Abstract
Efficient DNA repair is critical for cell survival and the maintenance of genome integrity. The homologous recombination pathway is responsible for the repair of DNA double-strand breaks within cells. Initiation of this pathway in bacteria can be carried out by either the RecBCD or the RecFOR proteins. An important regulatory player within the RecFOR pathway is the RecOR complex that facilitates RecA loading onto DNA. Here we report new data regarding the assembly of Deinococcus radiodurans RecOR and its interaction with DNA, providing novel mechanistic insight into the mode of action of RecOR in homologous recombination. We present a higher resolution crystal structure of RecOR in an ‘open’ conformation in which the tetrameric RecR ring flanked by two RecO molecules is accessible for DNA binding. We show using small-angle neutron scattering and mutagenesis studies that DNA binding does indeed occur within the RecR ring. Binding of single-stranded DNA occurs without any major conformational changes of the RecOR complex while structural rearrangements are observed on double-stranded DNA binding. Finally, our molecular dynamics simulations, supported by our biochemical data, provide a detailed picture of the DNA binding motif of RecOR and reveal that single-stranded DNA is sandwiched between the two facing oligonucleotide binding domains of RecO within the RecR ring.
INTRODUCTION
Homologous recombination (HR) is one of the primary pathways in all organisms by which double-stranded DNA (dsDNA) breaks and single-stranded DNA (ssDNA) gaps are repaired (1–4). This process is also essential for the restart of stalled replication forks, the maintenance of genomic integrity, the proper segregation of chromosomes and the generation of genomic diversity. HR requires extensive regions of DNA homology and repairs DNA damages accurately by using information from the intact homologous template. This pathway is initiated by the binding of a RecA-like recombinase to ssDNA resulting in the formation of a long nucleoprotein filament called the presynaptic complex that induces DNA strand invasion.
Recombinase function is highly regulated in all cells, and the regulation occurs at many levels. For example, ssDNA-binding proteins (SSB) inhibit the formation of the presynaptic complex when prebound to ssDNA, while another family of proteins, the recombination mediator proteins (RMPs), overcomes these inhibitory effects (5). RMPs include phage UvsY (6), prokaryotic RecBCD and RecFOR proteins (7–10) and numerous eukaryotic members (11). Mutations of human RMPs have been found to be associated with cancer predisposition, mental retardation, UV sensitivity and premature aging (12–15).
The proteins RecF, RecO and RecR are the most common RMPs in bacteria (16). Mutation of the recF, recO or recR genes results in a delay of SOS induction (17,18). This delay and the loss of DNA repair are suppressed by specific mutations in the recA gene, called srf (suppressors of recF mutation) mutations. The RecA803 protein is the product of one srf suppressor; it binds efficiently to ssDNA that is complexed with SSB protein, whereas the wild-type RecA protein does not (19,20). These, and other, in vivo and in vitro observations strongly suggest that the RecFOR proteins are all needed for assembly of the RecA filament on SSB-coated ssDNA (21,22). At present, however, there is little evidence for the existence of a stable heterotrimeric RecFOR complex.
The individual crystal structures of all three RecF, RecO and RecR proteins and of the RecOR complex have been reported from Deinococcus radiodurans (dr) (23–27), a bacterium that displays an extraordinary resistance to a wide range of DNA-damaging agents. In addition, the crystal structure of Escherichia coli RecO in complex with a peptide of the conserved SSB C-terminus has recently been published (28). However, up to now, little is known regarding the interactions of these proteins with DNA.
The crystal structure of drRecR revealed a tetrameric architecture, consisting of a dimer of dimers that has been suggested to act as a DNA clamp (23). These RecR tetramers exhibit two different RecR–RecR interfaces, involving domain swapping of either the N-terminal or the C-terminal domains. Residues lysine 23 and arginine 27 that are located inside of the ring-like structure have been reported to be important for DNA binding (23,27). However, other RecR homologs from Thermus thermophilus, E. coli and Thermoanaerobacter tengcongensis were described to be dimeric in solution (29–32). These RecR dimers assemble through the N-terminal domains of RecR and are referred to as RecR N-N dimers. The D.radiodurans and Bacillus subtilis RecR proteins reveal weak binding to DNA (23,33), while the E. coli and T. tengcongensis homologs have no known DNA-binding activity (32). RecR interacts with both RecF and RecO in vitro and both complexes show a significantly increased apparent affinity for ssDNA and dsDNA compared with the individual proteins (5,31,34). The RecF protein exhibits extensive structural similarity with the head domain of the eukaryotic Rad50 protein but lacks the long coiled-coil domain of Rad50. RecF belongs to the ATP binding cassette ATPase family of proteins and binds to DNA with increased affinity for dsDNA (5,31,34,35). ATP binding triggers RecF dimerization (26), and ATP hydrolysis triggers dissociation from DNA (36). The RecO protein contains an N-terminal oligonucleotide binding (OB) fold domain, a central helical domain and a zinc binding domain (24,25). RecO binds both ssDNA and dsDNA likely via its OB-fold domain and, potentially, charged surface areas of other domains (24). RecO interacts with SSB and promotes the annealing of complementary ssDNA strands (37,38).
The crystal structure of the drRecOR complex solved at 3.8 Å resolution revealed a four-to-two stoichiometry between RecR and RecO (27). RecR forms a tetrameric ring and the two RecO molecules are bound in trans. Unexpectedly, the amino acids in RecR that are important for DNA binding are occluded by RecO in the crystal structure. This structure led to the idea that the RecOR complex may open up on DNA binding by displacing at least one copy of RecO and possibly two copies of RecR. Recent work by Tang et al. (32) strongly indicates that the RecOR complex of T. tengcongensis (tteRecOR) displays a 2:1 stoichiometry in solution, consisting of a RecR N-N dimer and a single RecO molecule. In this study, tteRecOR was also shown to bind specifically to ssDNA and not to dsDNA and the authors propose a model for ssDNA binding to the heterotrimeric RecOR complex. However, so far there is no structural data available describing the precise mechanisms by which the RecOR complex interacts with DNA substrates.
Here we report a combined low- and high-resolution study, involving complementary structural biology approaches, of the drRecOR complex in the presence and absence of DNA. We show that drRecR is dimeric in solution and propose that binding of RecO triggers its tetramerization. A new crystal structure of the drRecOR complex was determined at higher resolution (3.3 Å) in a conformation where the inside of the RecR ring is partially accessible for ssDNA to bind. We demonstrate using small-angle neutron scattering (SANS) that the RecOR complex binds DNA substrates within this accessible cavity but with different binding modes for ssDNA and dsDNA. Our mutagenesis and biochemical data, together with molecular dynamics (MD) simulations carried out concomitantly on the RecOR complex with and without ssDNA, allow us to establish a molecular model for how this complex interacts with its DNA substrates.
MATERIALS AND METHODS
Expression, purification and crystallization of drRecOR
Expression and purification of drRecO (SwissProt: Q9RW50) and drRecR (SwissProt: Q9ZNA2) proteins for isolation of the drRecOR complex were largely performed as described previously (24,27). The various RecO and RecR mutants were generated by site-directed mutagenesis (Stratagene QuikChange site-directed mutagenesis kit) and the mutant complexes were purified using the same protocol as for the wild-type complex. Briefly, the wild-type or mutant RecO and RecR proteins were individually expressed in BL21 (DE3) RIL cells at 20°C for 16 h and afterward extracted together by sonication in lysis buffer (150 mM NaCl; 50 mM Tris, pH 7.5; 5 mM β-mercaptoethanol). Both proteins fused to an N-terminal hexa-histidine tag were purified by Ni-affinity chromatography. The proteins were eluted with lysis buffer containing 500 mM imidazole and immediately dialyzed against buffer A (100 mM NaCl; 50 mM Tris, pH 8.0; 5 mM β-mercaptoethanol). The protein complex was further purified on a MonoQ column equilibrated in buffer A and eluted by applying a linear gradient up to 1 M NaCl. The complex was finally loaded on a Superdex 200 10/300 column equilibrated in buffer A. Before crystallization experiments, the drRecOR complex was concentrated to a final concentration of 14 mg/ml using an Amicon Ultracel-10 K (Millipore). Several crystallization hits were obtained for the drRecORK23A/R27A mutant using the crystallization condition reported previously for wild-type drRecOR (0.1 M MES, pH 6.5, 10% dioxane, 1.6 M ammonium sulphate) in combination with the Silver Bullets optimization screen (Hampton Research). The crystal used for data collection resulted from the Silver Bullets condition N°8 (containing 0.25% w/v 1-Pentanesulfonic acid sodium salt monohydrate; 0.25% w/v 3,5-dinitrosalicylic acid; 0.25% 3-aminosalicylic acid; 0.25% w/v salicylamide; 0.02 M HEPES sodium, pH 6.8).
Data collection, structure determination and refinement
X-ray diffraction data were collected on ID14-4 beamline at the European Synchrotron Radiation Facility (ESRF) in Grenoble, France. The crystal used for data collection belonged to the space group P1 and had unit cell dimensions of a = 63.130 Å, b = 93.110 Å, c = 92.350 Å, α = 103.6°, β = 110.42° and γ = 106.22°. Diffraction was observed to a maximum resolution of 3.34 Å. Data were processed with the program iMosflm (39) and scaled with SCALA from the CCP4 program suite (40). The structure of drRecOR was determined by molecular replacement using the program PHASER (41) and drRecR (pdb1VDD) and drRecO (pdb1W3S) as search models. The model was built using the program Coot (42) and refinement was carried out with Phenix (43) and Refmac5 (44) using TLS refinement. The quality of the model was checked using the Molprobity server (45). Structural figures were generated with PYMOL (www.pymol.org).
Cloning, expression, purification of drRecOR and drRecR for small-angle scattering analysis
To obtain high-quality small-angle scattering profiles, the protein constructs were optimized. drRecO was cloned into a pET24d vector using the NcoI and XhoI restriction sites to code for a C-terminally hexa-histidine tagged protein. drRecR full-length protein was cloned into a pET24d vector with either an N-terminal or C-terminal hexa-histidine tag and into a pET151b vector for TEV-cleavable N-terminal hexa-histidine protein expression. The drRecR construct encoding for residues 1–197 was cloned into a pET24d with a C-terminal hexa-histidine tag. All constructs were expressed in BL21 RIL (DE3) cells for 18 h at 20°C. For the purification of the drRecR protein, cells were lysed and the soluble fraction was loaded on a 1 ml HisTrap column (GE Healthcare) equilibrated in lysis buffer (150 mM NaCl; 50 mM Tris, pH 8.0; 5 mM β-mercaptoethanol). The column was washed in lysis buffer containing 1 M NaCl and then with lysis buffer containing 50 mM imidazole. The proteins were eluted with buffer containing 500 mM imidazole and immediately dialyzed against 50 mM Tris, pH 8.0; 100 mM NaCl; 5 mM β-mercaptoethanol. For the drRecR construct containing the cleavable hexa-histidine tag, a Tobacco Etch Virus protease digestion was performed and the protein was applied to a second HisTrap column. All proteins were further purified on a MonoQ column. Fractions containing the drRecR complex were pooled and applied to a Superdex 200 10/300 column equilibrated in 10 mM Tris, pH 8.0; 100 mM NaCl; 5 mM β-mercaptoethanol. For the purification of the drRecOR, complex cells expressing drRecO and drRecR were mixed and lysed together and further purified using the same protocol as for the drRecR purification. No difference concerning the complex formation could be observed between the different constructs. For the SAXS and SANS data presented here, the drRecR protein containing a C-terminal hexa-histidine tag was used.
Fluorescence anisotropy
Equilibrium DNA-binding assays were performed on a Synergy H4 microplate reader (Biotek) fitted with polarization filters to measure fluorescence anisotropy. The following DNA substrates were used: a 50mer polydT ssDNA oligonucleotide labeled with fluorescein and a 50mer dsDNA consisting of a 5′ FAM-labeled strand 5′-GACTACGTACTGTTACGGCTCCATCTCTACCGCAATCAGGCCAGATCTGC-3′, annealed to its complementary strand 5′-GCAGATCTGGCCTGATTGCGGTAGAGATGGAGCCGT AACAGTACGTAGTC-3′. The binding assays were conducted at 25°C in 80 µl reaction volumes in 50 mM Tris, pH 8.0, 100 mM NaCl, 5 mM β-mercaptoethanol and 0.05% Tween 20. Zero to forty micromolar drRecR, drRecO [prepared as described previously (24)], drRecOR and drRecOR mutant complexes were titrated into 1 nM DNA. Samples were loaded into 384-well black microplates before reading. Each data point was the average of 100 readings of anisotropy. Averaged data from the triplicate measurements were fitted to a standard binding equation assuming a single binding site with Hill slope (h value) using GraphPad Prism6. The R2 values for the fits were all >0.99.
Small-Angle X-ray scattering
Small-angle X-ray scattering (SAXS) experiments were conducted at the ESRF ID14-3 beamline in Grenoble (France). All proteins used were purified by size-exclusion chromatography before SAXS measurements. The buffer of the size-exclusion chromatography step was used as reference for background subtraction for the SAXS measurement. For each sample, six different concentrations ranging between 0.5 and 10 mg/ml were measured. Protein solution (60 μl) for each sample (and buffer) was exposed to X-rays and scattering data were collected. Ten individual frames of 30 s were collected for every exposure using a Pilatus 1M detector (Dectris). Individual frames were processed automatically and independently using the software BsxCUBE, yielding individual radially averaged curves of normalized intensity versus scattering angle q = 4πSINθ/λ. Frames were combined, excluding any data points affected by aggregation induced by radiation damage, to give the average scattering curve for each measurement (measured before and after every sample and background subtracted).
Small-angle neutron scattering
SANS data were collected with the D22 instrument at the Institut Laue-Langevin in Grenoble, France. All drRecOR samples resulted from the same purification, which was divided to perform the different measurements with and without DNA assuring that observed changes are due to ligand binding and not to differences between protein purifications. The concentration of the RecOR complex was at 2 mg/ml. For the measurements in the presence of DNA, 50mer ssDNA and dsDNA (same oligonucleotide sequences as those used for the fluorescence anisotropy experiments) was added in a slight excess at a DNA:protein ratio of 1.2:1. Data were recorded using two detector distances at 2 and 10 m with a neutron wavelength of 6 Å, covering a q range of 0.007–0.35 Å−1, where q is the scattering vector (4πsinθ/λ). Data reduction was performed using the GRASansP software. After SANS data collection the binding of DNA to the drRecOR complex was verified by gel electrophoresis.
Small-angle scattering data processing
SAXS and SANS data were processed and analysis steps were performed using the ATSAS package (46) as described in (47). The final merged scattering data were further evaluated using PRIMUS. At low angle, the isotropic scattering data can be expressed as the Guinier approximation, I(q) = I(0) exp 1/3 Rg2q2. The isotropic scattering intensity I(q) was transformed to the distance distribution function P(r) using the program GNOM, which was also used to calculate the particle maximum dimensions Dmax. The optimum value of Dmax was found when the Rg obtained from the P(r) plot was equal that obtained from the Guinier analysis. For ab initio modeling of the SAXS data, 20 sets of independent models were calculated using Dammin (48), and then averaged and aligned using DAMAVER (49). Ab initio modeling of the drRecOR–DNA complexes were performed with the multi-phase dummy-atom modeling program MONSA (48). Theoretical scattering curves for known structures were calculated and compared with experimental data using CRYSOL (50) for X-ray data and CRYSON (51) for neutron data.
MD simulations
All MD simulations were carried out using the NAMD (52) package, in the isobaric-isothermal ensemble. The temperature and the pressure were kept constant at 300 K and 1 atm using, respectively, Langevin dynamics (53) and the Langevin piston method (54). The particle mesh Ewald algorithm (55) was used to account for long-range electrostatic interactions. Covalent bonds involving hydrogen atoms were constrained to their equilibrium length by means of the Rattle algorithm (56). The equations of motion were integrated by means of a multiple-time step algorithm (57) with a time step of 2 and 4 fs for short- and long-range interactions, respectively. The CHARMM27 (58,59) force field including CMAP (60) corrections was used to model proteins, DNA and counter-ions; the TIP3P (61) model was used to describe water.
Molecular assays were built starting from the crystal structure reported in this work (later called RecOR-‘open’) using the following procedure. The MODELLER package (62) was used to infer the position of the amino acids (G41, V42, K43, G44, P45, L46, S47) of the two RecO loop subunits missing in the crystal structure. After adding proper hydrogen atoms, the protein was fully solvated in a water box of 110 × 100 × 125 Å. An elongated single-strand of DNA composed of 10 thymine nucleotides was then inserted randomly in the central cavity of the complex, while preventing sterical clashes between ssDNA and proteins. The initial simulation box contains ∼110 000 atoms. Two different systems were considered involving, respectively, the K23A/R27A double-mutant and the wild-type RecOR complex. The latter was constructed by mutating residues A23 and A27 observed in the crystal structure of the four RecR subunits to K23 and R27. Proper positioning of the side chains of these lysine and arginine residues was made using the crystal structure of the wild type as a template (later called RecOR-‘closed’, PDB code 2V1C). All setups and analyses were made with VMD (63).
After 2000 steps of energy minimization, the two molecular assays were thermalized for 100 ns using the following protocol. During the first 5 ns, all the atoms of the system but water and counter-ions were kept fixed. For the next 40 ns, side chains and ssDNA were allowed to move while relaxing gently the position of the backbone atoms with stepwise-diminishing harmonic restraints. The overall system was then equilibrated further during 55 ns. Finally, data production and analysis was carried out all along the next 150 ns.
In a last step, starting from the wild-type and mutant equilibrated RecOR/ssDNA complexes, two apo-RecOR assays were built and subjected to the molecular-dynamics protocol described above. In a nutshell, four different assays were considered in this work, namely apo- and ssDNA complex of the wild-type and double-mutant RecOR complex. For each of them a trajectory of 250 ns was generated representing an overall effort of 1 µs. For the wild-type apo complex, a control simulation of 100 ns was produced starting from the ‘open’ crystal structure thermalized as described above.
RESULTS
Solution structure of the drRecR protein
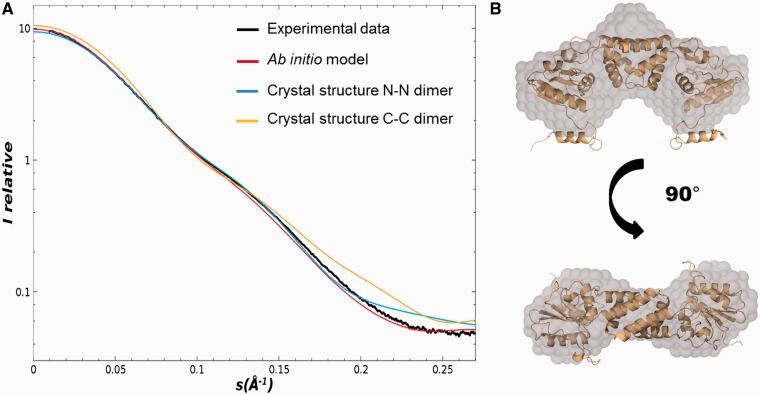
Despite many structural studies, the quaternary organization of drRecR in solution, in its free form remains unclear. Here, we performed SAXS experiments to determine the solution structure of drRecR. We compared various constructs of drRecR with N-terminal (RecR-NH) or C-terminal His-tag (RecR-CH) and without tag (RecR) to ensure that the oligomeric state is not dependent on the presence or absence of the tag. In addition, we tested a construct comprising drRecR residues 1–197 leaving out the C-terminal 23 residues (RecRdC) that are not conserved in other RecR homologs. All constructs tested in these experiments show similar scattering curves (Supplementary Figure S1) and the overall parameters of the constructs summarized in Table 1 clearly indicate that drRecR is dimeric in solution. The dimeric assembly of RecR in solution was additionally confirmed by static light scattering (SLS) measurements (Table 1). Moreover, calculated ab initio models derived from the SAXS curves are in good agreement with the structure of the drRecR dimer as shown in Figure 1A and B and could allow us to discriminate between N-N and C-C dimers. The calculated scattering curve of the N-N dimer displayed a much better fit to our experimental curve with a χ2 value of 4.16 than did the curve derived from the C-C dimer (χ2 value of 12.33). Our SAXS measurements also revealed the appearance of interparticle effects and the formation of larger particles at higher protein concentrations (above 5 mg/ml), which could be indicative of the formation of tetramers or higher order oligomers of drRecR.
Table 1.
In solution small-angle X-ray scattering data for drRecR and drRecOR
| Sample | Rg (Å) | Dmax (Å) | MM from I0 (kDa) | Average molecular mass (kDa) by SLS | ||
|---|---|---|---|---|---|---|
| RecR-NH | 33.6 ± 0.01 | 108 | 56 ± 0.1 (24.6) | 47.9 ± 0.5 | ||
| RecR-CH | 34.8 ± 0.01 | 110 | 58 ± 0.1 (24.8) | 48.4 ± 0.5 | ||
| RecR | 33.6 ± 0.01 | 108 | 56 ± 0.1 (24.3) | 48.8 ± 0.5 | ||
| RecRdC | 31 ± 0.1 | 100 | 54 ± 0.1 (22.3) | 45.2 ± 0.3 |
| Sample | Rg (Å) | Dmax (Å) | MM from I0 (kDa) | Porod Vol (Å3) | Excluded Vol (Å3) | MM (MMseq) (kDa) |
|---|---|---|---|---|---|---|
| RecOR | 40.0 ± 0.01 | 130 | 130 ± 5 | 234.5 | 242.7 | (153) 151.7 |
| RecOR K23A/R27A | 39.9 ± 0.01 | 130 | 130 ± 5 | n.d. | n.d. | n.d. |
RecR molecular masses (MM) obtained by SLS measurements are also presented.
Figure 1.
drRecR solution structure analyzed by SAXS. (A) Superposition of the experimental scattering curve obtained for drRecR (black) with the calculated scattering curves derived from the ab initio model of drRecR (red) and the crystal structures of the N-N (blue) and C-C (yellow) dimers of drRecR (pdb entry: 1vdd). (B) Overlay of the drRecR N-N dimer crystal structure with the envelope of the ab initio model calculated with dammin.
The structure of the drRecOR complex in an open conformation
Previously, the crystal structure of the drRecOR complex was reported to a maximum resolution of 3.8 Å (27). Owing to a limited observation-to-parameter ratio, refinement was terminated after a single round of rigid-body refinement (Rwork/Rfree of 45.9%/44.3%, respectively) and subsequent manual rebuilding of some residues. To improve data quality and to get more detailed information about the drRecOR complex, new crystallization trials were performed and we succeeded in the crystallization of the drRecOR complex using a RecR double-mutant (drRecORK23A/R27A), which still binds RecO and DNA. The mutations lie >20 Å away from the RecO–RecR interface. Using the RecOR mutant led to purer and highly homogenous material and facilitated its crystallization. SAXS experiments confirmed that the wild-type (drRecORWT) and the mutant complexes both consist of a hetero-hexameric complex and adopt a similar structure in solution displaying the same Rg and Dmax values (Table 1). The newly obtained crystals belong to the space-group P1 and diffracted X-rays to 3.34 Å resolution. The structure was determined by molecular replacement and subsequent refinement resulted in a Rwork/Rfree of 24.0%/27.8%, respectively (see Table 2 for complete refinement statistics).
Table 2.
Data collection and refinement statistics
| drRecOR-‘open’ | |||
|---|---|---|---|
| Data collection |
Refinement |
||
| Space group | P1 | PDB code | 4JCV |
| Cell dimensions | Resolution (Å) | 3.34 | |
| a, b, c (Å) | 63.1, 93.1, 92.3 | N° reflections | 22 815 |
| α, β, γ (°) | 103.6, 110.4, 106.2 | Rwork/Rfree (%) | 24.0/27.8 |
| Wavelength (Å) | 0.9793 | No. of atoms | |
| Resolution (Å) | 83.07–3.34 (3.52–3.34)a | Protein | 9156 |
| Rsym (%) | 6.1 (35.1)a | Zn | 6 |
| I/σI | 8.8 (2.0)a | Average B-factor | 52.6 |
| Completeness (%) | 94.2 (96.3)a | R.m.s deviations | |
| Redundancy | 1.8 (1.8)a | Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.329 | ||
aValues in parentheses are for highest-resolution shell.
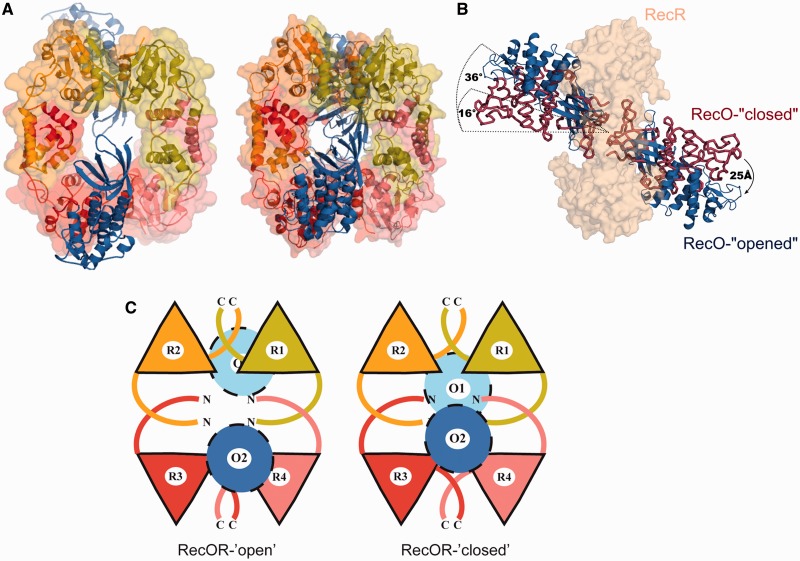
This new crystal structure reveals the same 4:2 stoichiometry as in the previous structure with RecR forming a tetrameric ring with one copy of RecO bound to each side of the ring (Figure 2A and C). Each asymmetric unit contained one full hetero-hexameric complex. Residues 200–220 of RecR and residues 41–47 and 227–244 of RecO could not be built owing to the absence or poor quality of the electron density for these residues. The root mean square deviation (rmsd) between the RecO molecules of the new and the previous structure is small, with a value of 1.256 Å (0.09 Å between the individual RecO molecules). The rmsd between the RecR rings of the two RecOR structures is slightly higher, with a value of 2.56 Å for all Cα atoms and of 2.76 Å compared with the RecR structure alone, indicating a certain flexibility of the RecR ring.
Figure 2.
Crystal structure of the RecOR complex in an open conformation. (A) Left: drRecOR-‘open’ crystal structure consisting of two RecO molecules shown in dark and light blue and four RecR chains (colored pale green, yellow, red and pink) forming a ring-like structure. Right: drRecOR-‘closed’ crystal structure shown in the same orientation and color scheme. (B) Comparison of the RecO orientation in the RecOR-‘open’ and RecOR-‘closed’ crystal structures. The RecR tetrameric ring, colored in beige and illustrated in surface representation, is viewed from the side. The crystal structures were superimposed on the RecR ring. The RecO from the RecOR-‘open’ structure is colored in blue, while the RecO from the RecOR-‘closed’ structure is colored in dark red. (C) Schematic representation of the hetero-hexameric RecOR complexes in their ‘open’ and ‘closed’ conformations. RecR tetrameric rings are made up of two RecR N-N dimers (R1-R4 and R2-R3) interacting via their C-terminal domains. The two RecO molecules (colored light and dark blue) largely obstruct the RecR ring in the RecOR-‘closed’ structure, whereas the opening of the RecO molecules in the RecOR-‘open’ structure creates a hole in the center of the hetero-hexameric complex.
This RecOR structure, however, shows significant differences compared with the previously reported RecOR regarding the localization and orientation of the RecO molecules (Figure 2B and C). In our new structure, the two RecO molecules are arranged at a 36° angle relative to an axis perpendicular to the center of the RecR tetrameric ring, and thus adopt a much more open conformation (Figure 2B); from here on, we will refer to this structure as RecOR-‘open’. In contrast, in the previous structure (now referred to as RecOR-‘closed’) this angle was only 16°. This 20° rotation results in an ∼25 Å movement of the position of the RecO helical domains (Figure 2C). In RecOR-‘closed’, the two facing OB domains interact tightly, leaving no gap between the RecO molecules, whereas in the ‘open’- RecOR structure, the distance between the two RecO molecules is significantly enlarged (closest contact is 13 Å), creating a large accessible area within the center of the RecR terameric ring and between the OB domains (Figure 2A and C and Supplementary Figure S2). In this conformation, the residues K23 and R27 of RecR that are implicated in DNA binding (23,27) and are located inside the RecR ring are now accessible for DNA binding.
drRecO–drRecR interaction area
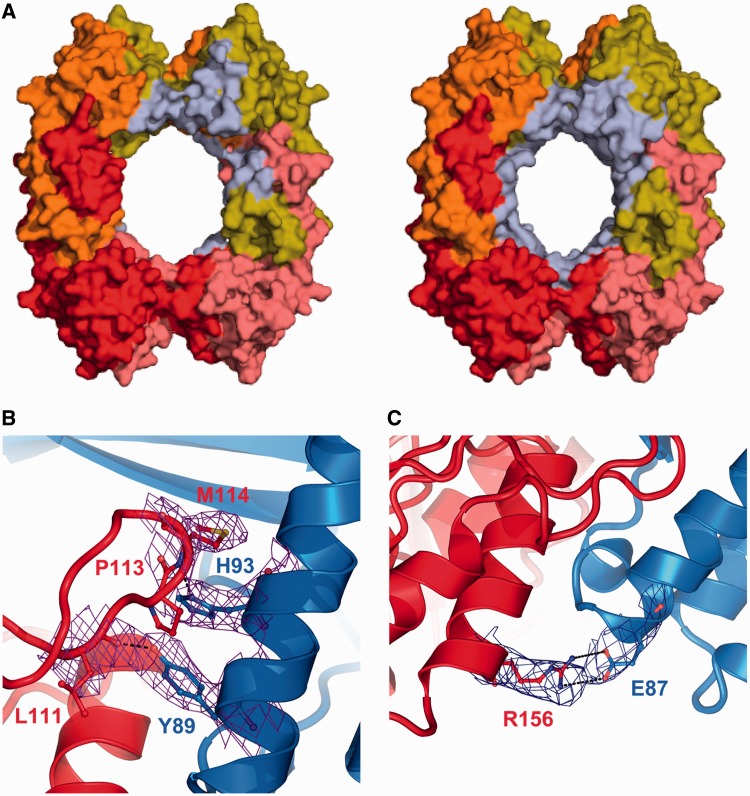
Because of the improved resolution of this new structure, a more detailed analysis of the protein–protein interfaces was possible. In the RecOR-‘closed’ structure, many of the RecR residues contributing to complex formation are located around its central hole (Figure 3A). In this conformation, the OB-fold domain of RecO forms extensive contacts with the N-terminal Helix-hairpin-Helix (HhH) (residues 1–30) motif, the C-terminal region, including the Walker B motif (residues 167–182) and the Toprim domain of RecR. In the RecOR-‘closed’ structure the surface area implicated in complex formation covers ∼3000 Å2. Within the RecOR-‘open’ structure, each RecO forms contacts with the Toprim and C-terminal domains of two of the RecR chains but shows fewer interactions with the HhH domain of RecR (Figure 3A). As a result, in this ‘open’ conformation the surface area implicated in complex formation is reduced to ∼2000 Å2. Two areas of the Toprim domain of RecR are involved in interactions with RecO (Figure 3B): (i) the loop following β-strand 4 (residues 110–115) and (ii) α-helix 6 (residues 144–156). Both of these regions interact with the α-helix 1 in the RecO molecule mostly through hydrophobic contacts, which are further stabilized by several ionic interactions involving notably RecO residues E87, Y89 and H93 and RecR residues L111, E146 and R156 (Figure 3B). RecR-E146 forms a salt bridge with RecO-R15 and as illustrated in Figure 3C, RecR-R156 forms a stable salt bridge with RecO-E87. RecO-H93 also interacts with RecR-P113 through π-stacking interactions. Mutational studies have previously shown that RecO-H93 is essential for RecOR complex formation (27). Finally, RecO contacts residues from both domain-swapped RecR C-terminal domains involved in the tetramerization interface and by this stabilizes the RecR tetramer. Despite the major conformational change observed in our new RecOR-‘open’ structure, 17 hydrophobic interactions, the π-stacking interaction between RecO-H93 and RecR-P113, and the salt bridge between RecO-R15 and RecR-E146 are observed in both RecOR crystal structures, underlining the importance of these residues in complex formation.
Figure 3.
drRecO-drRecR interface. (A) Surface representations of RecR (colored as in Figure 2), illustrating the regions involved in interactions with RecO (colored in pale blue) as observed in the RecOR-‘open’ (left) and RecOR-‘closed’ (right) structures. (B and C) Close-up views of the essential, electrostatic and ionic interactions formed between drRecR (red) and drRecO (blue) that contribute to the stability of the RecOR complex. The residues involved in these contacts are labeled and represented in sticks, and the electron density map (2Fo-Fc contoured at 1σ) corresponding to these residues is shown in purple.
DNA binding to drRecO, drRecR and the drRecOR complex
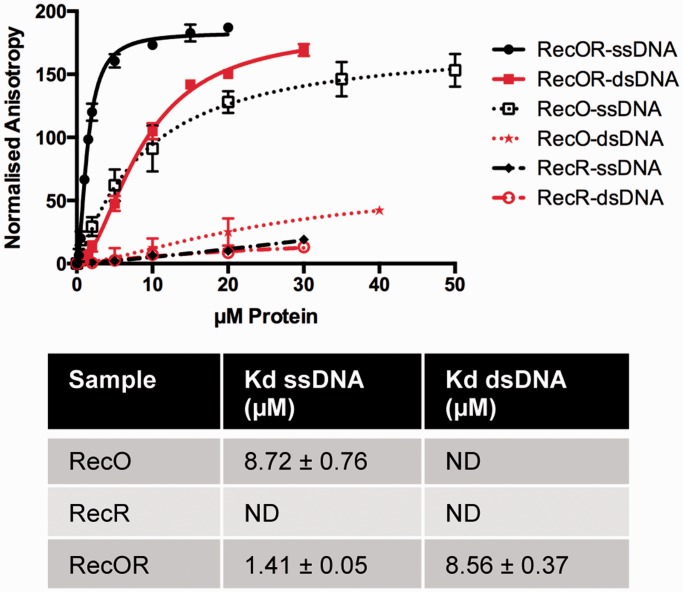
Binding of RecO, RecR and RecOR to 50mer fluorescently labeled ss- and dsDNA was assayed by fluorescence anisotropy measurements. In these experiments, no significant binding of RecR to either ssDNA or dsDNA could be detected (Figure 4) and no reliable dissociation constants could be derived from the fluorescence polarization measurements. RecO, as expected, showed a clear preference for ssDNA with a Kd of 8.7 µM. The RecOR complex in contrast bound to both ssDNA and dsDNA substrates with Kd values of, respectively, 1.4 and 8.5 µM (Figure 4).
Figure 4.
DNA binding of drRecOR, drRecO and drRecR. Fluorescence anisotropy measurements of drRecOR (solid lines), drRecO (dotted line) and drRecR (dashed lines) binding to ds- (red) and ssDNA (black) 50mer fluorescently labeled oligonucleotides. The data points corresponding to the average of three independent experiments were fitted to a standard binding equation assuming a single binding site with Hill slope using GraphPad Prism6 and standard deviations are depicted as vertical error bars. The estimated dissociation constants (Kd) derived from these fits are presented in the table below.
Solution structure of the drRecOR complex alone and with DNA
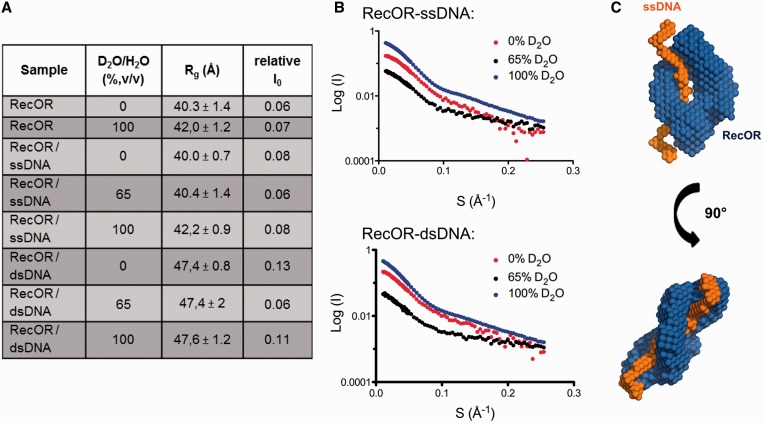
The results of SAXS and SANS experiments on RecOR are summarized in Table 1. The measurements revealed an Rg of ∼40 Å, which is in good agreement with the calculated ones of 37.6 Å for RecOR-‘open’ and 35.1 Å for RecOR-‘closed’. Both crystal structures are lacking several residues of the full-length proteins, especially the flexible C-terminal region of RecR, which explains the slightly smaller calculated Rg values compared with the experimental ones. The molecular weight determined from the I0 was ∼130 kDa compared with the theoretical value of 153 kDa. To verify the molecular weight of the complex in solution we calculated the excluded volume from 10 independent ab initio models. For globular proteins division of the excluded volume by 1.6 gives a good estimation for the molecular weight of the scattering object. The calculated molecular weight of 151.7 kDa matches well the theoretical one, indicating that RecOR has the same stoichiometry in solution as seen in the crystal structure. Fitting of the two crystal structures to the scattering curves results in a slightly better fit for the RecOR-‘open’ structure with a χ2 value of 1.63 compared with the RecOR-‘closed’ structure with a χ2 value of 2.23. Taken together these results suggest that the solution structure of the RecOR complex is compatible with both crystal structures.
To test how the drRecOR complex interacts with DNA substrates we performed SANS experiments. The use of SANS with contrast variation allows to separate the information about shape and internal structure by changing solvent density. This is possible because DNA has a different scattering length density than proteins. We measured the scattering of the RecOR complex bound to ssDNA and dsDNA in 0%, 65% (matching point of DNA) and 100% deuterium (Figure 5). The Rg values derived from the data recorded for the RecOR complex bound to ssDNA are similar to these values obtained for the RecOR complex alone, indicating that the solution structure of the complex remains the same in the presence of ssDNA (Figure 5A and B). Ab initio modeling of the different phases using the program MONSA resulted in structural models where the DNA substrate is always bound in the center of the RecOR complex and located in the center of the RecR tetrameric ring (Figure 5C). In contrast, the measurements of drRecOR bound to dsDNA resulted in larger Rg values approaching ∼47 Å (Figure 5A), indicating that the complex undergoes larger conformational changes to bind dsDNA. The I0 values, however, show that the overall stoichiometry of the complex remains the same. Analysis of the derived Rg values and ab initio models suggests that the complex binds to dsDNA in a more open conformation than to ssDNA, but here again the data strongly indicates that dsDNA is bound within the center of the RecR ring.
Figure 5.
SANS analysis of drRecOR-ssDNA and drRecOR–dsDNA complexes. (A) Table summarizing the neutron scattering data for drRecOR and drRecOR–DNA complexes. (B) Scattering profiles obtained for drRecOR-ssDNA (upper) and drRecOR-dsDNA (lower), which are displaced along the logarithmic axis for visualization, are shown as the logarithm of the scattering intensity, I (black dots), as a function of the momentum transfer, q ¼ 4π sin(θ) / λ, where 2θ is the scattering angle and λ is the X-ray wavelength. (C) Ab initio model calculated with Monsa for the drRecOR–ssDNA complex. The protein phase (drRecOR) is represented in blue, the ssDNA phase in orange.
MDs simulations
The drRecOR-‘open’ crystal structure exhibits a markedly open conformation compatible with the binding of ssDNA inside the RecR ring as suggested by our SANS experiments. To further assess the validity of this hypothesis, all-atom simulations were systematically carried out and four independent MD trajectories were generated for the wild-type and mutant drRecORK23A/R27A complexes with and without ssDNA, respectively. In the absence of structural information regarding the initial position of the substrate in the complex, an extended 10-nucleotide poly-dT ssDNA was arbitrarily threaded all the way through the RecR ring (Figure 6A and B). In all cases, the overall structure of the RecOR-‘open’ complex is retained over 250 ns, irrespective of the presence of ssDNA, and time-evolution functions of the backbone atom-positional rmsd fluctuate around ∼6.5 Å (Supplementary Figure S3) consistent with the size of this molecular assembly. The same quantity computed independently for all the complex subunits (Supplementary Figure S4) shows that the structure of each individual chain evolves marginally with respect to its initial conformation in the crystal. Fluctuations of the rmsd proved to be slightly larger for the RecR chains (∼2–4 Å) compared with those of RecO (∼2–3 Å) in agreement with the higher plasticity of each single RecR entity observed at the experimental level.
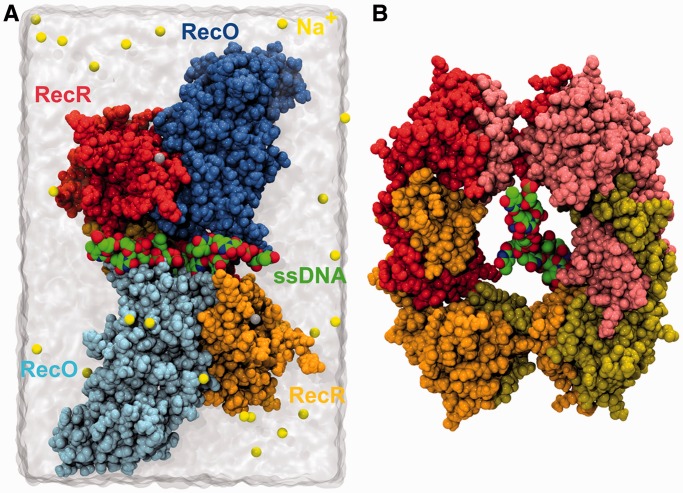
Figure 6.
MDs simulations set-up. (A) Initial setup of the drRecOR/ssDNA complex immerged in a water box. Yellow and gray spheres depict sodium counter-ions and zinc ions, respectively. For clarity purposes, only two RecR subunits are shown. (B) View of the initial position of the 10-thymine ssDNA in the central cavity of the drRecR tetrameric ring. The RecO and RecR chains are colored as in Figure 2.
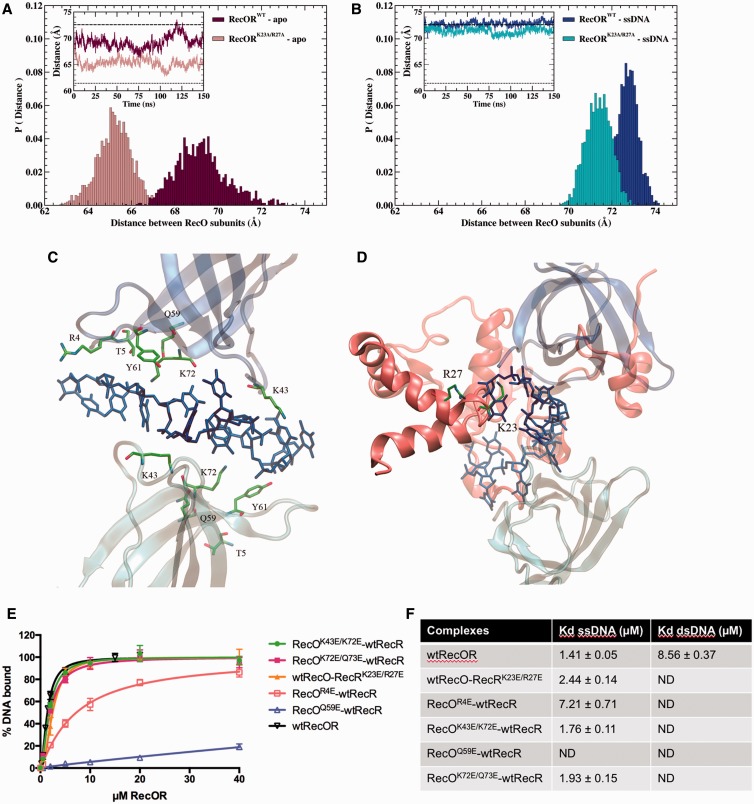
Although the insertion of ssDNA does not alter the overall structure of the complex, it modifies the positional dynamics of the two RecO subunits with respect to each other (Figure 7A and B). When ssDNA is bound to the complex, the distance between the center of mass of the two RecO proteins remains essentially near 71.4 Å (K23A/R27A) and 72.8 Å (wild-type), in line with the corresponding distance, 71.6 Å, observed in the ‘open’ crystal structure (Figure 7B). For the apo systems, the latter distance is markedly shorter, with the mean values of 65.4 Å (K23A/R27A) and 69.1 Å (wild-type) and navigates continuously between values measured in RecOR-‘closed’ (61.4 Å) and RecOR-‘open’ (71.6 Å) assemblies, respectively (Figure 7A and Supplementary Figure S5). The tilt angle between the normal to RecR tetrameric ring and the axis connecting the center of mass of the two RecO subunits follows a similar trend (Supplementary Figure S6), its mean value switches from ∼38° in the presence of ssDNA to ∼29° for the wild type apo system. The Cryson program was used to fit the experimental SANS data acquired for RecOR with and without ssDNA against structures sampled all along the MD trajectories. Rg values extracted from the fitting procedure (Figures S7) are distributed around ∼36.2 Å and ∼36.8 Å for apo and holo complexes, respectively, consistent with the value (37.6 Å) computed for the ‘open’ crystal structure. Taken together these results demonstrate that the RecOR-‘open’ structure can accommodate ssDNA in its central cavity without any major structural rearrangements.
Figure 7.
MD simulations analysis of drRecOR in the presence and absence of ssDNA. (A and B) Probability distribution and time evolution (insets) of the distance between the center of mass of the two RecO subunits in (A) drRecOR wild-type–apo and drRecORK23A/R27A–apo, and (B) drRecOR wild-type–ssDNA and drRecOR K23A/R27A–ssDNA. Dotted and dashed lines (insets) depict the corresponding distances observed in RecOR-‘closed’ (61.4 Å) and RecOR-‘open’ (71.6 Å), respectively. (C and D) Detailed view of the interaction between ssDNA and the wild type drRecOR complex predicted by MDs simulation. (C) Residues R4, T5, K43, Q59, Y61 and K72 of both drRecO subunits interacting with ssDNA. (D) Residues K23 and R27 of drRecR interacting with ssDNA. (E) Fluorescence anisotropy measurements of wild-type and mutant drRecOR complexes binding to 50mer fluorescently labeled ssDNA. The data points are the average of three independent experiments and were fitted to a standard binding equation assuming a single binding site with Hill slope using GraphPad Prism6. Standard deviations are depicted as vertical error bars. (F) Table summarizing the binding affinities (Kd values in µM) of wild-type and mutant drRecOR complexes for 50mer ss- and dsDNA derived from our fluorescence anisotropy measurements.
Molecular-dynamics trajectories further provide the atomistic view of ssDNA recognition by RecOR. Starting from an extended conformation, ssDNA folds in the middle of the ring cavity and is rapidly sandwiched between the two facing OB domains of RecO, forming long-lived electrostatic interactions. The stable binding motif, monitored for both the wild type and the double-mutant complex, involves polar and basic residues R4, T5, K43, Q59, Y61 and K72 symmetrically distributed over the two RecO subunits (Figure 7C). These residues form electrostatic interactions with six consecutive nucleotides in the ssDNA chain. Outside this intercalated region, the interaction of ssDNA with RecOR appears to be less specific and the terminal nucleotides are particularly flexible and freely accessible to the solvent. In the wild-type complex, an equally stable electrostatic interaction is formed with residues K23 and R27 of one of the four RecR subunits (Figure 7D) congruent with what has been described experimentally.
Mutagenesis analysis
To validate our SANS and MD data, we mutated residues R4, K43, Q59, K72 and Q73 of RecO and residues K23 and R27 of RecR to glutamates and measured the binding of these mutant RecOR complexes to either ss- or dsDNA (Figure 7E and F). All the mutants displayed impaired binding to dsDNA, clearly indicating that dsDNA binds through the center of the RecOR ring. RecO mutants K43E/K72E and K72E/Q73E retained wild-type ssDNA binding, while RecO mutant R4E showed a significantly weaker ssDNA binding ability. The RecOR complexes containing either RecO mutant Q59E or RecR mutant K23E/R27E showed no detectable binding to either ss- or dsDNA. Taken together, these results confirm that ssDNA also binds within the center of the RecOR complex and interacts with the OB domains of RecO, as suggested by our SANS and MD data.
DISCUSSION
Over the past years, a lot of structural information of RMPs belonging to the RecFOR pathway has become available. However, there are still a number of important questions that remain unanswered. The objective of this study was to gain insight into the assembly of the RecOR complex and improve our understanding of how it interacts with its DNA substrates.
The RecR protein of D. radiodurans has been reported to assemble as a tetramer and to act as a DNA clamp (23), while other RecR homologs have been shown to be dimeric in solution (29–32). The steps leading to assembly of RecR on DNA depend to a large extent on the quaternary structure of RecR in solution. As a tetramer, the RecR ring would have to open to encircle the DNA, whereas a RecR dimer may assemble on DNA as a dimer and then form a tetramer around the DNA duplex. Here SAXS and SLS analyses carried out on drRecR show that the protein is dimeric in solution, which is in agreement with recently published data (64) and strongly suggests that the tetrameric structures of RecR observed in crystals are favored by the high protein concentration and protein–protein contacts in the crystal lattice. All RecR dimers described so far consist of N-N dimers in which the HhH domain of RecR forms the principal interaction interface (29–32,64). In the available tetrameric structures of RecR (23,32), interactions between the C-terminal regions of two RecR monomers are also observed and the interaction surfaces involved in either N-N or C-C dimer interfaces are similar (∼2000 Å2). In the case of drRecR, our SAXS data clearly indicate that the stable dimer observed in solution is also the N-N dimer indicating that C-C dimers of drRecR do not form alone in solution.
Formation of RecR tetramers, consisting of two N-N dimers interacting via their C-terminal regions, is most likely induced by the interaction of RecR with one of its cellular partners (RecO and/or RecF) and/or with DNA. Our DNA binding data, in agreement with previous measurements, show that drRecR binds poorly to both ss- and dsDNA. In our fluorescence anisotropy measurements, the binding was so weak that we could not determine an accurate Kd value. DNA binding is therefore unlikely to be the inducer of tetramerization. In contrast, there is increasing evidence that binding of RecR to either RecO or RecF strongly favors RecR tetramerization. Our high- and low-resolution structural studies of the drRecOR complex and size-exclusion chromatography and SAXS analyses of T. thermophilus RecFR complex (30) indicate that both types of complexes contain a RecR tetramer. Single-molecule Förster resonance energy transfer measurements also recently showed that addition of RecO significantly stimulated the formation of drRecR tetramers (64). In the RecOR complex, over one-third of the RecR residues interacting with RecO are also implicated in the tetramerization interface of two RecR dimers. A given RecO molecule thus interacts with RecR molecules belonging to both dimers and thereby stabilizes the RecR ring-like structure. The RecO molecules also interact, but to a lesser extent, with several N-terminal residues of RecR implicated in N-N dimer formation.
The higher resolution crystal structure of drRecOR presented here allowed us to provide a more complete and reliable description of the RecO–RecR interface. In addition to its C-terminal region, two areas of the Toprim domain of drRecR are involved in interactions with drRecO. These regions and residues are conserved to varying extents in RecR and RecO proteins (Supplementary Figures S9 and S10). In RecR proteins, other than Helicobacter pylori RecR, the regions involved in complex formation are highly conserved. Only RecR-R156 that interacts strongly with E87 from RecO is not conserved. For RecO proteins, the overall sequence conservation is low and the regions involved in interactions with RecR are also only poorly conserved, indicating that these protein–protein interfaces may be species-specific and that the nature and stability of the RecOR complex may differ from one organism to another.
The two conformations observed in the RecOR crystal structures may reflect the intrinsic flexibility of the complex and an inherent ability of the RecO molecule to open and close the access to the RecR ring. The solution structure of drRecOR is in agreement with both of these crystal structures, thus confirming the four-to-two stoichiometry and overall assembly of RecOR as a tetrameric RecR ring with one RecO on either side also in solution. Each RecR molecule shows intrinsic flexibility that is in part stabilized by domain swapping at its N- and C-termini and RecO in contrast is a more rigid molecule but its orientation and interaction with RecR fluctuates as shown by MD simulations. When ssDNA is not bound to RecOR, MD reveals that the conformation of RecOR travels continuously between its ‘open’ and ‘closed’ states, although the limited sampling time does not allow capture of the complete transition. On the contrary, this positional dynamics is hampered by ssDNA, locking the protein complex in its ‘open’-structure.
The previously published drRecOR structure shows the complex with a four-to-two stoichiometry in a conformation where the inside of the RecR tetrameric ring is occluded by the two copies of RecO bound to it. In this conformation RecOR cannot bind DNA in the inside of the RecR ring. It was therefore proposed that the complex might undergo large conformational rearrangements and change its stoichiometry to interact with DNA (27). In this work the crystal structure of the RecOR complex was solved in an open conformation where the inside of the RecR tetrameric ring is partially accessible, providing sufficient space for ssDNA to bind.
Fluorescence anisotropy experiments show that RecOR, and not RecR alone, interacts with ss- and dsDNA. This differs from tteRecOR, which has been shown to specifically interact with ssDNA and not with dsDNA (32). Interestingly, tteRecOR has also been suggested to form a hetero-trimeric complex consisting of a RecR dimer and a RecO monomer (32). Taken together, these observations suggest that the ring-like tetrameric assembly of RecR may be required to enable RecR to bind to duplexed DNA.
SANS experiments of the drRecOR complex bound to ssDNA or dsDNA revealed that the RecOR complex maintains its four-to-two stoichiometry in the presence of DNA and indicate that the DNA locates to the inside of the complex. When bound to ssDNA, the overall conformation of the complex is the same as in its apo form, whereas when bound to dsDNA the complex behaves differently and significant conformational changes are observed. The increased Rg of the protein part of the complex indicates that binding of the complex to dsDNA induces a more open conformation than when binding to ssDNA. This could be explained by the larger diameter and reduced flexibility of dsDNA compared with ssDNA. In RecOR-‘open’, the accessible area within the RecR tetrameric ring is not large enough to harbor dsDNA, and larger conformational changes would be needed to accommodate a duplexed DNA within the center of the RecOR complex.
To gain further insight into the RecOR structure and dynamics in solution, our structural studies were combined with a MD simulation analysis of RecOR in the absence and presence of ssDNA. Both the previously reported RecOR-‘closed’ structure and the RecOR-‘open’ structure reported here are consistent with the calculated trajectories. In the presence of ssDNA, the distance between the centers of mass of the two RecO molecules remains larger than in the apo RecOR-‘closed’ complex structure and close to the value seen in the RecOR-‘open’ structure. Furthermore, on the time-scale explored in this work, the structure of the central cavity is not distorted on insertion of ssDNA. These results demonstrate that the RecOR-‘open’ structure is in a conformation compatible with the binding of ssDNA in its core.
We used MD simulations together with biochemical experiments to investigate the detailed interactions of drRecOR with ssDNA. The analysis revealed that ssDNA is sandwiched between the two copies of RecO forming extensive contacts with the OB domains of RecO. RecO residues interact in a nonspecific manner with the ssDNA, forming mostly ionic interactions with the phosphate backbone (Supplementary Figure S11). Our mutagenesis and DNA-binding studies reveal that RecO residues R4 and Q59 and RecR residues K23 and R27 are particularly important for RecOR binding to DNA.
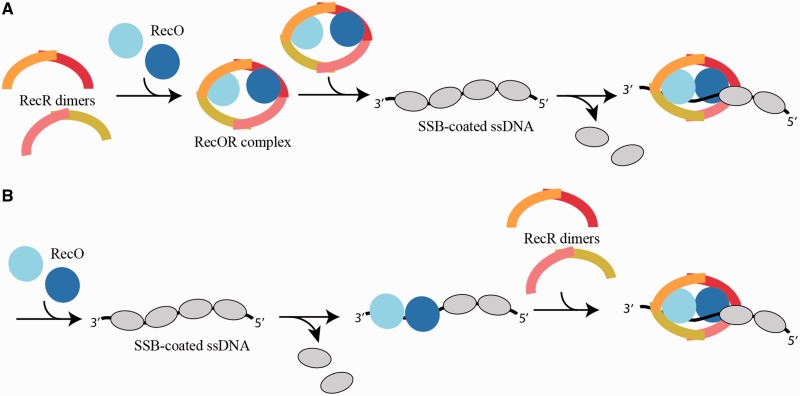
Altogether our data allow us to propose a model for the assembly of the drRecOR complex on DNA (Figure 8). In the cell as in solution, RecO most likely occurs as a monomer and RecR as a dimer (N-N dimer). Our data indicates that the interaction of RecO with RecR strongly favors the formation of RecR tetramers. The assembly of this hetero-hexameric complex may form in the cell before interaction with its SSB-coated ssDNA substrate (model A) or may alternatively assemble directly on SSB-coated ssDNA (model B). At present our data and the available literature do not allow us to discriminate between these two models. In both cases, RecO is believed to be involved in binding and perhaps also removal of SSB from the DNA. In model A, the threading of ssDNA through the RecOR complex would result in removal of the bound SSB and facilitate the assembly of RecA. In model B, RecO alone may bind to DNA and remove SSB before assembly of the RecOR complex. While our data clearly indicate that RecO triggers the tetramerization of RecR and the subsequent formation of the stable hetero-hexameric RecOR assembly and that ssDNA can bind inside such a complex, further studies will be needed to distinguish between these two models.
Figure 8.
Model for the assembly of the drRecOR complex on DNA. In solution, drRecO is a monomer and drRecR is a dimer (N-N). Tetramerization of drRecR is stabilized by the formation of a hetero-hexameric complex with drRecO. The assembly of this complex may form in the cell before interaction with its DNA substrates (A) or may alternatively assemble directly on the DNA (B). The binding of drRecOR to SSB-coated DNA would then disrupt SSB-ssDNA interactions, thereby facilitating the loading of RecA onto the DNA.
ACCESSION NUMBERS
pdb code: 4JCV.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–11 and References [65,66].
FUNDING
ESRF in-house research program and an ATIP-AVENIR-Centre National de la Recherche Scientifique-Ligue contre le Cancer program (to J.T.). Funding for open access charge: ATIP-AVENIR grant from the Centre National de la Recherche Scientifique and Ligue contre le Cancer.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The super computer center of the University of Aix-Marseille is gratefully acknowledged for the generous provision of Cpu time (project selected for testing the architecture). The authors thank Dr Phil Callow and Dr Giuseppe Zaccai (ILL) for providing local support on the instrument D22 and help for data processing.
REFERENCES
- 1.Cox MM. Recombinational DNA repair in bacteria and the RecA protein. Prog. Nucleic Acid Res. Mol. Biol. 1999;63:311–366. doi: 10.1016/s0079-6603(08)60726-6. [DOI] [PubMed] [Google Scholar]
- 2.Cox MM, Goodman MF, Kreuzer KN, Sherratt DJ, Sandler SJ, Marians KJ. The importance of repairing stalled replication forks. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 3.Kowalczykowski SC. Initiation of genetic recombination and recombination-dependent replication. Trends Biochem. Sci. 2000;25:156–165. doi: 10.1016/s0968-0004(00)01569-3. [DOI] [PubMed] [Google Scholar]
- 4.Kuzminov A. DNA replication meets genetic exchange: chromosomal damage and its repair by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8461–8468. doi: 10.1073/pnas.151260698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Umezu K, Kolodner RD. Protein interactions in genetic recombination in Escherichia coli. Interactions involving RecO and RecR overcome the inhibition of RecA by single-stranded DNA-binding protein. J. Biol. Chem. 1994;269:30005–30013. [PubMed] [Google Scholar]
- 6.Sweezy MA, Morrical SW. Biochemical interactions within a ternary complex of the bacteriophage T4 recombination proteins uvsY and gp32 bound to single-stranded DNA. Biochemistry. 1999;38:936–944. doi: 10.1021/bi9817055. [DOI] [PubMed] [Google Scholar]
- 7.Fujii S, Isogawa A, Fuchs RP. RecFOR proteins are essential for Pol V-mediated translesion synthesis and mutagenesis. EMBO J. 2006;25:5754–5763. doi: 10.1038/sj.emboj.7601474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lloyd RG, Thomas A. On the nature of the RecBC and RecF pathways of conjugal recombination in Escherichia coli. Mol. Gen. Genet. 1983;190:156–161. doi: 10.1007/BF00330339. [DOI] [PubMed] [Google Scholar]
- 9.Wang TC, Smith KC. Mechanisms for recF-dependent and recB-dependent pathways of postreplication repair in UV-irradiated Escherichia coli uvrB. J. Bacteriol. 1983;156:1093–1098. doi: 10.1128/jb.156.3.1093-1098.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kolodner R, Fishel RA, Howard M. Genetic recombination of bacterial plasmid DNA: effect of RecF pathway mutations on plasmid recombination in Escherichia coli. J. Bacteriol. 1985;163:1060–1066. doi: 10.1128/jb.163.3.1060-1066.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Symington LS. Role of RAD52 epistasis group genes in homologous recombination and double-strand break repair. Microbiol. Mol. Biol. Rev. 2002;66:630–670. doi: 10.1128/MMBR.66.4.630-670.2002. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ouyang KJ, Woo LL, Ellis NA. Homologous recombination and maintenance of genome integrity: cancer and aging through the prism of human RecQ helicases. Mech. Ageing Dev. 2008;129:425–440. doi: 10.1016/j.mad.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Tal A, Arbel-Goren R, Stavans J. Cancer-associated mutations in BRC domains of BRCA2 affect homologous recombination induced by Rad51. J. Mol. Biol. 2009;393:1007–1012. doi: 10.1016/j.jmb.2009.09.011. [DOI] [PubMed] [Google Scholar]
- 14.Powell SN, Willers H, Xia F. BRCA2 keeps Rad51 in line. High-fidelity homologous recombination prevents breast and ovarian cancer? Mol. Cell. 2002;10:1262–1263. doi: 10.1016/s1097-2765(02)00789-x. [DOI] [PubMed] [Google Scholar]
- 15.Thompson LH, Schild D. Recombinational DNA repair and human disease. Mutat. Res. 2002;509:49–78. doi: 10.1016/s0027-5107(02)00224-5. [DOI] [PubMed] [Google Scholar]
- 16.Rocha EP, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:e15. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Volkert MR, Hartke MA. Effects of the Escherichia coli recF suppressor mutation, recA801, on recF-dependent DNA-repair associated phenomena. Mutat. Res. 1987;184:181–186. doi: 10.1016/0167-8817(87)90015-0. [DOI] [PubMed] [Google Scholar]
- 18.Hegde S, Sandler SJ, Clark AJ, Madiraju MV. recO and recR mutations delay induction of the SOS response in Escherichia coli. Mol. Gen. Genet. 1995;246:254–258. doi: 10.1007/BF00294689. [DOI] [PubMed] [Google Scholar]
- 19.Wang TC, Smith KC. recA (Srf) suppression of recF deficiency in the postreplication repair of UV-irradiated Escherichia coli K-12. J. Bacteriol. 1986;168:940–946. doi: 10.1128/jb.168.2.940-946.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang TC, Chang HY, Hung JL. Cosuppression of recF, recR and recO mutations by mutant recA alleles in Escherichia coli cells. Mutat. Res. 1993;294:157–166. doi: 10.1016/0921-8777(93)90024-b. [DOI] [PubMed] [Google Scholar]
- 21.Madiraju MV, Templin A, Clark AJ. Properties of a mutant recA-encoded protein reveal a possible role for Escherichia coli recF-encoded protein in genetic recombination. Proc. Natl Acad. Sci. USA. 1988;85:6592–6596. doi: 10.1073/pnas.85.18.6592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bell JC, Plank JL, Dombrowski CC, Kowalczykowski SC. Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature. 2012;491:274–278. doi: 10.1038/nature11598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee BI, Kim KH, Park SJ, Eom SH, Song HK, Suh SW. Ring-shaped architecture of RecR: implications for its role in homologous recombinational DNA repair. EMBO J. 2004;23:2029–2038. doi: 10.1038/sj.emboj.7600222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leiros I, Timmins J, Hall DR, McSweeney S. Crystal structure and DNA-binding analysis of RecO from Deinococcus radiodurans. EMBO J. 2005;24:906–918. doi: 10.1038/sj.emboj.7600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makharashvili N, Koroleva O, Bera S, Grandgenett DP, Korolev S. A novel structure of DNA repair protein RecO from Deinococcus radiodurans. Structure. 2004;12:1881–1889. doi: 10.1016/j.str.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 26.Koroleva O, Makharashvili N, Courcelle CT, Courcelle J, Korolev S. Structural conservation of RecF and Rad50: implications for DNA recognition and RecF function. EMBO J. 2007;26:867–877. doi: 10.1038/sj.emboj.7601537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Timmins J, Leiros I, McSweeney S. Crystal structure and mutational study of RecOR provide insight into its mode of DNA binding. EMBO J. 2007;26:3260–3271. doi: 10.1038/sj.emboj.7601760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ryzhikov M, Koroleva O, Postnov D, Tran A, Korolev S. Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein. Nucleic Acids Res. 2011;39:6305–6314. doi: 10.1093/nar/gkr199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Honda M, Inoue J, Yoshimasu M, Ito Y, Shibata T, Mikawa T. Identification of the RecR Toprim domain as the binding site for both RecF and RecO. A role of RecR in RecFOR assembly at double-stranded DNA-single-stranded DNA junctions. J. Biol. Chem. 2006;281:18549–18559. doi: 10.1074/jbc.M512658200. [DOI] [PubMed] [Google Scholar]
- 30.Honda M, Fujisawa T, Shibata T, Mikawa T. RecR forms a ring-like tetramer that encircles dsDNA by forming a complex with RecF. Nucleic Acids Res. 2008;36:5013–5020. doi: 10.1093/nar/gkn471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Webb BL, Cox MM, Inman RB. An interaction between the Escherichia coli RecF and RecR proteins dependent on ATP and double-stranded DNA. J. Biol. Chem. 1995;270:31397–31404. doi: 10.1074/jbc.270.52.31397. [DOI] [PubMed] [Google Scholar]
- 32.Tang Q, Gao P, Liu YP, Gao A, An XM, Liu S, Yan XX, Liang DC. RecOR complex including RecR N-N dimer and RecO monomer displays a high affinity for ssDNA. Nucleic Acids Res. 2012;40:11115–11125. doi: 10.1093/nar/gks889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alonso JJ, Marro J, Gonzalez-Miranda JM. Reaction-diffusion lattice gas: theory and computer results. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 1993;47:885–898. doi: 10.1103/physreve.47.885. [DOI] [PubMed] [Google Scholar]
- 34.Webb BL, Cox MM, Inman RB. Recombinational DNA repair: the RecF and RecR proteins limit the extension of RecA filaments beyond single-strand DNA gaps. Cell. 1997;91:347–356. doi: 10.1016/s0092-8674(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 35.Inoue J, Honda M, Ikawa S, Shibata T, Mikawa T. The process of displacing the single-stranded DNA-binding protein from single-stranded DNA by RecO and RecR proteins. Nucleic Acids Res. 2008;36:94–109. doi: 10.1093/nar/gkm1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Webb BL, Cox MM, Inman RB. ATP hydrolysis and DNA binding by the Escherichia coli RecF protein. J. Biol. Chem. 1999;274:15367–15374. doi: 10.1074/jbc.274.22.15367. [DOI] [PubMed] [Google Scholar]
- 37.Sugiyama T, New JH, Kowalczykowski SC. DNA annealing by RAD52 protein is stimulated by specific interaction with the complex of replication protein A and single-stranded DNA. Proc. Natl Acad. Sci. USA. 1998;95:6049–6054. doi: 10.1073/pnas.95.11.6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mazloum N, Zhou Q, Holloman WK. DNA binding, annealing, and strand exchange activities of Brh2 protein from Ustilago maydis. Biochemistry. 2007;46:7163–7173. doi: 10.1021/bi700399m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Battye TG, Kontogiannis L, Johnson O, Powell HR, Leslie AG. iMOSFLM: a new graphical interface for diffraction-image processing with MOSFLM. Acta Crystallogr. D Biol. Crystallogr. 2011;67:271–281. doi: 10.1107/S0907444910048675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Winn MD, Ballard CC, Cowtan KD, Dodson EJ, Emsley P, Evans PR, Keegan RM, Krissinel EB, Leslie AG, McCoy A, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D, Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J. Appl. Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr. D Biol. Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 45.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, III, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: defining accurate macromolecular structures, conformations and assemblies in solution. Q. Rev. Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 48.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003;36:860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- 51.Svergun DI, Richard S, Koch MH, Sayers Z, Kuprin S, Zaccai G. Protein hydration in solution: experimental observation by X-ray and neutron scattering. Proc. Natl Acad. Sci. USA. 1998;95:2267–2272. doi: 10.1073/pnas.95.5.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K. Scalable molecular dynamics with NAMD. J. Comput. Chem. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feller SE, Zhang YH, Pastor RW, Brooks BR. Constant pressure molecular dynamics simulation: the Langevin piston method. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 54.Martyna GJ, Tobias DJ, Klein ML. Constant-pressure molecular-dynamics algorithms. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 55.Darden T, York D, Pedersen L. Particle mesh Ewald: an N.Log(N) Method for Ewald sums in large systems. J. Chem. Phys. 1993;98:10089–10092. [Google Scholar]
- 56.Andersen HC. Rattle: a velocity version of the shake algorithm for molecular-dynamics calculations. J. Comput. Phys. 1983;52:24–34. [Google Scholar]
- 57.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular-dynamics. J. Chem. Phys. 1992;97:1990–2001. [Google Scholar]
- 58.MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 59.Foloppe N, MacKerell AD. All-atom empirical force field for nucleic acids: I. Parameter optimization based on small molecule and condensed phase macromolecular target data. J. Computat. Chem. 2000;21:86–104. [Google Scholar]
- 60.Mackerell AD, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Computat. Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 61.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 62.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 63.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. Model. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 64.Kim C, Kim JY, Kim SH, Lee BI, Lee NK. Direct characterization of protein oligomers and their quaternary structures by single-molecule FRET. Chem. Commun. 2012;48:1138–1140. doi: 10.1039/c2cc16528g. [DOI] [PubMed] [Google Scholar]
- 65.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res. 2003;31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.