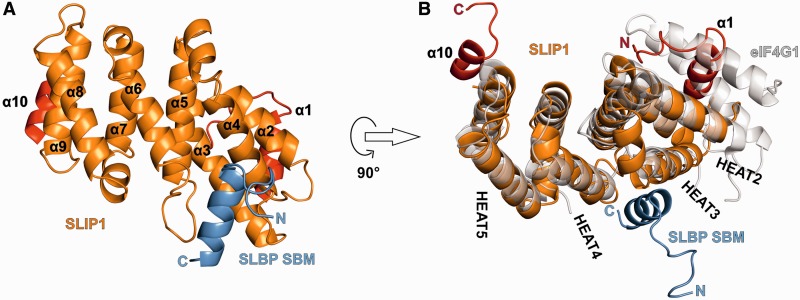
SLIP1 is an all α-helical protein (Figure 1A). The secondary structure includes 10 α-helices (α1–α10) consisting of 11–22 residues each and arranged in an antiparallel fashion. Eight helices (α2–α9) have the characteristic arrangement found in HEAT-repeat proteins. HEAT repeats are ∼40 amino acids motifs that fold into helical hairpins formed by two antiparallel α-helices, known as A and B helices (33). In HEAT-repeat proteins, multiple helical hairpins pack against each other consecutively and in a parallel fashion, giving rise to solenoid structures [reviewed in (34)]. The regular topology of secondary structure elements results in the A and B helices lining the outer convex surface and the inner concave surface of the solenoid, respectively. Database searches for structural similarities to SLIP1 using the program Dali (35) identified the HEAT-repeat fold of MIF4G. In contrast to MIF4G-like domains, however, SLIP1 contains only four of the canonical five HEAT repeats (HEAT 2–5, between α2–α9) (Figure 1B). Instead of a parallel helical hairpin as HEAT 1, SLIP1 features a single helix that packs perpendicular to the helices of HEAT 2. Another unusual feature of SLIP1 is the C-terminal helix (α10). Helix α10 deviates from the HEAT-repeat architecture and forms a C-terminal extension that interacts intra-molecularly with the concave surface of the solenoid, against the side of HEAT 5 (α9, Figure 1). In addition, helix α10 interacts inter-molecularly with helix α8 in the second monomer of SLIP1 present in the crystal lattice.
Figure 1.
Structure of SLIP1 bound to the SBM of SLBP. (A) Structure of the D.r. SLIP1–SLBP complex. SLBP is in blue. SLIP1 is shown with the 4 HEAT repeats in orange and the N- and C-terminal helices in red. The N- and C-termini of SLBP are labeled and the helices of SLIP1 are numbered. (B) The structure of D.r. SLIP1–SLBP is shown in an orientation related by a 90° rotation around a horizontal axis with respect to panel A. It is superposed on the MIF4G domain of eIF4G (in gray, PDB code 2VSO). The HEAT repeats of SLIP1 are numbered.
A conserved convex surface of SLIP1 interacts with SLBP
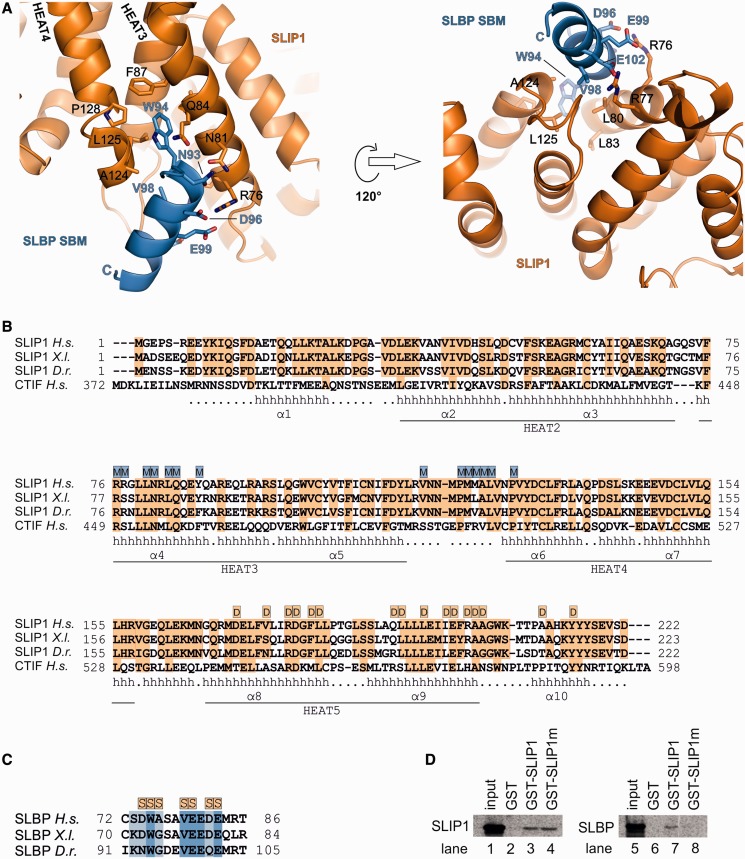
Zebrafish SLBP89–105 folds into an α-helix encompassing residues 94–105 and flanked by an N-terminal segment. The helix docks at the convex surface of SLIP1, binding between the outer helices of HEAT 3 and 4 (α4 and α6, Figures 1 and 2A). The interaction is centered at the highly conserved Trp94 and Val98 of D.r. SLBP (Figure 2C), which are recognized at a shallow hydrophobic pocket on SLIP1 lined by Leu80, Leu83, Phe87, Ala124, Leu125 and Pro128 (Figure 2A). The conserved acidic residues Asp96, Glu99 and Glu102 in SLBP interact with SLIP1 Arg77 and Arg76. Polar contacts engage SLBP Asn93 with SLIP1 Asn81 and Gln84. SLIP1 Asn81 also contacts the SLBP main chain at Gly95. These interactions are highly conserved in metazoans (Figure 2B and C). Consistently, a single point mutation of Xenopus SLBP at Trp75 (corresponding to Trp94 in the D.r. orthologue) has been shown to abolish the interaction with SLIP1 and the translation stimulation activity of SLBP (23). Conversely, mutation of exposed polar residues lining in the Trp-binding pocket of human SLIP1 (Arg77, Asn81 and Gln84 to alanine residues) resulted in loss of SLBP binding in pull-down assays (Figure 2D).
Figure 2.
Determinants and conservation of the SLIP1–SLBP interaction. (A) Close-up view of the interactions between SLIP1 (in orange) and the SBM of SLBP (in blue). The molecules are in a similar orientation as in Figure 1A. Selected residues are shown in stick representation and labeled. (B) Alignment of SLIP1 orthologues from Homo sapiens (H.s., Uniprot accession code A9UHW6), Xenopus laevis (X.l., Uniprot accession code Q801N6) and Danio rerio (D.r., Uniprot accession code Q5EAQ1). The secondary-structure elements observed in the D.r. SLIP1–SLBP89–105 crystal structure are shown below the sequences (h = helix). The HEAT repeats are labeled according to a superposition with MIF4G structure (PDB code 2VSO). Also labeled are the antiparallel helices of the MIF4G-like fold of SLIP1 (α1–α10). Highlighted in orange are conserved residues. The residues of SLIP1 interacting with the corresponding motif of SLBP are highlighted above the sequences (with M, in blue). The residues involved in dimerization are also highlighted (with D, in orange). The alignment also includes the C-terminal portion of the related human protein CTIF (Uniprot accession code O43310). (C) Alignment of the SBM of SLBP orthologues (H.s., Uniprot accession code Q14493; X.l., Uniprot accession code P79943; D.r., Uniprot accession code Q7SXI8) is shown with a similar representation as in panel B. The residues of SLBP involved in the interaction with SLIP1 are highlighted above the sequences (with S, in orange). (D) SLIP1 (lanes 1–4) or SLBP (lanes 5–8) was labeled with 35S-methionine by in vitro translation in rabbit reticulocyte lysate. The lysates were incubated with GST, GST–SLIP1 or GST–SLIP1m (a mutant with the R77A/N81A/Q84A substitutions at the SBM-binding site). The complexes were bound to glutathione sepharose and analyzed by SDS gel electrophoresis and the bound proteins detected by autoradiography.
It has recently been shown that SLBP interacts with the protein CTIF (CBP80/20-dependent translation initiation factor) and that the interaction is impaired when human SLBP Trp75 is mutated (36). CTIF contains an N-terminal CBP80-binding domain and a C-terminal MIF4G domain (37), which shares 33% sequence identity with SLIP1. Structure-based sequence analysis shows that the SLBP-binding residues we identified in SLIP1 are conserved in the MIF4G domain of CTIF (Figure 2B). CTIF may bind SLBP in a similar manner as observed for SLIP1.
SLIP1-binding motifs are present in additional proteins
To determine what additional proteins might interact with human SLIP1, we carried out a yeast two-hybrid screen using a mating library approach that allowed us to screen ∼10 million clones. We obtained 220 positive clones. Among these, we identified multiple independent clones encoding the proteins DBP5, eIF3g and c-myc (Figure 3A). In addition, we obtained 3′hExo (a 3′ to 5′ exonuclease that forms a specific complex with SLBP and the stem-loop RNA (38), as well as SLIP1 itself, consistent with the report that SLIP1 is a dimer (39). To validate the yeast two-hybrid results, we analyzed the interactions by in vitro co-precipitation assays with a similar strategy used previously (23). We synthesized the candidate SLIP1-binding proteins using an in vitro rabbit reticulocyte lysate system, labeled them with 35S-methionine and carried out pull-down assays with purified GST–SLIP1. All the proteins identified in the two-hybrid assay co-precipitated with GST–SLIP1 on glutathione beads, with a similar efficiency as compared with SLBP (Figure 3B).
Figure 3.
SBMs are present in eIF3g and DBP5. (A) A two-hybrid screen was carried out using SLIP1 as bait. Over 200 positive clones were characterized by isolating the insert by polymerase chain reaction and digesting the fragment with AluI to identify clones that contained overlapping inserts. Over 120 of these clones encoded DBP5, eIF3g or myc. A number of clones of different sizes from each were sequenced (indicated in parentheses). The length of the protein in amino acids is shown, together with the smallest positive clone isolated. Smaller numbers of clones for several other proteins were obtained. (B) To validate the two-hybrid results, each of the proteins was labeled with 35S-methionine by in vitro translation and analyzed in a pull-down assay as in Figure 2D. (C) Alignment of the SBM of H.s. SLBP with the putative SBMs we identified in eIF3g and DBP5 in several vertebrates. (D) Schematic representation of the domain organization of human DBP5 and eIF3g. The folded domains of DBP5 and eIF3g are boxed and labeled. In each protein, the putative SBMs reside in regions of low complexity that are predicted to be unstructured.
Sequence analysis of eIF3g and DBP5 revealed the presence of a stretch of amino-acid residues that is remarkably similar to the SLIP1-binding motif (SBM) of SLBP (Figure 3C), while no such sequence motif could be identified in c-myc or 3′-hExo. The binding of c-myc to SLIP1 is not affected by the mutants in the SBM in Figure 2D, consistent with the idea that c-myc binds to another region of SLIP1 (R.L., A.R, W.F.M., not shown). In both eIF3g and DBP5, the putative SBM sequences are located in intrinsically unstructured regions at the N-terminus of the proteins, separate from the folded domains that are either known or expected from structural prediction programs (Figure 3D). In both proteins, these sequences are conserved in vertebrates (Figure 3C) and are not present in yeast, consistent with the notion that the yeast genome does not contain a SLIP1 orthologue and that yeast histone mRNAs feature a canonical poly(A) tail rather than a specialized stem-loop structure at their 3′ end. EIF3g is an outer subunit of the translation initiation complex that is recruited to the 5′ end of the mRNA (40). Recently, eIF3g has also been shown to interact directly with the SLIP1-paralogue CTIF (36). DBP5 is a DEAD-box protein essential for mRNA export [reviewed in (41)], but not known to connect functionally to either SLIP1 or SLBP.
SLIP1 interacts with DBP5 in vivo and in vitro
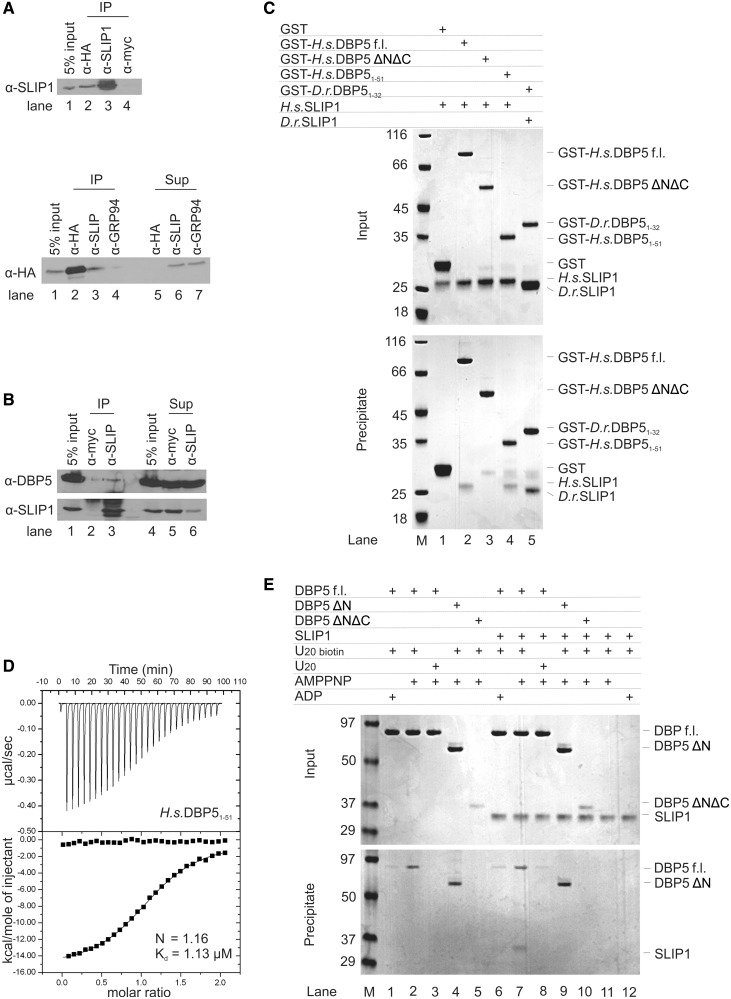
To assess whether SLIP1 interacts with DBP5 in vivo, we created a stable cell line expressing HA-tagged DBP5 and carried out immunoprecipitation experiments with an anti-HA, anti-SLIP1 or a control antibody (anti-myc). There is not an anti-DBP5 antibody that efficiently immunoprecipitated endogenous DBP5. The immunoprecipitates were resolved by gel electrophoresis and the proteins were detected by western blotting. We tested the interaction of HA-tagged-DBP5 with endogenous SLIP1 after immunoprecipitation with anti-SLIP1 (Figure 4A, top panel): we detected SLIP1 when immunoprecipitating with the anti-HA antibody (lane 2) but not with the control anti-myc antibody (lane 4). We then tested the interaction of endogenous SLIP1 with HA-DBP5 after immunoprecipitation with anti-SLIP1 (Figure 4A, bottom panel): we detected HA-tagged DBP5 when immunoprecipitating with the anti-SLIP1 antibody (lane 3) but not with the control antibody (lane 4). The anti-HA efficiently depleted the HA-DBP5 from the extract (Figure 4A, bottom, lanes 2 and 5).
Figure 4.
DBP5 binds SLIP1 in vivo and in vitro. (A) Top panel: lysates from HeLa cells stably expressing N-terminally HA-tagged DBP5 were treated with anti-HA (lane 2), anti-SLIP1 (lane 3) or anti-Myc (lane 4) and the immunoprecipitates resolved by SDS gel electrophoresis and probed by western blotting for SLIP1. Lane 1 is 5% of the input. Note that the anti-SLIP1 efficiently immunoprecipitated endogenous SLIP1. Bottom panel: lysates from the cells expressing HA-DBP5 were treated with anti-HA (lanes 2, 5), anti-SLIP1 (lanes 3, 6) or anti-GRP94 (as control, lanes 4, 7). The immunoprecipitates were analyzed for HA–DBP5 by western blotting (lanes 1–4). Five percent of the input is in lane 1. Lanes 5–7 are 5% of the supernatants after the immunoprecipitation. (B) Lysates were prepared from exponentially growing HeLa cells and treated with either antisera against the myc epitope (lanes 2, 5) or anti-SLIP1 antisera (lanes 3, 6). The immunoprecipitates were resolved by gel electrophoresis and probed by western blotting with anti-DBP5 (top) or anti-SLIP1 (bottom). Five percent of the supernatant remaining after immunoprecipitation was analyzed by western blotting (lanes 6, 7). Lanes 1 and 5 are 5% of the input. (C) Qualitative in vitro pull-down assays of SLIP1 and DBP5. Human GST–DBP5 f.l. and a fragment containing GST–DBP5 residues 1–51 efficiently precipitated SLIP1 (lanes 2, 4), while no SLIP1 precipitation was observed when using the N-terminal RecA domain of DBP5 (GST DBP5ΔNΔC, lane 3). In this assay, D.r. GST–DBP5 residues 1–32 precipitated D.r. SLIP1 (lane 5). Note that GST–D.r. DBP51–32 migrates slower on SDS PAGE than H.s. GST-DBP51–51. Bands and samples were confirmed by mass spectrometry. (D) Quantitative measurements of the H.s. SLIP1 and DBP51–51 interaction by ITC. Both the raw data and the integrated heat data are shown. (E) Protein precipitations with biotinylated single-stranded RNA. Purified proteins were mixed with 5′ end-biotinylated 20-mer single-stranded RNA and incubated with or without AMPPNP or ADP, as indicated. Proteins mixtures before (input, 17% of the total) and after coprecipitation (precipitate) were separated on a 15% (w/v) acrylamide SDS PAGE and visualized using Coomassie stain. The molecular weight standards are shown.
To assess whether the endogenous SLIP1 and DBP5 interacted, we immunoprecipitated using the anti-SLIP1 antibody, and analyzed the immunoprecipitates for DBP5 by western blotting with an anti-DBP5 antibody. The supernatants were also analyzed. The anti-SLIP1 antibody efficiently precipitated the endogenous SLIP1 (Figure 4B, bottom, lanes 3 and 6). A small amount of DBP5 was co-immunoprecipitated with SLIP1 (Figure 4B, lane 3) and not with the control antibody (Figure 4B, lane 2), indicating that the endogenous proteins also interacted.
DBP5 binds RNA and ATP using its two RecA domains, which are also the site for the interaction with the nucleoporins NUP214 and GLE1 (31,42,43). No protein has yet been reported to interact with the N-terminal domain of DBP5, which contains the putative SBM (at residues 1–20). In GST-pull-down assays with purified proteins, DBP5 f.l. and DBP51–51 precipitated SLIP1 (Figure 4C, lane 2 and lane 4). We analyzed the binding quantitatively using ITC. DBP51–51 bound SLIP1 with a Kd of ∼1.1 µM (Figure 4D). Full-length DBP5 bound SLIP1 with a Kd of 0.1 µM (Supplementary Figure S1A), suggesting that the RecA core also contributes to binding. However, the RecA core in isolation (DBP5 ΔN) showed no detectable binding to SLIP1 by ITC (Supplementary Figure S1B), indicating that the major SLIP1-binding site resides in the N-terminal region of DBP5. We tested whether SLIP1 and RNA compete for binding to DBP5. We incubated a single-stranded RNA biotinylated at the 5′ end with recombinant proteins and incubated the samples with AMPPNP before precipitation with streptavidin magnetic beads. As expected, both DBP5 f.l. and the DEAD-box core (DBP5 ΔN) precipitated in the presence of AMPPNP (Figure 4E, lanes 2 and 4), while a single RecA domain did not (DBP5 ΔNΔC, lane 5) (31). In the RNA-pull-down assay, SLIP1 co-precipitated with DBP5 f.l. (Figure 4E, lane 7) but not with DBP5 ΔN (lane 9). These results indicate that DBP5 binds SLIP1 mainly via the N-terminal region and can concomitantly bind RNA via the RecA domains.
Structure of SLIP1 bound to the N-terminus of DBP5
To obtain the crystal structure of a SLIP1–DBP5 complex, we purified the corresponding zebrafish orthologues. In pull-down assays, D.r. SLIP1 interacted with D.r. DBP51–32 (corresponding to human DBP51–32) (Figure 4C, lane 5). In ITC experiments, D.r. SLIP1 bound D.r. DBP51–32 with a Kd of 2 μM (Supplementary Figure S1C), comparable with the binding affinities measured for the human proteins (Figure 4D). The complex between zebrafish SLIP1 and DBP51–32 yielded crystals that contain two complexes in the asymmetric unit. We determined the structure by molecular replacement using the SLIP1 coordinates and refined it to 3.25 Å resolution. The final atomic model of the SLIP1–DBP5 complex has an Rfree of 27.2%, Rwork of 24.5% and good stereochemistry (Table 1). The complex contains residues 7–222 of SLIP1 and residues 2–20 of DBP5. No ordered electron density was observed for residues 21–32 of DBP5, suggesting that this region is flexible in the crystals we obtained.
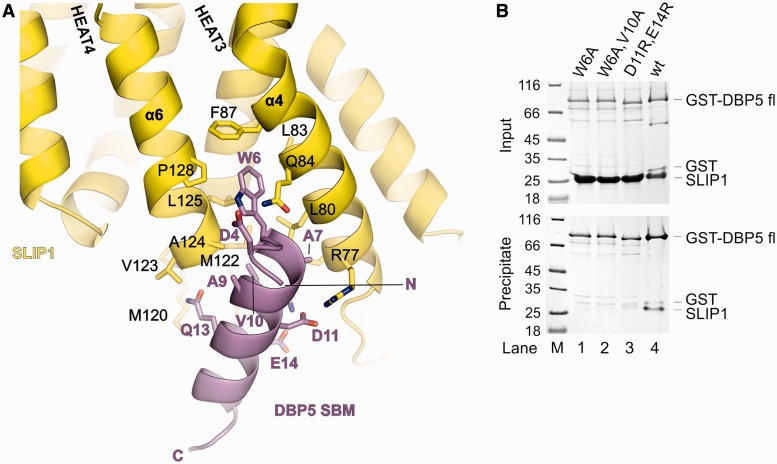
The DBP5 SBM forms an α-helix engaged in similar interactions with SLIP1 as described above for the SBM of SLBP (Figure 5A). Briefly, Trp6, Val10, Asp11 and Glu14 of zebrafish DBP5 are at the equivalent structural positions of Trp94, Val98, Glu99 and Glu102 of zebrafish SLBP. The small side chain of D.r. DBP5 Ala7 is accommodated in the shallow pocket of SLIP1 without significant rearrangements as compared with D.r. SLBP Gly95. Minor changes occur in the peripheral interactions. For example, DBP5 Ser5 does not contact SLIP1 Asn81 as was the case for SLBP Asn93, but a compensatory interaction is formed between DBP5 Asp4 and SLIP1 Lys88. Based on the structural analysis, we tested the effect of mutations in the interaction of the human f.l. proteins. In in vitro pull-down experiments, the interaction with SLIP1 was abolished with a Trp6Ala DBP5 mutant (Figure 5B, lane 1), homologous to a critical residue for binding in SLBP and SLIP1, and was drastically reduced with an Asp11Arg, Glu14Arg DBP5 mutant (Figure 5B, lane 3, compare with the wild type in lane 4). We conclude that DBP5 is a bona fide direct binding partner of SLIP1.
Figure 5.
Structure of SLIP1 bound to the SBM of DBP5. (A) Close-up view of the interactions between SLIP1 (in yellow) and the SBM of DBP5 (in violet). The molecules are in a similar orientation as in Figure 2A. Selected residues are shown in stick representation and labeled. (B) Protein co-precipitations by GST pull-down assays. GST-tagged human DBP5 wild type or mutants were incubated with SLIP1 in a buffer containing 100 mM NaCl. One-sixth of the sample was kept as input control (upper panel), and the rest was co-precipitated with glutathione sepharose beads (lower panel). Both input and pull-down samples were analyzed on Coomassie stained 15% SDS PAGE. The lane on the left shows a molecular weight marker.
The SLIP1 homodimer binds two SBMs simultaneously
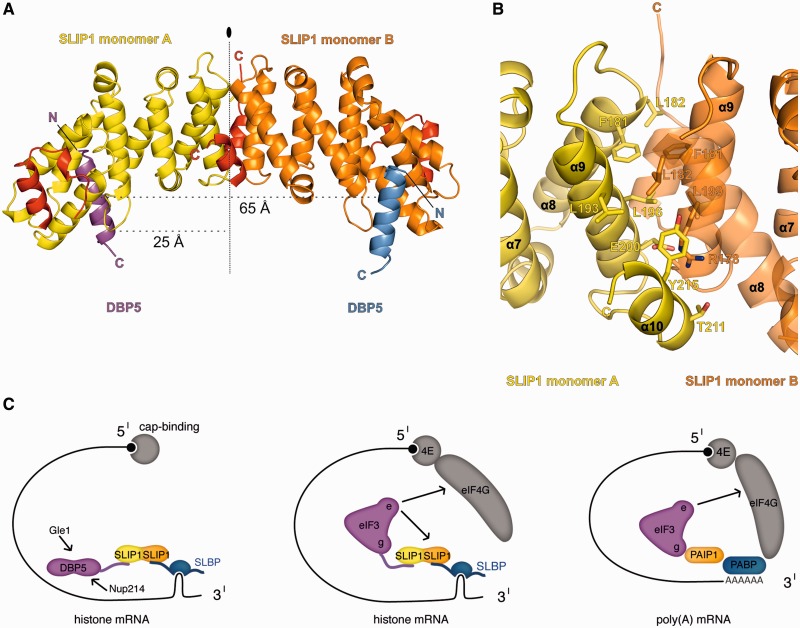
SLIP1 is a dimer in solution by ultracentrifugation experiments (39) and by static light scattering (Supplementary Figure S2A). The SLIP1–DBP5 crystals contain a SLIP1 dimer in the asymmetric unit. The two SLIP1 monomers are related by a non-crystallographic 2-fold axis (located between the α9 helices) and interact with the three C-terminal helices (α8–α10, Figure 6A and B). The dimerization interface buries 8.5% of the surface of each monomer (984 Å2 of surface area buried out of 11 635 Å2), as calculated using the PISA server (44). The same tail-to-tail dimer is also observed in the structure of apo SLIP1 (2I2O), independently of the fact that the crystals belong to a different space group and have different lattice contacts (Supplementary Figure S2B). This dimer is also generated by a crystallographic 2-fold axis in the case of the SLIP1–SLBP crystals (Supplementary Figure S2). The SLIP1–SLIP1 dimerization interface is formed by evolutionary conserved residues (Figures 2B and 6B). The intermolecular interactions are mainly hydrophobic, and involve Phe181, Leu182, Leu193, Leu196 and Tyr215 of each monomer (Figure 6B). In addition, there is a conserved salt bridge between Glu200 of one monomer and Arg178 of the other monomer.
Figure 6.
Concurrent binding of two SBMs to a SLIP1 homodimer. (A) Structure of the dimeric SLIP1–SLBP complex observed in the crystal lattice. The two monomers (A and B) are labeled. The vertical line indicates the 2-fold symmetry of the dimer. (B) Close-up view of the dimerization interface, shown in the same orientation as in panel A. Selected conserved residues are shown in stick representation and labeled. For clarity, helix 8 is shown with 50% transparency. (C) Left panel: model for the involvement of SLBP in histone mRNA export. The SLIP1 heterodimer is shown in yellow and orange, as in panel A. The model does not include additional levels of regulation, such as phosphorylation (39). Central panel: model for the mechanisms of translation stimulation of histone mRNAs as compared with that of poly(A)-containing mRNAs (right panel). Arrows indicate additional interactions reported in the literature.
The SLIP1 homodimer has two SBM-binding sites (Figure 6A). The SBM helices are more than 25 Å away from the dimerization interface. Indeed, mutation of Glu200 in the human orthologue has been shown to impair SLIP1 dimerization and to maintain SLBP binding (39). The two parallel SBM helices are positioned with their C-termini at a 65 Å distance from each other. SLIP1 can thus in principle bind to two SBM-containing proteins simultaneously, provided the architecture and properties of the flanking domains C-terminal to the SBM do not give rise to steric or electrostatic clashes. Interestingly, the dimerization interface of SLIP1 is conserved in CTIF (Figure 2B), suggesting the possible formation of CTIF–CTIF homodimers and/or SLIP1–CTIF heterodimers. Indeed, a recent study has shown that SLIP1, CTIF, SLBP and CBP80 co-immunoprecipitate in an RNA-independent manner, supporting the existence of SLIP1–CTIF heterodimers in vivo (36).
CONCLUSIONS
SLIP1 is a MIF4G-like protein that contains the binding site for a short linear sequence motif (SLiM) that we named SBM. The SBM is present in SLBP, the protein that binds the specialized stem-loop structure at the 3′ end of replication-dependent histone mRNAs in metazoans. The vertebrate orthologues of eIF3g and DBP5 also contain a SBM sequence, which thus appears to have evolved convergently in different proteins. In these three proteins, the SBM has other typical SLiM features (45). First, it encompasses a sequence region of strong evolutionary conservation amid variable neighboring regions. Second, it is present in an intrinsically unfolded domain and becomes folded on binding. Third, it binds SLIP1 with few direct contacts and with low micromolar affinity. Such a relatively low affinity is typical of transient and dynamic complexes, and can often be modulated by posttranslational modifications (45). The regions flanking the SBMs of both SLBP and eIF3g contain phosphorylation sites (19,46), which might influence the interaction in the context of specific cellular conditions.
SLIP1 is a non-canonical MIF4G-like protein, not only in terms of the fold (i.e. the number of its HEAT repeats) but also in terms of the oligomeric state. The existence of SLIP1 as a homodimer raises the possibility that its two SBM-binding sites can be occupied either by the same or by different SBM-containing proteins. In the latter case, it would allow SLIP1 to act as a platform for bridging DBP5 or eIF3g to SLBP. The relevance of these simultaneous interactions and the emerging models (Figure 6C) are discussed below. We note that it is also possible that SLIP1 might bridge DBP5 and eIF3g, with an interaction that would be independent of histone mRNA metabolism. Indeed, SLIP1 is expected to have additional functions because knockdown of SLIP1 affects cellular viability while knockdown of the histone-specific factor SLBP only affects cell-cycle progression (16,23,47). SLIP1 could also bridge analogous interactions in the context of a SLIP1–CTIF heterodimer, which would in addition be able to connect to the cap-binding protein CBP80 (37).
DBP5 (also known as DDX19 in metazoans) belongs to the DEAD-box family of RNA-dependent ATPases. DBP5 is essential for the export of poly(A)-containing mRNPs: it is believed to remodel the mRNP at the cytoplasmic side of the nuclear pore complex, removing the transport factor before the mRNA is released to the cytoplasm [reviewed in (41)]. Two cytoplasmic nucleoporins, GLE1 and NUP214, bind DBP5 directly and activate the ATPase cycle that likely underlies mRNP remodeling. No other factor is known to direct DBP5 to its specific RNA targets. A DBP5–SLIP1/SLIP1–SLBP complex could specifically target the ATPase to histone-containing mRNPs (Figure 6C, left panel). Importantly, the loading of DBP5 onto the histone mRNP would not physically interfere with the domains of DBP5 responsible for RNA binding and GLE1/NUP214 regulation. This interaction would rationalize the active role of SLBP in nuclear export in mammalian cells (14).
It is possible that after export, the SLIP1–SLBP complex dissociates from DBP5 and then interacts with eIF3g to stimulate translation of histone mRNA. EIF3g is a peripheral component of the mammalian eIF3 complex and is involved in translational regulation (48). In poly(A)-containing mRNPs, eIF3g interacts directly with PAIP1 and stimulates translation (49). PAIP1 is a MIF4G-containing protein with a multidomain architecture, allowing it to bind eIF3g and PABP simultaneously (Figure 6C, right panel). PABP in turn binds the 3′ poly(A) tail and eIF4G, a scaffolding protein that connects to the 5′ cap structure and to the eIF3e/INT6 subunit (50,51). Mechanistically, eIF3g–PAIP1 is believed to stabilize the interactions that bring together the 3′ and 5′ ends of the message, circularizing it and facilitating multiple initiation-termination-reinitiation cycles (49). The eIF3g-PAIP1-PABP interaction can be readily compared with eIF3g–SLIP1/SLIP1–SLBP (Figure 6C, right and central panels). In the case of histone mRNPs, there is no known direct, RNA-independent SLBP–eIF4G contact similar to that of PABP–eIF4G. However, eIF3e/INT6 contributes an alternative interaction with SLIP1–SLBP that is necessary for efficient histone mRNA translation (24) (Figure 6C, central panel). This model rationalizes the finding that both the 5′ cap and the 3′ stem-loop are required for histone translation in vivo in mammalian cells (52). It suggests that histone mRNPs and poly(A)-containing mRNPs might establish an equivalent network of multiple and reinforcing interactions to promote efficient translation initiation.
ACCESSION NUMBERS
Atomic coordinates and structure factors have been deposited in the Protein data Bank with accession codes 4JHK for the SLIP1–SLBP complex and 4JHJ for the SLIP1–DBP5 complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2.
FUNDING
Max Planck Gesellschaft; European Research Council Advanced Investigator Grant [294371]; Marie Curie Initial Training Network RNPnet and the Deutsche Forschungsgemeinschaft [SFB646, SFB1035, FOR1680, GRK1721 and CIPSM to E.C.]; National Institute of Health (NIH) [GM29832 to W.F.M.]; EMBO Long-Term Fellowship [ALTF 1557-2010 to H.v.M.]; NIH postdoctoral fellowship [F32 GM080007 to R.L.]. Funding for open access charge: Max Planck Gesellschaft.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank the Max Planck Institute of Biochemistry Core Facility and Crystallization Facility; the staff members at the beamlines PXII and PXIII of the Swiss Light Source; Monika Krause for the drawings in Figure 6C. The authors also thank Fabien Bonneau and members of our labs for useful discussions and critical reading of the manuscript and Markus Wahl for generous support. H.v.M. and E.C. designed experiments and wrote the manuscript. H.v.M. cloned, purified, crystallized the complexes, solved the structures and carried out in vitro binding assays and ITC experiments. C.B. performed and analyzed ITC and static light scattering experiments. R.L., A.R. and W.F.M. designed and carried out the yeast two-hybrid analysis, the in vivo experiments and the pull-downs with in vitro translated proteins.
REFERENCES
- 1.Moore MJ. From birth to death: the complex lives of eukaryotic mRNAs. Science. 2005;309:1514–1518. doi: 10.1126/science.1111443. [DOI] [PubMed] [Google Scholar]
- 2.Fortes P, Inada T, Preiss T, Hentze MW, Mattaj IW, Sachs AB. The yeast nuclear cap binding complex can interact with translation factor eIF4G and mediate translation initiation. Mol. Cell. 2000;6:191–196. [PubMed] [Google Scholar]
- 3.Imataka H, Gradi A, Sonenberg N. A newly identified N-terminal amino acid sequence of human eIF4G binds poly(A)-binding protein and functions in poly(A)-dependent translation. EMBO J. 1998;17:7480–7489. doi: 10.1093/emboj/17.24.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 5.Jaeger S, Barends S, Giege R, Eriani G, Martin F. Expression of metazoan replication-dependent histone genes. Biochimie. 2005;87:827–834. doi: 10.1016/j.biochi.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Marzluff WF, Wagner EJ, Duronio RJ. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 2008;9:843–854. doi: 10.1038/nrg2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang ZF, Whitfield ML, Ingledue TC, 3rd, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- 8.Martin F, Schaller A, Eglite S, Schumperli D, Muller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem-loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol. Cell. Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Erkmann JA, Wagner EJ, Dong J, Zhang Y, Kutay U, Marzluff WF. Nuclear import of the stem-loop binding protein and localization during the cell cycle. Mol. Biol. Cell. 2005;16:2960–2971. doi: 10.1091/mbc.E04-11-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dominski Z, Zheng LX, Sanchez R, Marzluff WF. Stem-loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol. Cell. Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang Y, Carmichael GG. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc. Natl Acad. Sci. USA. 1997;94:10104–10109. doi: 10.1073/pnas.94.19.10104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol. Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 14.Sullivan KD, Mullen TE, Marzluff WF, Wagner EJ. Knockdown of SLBP results in nuclear retention of histone mRNA. RNA. 2009;15:459–472. doi: 10.1261/rna.1205409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wagner EJ, Berkow A, Marzluff WF. Expression of an RNAi-resistant SLBP restores proper S-phase progression. Biochem. Soc. Trans. 2005;33:471–473. doi: 10.1042/BST0330471. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X, McKillop-Smith S, Muller B. The human histone gene expression regulator HBP/SLBP is required for histone and DNA synthesis, cell cycle progression and cell proliferation in mitotic cells. J. Cell Sci. 2004;117:6043–6051. doi: 10.1242/jcs.01523. [DOI] [PubMed] [Google Scholar]
- 17.Gorgoni B, Andrews S, Schaller A, Schumperli D, Gray NK, Muller B. The stem-loop binding protein stimulates histone translation at an early step in the initiation pathway. RNA. 2005;11:1030–1042. doi: 10.1261/rna.7281305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanchez R, Marzluff WF. The stem-loop binding protein is required for efficient translation of histone mRNA in vivo and in vitro. Mol. Cell. Biol. 2002;22:7093–7104. doi: 10.1128/MCB.22.20.7093-7104.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koseoglu MM, Graves LM, Marzluff WF. Phosphorylation of threonine 61 by cyclin a/Cdk1 triggers degradation of stem-loop binding protein at the end of S phase. Mol. Cell. Biol. 2008;28:4469–4479. doi: 10.1128/MCB.01416-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Battle DJ, Doudna JA. The stem-loop binding protein forms a highly stable and specific complex with the 3′ stem-loop of histone mRNAs. RNA. 2001;7:123–132. doi: 10.1017/s1355838201001820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Lam TT, Tonelli M, Marzluff WF, Thapar R. Interaction of the histone mRNA hairpin with stem-loop binding protein (SLBP) and regulation of the SLBP-RNA complex by phosphorylation and proline isomerization. Biochemistry. 2012;51:3215–3231. doi: 10.1021/bi2018255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dominski Z, Marzluff WF. Three-hybrid screens for RNA-binding proteins. Proteins binding 3′ end of histone mRNA. Methods Mol. Biol. 2001;177:291–318. doi: 10.1385/1-59259-210-4:291. [DOI] [PubMed] [Google Scholar]
- 23.Cakmakci NG, Lerner RS, Wagner EJ, Zheng L, Marzluff WF. SLIP1, a factor required for activation of histone mRNA translation by the stem-loop binding protein. Mol. Cell. Biol. 2008;28:1182–1194. doi: 10.1128/MCB.01500-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neusiedler J, Mocquet V, Limousin T, Ohlmann T, Morris C, Jalinot P. INT6 interacts with MIF4GD/SLIP1 and is necessary for efficient histone mRNA translation. RNA. 2012;18:1163–1177. doi: 10.1261/rna.032631.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 26.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nat. Struct. Mol. Biol. 2009;16:247–254. doi: 10.1038/nsmb.1561. [DOI] [PubMed] [Google Scholar]
- 32.Thapar R, Mueller GA, Marzluff WF. The N-terminal domain of the Drosophila histone mRNA binding protein, SLBP, is intrinsically disordered with nascent helical structure. Biochemistry. 2004;43:9390–9400. doi: 10.1021/bi036314r. [DOI] [PubMed] [Google Scholar]
- 33.Andrade MA, Petosa C, O'Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J. Mol. Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- 34.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr. Opin. Struct. Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 35.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choe J, Kim KM, Park S, Lee YK, Song OK, Kim MK, Lee BG, Song HK, Kim YK. Rapid degradation of replication-dependent histone mRNAs largely occurs on mRNAs bound by nuclear cap-binding proteins 80 and 20. Nucleic Acids Res. 2013;41:1307–1318. doi: 10.1093/nar/gks1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim KM, Cho H, Choi K, Kim J, Kim BW, Ko YG, Jang SK, Kim YK. A new MIF4G domain-containing protein, CTIF, directs nuclear cap-binding protein CBP80/20-dependent translation. Genes Dev. 2009;23:2033–2045. doi: 10.1101/gad.1823409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tan D, Marzluff WF, Dominski Z, Tong L. Structure of histone mRNA stem-loop, human stem-loop binding protein, and 3'hExo ternary complex. Science. 2013;339:318–321. doi: 10.1126/science.1228705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bansal N, Zhang M, Bhaskar A, Itotia P, Lee E, Shlyakhtenko LS, Lam TT, Fritz A, Berezney R, Lyubchenko YL, et al. Assembly of the SLIP1-SLBP Complex on Histone mRNA Requires Heterodimerization and Sequential Binding of SLBP Followed by SLIP1. Biochemistry. 2013;52:520–536. doi: 10.1021/bi301074r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat. Rev. Mol. Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Valkov E, Dean JC, Jani D, Kuhlmann SI, Stewart M. Structural basis for the assembly and disassembly of mRNA nuclear export complexes. Biochimica Biophys. Acta. 2012;1819:578–592. doi: 10.1016/j.bbagrm.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 42.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472:238–242. doi: 10.1038/nature09862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. Proc. Natl Acad. Sci. USA. 2009;106:3089–3094. doi: 10.1073/pnas.0813267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 45.Davey NE, Van Roey K, Weatheritt RJ, Toedt G, Uyar B, Altenberg B, Budd A, Diella F, Dinkel H, Gibson TJ. Attributes of short linear motifs. Mol. Biosyst. 2012;8:268–281. doi: 10.1039/c1mb05231d. [DOI] [PubMed] [Google Scholar]
- 46.Damoc E, Fraser CS, Zhou M, Videler H, Mayeur GL, Hershey JW, Doudna JA, Robinson CV, Leary JA. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol. Cell. Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 47.Townley-Tilson WH, Pendergrass SA, Marzluff WF, Whitfield ML. Genome-wide analysis of mRNAs bound to the histone stem-loop binding protein. RNA. 2006;12:1853–1867. doi: 10.1261/rna.76006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martineau Y, Derry MC, Wang X, Yanagiya A, Berlanga JJ, Shyu AB, Imataka H, Gehring K, Sonenberg N. Poly(A)-binding protein-interacting protein 1 binds to eukaryotic translation initiation factor 3 to stimulate translation. Mol. Cell. Biol. 2008;28:6658–6667. doi: 10.1128/MCB.00738-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeFebvre AK, Korneeva NL, Trutschl M, Cvek U, Duzan RD, Bradley CA, Hershey JW, Rhoads RE. Translation initiation factor eIF4G-1 binds to eIF3 through the eIF3e subunit. J. Biol. Chem. 2006;281:22917–22932. doi: 10.1074/jbc.M605418200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gingras AC, Raught B, Sonenberg N. eIF4 initiation factors: effectors of mRNA recruitment to ribosomes and regulators of translation. Ann. Rev. Biochem. 1999;68:913–963. doi: 10.1146/annurev.biochem.68.1.913. [DOI] [PubMed] [Google Scholar]
- 52.Gallie DR, Lewis NJ, Marzluff WF. The histone 3′-terminal stem-loop is necessary for translation in Chinese hamster ovary cells. Nucleic Acids Res. 1996;24:1954–1962. doi: 10.1093/nar/24.10.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.