Abstract
Left-handed Z-DNA has been identified as a transient structure occurred during transcription. DNA B-Z transition has attracted much attention because of not only Z-DNA biological importance but also their relation to disease and DNA nanotechnology. Recently, photoluminescent carbon dots, especially highly luminescent nitrogen-doped carbon dots, have attracted much attention on their applications to bioimaging and gene/drug delivery because of carbon dots with low toxicity, highly stable photoluminescence and controllable surface function. However, it is still unknown whether carbon dots can influence DNA conformation or structural transition, such as B-Z transition. Herein, based on our previous series work on DNA interactions with carbon nanotubes, we report the first example that photoluminescent carbon dots can induce right-handed B-DNA to left-handed Z-DNA under physiological salt conditions with sequence and conformation selectivity. Further studies indicate that carbon dots would bind to DNA major groove with GC preference. Inspired by carbon dots lighting up Z-DNA and DNA nanotechnology, several types of DNA logic gates have been designed and constructed based on fluorescence resonance energy transfer between photoluminescent carbon dots and DNA intercalators.
INTRODUCTION
DNA is polymorphic and exists in a variety of distinct conformations in vivo (1–3). In addition to the canonical right-handed B-form, left-handed Z-DNA has been identified under physiological conditions as a transient structure occasionally induced by biological process, such as transcription, methylation of cytosine and DNA supercoiling (4,5). The high-binding affinity and specificity of Z-DNA-binding proteins or enzymes to Z-DNA domain indicate that the formed transient structure is novel and important regulators of several genes, such as c-myc, colony stimulating factor 1(CSF-1) and a disintegrin and metalloproteinase domain-containing protein 12 (human ADAM-12) (6,7). The biological relevance of Z-DNA with Alzheimer’s disease (AD) risk factors has also been recently reported (8). Besides, DNA B-Z transition has been used in DNA nanotechnology for design and manufacture of artificial nucleic acid structures (9,10) for technological uses and even for synthetic biology (11). Therefore, DNA B-Z transition has attracted much attention because of not only Z-DNA biological importance but also their relation to disease and DNA nanotechnology. From numerous experimental and theoretical approaches, it is known that GC-rich B-DNA, which usually contains alternating syn-G and anti-C nucleosides, is favored to convert to Z-DNA by several external stimuli, such as high ionic strength (over 2 M NaCl) (12,13), negative supercoiling (14), solvent condition (8), protein binding (15) and chemical modification (16). Few small molecules, cobalt hexamine, polyamines, cationic copolymer and polynuclear platinum complexes (17–21) and two pairs of enantiomers (13,22) have been reported to stabilize Z-DNA and induce DNA allosteric transition. Considering the biological importance of Z-DNA and application of B-Z transition in DNA nanotechnology, greater efforts are urgently needed for developing new Z-DNA-binding agents that can promote B-Z transition under physiological conditions and provide potential for DNA nanotechnology.
Carbon nanomaterials have shown potential applications ranging from gene therapy, novel drug delivery to biosensing. Among them, the interactions between single-walled carbon nanotubes (SWNTs) and DNA have received great attention, and significant progress has been made. Zheng et al. (23) have shown that a particular sequence of single-stranded DNA self-assembles into a helical structure around individual carbon nanotubes. Strano et al. (24) have reported that DNA B-Z transition induced by metal ions can still occur on the surface of SWNTs. Our group has carried out a series of DNA interactions with SWNTs (25–32) and found that SWNTs can destabilize duplex DNA, induce a sequence-dependent DNA B–A transition (29) and can facilitate self-structuring of ssRNA poly(rA) to form A·A+ duplex structure (30). Intriguingly, SWNTs, not multi-walled carbon nanotubes, can selectively stabilize and induce human telomeric i-motif DNA formation and cause telomere dysfunction (25–28). All these works indicate that the intrinsic properties of material itself have a great influence on its applications, such as the surface functionalization, the degree of match between the size and DNA structure (25–32). As a new emerging member of carbon nanomaterials, photoluminescent carbon dots (C-dots, sub-10 nm), especially nitrogen-doped C-dots, have attracted growing interests in recent years owing to their great potential in biological labeling, bioimaging, drug delivery and optoelectronic device applications with stable photoluminescence, broad excitation spectrum, tunable emission wavelength and excellent biocompatibility (33–35). Recently, polyethylenimine-passivated C-dots as a nanogene vector has been reported to condense double-stranded plasmid DNA and mediate its transfection and exhibit upregulation of marker gene expression over naked DNA using COS-7 and HepG2 cells (36). Inspired by the intriguing enhanced photoluminescence of nitrogen-doped C-dots and progress on carbon nanotube–DNA interactions, we attempted to explore whether functionalized C-dots can change DNA structure and facilitate DNA structure transition, especially B-Z transition.
We report here for the first time that positively charged spermine-functionalized C-dots (SC-dots) can induce DNA B-Z transition, even under physiological (low salt concentration) condition [10 mM PBS, 100 mM NaCl (pH = 7.2)] with sequence selectivity (Scheme 1A). The binding mode, binding preference and the impact on DNA conformation transition were carefully studied. Contrasting changes for SC-dots binding to poly[dGdC]:poly[dGdC] (GC-DNA) and poly[dAdT]:poly[dAdT] (AT-DNA) were observed. For GC homopolymer and calf thymus DNA (ct-DNA), on SC-dots binding, typical circular dichroism (CD) spectra of Z-DNA were observed, but no such change was observed for AT-DNA. Our spectral and competitive binding studies provided direct evidence that SC-dots could induce DNA B to Z transition under low salt condition, and they bound to the DNA major groove with GC preference. Furthermore, using this unique photoluminescent SC-dots to lighten up Z-DNA structure, several optical logic gates were designed (Scheme 1B), which might provide new insights into the applications of C-dots under physiological conditions and the novel use of Z-DNA in nanoscience and DNA nanotechnology.
Scheme 1.
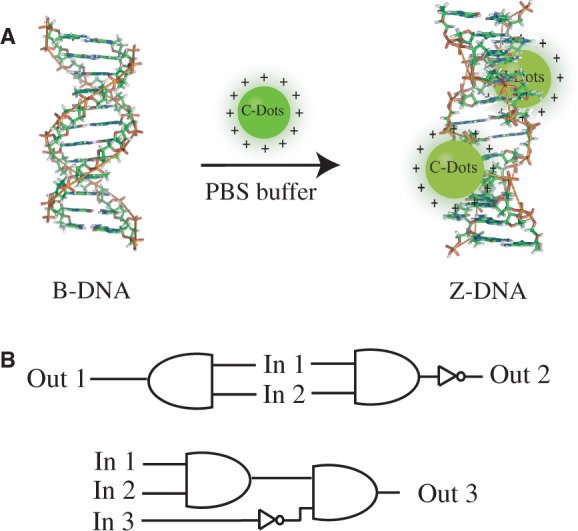
Schematic representation of the B-Z DNA transition induced by SC-dots (A) and designed logic gates (B).
MATERIALS AND METHODS
Materials
D-(+)-glucose (99.995%), spermine (97%) and NaCl (99.999%) were purchased from Aldrich. The ct-DNA was obtained from Sigma and purified before use. Poly[dGdC]:poly[dGdC] (GC-DNA), poly[dAdT]:poly[dAdT] (AT-DNA), polydA and polydT were purchased from Pharmacia as shown in Supplementary Table S1. The concentration of ct-DNA, GC-DNA and AT-DNA were determined by ultraviolet (UV) absorbance measurements using the extinction coefficient: Ɛ260 = 12 824 M−1 cm−1, Ɛ260 = 16 800 M−1 cm−1, Ɛ260 = 13 200 M−1 cm−1, respectively. Oligonucleotides were stored frozen −20°C before use, and the concentration of stock solutions was determined by UV absorbance. Ethidium bromide (EB), Hoechst 33258 and daunomycin (DM) were purchased from Sigma, and methyl green (MG) was purchased from Aldrich and were used without further purification. Their concentrations were determined by absorbance measurements using the extinction coefficient: Ɛ480 = 5600 M−1 cm−1, Ɛ338 =42 000 M−1 cm−1, Ɛ480 = 11 500 M−1 cm−1 for EB, Hoechst 33258 and DM, respectively. All the experiments were carried out in phosphate buffer [10 mM, 100 mM NaCl (pH = 7.2)] unless stated otherwise. In sodium iodide fluorescence quenching experiments, the ionic strength was kept constant. All aqueous solutions were prepared using ultra-pure water (18.2 MΩ, Milli-Q, Millipore).
Measurements
FT-IR characterization was carried out on a BRUKE Vertex 70 FT-IR spectrometer. Sample was thoroughly ground with exhaustively dried KBr. Atomic force microscope (AFM) measurements were performed using Nanoscope V multimode atomic force microscope (Veeco Instruments, USA). Transmission electron microscopy (TEM) images were recorded using a FEI TECNAI G2 20 high-resolution transmission electron microscope operating at 200 kV. Powder X-ray diffraction data were collected on a Bruker D8-ADVANCE diffractometer equipped with Cu Kα at a scan speed of 5 min−1. The ζ-potentials of the carbon dots were measured in a Zetasizer 3000HS analyzer. Energy Dispersive Spectrometer (EDX) for element analysis was carried out on a Hitachi S-4500 instrument. X-ray photoelectron spectroscopy (XPS) analysis was measured on an ESCALAB MK II X-ray photoelectron spectrometer using Mg as the exciting source.
CD spectra were measured at 25°C on a JASCO J-810 spectropolarimeter with a computer-controlled water bath. The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen (99.99%) for 45 min before use and kept the nitrogen atmosphere during experiments. Three scans were accumulated and automatically averaged. Absorbance measurements and melting experiments were made on a Cary 300 UV/Vis spectrophotometer, equipped with a Peltier temperature control accessory. All UV/Vis spectra were measured in 1.0 cm path length quartz cuvettes with the same concentration of SC-dots aqueous solution as the reference. Primary data were transferred to the graphics program Origin for plotting and analysis. Fluorescence experiments were carried out on a JASCO FP-6500 spectrofluorometer at 25°C.
Preparation of carbon dots
According to our described procedures on carbon dots synthesis (35), the mixture of 0.1 g D-(+)-glucose, 0.1 g spermine and 0.01 g NaCl was mixed thoroughly in 1 ml H2O under lower power, then irradiated with intermittent heating (∼30 s/time) and mixing in a domestic microwave oven (750 W) for a total of 3 min. The color-changed solution was subsequently dialyzed through the molecularporous memebrane tubing (MWCO:1000, Spectrum Laboratories, Inc. US) for further characterization and use. G-dots solution was obtained as control by the similar procedure, just no spermine containing.
Logic operations
AND gate: Four separate Eppendorf tubes corresponding to the four possible states of the AND gate (0,0; 1,0; 0,1; 1,1) were prepared by addition of solution of the AND gate (5 µg/ml SC-dots solution), 2 µM ct-DNA (input-1, In 1) and 1 µM EB (input-2, In 2) in 10 mM phosphate buffer, 100 mM NaCl (pH 7.2). These samples were incubated at 25°C for 2 min. Fluorescence spectra were then measured individually be excitation at 400 nm and recording the fluorescence emission at 585 nm (Out 1).
NAND gate: For NAND gate, the same conditions were used as AND gate. Fluorescence spectra were then measured individually being excited at 400 nm and recording the fluorescence emission at 465 nm (Out 2).
AND + INH and NAND + INH gates: Two gates were constructed based on whether the NaI can quench the fluorescence of SC-dots, and then they influenced the fluorescence resonance energy transfer (FRET) process. Based on the aforementioned logic gates, iodide ion was further adopted as the third input (input-3, In 3). These samples were incubated at 25°C for 2 min. Fluorescence spectra were then measured individually be excitation at 400 nm and recording the fluorescence emissions at 585 nm (Out 3) and 465 nm (Out 4).
RESULTS AND DISCUSSION
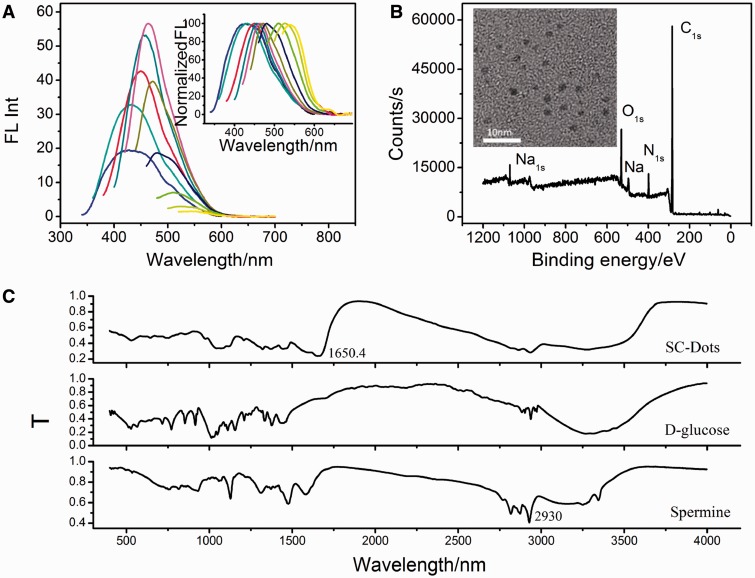
To perform DNA B-Z transition, water-soluble positively charged SC-dots were first prepared through microwave assisted synthesis, which would provide a facile, economic and green one-step route for large-scale synthesis of C-dots (Supplementary Scheme S1) (37). The mixture of D-glucose, spermine and a tiny amount of NaCl (10:10:1, weight ratio) was irradiated with intermittent heating and mixing in a domestic microwave oven (750 W) for a total of 3 min. As shown in Figure 1A and inset, the emission spectra of the as-prepared SC-dots were broad, ranging from ∼425 (blue) to ∼540 nm (yellow), with a dependence on the excitation wavelength. TEM (Figure 1B inset) showed that the as-prepared SC-dots had a diameter distribution of 2.26 ± 0.9 nm (based on statistical analysis of >100 dots, Supplementary Figure S2). The XPS spectra (Figure 1B) of SC-dots showed three peaks at 284.5, 400.0 and 530.8 eV, which were attributed to C1s, N1s and O1s, respectively. The C1s spectrum (Supplementary Figure S3B) showed two peaks at 284.6 and 288.1 eV, which were attributed to C–C and C = N/C = O, respectively. The N1s peak around 400.8 eV also indicated the presence of C = N−C formation (38). Elemental analysis revealed that the composition of SC-dots was C 51.73, O 24.06, N 21.96 and H (calculated) 2.25%. FT-IR spectra (Figure 1C) were further used to identify the functional groups presented on SC-dots. The broad bands ∼2930 cm−1 and 3345 cm−1 suggested the existence of N-H and –OH groups. A significant new band ∼1650.4 cm−1 was attributed to the stretching vibration peak of C = N bond, i.e. Schiff’s base structure by the reaction of aldehyde ends of the D-glucose and amine moieties of spermine with the imine linkages (39,40). X-ray diffraction pattern displayed a broad diffraction peak at 2θ = 20.8°, indicating an amorphous nature (Supplementary Figure S4) (36). The SC-dots were positively charged owing to the presence of amino groups outside, which was consistent with the zeta potential value (18.24 mV).
Figure 1.
(A) Emission spectra (with progressively longer excitation wavelengths from 300 to 480 nm in 20 nm increments) of SC-dots. Inset: the emission intensity was normalized. (B) XPS spectra of SC-dots. Inset: TEM image of SC-dots. (C) FT-IR spectra SC-dots, D-glucose and spermine, respectively.
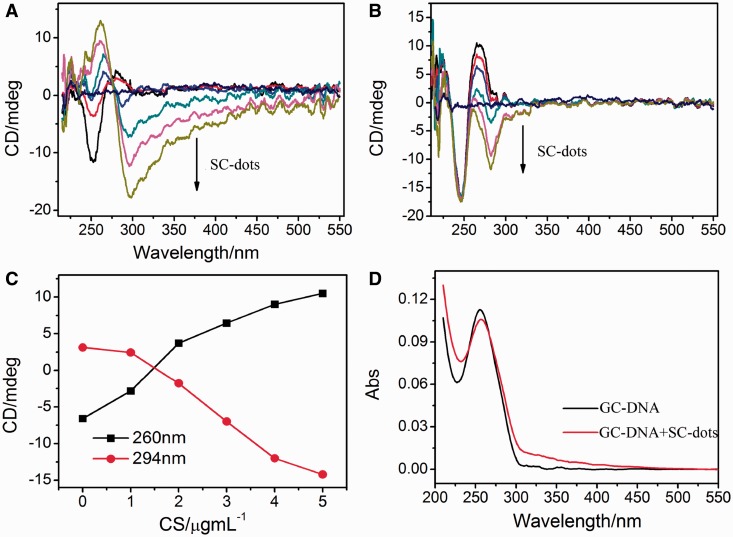
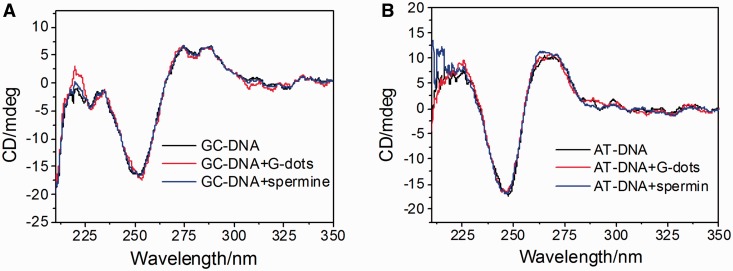
Next, we studied the interactions between DNA and SC-dots. CD spectra showed that GC- and AT-DNA DNA were in B-form with a positive band near 270 nm and a negative band near 250 nm in the absence of SC-dots under physiological conditions (Figure 2) (29). On addition of SC-dots, the canonical B-form of GC-DNA was changed, at 5 µg/ml SC-dots, Z-DNA CD characteristics with a positive band near 260 nm and a negative band at 294 nm were observed (17–22), indicating that B-Z transition occurred (Figure 2A) (12–21,41). The transition was cooperative and the transition midpoint was ∼2 µg/ml SC-dots. As for AT-DNA, it persisted in B-form on addition of SC-dots under the same conditions. The negative band of B-DNA near 250 nm was hardly changed even at 5 µg/ml SC-dots (Figure 2B and Supplementary Figure S5A). The Z-DNA peaks of GC-DNA increased after gradually addition of SC-dots (Figure 2C). As the B-Z transition was accompanied by a change in the UV spectrum, a marked hypochromism and bathrochromic shift was also observed (Figure 2D) (17,20). UV spectral change of AT-DNA was also different from GC-DNA, and there was no obvious shift on SC-dots binding (Supplementary Figure S5B). Moreover, carbon dots prepared by using glucose alone, named as G-dots (zeta potential of −0.13 mV, Supplementary Figure S6), and spermine itself were used as control agents; neither of them could induce B-Z transition under the same experimental conditions (Figure 3). As indicated previously (17–22), two factors are mainly required to trigger B-Z transition, shielding of the electrostatic repulsion from phosphate groups of DNA and reducing water activity in Z-DNA stabilization (17–22). As DNAs bind to positively charged SC-dots, various interactions of DNA bases and backbone with SC-dots, such as hydrophobic interactions, electrostatic interaction, van der Waals, can take place. The strong interaction between SC-dots and DNA can disturb DNA hydration layer, even DNA structure, which was similar to SWNTs (25–32). Previous studies showed that GC homopolymer has a strong tendency for aggregation than AT homopolymer, and GC homopolymer could undergo B-Z transition by reducing water activity (42). Under the usual experimental condition for the transition, GC homopolymer will aggregate, whereas AT homopolymer remains in B-form (17–22). We further examined the change in 2 M NaCl solution; however, the transition was not induced by SC-dots, suggesting that the electrostatic interaction between SC-dots and DNA was important for the B-Z transition to occur (Supplementary Figure S7). Surface charge of a nanoparticle plays a crucial role for its potential capability to alter DNA structure. It has been reported recently that cationic gold nanoparticle (∼1.5 nm) can induce DNA bending and strand separation, by tuning and balancing charge and hydrophobicity one could envision nanoparticles engineered to evoke a specific structural response from DNA (43).
Figure 2.
CD spectra of 2 µM GC-DNA (A) and AT-DNA (B) titrated with different SC-dots: 0, 1, 2, 3, 4, 5 µg/ml. The straight line in middle is CD spectrum of 5 µg/ml SC-dots alone. (C) Plot of CD intensity at the 260 nm (black square) or at 294 nm (red circles) as a function of the SC-dots concentration. The data were adopted from (A). (D) UV spectra of GC-DNA in the absence or presence of 2µg/ml SC-dots.
Figure 3.
CD spectra of (A) GC-DNA and (B) AT-DNA (black), DNA solutions after addition with 5 µg/ml G-dots (red) and after addition with 5 mM spermine (blue).
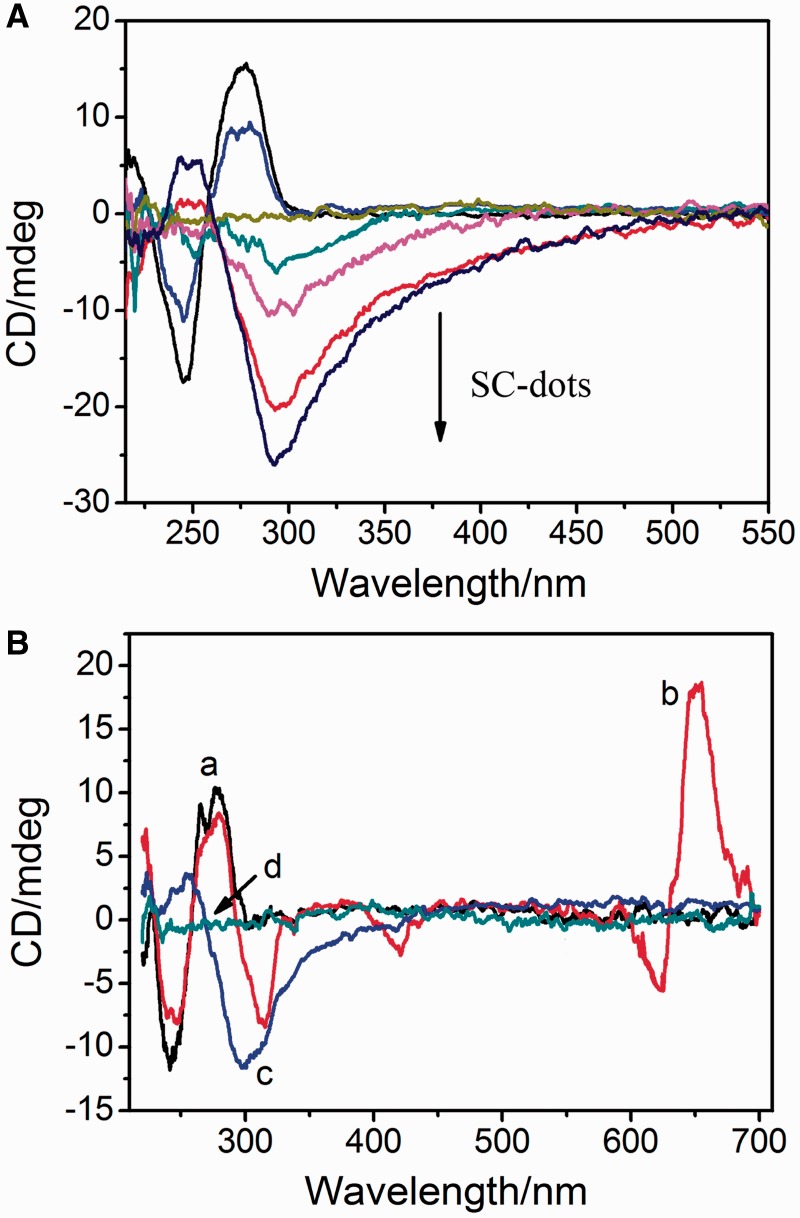
Although SC-dots could induce natural ct-DNA (42% GC and 58%AT) (29) to Z-form, it was not as easily as that for GC-DNA (Figure 4A). This indicated that SC-dots-induced B-Z transition was dependent on DNA GC content. One comparably minor increase at near 250 nm and a decrease near 295 nm were observed when SC-dots were added to ct-DNA solution (Figure 4A and Supplementary Figure S8). However, the G-dots and spermine also could not induce ct-DNA to occur B-Z transition (Supplementary Figure S9A). Under high salt condition, the CD spectra of ct-DNA were compared before or after addition of SC-dots (Supplementary Figure S9B), the transition was not induced by SC-dots, further indicating that the electrostatic interaction between SC-dots and DNA was important for the B-Z transition to occur. Supplementary Figure S10 showed the results of 1% agarose gel electrophoresis of ct-DNA with SC-dots. With the increase of amount of SC-dots, the intensity of ct-DNA bands decreased, and the migration was retarded. As water activity is an important apparent driving force for B-Z transition and the water activity of GC-rich region is lower than AT-rich region (29), GC-DNA undergoes the B to Z transition most easily, whereas AT-DNA resists the B-Z transition. It speculates that the cationic surface of SC-dots serves to reduce electrostatic repulsion among DNA phosphate backbone, and the hydrophilic sugar groups, which reduce activity of water; these two factors can act cooperatively to unwind DNA helix and lead to destabilization of double-stranded DNA (17–22,25–29).
Figure 4.
(A) CD spectra of ct-DNA titrated with different SC-dots: 0, 1, 2, 3, 5, 8 µg/ml. The straight lint in middle is CD spectrum of 5 µg/ml SC-dots alone. (B) CD spectra of ct-DNA alone (a, black); ct-DNA with MG molecule (b, red); and addition of SC-dots to the above solution (c, blue); SC-dots alone (d, green), ct-DNA: 30 µg/ml; MB: 25 µM; SC-dots: 5 µg/ml.
CD spectra showed that the DNA ellipticity changed following the order GC-DNA>ct-DNA>AT-DNA>polydApolydT (Supplementary Figure S11). It is well-known that GC-DNA, AT-DNA and ct-DNA are in the B-form, whereas polydApolydT has distinct structural and functional properties and adopts a non-B conformation (17,44). The CD spectrum of polydApolydT displayed an unusual shape with split bands in the region of 260–300 nm as a result of the stacking of bases with relatively large propeller twist (18–24°) (17). On addition of SC-dots, UV spectrum of polydApolydT showed a cross point at 284 nm, and the CD signal was decreased slightly while no obvious change occurred at 5 µg/ml SC-Dots, which indicated that SC-dots could not facilitate polydApolydT to occur structure transition (Supplementary Figure S11).
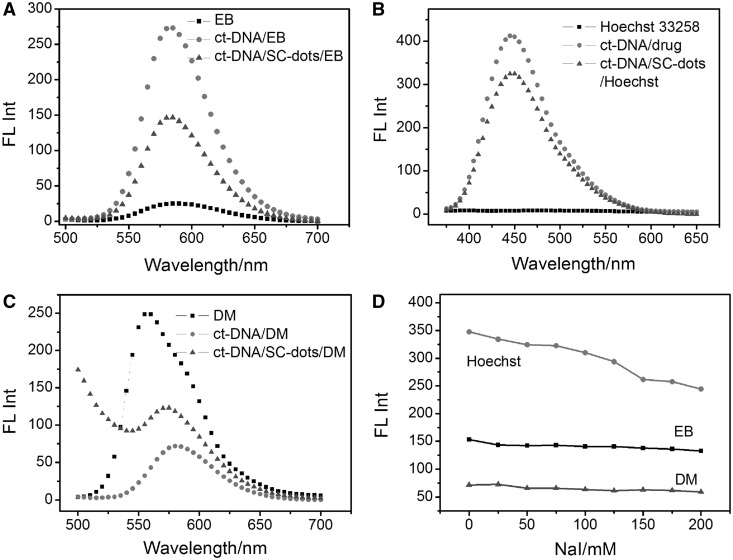
To further establish SC-dots DNA binding mode, the commonly used competitive binding assay was carried out (29). Our results clearly indicated that SC-dots could influence the interactions between DNA and MG, a proven DNA major groove binder (29,45,46). Figure 4B showed CD spectral changes in the presence of SC-dots. For MG, there was no CD signal under our experimental conditions. When it bound to ct-DNA, three induced CD signals characteristic of bound MG ∼310, 430 and 650 nm were observed. With addition of SC-dots, the induced CD intensity was disappeared; typical data were shown in Figure 4B, which was consistent with our previous reports that SWNTs would exclude MG out of DNA major groove (29). Then, the ct-DNA CD spectrum became like the one of DNA/SC-dots in the absence of MG, further supporting that SC-dots could bind to DNA major groove. It is well-known that EB and DM can intercalate into DNA through the minor groove, and Hoechst 33258 is a classical DNA minor groove binder (47–49). When bound to DNA, the fluorescence of EB or Hoechst is greatly enhanced, and DM fluorescence is strongly quenched. With this in mind, if SC-dots competitively bind to the same site of DNA as EB, Hoechst or DM, the fluorescence of EB and Hoechst would strongly decrease, and the fluorescence of DM would significantly increase because the strong binding of SC-dots with DNA should exclude these DNA binders out of their minor groove binding sites. The fluorescence competitive binding assay has been widely used to establish DNA-binding mode (26,50–52). As shown in Figure 5A–C, when these molecules were added to ct-DNA/SC-dots solution, DM fluorescence was also well quenched, and the fluorescence of EB or Hoechst was still enhanced, although the emission intensity was lower than that of EB or Hoechst bound to B-DNA alone (8). It indicated that EB, Hoechst and DM could still bind to DNA through minor groove even in the presence of SC-dots binding. In combination with CD competitive binding results (Figure 4) of DNA major groove binder, MG, fluorescence experiments further supported that SC-dots could bind to DNA major groove, not to minor groove.
Figure 5.
Fluorescence emission spectra of (A) 1 µM EB (Excitation wavelength: 480 nm); (B) 1 µM Hoechst 33258 (Excitation wavelength: 355 nm); (C) 1 µM DM (Excitation wavelength: 480 nm): 1 µM fluorophore (EB, Hoechst 33258, DM) alone (squares), 30 µg ml−1 ct-DNA+ 1 µM fluorophore (dots), 30 µg ml−1 ct-DNA+5 µg ml−1 SC-dots+ 1 µM fluorophore (uptriangles). (D) Plot of fluorescence intensity of fluorephore-DNA–SC-dots versus NaI concentrations from 0 to 200 mM. ct-DNA: 30 µg ml−1; SC-dots: 5 µg ml−1; 1 µM different fluorephore drugs.
We also carried out NaI quenching experiments. Iodide ions cannot quench the fluorescence of these dye molecules when they are bound to DNA (53). Figure 5D showed that the fluorescence was not quenched by iodide for EB or DM. For Hoechst 33258, the whole intensity decreased because SC-dots also had strong emission ∼460 nm as the same as Hoechst 33258, and iodide ions could well quench the fluorescence of SC-dots, as shown in Supplementary Figure S12 and inset. All the aforementioned results indicated that SC-dots could bind to DNA major groove. Spermine molecule could neutralize repulsive phosphate interactions between DNA helices and bridged them by a hydrogen-bonding network and formed hydrogen bonds exclusively with convex surface (the Z-DNA equivalent of the major groove) of duplex and with phosphate oxygens (53,54). The convex surface of Z-DNA is ∼2 nm, which is more suitable for the SC-dots binding. Furthermore, one spermine molecule has been reported to interact simultaneously with three Z-DNA duplex (53), which would supply another clue for the possibility of condensed mechanism between DNA and SC-dots. AFM studies showed that ct-DNA was condensed when it bound with SC-dots, and the condensation was depended on the amount of SC-dots (Supplementary Figure S13).
Recently, the B–Z transition has been explored from nanotechnological points of view and used as the driving machinery of a nanomechanical DNA device (55,56). DNA molecules are versatile building blocks for construction of synthetic logic gates to conduct computing and logic operations at literally the nanosized level (57–60). Here, based on occurrence of FRET between SC-dots and DNA intercalater, EB, several optical Z-DNA-based logic gates have been designed and constructed.
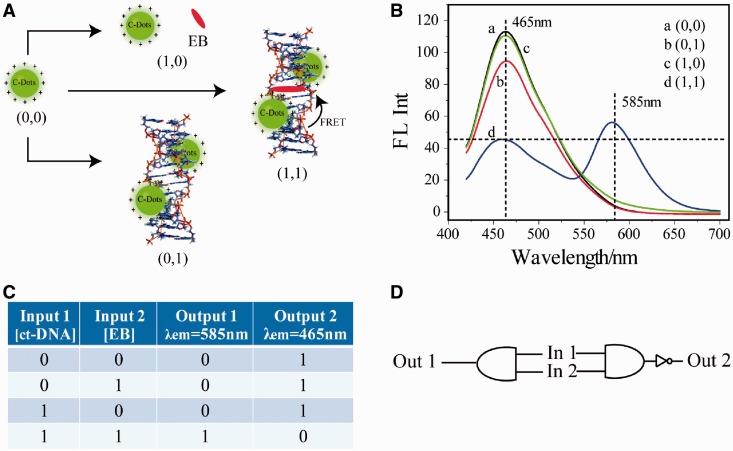
To create a Z-DNA-based AND gate, we coupled two molecular recognition events in series to cause a change in the photonic output of the gate in solution (Figure 6A). AND logic is represented by the situation where the output of an AND gate is true only if both inputs are true. SC-dots solution showed highest emission near 465 nm on excitation at 400 nm, and the two inputs were ct-DNA (In 1) and the DNA intercalater, EB (In 2). EB had absorption ∼480 nm, which overlaped with the emission wavelength of SC-dots (Figure 1A). The output of the AND gate was measured on excitation at 400 nm by fluorescence output at 585 nm (Out 1, Figure 6), the emission wavelength of EB. After interacting with one of the inputs (ct-DNA or EB), there was nearly no fluorescence ∼585 nm. Only both of the inputs existed, the fluorescence increased, indicating that the SC-Dots and EB came close enough and an efficient FRET occurred (61,62). NAND logic is the result of sending an AND gate through an inverter, which causes all 0 states to switch to 1 and vice versa. Thus, NAND logic has an output of 1 for all combinations of binary inputs except the (1,1) situation where the output now becomes 0. The Z-DNA-based AND gate was converted to a NAND gate by simply switching the emission wavelength to 465 nm (emission of SC-dots, Out 2, Figure 6) instead of 585 nm.
Figure 6.
(A) Scheme of AND and NAND logic gates principle. (B) Fluoresent spectra of (a) SC-dots only, (b) SC-dots/ct-DNA, (c) SC-dots/ct-DNA/EB, (d) SC-dots/EB, excitation wavelength: 400 nm. On excitation at 400 nm, fluorescence output at 585 and 465 nm were recorded in AND and NAND gates, respectively. (C) Truth table. (D) AND/NAND logic scheme.
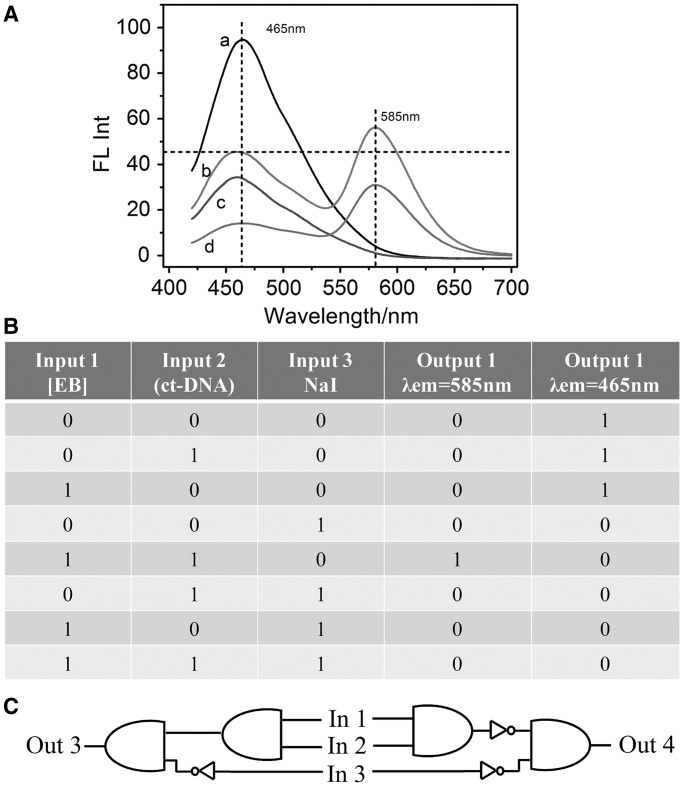
Iodide ion has been proved to quench the fluorescence of SC-dots (Supplementary Figure S12 and inset) but cannot quench EB fluorescence on its binding to DNA as discussed earlier in the text. This was adopted as the third input (In 3). In the induced Z-DNA system, the FRET occurred with the excitation of 400 nm, and the gradual addition of NaI could quench the fluorescence of SC-dots at 465 nm, then induce the fluorescence of EB at 585 nm decreased (Supplementary Figure S14A). If excited at 480 nm directly, the EB emission was almost unchangeable as shown in Supplementary Figure S14B. Therefore, the AND + NIH, NAND + NIH logic gates were constructed with fluorescence outputs at 585 nm (Out 3) and 465 nm (Out 4), respectively, by using the specific system induced by SC-dots through lightening up Z-DNA and quenched by iodide ion (Figure 7 and Supplementary Figure S15).
Figure 7.
(A) Fluoresent spectra of (a) SC-dots/ct-DNA, (b) SC-dots/ct-DNA/EB, (c) SC-dots/ct-DNA/NaI, (d) SC-dots/ct-DNA/EB/NaI. (B) Truth table. (C) AND + INH/NAND + INH logic scheme.
CONCLUSION
In summary, photoluminescent SC-dots can induce DNA B-Z transition under physiological salt condition with sequence selectivity. Contrasting change for SC-dots binding to GC-DNA and AT-DNA are observed. Further studies indicate that SC-dots bind to DNA major groove with GC preference. Based on photoluminescent SC-dots-induced DNA B-Z transition and FRET, several DNA logic gates are constructed. To the best of our knowledge, this is the first example that functionalized C-dots can influence DNA structure and induce DNA B-Z transition under physiological low salt condition. As C-dots have shown potential applications that range from nanodevices, gene therapy and drug delivery, our studies would provide new insight into understanding the biomedical effects of C-dots on Z-DNA and shed light on their applications to DNA nanotechnology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–15.
FUNDING
Funding for open access charge: The National Basis Research Program of China [2011CB936004, 2012CB720602]; National Natural Science Foundation of China [21210002, 91213302, 21202158].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Phan AT, Kuryavyi V, Patel DJ. DNA architecture: from G to Z. Curr. Opin. Struct. Biol. 2006;16:288–298. doi: 10.1016/j.sbi.2006.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Choi J, Majima T. Conformational changes of non-B DNA. Chem. Soc. Rev. 2011;40:5893–5909. doi: 10.1039/c1cs15153c. [DOI] [PubMed] [Google Scholar]
- 3.Mergny JL, Helene C. G-quadruplex DNA: a target for drug design. Nat. Med. 1998;4:1366–1367. doi: 10.1038/3949. [DOI] [PubMed] [Google Scholar]
- 4.Rich A, Zhang SG. Z-DNA: the long road to biological function. Nat. Rev.Genet. 2003;4:566–572. doi: 10.1038/nrg1115. [DOI] [PubMed] [Google Scholar]
- 5.Cer RZ, Bruce KH, Mudunuri US, Yi M, Volfovsky N, Luke BT, Bacolla A, Collins JR, Stephens RM. Non-B DB: a database of predicted non-B DNA-forming motifs in mammalian genomes. Nucleic Acids. Res. 2011;39:D383–D391. doi: 10.1093/nar/gkq1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu H, Mulholland N, Fu HQ, Zhao F. Cooperative activity of BRG1 and Z-DNA formation in chromatin remodeling. Mol. Cell. Biol. 2006;26:2550–2559. doi: 10.1128/MCB.26.7.2550-2559.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ray BK, Dhar S, Shakya A, Ray A. Z-DNA-forming silencer in the first exon regulates human ADAM-12 gene expression. Proc. Nat. Acad. Sci. USA. 2011;108:103–108. doi: 10.1073/pnas.1008831108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geng J, Zhao C, Ren J, Qu XG. Alzheimer’s disease amyloid beta converting left-handed Z-DNA back to right-handed B-form. Chem. Commun. 2010;46:7187–7189. doi: 10.1039/c0cc02049d. [DOI] [PubMed] [Google Scholar]
- 9.Mao C, Sun W, Shen Z, Seeman NC. A nanomechanical device based on the B-Z transition of DNA. Nature. 1991;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 10.Lee JB, Peng S, Yang DY, Roh YH, Funabashi H, Park N, Rice E, Chen L, Long R, Wu M, et al. A mechaical metamaterial made from a DNA hygrogel. Nat. Nanotechnol. 2012;7:816–820. doi: 10.1038/nnano.2012.211. [DOI] [PubMed] [Google Scholar]
- 11.Pinherio AV, Han D, Shih WM, Yan H. Challenges and opportunities for structural DNA. Nat. Nanotechol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suh D, Sheardy RD, Chaires JB. Unsual binding of ethidium to a deoxyoligonucleotide containing a B-Z junction. Biochemistry. 1991;30:8722–8726. doi: 10.1021/bi00100a002. [DOI] [PubMed] [Google Scholar]
- 13.Qu XG, Trent JO, Fokt I, Priebe W, Chaires JB. Allosteric, chiral-selective drug binding to DNA. Proc. Natl Acad. Sci. USA. 2000;97:12032–12037. doi: 10.1073/pnas.200221397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nordheim A, Lafer EM, Peck LJ, Wang JC, Stllar BD, Rich A. Negatively supercoiled plasmids contain left-handed Z-DNA segments as detected by specific antibody binding. Cell. 1982;31:309–318. doi: 10.1016/0092-8674(82)90124-6. [DOI] [PubMed] [Google Scholar]
- 15.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zα domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 16.Hebe M, Felsenfeld G. Effects of methylation on a synthetic polynucleotide: the B-Z transition on poly (dG-m5dC).poly (dG-m5dC) Proc. Natl Acad. Sci. USA. 1981;78:1619–1623. doi: 10.1073/pnas.78.3.1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang HY, Yu HJ, Ren JS, Qu XG. Reverible B/Z-DNA transition under the low salt condition and non-B-form polydApolydT selectivity by a cubane-like europium-L-aspartic acid complex. Biophys. J. 2006;90:3203–3207. doi: 10.1529/biophysj.105.078402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parkinson A, Hawken M, Hall M, Sanders KJ, Rodger A. Amine iduced Z-DNA in poly (dG-dC) poly (dG-dC): circular dichrosim and gel electrophoresis study. Phys. Chem.Comm. Phys. 2000;2:5469–5478. [Google Scholar]
- 19.Wu Z, Tian T, Yu J, Weng X, Liu Y, Zhou X. Formation of sequence-independent Z-DNA induced by a ruthenium complex at low salt concentrations. Angew. Chem. Int. Ed. 2011;50:11962–11967. doi: 10.1002/anie.201104422. [DOI] [PubMed] [Google Scholar]
- 20.Shimada N, Kano A, Maruyama A. B-Z DNA transition triggered by a cationic comb-type copolymer. Adv. Funct. Mater. 2009;19:3590–3595. [Google Scholar]
- 21.Johnson A, Qu Y, Houten BV, Farrell N. B-Z DNA conformational changes induced by a family of dinuclear bis(platinum) complexes. Nucleic Acids. Res. 1992;20:1697–1703. doi: 10.1093/nar/20.7.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu Y, Zhang YX, Sugiyama H, Umano Y, Osuga H, Tanaka K. (P)-Helicene displays chiral selection in binding to Z-DNA. J. Am. Chem. Soc. 2004;126:6566–6567. doi: 10.1021/ja0499748. [DOI] [PubMed] [Google Scholar]
- 23.Zheng M, Jagota A, Strano MS, Santos AP, Barone P, Chou SG, Diner BA, Dresselhaus MS, Mclean RS, Onoa GB, et al. Structure-based carbon nanotube sorting by sequence-dependent DNA assembly. Science. 2003;28:1545–1548. doi: 10.1126/science.1091911. [DOI] [PubMed] [Google Scholar]
- 24.Heller DA, Jeng ES, Yeung TK, Martinez BM, Moll AE, Gastala JB, Strano MS. Optical detection of DNA conformational polymorphism on single-walled carbon nanotubes. Science. 2006;311:508–511. doi: 10.1126/science.1120792. [DOI] [PubMed] [Google Scholar]
- 25.Chen Y, Qu K, Zhao C, Wu L, Ren J, Wang J, Qu X. Insights into the biomedical effects of carboxylated single-wall carbon nanotubes on telomerase and telomeres. Nat. Commun. 2013;3:1074. doi: 10.1038/ncomms2091. 1–13. [DOI] [PubMed] [Google Scholar]
- 26.Li X, Peng Y, Ren J, Qu X. Carboxyl-modified single-walled carbon nanotubes selectively induce human telomeric i-motif formation. Proc. Natl Acad. Sci. USA. 2006;103:19658–19663. doi: 10.1073/pnas.0607245103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng Y, Wang X, Xiao Y, Feng L, Zhao C, Ren J, Qu X. i-motif quadruplx DNA-based biosensor for distinguishing single- and multiwalled carbon nanotubes. J. Am. Chem. Soc. 2009;131:13813–13818. doi: 10.1021/ja9051763. [DOI] [PubMed] [Google Scholar]
- 28.Zhao C, Song Y, Ren J, Qu X. A DNA nanomachine induced by single-walled carbon nanotubes on gold surface. Biomaterials. 2009;30:1739–1745. doi: 10.1016/j.biomaterials.2008.12.034. [DOI] [PubMed] [Google Scholar]
- 29.Li X, Peng Y, Ren J, Qu X. Carbon nanotubes selective destabilization of duplex and triplex DNA and inducing B-A transition in solution. Nucleic Acids. Res. 2006;34:3670–3676. doi: 10.1093/nar/gkl513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao C, Peng Y, Song Y, Ren J, Qu X. Self-assembly of single-stranded RNA on carbon nanotube: polyadenylic acid to form a duplex structure. Small. 2005;4:656–661. doi: 10.1002/smll.200701054. [DOI] [PubMed] [Google Scholar]
- 31.Song Y, Feng L, Ren J, Qu X. Stabilization of unstable CGC+ triplex DNA by single-walled carbon nanotubes under physiological conditions. Nucleic Acids. Res. 2011;39:6835–6843. doi: 10.1093/nar/gkr322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao C, Ren J, Qu X. Single-walled carbon nanotubes binding to human telomeric i-motif DNA under molecular-crowding conditions: more water molecules released. Chem. Eur. J. 2008;14:5435–5439. doi: 10.1002/chem.200800280. [DOI] [PubMed] [Google Scholar]
- 33.Baker SN, Baker JN. Luminescent carbon nanodots: emergent nanolights. Angew. Chem. Int. Ed. 2010;49:6726–6744. doi: 10.1002/anie.200906623. [DOI] [PubMed] [Google Scholar]
- 34.Li H, He X, Kang Z, Huang H, Liu Y, Liu J, Lian S, Tsang CH, Yang X, Lee S. Water-soluble fluorescent carbon quantum dots and photocatalyst design. Angew. Chem. Int. Ed. 2010;49:4430–4434. doi: 10.1002/anie.200906154. [DOI] [PubMed] [Google Scholar]
- 35.Wang X, Qu K, Xu B, Ren J, Qu X. Multicolor luminescent carbon nanoparticles: synthesis, supramolecular assembly with porphyrin, intrinsic peroxidase-like catalytic activity and applications. Nano Res. 2011;4:908–920. [Google Scholar]
- 36.Liu C, Zhang P, Zhai X, Tian F, Li W, Yang J, Liu Y, Wang H, Liu W. Nano-carrier for gene delivery and bioimaging based on carbon dots with PEI-passivation enhanced fluorescence. Biomaterials. 2012;33:3604–3613. doi: 10.1016/j.biomaterials.2012.01.052. [DOI] [PubMed] [Google Scholar]
- 37.Wang X, Qu K, Xu B, Ren J, Qu X. Microwave assisted one-step green synthesis of cell-permeable multicolor photoluminescent carbon dots without surface passivation reagents. J. Mater. Chem. 2011;21:2445–2450. [Google Scholar]
- 38.Liu S, Tian J, Wang L, Zhang Y, Qin X, Luo Y, Asiri AM, Al-Youbi AO, Sun X. Hydrothermal treatment of grass: a low-cost, green route to nitrogen-doped, carbon-rich, photoluminescent polymer nanodots as an effective fluorescent sensing platform for label-free detection of Cu(II) ions. Adv. Mater. 2012;24:2037–2041. doi: 10.1002/adma.201200164. [DOI] [PubMed] [Google Scholar]
- 39.Yalpani M, Hall LD. Some chemical and analytical aspects of polysaccharide modifications. III. Formation of branched-chain, soluble chitosan derivatives. Macromolecules. 1984;17:272–281. [Google Scholar]
- 40.Wang T, Turhan M, Gunasekaran S. Selected properties of pH-sensitive, biodegradable chitosan–poly(vinyl alcohol) hydrogel. Polym. Int. 2004;53:911–918. [Google Scholar]
- 41.Ha SC, Lowenhaupt K, Rich A, Kim YG, Kim KK. Crystal structure of a junction between B-DNA and Z-DNA reveals two extruded bases. Nature. 2005;437:1183–1186. doi: 10.1038/nature04088. [DOI] [PubMed] [Google Scholar]
- 42.Pohl FM. Polymorphism of a synthetic DNA in solution. Nature. 1976;12:365–366. doi: 10.1038/260365a0. [DOI] [PubMed] [Google Scholar]
- 43.Railsback JG, Singh A, Pearce RC, Mcknight TE, Collazo R, Sitar Z, Yingling YG, Melechko AV. Weakly charged cationic nanoparticles induce DNA bending and strand separation. Adv. Mater. 2006;45:4261–4265. doi: 10.1002/adma.201104891. [DOI] [PubMed] [Google Scholar]
- 44.Brahms S, Brahms JG. DNA with adenine tracts contains poly(dA).poly(dT) conformational features in solution. Nucleic Acids. Res. 1990;18:1559–1564. doi: 10.1093/nar/18.6.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nitta Y, Kuroda R. Quantitative analysis of DNA–porphyrin interactions. Biopolymer. 2006;81:376–391. doi: 10.1002/bip.20430. [DOI] [PubMed] [Google Scholar]
- 46.Tuite E, Sehlstedt U, Hagmar P, Norden B, Takahashi M. Effects of minor and major groove-binding drugs and intercalators on the DNA association of minor groove-binding proteins RecA and deoxyribonuclease I detected by flow linear dichroism. Eur. J. Biochem. 1997;49:5665–5667. doi: 10.1111/j.1432-1033.1997.0482a.x. [DOI] [PubMed] [Google Scholar]
- 47.Chaires JB. Allosteric conversion of Z DNA to an intercalated right-handed conformation by daunomycin. J. Bio.Chem. 1986;261:8899–8907. [PubMed] [Google Scholar]
- 48.Chaires JB. Daunomycin inhibits the B leads to Z transition in poly d(G-C) Nucleic Acids Res. 1983;11:8485–8494. doi: 10.1093/nar/11.23.8485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wu PK, Kharatishivili M, Qu Y, Farrell N. A circular dichroism study of ethidium bromide binding to Z-DNA induced by dinuclear platinum complexes. J. Inorg. Biochem. 1996;63:9–18. doi: 10.1016/0162-0134(95)00163-8. [DOI] [PubMed] [Google Scholar]
- 50.Roger DL, Fink BE, Brunette SR, Tse WC, Hedrick MP. A simple, high resolution method for establishing DNA binding affinity and sequence selectivity. J. Am. Chem. Soc. 2001;123:5878–5891. doi: 10.1021/ja010041a. [DOI] [PubMed] [Google Scholar]
- 51.Shangguan G, Xing F, Qu X, Mao J, Zhao D, Zhao X, Ren J. DNA binding specificity and cytotoxicity of novel antitumor agent Ge132 derivatives. Bioorg. Med. Chem. Lett. 2005;15:2962–2965. doi: 10.1016/j.bmcl.2005.04.053. [DOI] [PubMed] [Google Scholar]
- 52.Suh D, Chaires JB. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995;3:723–728. doi: 10.1016/0968-0896(95)00053-j. [DOI] [PubMed] [Google Scholar]
- 53.Bancroft D, Willliams LD, Rich A, Egli M. the low-temperature crystal structure of the pure-spermine form of Z-DNA reveals binding of a spermine molecule in the minor groove. Biochemistry. 1994;33:1073–1086. doi: 10.1021/bi00171a005. [DOI] [PubMed] [Google Scholar]
- 54.Doi I, Tsuji G, Kawakami K, Nakagawa O, Taniguchi Y, Sasaki S. The spermine–bisaryl conjugate as a potent inducer of B- to Z-DNA transition. Chem. Eur. J. 2010;16:11993–11999. doi: 10.1002/chem.201000947. [DOI] [PubMed] [Google Scholar]
- 55.Niemeyer C, Alder M. Nanomechanical devices based on DNA. Angew. Chem. Int. Ed. 2002;41:3779–3783. doi: 10.1002/1521-3773(20021018)41:20<3779::AID-ANIE3779>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 56.D’Urso A, Mammana A, Balaz M, Holmes AE, Berova N, Lauceri R, Purrello R. Interactions of a tetraanionic porphyrin with DNA: from a Z-DNA sensor to a versatile supramolecular device J. Am. Chem. Soc. 2009;131:2046–2047. doi: 10.1021/ja808099u. [DOI] [PubMed] [Google Scholar]
- 57.Benenson Y, Gil B, Uri Ben-Dor RA, Shapiro E. An autonomous molecular computer for logical control of gene expression. Nature. 2004;429:423– 429. doi: 10.1038/nature02551. [DOI] [PubMed] [Google Scholar]
- 58.Seelig G, Soloveichik D, Zhang DY, Winfree E. Enzyme-free nucleic acid logic circuits. Science. 2006;314:1585–1588. doi: 10.1126/science.1132493. [DOI] [PubMed] [Google Scholar]
- 59.Huang Z, Tao Y, Pu F, Ren J, Qu X. Versatile logic devices based on programmable DNA-regulated silver-nanocluster signal transducers. Chem. Eur. J. 2012;18:6663–6669. doi: 10.1002/chem.201103859. [DOI] [PubMed] [Google Scholar]
- 60.Feng L, Huang Z, Ren J, Qu X. Toward site-specific, homogeneous and highly stable fluorescent silver nanoclusters fabrication on triplex DNA scaffolds. Nucleic Acids Res. 2012;40:e122. doi: 10.1093/nar/gks387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Patalsky F, Gill R, Weizmann Y, Mokari T, Banin U, Willner I. Lighting-up the dynamics of telomerization and DNA replication by CdSe−ZnS quantum dots. J. Am. Chem. Soc. 2003;125:13918–13919. doi: 10.1021/ja035848c. [DOI] [PubMed] [Google Scholar]
- 62.Gill R, Zayats M, Willner I. Semiconductor quantum dots for bioanalysis. Angew. Chem. Int. Ed. 2008;47:7602–7625. doi: 10.1002/anie.200800169. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.