Abstract
The mouse embryo hindbrain is a robust and adaptable model to study sprouting angiogenesis. It permits the spatiotemporal analysis of organ vascularisation in normal mice and in mouse strains with genetic mutations that result in late embryonic or perinatal lethality. Unlike postnatal models such as retinal angiogenesis or Matrigel implants, there is no requirement for breeding of conditional knockout mice. The unique architecture of the hindbrain vasculature allows whole-mount immunolabelling of blood vessels and high-resolution imaging as well as easy quantitation of angiogenic sprouting, network density and vessel calibre. The hindbrain model also permits the visualisation of ligand binding to blood vessels in situ and the analysis of blood vessel growth within a natural multicellular microenvironment, where endothelial cells (ECs) interact with non-ECs to refine the 3D organ architecture. The entire procedure, from embryo isolation to imaging through to result analysis, takes approximately 4 d.
INTRODUCTION
In this protocol we describe examples of studies that have used the mouse embryo hindbrain to elucidate the molecular and cellular mechanisms of sprouting angiogenesis and present an experimental protocol to image and quantitate sprouting angiogenesis in this organ at high spatiotemporal resolution.
THE MOUSE EMBRYO HINDBRAIN
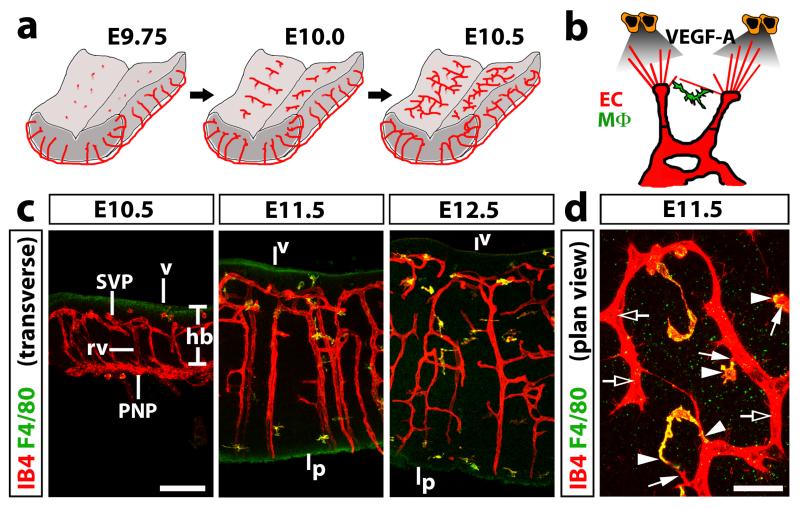
Owing to its unique architecture and vascularisation early in development, the mouse embryo hindbrain has become a key model system to study sprouting angiogenesis in vivo. Similar to other parts of the vertebrate central nervous system, the hindbrain is vascularised early in development to support the growth and function of the rapidly proliferating neural progenitors and their progeny. Vessels first sprout from the perineural vascular plexus into the mouse hindbrain at around embryonic day (E) 9.75 and then grow centripetally towards the ventricular zone (Fig. 1a, stage 1) 1. From E10 onwards, these radial vessels extend sprouts at near right angles to extend parallel to, but beneath the ventricular hindbrain surface (Fig. 1a, step 2) 1. As neighbouring sprouts meet and anastomose, a subventricular vascular plexus (SVP) begins to form (Fig. 1a, step 3) 1,2. Arteriovenous differentiation in the hindbrain appears to take place well after capillary networks are established. Thus, perfused vascular networks in the hindbrain appear from E10.5 onwards 1, but the arterial marker smooth muscle alpha actin (SMA) is expressed only from around E14.5 onwards in the hindbrain (A. Plein and C. Ruhrberg, unpublished observations).
Fig. 1. Time course of blood vessel growth in the mouse embryo hindbrain.
(a) Schematic representation of hindbrain vascularisation. Vessels begin to sprout from the perineural vascular plexus to invade the hindbrain at around E9.75 and grow radially towards the ventricular zone (stage 1). From around E10.0 onwards, radial vessels arrest below the ventricular surface and begin to sprout laterally, turning at near right angles (stage 2). At around E10.5, neighbouring sprouts begin to anastomose to form the SVP (stage 3).
(b) Schematic representation of the mechanism of sprouting angiogenesis in the hindbrain. Sprouting blood vessels (red) are led by filopodia-studded tip cells; the filopodia detect VEGF gradients (grey) that are secreted by neuronal progenitors (orange) to guide the sprouting vessels. Anastomosis of neighbouring tip cells is initiated by filopodia contact and optimised by yolk sacderived tissue macrophages (MΦ, green), which interact with tip cells and tip cell filopodia.
(c,d) Fluorescent labelling of hindbrain tissue of the indicated developmental stages with IB4 (red) to highlight blood vessels and F4/80 (green) to identify macrophages; transverse sections are shown in (c), a plan view of a flatmounted hindbrain, ventricular side facing up, is shown in (d). Clear arrows indicate examples of areas with endothelial stalk cells, solid arrows filopodia-studded endothelial tip cells and solid arrowheads tissue macrophages interacting with tip cells. Scale bars: 100 μm (c) and 25 μm (d). Abbreviations: hb, hindbrain; v, ventricular brain surface; p, pial brain surface; PNP, perineural vascular plexus; rv, radial vessel; SVP, subventricular vascular plexus. All animal procedures were performed in accordance with institutional and UK Home Office guidelines.
THE MOUSE EMBRYO HINDBRAIN AS AN ANGIOGENESIS MODEL
We pioneered the mouse embryo hindbrain as a model with the view to study growth factor signalling in sprouting angiogenesis 2. Specifically, our analysis of hindbrains from mouse mutants expressing single isoforms of soluble versus matrix-binding vascular endothelial growth factor A (VEGF-A) showed that the isoform balance regulates blood vessel branching and calibre 2. Accordingly, loss of the matrix-binding isoforms disrupted the formation of VEGF-A gradients in the extracellular environment of growing vessels, thereby decreasing the formation of filopodia-led endothelial sprouts whilst increasing luminal growth. Vice versa, expression of the VEGF-A isoform that binds the matrix most avidly at the expense of the soluble isoforms led to hyperbranched and thin vessels 2. These findings formed the basis of further studies in the mouse retina, which demonstrated that filopodia-studded endothelial tip cells at the front of the growing vessels sense VEGF-A isoform gradients to direct vessel growth (Fig. 1B) 3.
Using Cre/Lox technology to create cell type specific deletions of Vegfa, it was subsequently demonstrated that neural progenitors are the source of VEGF-A for proper hindbrain vascularisation 4, whereas the heterozygous targeting of one Vegfa allele in these cells showed that the VEGF dose is critically important to regulate vessel sprouting during SVP formation 1. The hindbrain model was also used to show that the shared VEGF-A/class 3 semaphorin receptor neuropilin 1 (NRP1) is essential for normal filopodia guidance in the hindbrain 5, independently of its ability to bind semaphorins 6. Moreover, the hindbrain model featured in seminal studies that demonstrated an essential role for the p110alpha isoform of phosphatidylinositol 3-kinase (PI3K) in endothelial cell migration 7 and an important role for netrin as a negative regulator of sprouting angiogenesis 8. In addition, the hindbrain model was used to show that heparin sulfate proteoglycans are required for pericyte recruitment to growing blood vessels 9 and that VEGFR3 promotes endothelial tip-to-stalk conversion during vascular development 10. Recently, we have used the mouse embryo hindbrain to identify a role for tissue macrophages in vascular anastomosis 1. These cells invade the embryonic brain independently of blood vessels and interact with opposing endothelial tip cells to promote sprout anastomosis (Fig. 1b-d) 1.
COMPARISON WITH OTHER METHODS: ADVANTAGES AND LIMITATIONS
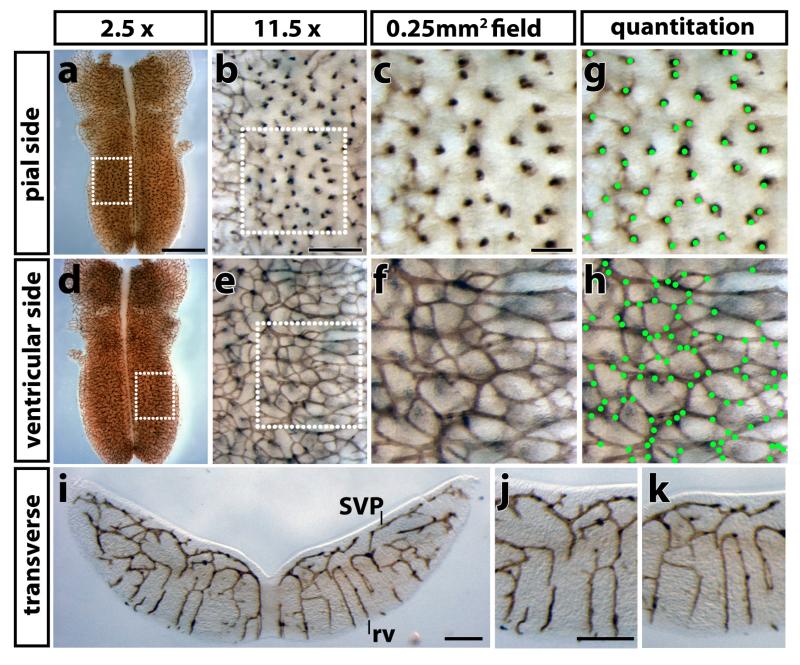
The mouse embryo hindbrain provides several advantages over other in vivo models to study angiogenesis. Firstly, it is ideally suited to quantitate angiogenic sprouting and vascular complexity, as it forms vessel networks of simple geometry. Thus, after flatmounting, three-dimensional vessel sprouting into the brain can be visualised as a one-dimensional process from its pial side, whilst sprouting within the brain appears as a near two-dimensional process on its ventricular side (Fig. 2, Fig. S1). The vascularisation of other organs, such as the developing kidney, heart or lung, as well as the angiogenic response in subcutaneous matrigel plugs, also proceeds in three dimensions; however, these organs are not suitable for flatmounting to obtain a simpler geometric representation of whole vessel networks. Secondly, quantitation in the hindbrain is easier and more accurate than in other vascular beds, because the temporal separation of arteriovenous specialisation from the earlier phase of sprouting and fusion results in the formation of relatively homogenous and extensive capillary networks. For example, the primary retinal vascular plexus can also be visualised in two dimensions like the hindbrain subventricular plexus 11, but arteriovenous remodelling occurs just behind the vascular front of the radially expanding vessel plexus 12, decreasing the size of areas containing capillaries suitable for quantitative analysis.
Fig. 2. Immunolabelling to visualise developing blood vessels in the mouse embryo hindbrain.
An E12.5 hindbrain was labelled by PECAM immunohistochemistry, flatmounted and imaged at the indicated magnifications. (a-c) Flatmounting the hindbrain with the pial side up allows visualisation of radial vessels entering the brain. (d-f) Flatmounting the hindbrain with the ventricular side up allows visualisation of the subventricular vascular plexus. The dotted boxes in (a,d) indicate the areas shown at higher magnification in (b,e), the dotted boxes in (b,e) those shown at higher magnification in (c,f), respectively; the size of each field in (c,f) is 500 μm × 500 μm, i.e. 0.25 mm2. (g,h) Counting of radial vessels and vascular intersections in the fields shown in (c,f); green dots were used to track vessels that have been counted. (i-k) A 100 μm transverse vibratome section through the E12.5 hindbrain shown in (a); radial vessels (rv) extend from the pial side of the hindbrain and then branch to form the SVP below the ventricular side of the hindbrain; (j,k) higher magnification images from both sides of the hindbrain shown in panel (i). Scale bars: (a,d) 1 mm; (b,e,i-k) 200 μm, (c,f) 100 μm. All animal procedures were performed in accordance with institutional and UK Home Office guidelines.
The mouse hindbrain can be used at earlier developmental stages than other angiogenesis models. For example, the mouse retina is suitable to analyse angiogenesis in the first two weeks after birth, whilst the neural tube is one of the earliest organs to develop and is vascularised earlier than kidneys, heart or lung. The primitive skin is vascularised early in development like the hindbrain, but the hindbrain forms a more homogenous capillary network. Owing to its vascularisation early in development, the hindbrain model is suitable to study mouse mutants that survive the period of vasculogenesis, but are lethal after E12.5. For instance, mice expressing only the soluble VEGF120 isoform of VEGF-A, but lacking the heparin/neuropilin binding VEGF164 isoform, die soon after birth 13, whilst mice lacking the VEGF164 receptor neuropilin (NRP) 1 die at around E12.5 14. In both cases, mechanistic detail of their angiogenic defects could be determined with the hindbrain model 2,5.
When specific mutations such as loss of VEGF-A, VEGFR2 or VEGFR1 (reviewed in ref. 15) cause embryonic lethality prior to E10.5 in mice, the hindbrain model cannot be used to study angiogenesis. However, this limitation can be overcome by modern genetic techniques, such as constitutive or tamoxifen-inducible Cre-Lox technology 16. For example, a constitutive Cre-Lox approach was used to inactivate Vegfa in neural progenitors in the hindbrain 1 to circumvent the early embryonic lethality caused by ablating one Vegfa allele in the germline 17,18.
A disadvantage of the mouse embryo hindbrain is that it does not afford similar accessibility to experimental manipulation as the retina, such as injection of growth factors or function-blocking reagents 11. In addition, vessel networks rapidly degenerate in organotypic culture 19, precluding live imaging or ex vivo pharmacological manipulation, as described for retinal explants 20. Finally, the increasing thickness of the mouse hindbrain from E13.5 onwards inhibits the penetration of antibodies into the deeper tissue parts, and changes in organ shape prevent flat-mounting for wholemount imaging. Therefore, the hindbrain vasculature at later developmental or postnatal stages is best visualised in immunolabelled tissue sections.
EXPERIMENTAL DESIGN
Fluorescence or histochemical staining of transverse sections with isolectin B4 (IB4) provides a robust and widely used method to visualise blood vessels in the central nervous system, including brain, spinal cord and retina 21. This method labels the endothelium in both the perineural and intraneural vascular plexi, including endothelial stalk cells and tip cell filopodia (Fig. 1c,d; Fig. S1; e.g. refs. [1,5]). In addition, IB4 labels tissue macrophages that can be double labelled with the F4/80 antibody to distinguish them from endothelial cells (Fig. 1c,d; Fig. S1) 22. Alternatively, the endothelial marker platelet endothelial cell adhesion molecule (PECAM, also known as CD31) 23 can be used to visualise endothelial cells in the hindbrain (Fig. 2).
In the case of fluorescent staining, multilabelling with primary antibodies raised in different species allows the simultaneous analysis of several proteins or cell populations and can be combined with IB4 staining. Staining for robust epitopes, such as IB4, can also be performed following in situ hybridisation 19. BrdU or equivalent compounds for proliferation studies may be administered to the pregnant dam prior to hindbrain isolation, allowing for homogenous labelling of the entire litter for comparison of littermate mutant and controls in mouse strains carrying recessive genetic mutations 2. When analysing hindbrains from genetically modified mouse embryos to identify alterations in vascular growth and patterning, it is essential to include appropriate controls, including hindbrains from wild type littermates or, in the case of conditional null mutants, hindbrains from littermates expressing Cre, but lacking the floxed target gene, and hindbrains lacking Cre, but carrying the floxed target genes.
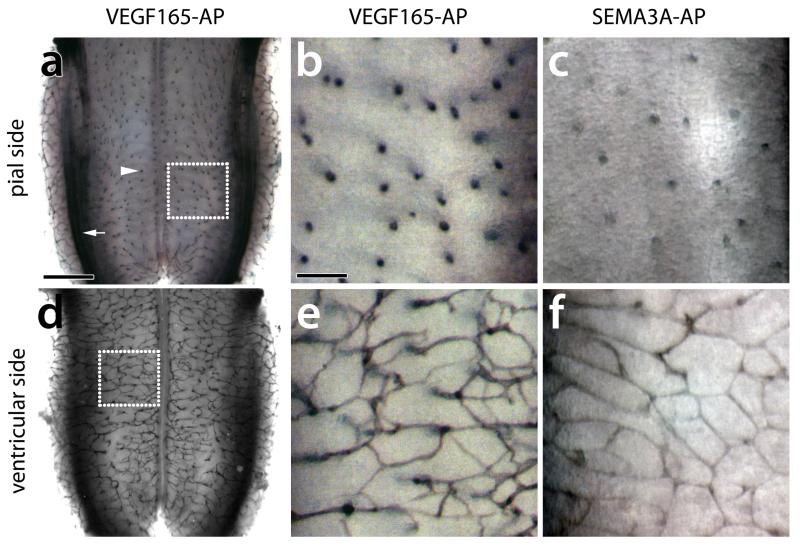
After staining with vascular markers such as PECAM or IB4, the radial vessels entering the brain can be visualised on the pial aspect of the flatmounted hindbrain (Fig. 2a-c, Fig. S1a-c), whilst the SVP vessels are apparent in a ventricular view (Fig. 2d-f, Fig. S1d-f) 1,2,6. Whilst histochemical methods are particularly suitable to provide overview images of the entire hindbrain vasculature after flatmounting (Fig. 2a,d, Fig. S1a,d), vibratome sectioning of the dissected hindbrains highlights the relationship of pial and SVP vessel segments (Fig. 2i-k, Fig. S1i,j) 1. The hindbrain also provides a useful platform for qualitative binding assays with alkaline phosphatase (AP)-tagged ligands to evaluate the suited to assess the binding of ligands to their vascular receptors in situ 6. For example, it can be used to show binding of VEGF165 and the alternative NRP1 ligand SEMA3A to growing hindbrain vessels (Fig. 3) 6.
Fig. 3. AP-fusion protein binding to the mouse embryo hindbrain.
Freshly dissected E12.5 hindbrains were reacted with the AP-tagged VEGF165 (a,b,d,e) and SEMA3A (c,f). (a-c) Flatmounting the hindbrain with the pial side up allows visualisation of radial vessels entering the brain (arrowhead); note that VEGF165 also detects axon tracts in the dorsolateral hindbrain (arrow). (d-f) Flatmounting the hindbrain with the ventricular side up allows visualisation of the subventricular vascular plexus. The dotted boxes in (a,d) indicate the areas shown at higher magnification in (b,e). Scale bars: (a,d) 500 μm, (b,c,e,f) 100 μm. All animal procedures were performed in accordance with institutional and UK Home Office guidelines.
Here we provide protocols for the hindbrain dissection (Fig. 4, Fig. S2 and supplementary videos 1 and 2), generic protocols for labelling with rat antisera and IB4 (Fig. 2, Fig. S1), a table of useful commercial antibodies (Table 1, 2), a protocol for AP fusion protein binding to whole hindbrains (Fig. 3), a table that details various angiogenic parameters that can be quantified in the hindbrain model and provides an overview of embryonic ages suitable for such quantitative analyses (Table 3) and a troubleshooting guide (Table 4) .
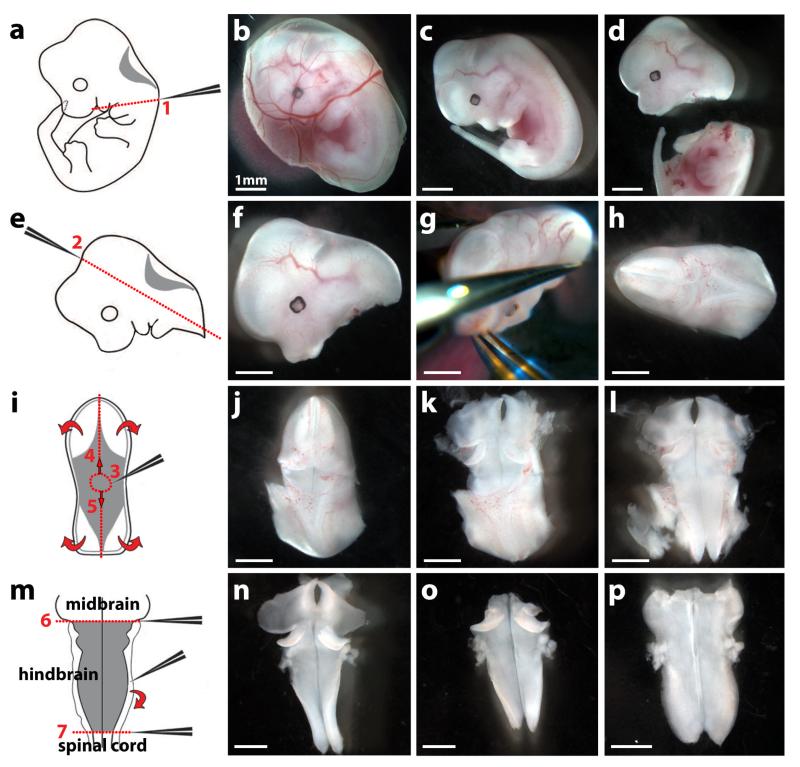
Fig. 4. Dissection of an E12.5 mouse embryo hindbrain.
The left hand side of the figure shows schematics and the right hand side dissected tissues.
(a-d) Remove the embryo and its yolk sac from the uterus with forceps (b); separate the embryo from the yolk sac (c); remove the head from the body with forceps by applying pressure along line 1 (a,d).
(e-h) Position the head (f) in a convenient orientation (g) and apply pressure along line 2 with forceps (e,g) to obtain the hindbrain with surrounding mesenchymal tissue (h).
(i-l) Position the hindbrain with surrounding mesenchymal tissue in a convenient orientation (j) and make a hole within the roofplate by inserting the forceps in position 3 (i,j). Use forceps to tear the roofplate along the dotted lines 4 and 5 (i) to expose the hindbrain (k) and allow it to unfurl (curved arrows in i; l).
(m) Tease away the pial membranes and surrounding mesenchyme from the neural tissue using forceps (curved arrow in m,n). Squeeze with forceps along the dotted lines 6 and 7 to remove midbrain and spinal cord tissue, respectively (m,o). The hindbrain now unfurls completely, ready for formaldehyde fixation; it can be easily visualised after carefully covering it with a glass coverslip (p).
Scale bars: 1 mm. All animal procedures were performed in accordance with institutional and UK Home Office guidelines. Adapted with permission from ref. 28.
Table 1. Examples of monoclonal antibodies specific for endothelial cells and vessel-associated cell types.
| cell type | Antibody | dilution |
|---|---|---|
| all EC | rat anti-mouse PECAM (CD31, MEC13.3) BD Pharmingen, cat. no. 553370 |
1:200 |
| capillary/venous EC | rat anti-mouse endomucin (clone V.7C7) Santa Cruz, cat. no. sc65495 |
1:200 |
| smooth muscle cells | mouse anti-SMA Sigma, cat. no. A5228 |
1:200 |
| macrophages | rat anti-mouse F4/80 AbD Serotec, cat. no. MCA497G |
1:500 |
Table 2. Examples of polyclonal antibodies specific for vascular extracellular matrix, vessel associated cell types, mitotic cells and proteins regulating angiogenesis.
| cell type | Antibody | dilution |
|---|---|---|
| basement membrane | rabbit anti-mouse collagen IV AbD Serotec, cat. no. 21501470 |
1:200 |
| pericytes | rabbit anti-mouse NG2 chondroitin sulfate proteoglycan Millipore, cat. no. AB5320 |
1:200 |
| cell proliferation | rabbit anti-mouse phosphorylated histone H3 (pH3, Ser10) Millipore, cat. no. 06-570 |
1:200 |
| various cell types, incl. EC | goat anti-mouse/rat NRP1 R&D Systems, cat. no. AF566 |
1:200 |
| EC | goat anti-mouse VEGFR2 R&D Systems, cat. no. AF644 |
1:200 |
Table 3. Examples of embryonic ages suitable to quantify various angiogenic parameters.
| side | age | angiogenic parameter | antibody | magnification |
|---|---|---|---|---|
|
| ||||
| ventricular | E10.5-11.5 | tip cell and filopodia number | IB4 (IF) | 40×, 63× |
| E10.5-12.5 | branchpoints or intersections | IB4, PECAM (IF or HRP) | 10× | |
| E10.5-12.5 | vessel diameter | IB4, PECAM (IF) | 63× | |
| E10.5-12.5 | endothelial cell proliferation | pH3 + IB4 (IF) | 10× | |
| E10.5-12.5 | tissue macrophage number | F4/80 ± IB4 (IF) | 10×, 20× | |
| E10.5-12.5 | pericyte density | NG2 + IB4 (IF) | 10×, 20× | |
|
| ||||
| pial | E11.5-12.5 | density of radial vessels | IB4, PECAM (IF or HRP) | 10× |
| E10.5-12.5 | vessel diameter | IB4, PECAM (IF) | 63× | |
| E10.5-12.5 | endothelial cell proliferation | pH3 + IB4 (IF) | 10× | |
Table 4. Troubleshooting guide.
| Step | Problem | Possible Reasons | Solution |
|---|---|---|---|
| 13A | No staining | Inadequate fixation; suboptimal antibody concentration. |
Use fresh 4 % formaldehyde solution; fix samples for 2 h on ice with gentle agitation; use fixative at 10× volume of tissue to be fixed; alternatively, test different fixative reagents; optimise antibody concentration; test alternative antibodies. |
| fluorescent speckles |
Secondary antibody forms precipitates. |
Spin secondary antibody in a refrigerated benchtop centrifuge at top speed before use. |
|
| Weak staining | Insufficient tissue penetration of antibodies; antibody concentration suboptimal; photobleaching. |
Increase incubation time; optimise antibody concentration; keep samples in the dark (e.g. wrap containers in tin foil); mount samples in antifade solution. |
|
| 13Bviii | No staining | Inadequate fixation; suboptimal antibody concentration. |
Use fresh 4 % formaldehyde solution, fix hindbrains for 2 hours on ice with gentle agitation; use fixative at 10× volume of tissue to be fixed; optimise antibody concentration; test fresh batches of primary or secondary antibodies. |
| High background |
Residual endogenous peroxidase activity; insufficient washing after antibody incubation; overdeveloped reaction. |
Check if correct bleaching with hydrogen peroxide was performed before staining; increase frequency of antibody washes; observe HRP reaction under a stereomicroscope every 5 minutes. |
|
| Weak staining | Insufficient tissue penetration of antibodies (E12.5 onwards); not enough time for HRP reaction; suboptimal antibody concentration. |
From E12.5 onwards, use vibratome sectioning followed by immunolabelling; allow at least 30 min for HRP reaction, observe reaction under a stereomicroscope every 5 minutes; incubate with new developing solution; optimise antibody concentration. |
|
| 13C | Hindbrain separates from agarose |
Excess liquid attached to hindbrain at embedding; incorrect settings for sectioning (e.g. alter sectioning speed). |
Carefully blot liquid from samples with clean tissue paper before embedding; optimise speed and vibration amplitude of vibratome. |
| 13D(i,ii) | Tissue degeneration |
PBT was used instead of PBS before tissue fixation. |
Use 10% FBS in PBS to block non-specific interactions before adding conditioned medium. Use PBS to wash unbound AP-fusion proteins before tissue fixation. |
| 13Dviii | Weak staining | Not enough time for AP reaction; suboptimal AP-fusion protein concentration. |
Observe under a stereomicroscope at 5 minute intervals; allow at least 30 min for AP reaction; incubate with fresh developing solution; spot conditioned medium on nitrocellulose filters and develop to verify AP-fusion protein expression. |
| 14C | Air bubbles in mounted sections. |
Trapping of air bubbles when adding the coverslip. |
Carefully lower the coverslip onto the section to avoid trapping air bubbles. |
MATERIALS
REAGENTS
4-nitro blue tetrazolium chloride (NBT, Roche, cat. no. 1087479).
5-bromo-4-chloro-3-indolyl-phosphate (BCIP, Roche, cat. no. 1585002).
Absolute methanol (Fisher Scientific, cat. no. M-4000-PC17). !CAUTION. It is irritating to eyes and skin. Danger of various serious irreversible effects through inhalation, in contact with skin and if swallowed. Wear protective goggles, clothing and gloves as appropriate.
Agarose (e.g. from BDH Electran, cat no. 438792U).
Diaminobenzidine and urea hydrogen peroxide tablets (Sigma-Aldrich, cat. no. D4293-50SET).
Glycerol (Acros Organics, cat no. AC15892-2500).
Heat inactivated normal goat serum (NGS, Sigma-Aldrich, cat. no. G9023).
Heat inactivated normal rabbit serum (NRS, Sigma-Aldrich, cat. no. R9133).
Hydrogen peroxide (Sigma-Aldrich, cat. no. H1009). !CAUTION. It is harmful if ingested and can cause serious damage to the eyes. Wear protective goggles, clothing and gloves as appropriate. Magnesium chloride 1 M (Sigma, cat. no. M1028).
Paraformaldehyde (PFA, Sigma-Aldrich. cat. no. P6148). !CAUTION. It is harmful if inhaled and swallowed. It is irritating to eyes, respiratory system and skin. It may cause sensitization of skin upon contact. Wear protective goggles, clothing and gloves as appropriate. Use it in a fume hood.
Phosphate buffered saline tablets (PBS, Sigma-Aldrich, cat. no. P4417).
Ready-to-use serum-free protein block solution (Dako, cat. no. X0909).
SlowFade Antifade reagent kit (Life Technologies, cat. no. S2828).
Sodium chloride (Sigma, cat. no. S3014)
Triton X100 (Sigma-Aldrich, cat. no. T8787). !CAUTION. It is harmful if ingested and can cause serious damage to the eyes. Wear protective goggles, clothing and gloves as appropriate.
Tris(hydroxymethyl)aminomethane (Tris, VWR, cat. no. 443864E)
AlexaFluor488-conjugated goat anti-rat secondary antibody (Life Technologies, cat. no. A11006). CRITICAL STEP: The protocol has been optimised with this fluorophore, but it is possible to choose an alternative fluorophore).
AlexaFluor488-conjugated streptavidin (Life Technologies, cat. no. S32354). CRITICAL STEP: The protocol has been optimised with this fluorophore, but it is possible to choose an alternative streptavidin-conjugated fluorophore.
Biotinylated lectin from Bandeiraea simplicifolia BS-I isolectin B4 (IB4; Sigma, cat. no. L2140). CRITICAL STEP: The protocol has been optimised for this lectin and its use is therefore recommended.
HRP-tagged rabbit anti-rat secondary antibody (Dako, cat. no. P045).
HRP-tagged streptavidin (Dako, cat. no. P0397).
Rat anti-mouse PECAM monoclonal antibody (CD31, MEC13.3; BD Pharmingen, cat. no. 553370). CRITICAL STEP: The protocol has been optimised for this antibody and its use is therefore recommended.
Additional primary and secondary antibodies may be required to label macrophages, smooth muscle cells, capillary and venous endothelia, vascular basement membrane, pericytes, mitotic cells or VEGF-A receptors. See Tables 1 and 2 for details.
Pregnant Dam. Adult male and female mice of any fertile mouse strain can be paired to obtain pregnant dams. For timed matings, mice are mated in the evening, and the morning of vaginal plug formation is counted as 0.5 days post coitum (dpc). !CAUTION: Institutional and governmental ethics regulations concerning rodent use must be followed.
DMEM (Life Technologies, cat. no. 31966-021).
FBS (Life Technologies, cat. no. 10437-028).
Lipofectamine (Life Technologies, cat. no. 18324012).
ddH2O.
Plasmid DNA.
EQUIPMENT
2.0 ml round-bottomed reagent tubes (safe-lock; e.g. Eppendorf, cat. no. 022363352).
24-well cell culture plates (e.g. BD Falcon, cat. no. 353047).
50 ml falcon tubes (e.g. BD Falcon, cat. no. 352098).
Benchtop tube roller (e.g. Stuart SRT9, Stuart).
Confocal laser scanning microscope (e.g. Zeiss LSM710, Zeiss).
Data acquisition software (e.g. OpenLab 3.5.1, Improvision Ltd.).
Disposable plastic moulds (e.g. Electron Microscopy Sciences, cat. no. 62352).
Epifluorescence stereomicroscope (e.g. Olympus, SZX16) equipped with a digital camera (e.g. Hamamatsu C4742-95-12HR).
Glass bottles (e.g. Schott Duran).
Glass coverslips (22×55mm; VWR international, cat. no. 631-0137).
Glass slides (VWR international, cat. no. 631-0111).
Gooseneck lamps (e.g. cold light source KL 2500 LCD, Schott).
Image editor software (e.g. Photoshop CS3, Adobe Inc.).
Plastic cell culture dishes (60 mm diameter; e.g. from BD Falcon, cat. no. 353002).
Plastic Pasteur pipettes (e.g. from Alpha Laboratories, cat. no. LW4003).
Stereomicroscope (e.g. Leica MZ16, Leica) equipped with a digital camera (e.g. QImaging Micropublisher 3.3 RTV).
Tabletop balance (e.g. ED Precision balance, Sartorius).
Vibratome (e.g. Vibratome 1000Plus Sectioning System, Intracel).
Watchmaker forceps no. 5 (e.g. from Dumont, cat. no. 91150-20).
Watchmaker forceps no. 55 (e.g. from Dumont, cat. no. 11255-20).
Water bath (e.g. Stuart SWB15D, Stuart).
REAGENT SETUP
Blocking buffer (10%, vol/vol): For 1 ml, mix 100 μl of serum with 900 μl of PBT. Prepare fresh for each immunostaining experiment.
Developing buffer
Dissolve 6 g of Tris base in 50 ml of ddH2O and adjust to pH 9.5 to yield 1 M Tris buffer. Dissolve 2.9 g of sodium chloride in 10 ml of ddH2O to yield 5 M sodium chloride. Mix 1 ml of 1 M Tris buffer with 200 μl of 5 M sodium chloride and 50 μl of 1 M magnesium chloride. Add ddH20 to 10 ml. Store at room temperature (21°C) indefinitely. Developing solution: Add 3.5 μl of NBT and 3.5 μl of BCIP to 1 ml of developing buffer. Protect from light. Prepare fresh for each experiment.
DAB solution
Dissolve 1 DAB tablet in 5 ml of ddH2O at room temperature on bench-top roller. Adjust tablet amount to final volume required. Protect from light. Prepare fresh for each immunostaining experiment.
DAB and urea/hydrogen peroxide solution
Dissolve 1 DAB and 1 urea hydrogen peroxide tablet in 5 ml of ddH2O at room temperature on bench-top roller. Protect from light. Prepare fresh for each immunostaining experiment.
Formaldehyde solution (4%, wt/vol)
Under a chemical fume hood, weigh 2 g PFA into a 50 ml Falcon tube and add 50 ml PBS. Dissolve PFA by heating the solution to 60°C in a water bath, mixing frequently until the PFA powder has dissolved. Store in aliquots at −20 °C for up to 1 year. Thaw at room temperature or at 37°C in a water bath and cool in ice prior to use.
Methanol gradient
To prepare 50 ml of 25%, 50% or 75% methanol in PBT, mix 12.5, 25 or 37.5 ml of absolute methanol with 37.5, 25 or 12.5 ml of PBT in 50 ml Falcon tubes. Store at room temperature indefinitely.
PBS
Dissolve 5 tablets of PBS in 1 l of ddH20. Store at room temperature indefinitely. CRITICAL STEP: Use PBS for non-permeabilised tissue staining, for example for visualisation of extracellular epitopes rather than cytoplasmic epitopes.
PBT
Add 50μl Triton X-100 to 50 ml PBS in a 50 ml Falcon tube; store at room temperature indefinitely. CRITICAL STEP: Use PBT for permeabilisation of the tissue to detect both cell surface and cytoplasmic epitopes.
Expression of AP fusion proteins
Expression vectors encoding a secreted form of AP suitable to insert cDNA sequences to be tested as AP fusion proteins can be obtained from various sources (e.g., pAPtag-4, GenHunter Corporation, or APTag-124). To generate fusion proteins, grow HEK-293T cells to 80% confluence in 10-cm culture dishes in growth medium (high glucose DMEM with 10% (vol/vol) FBS and antibiotics) and transfect the cells as follows: incubate 25 μl of Lipofectamine and 10 μg of plasmid DNA with two separate 500-μl aliquots of prewarmed serum- and antibiotics-free DMEM for 15 min at room temperature. Next, combine Lipofectamine and DNA solution and incubate the mixture for 15 min at room temperature; next, remove the medium from the HEK-293T cells and add the Lipofectamine/DNA solution to the cells in a dropwise manner. Add 4 ml of prewarmed serum- and antibiotics-free DMEM and place the cells in a 37 °C 5% CO2 humified incubator for 6 h; next, replace the medium with prewarmed growth medium and put the cells back in the incubator for 42 h. Finally, collect the supernatant and pass it through a 0.22-μm filter to remove detached cells; divide the supernatant into aliquots and store them at −80°C. This process takes ~2 d. Alternative methods have been published24.
EQUIPMENT SETUP
Plastic Pasteur pipettes
Use to transfer embryos and hindbrains between dishes: Cut off the tip with a clean scalpel or scissors to create an appropriately sized opening. Alternatively, slice the bulb of the pipette resulting in a spoon shaped opening suitable to transfer large embryos.
Benchtop tube roller
Should be placed in a cold room or cold cabinet, because antibody incubations should be performed in tubes that are gently rolling at 4 °C.
PROCEDURE
EMBRYO ISOLATION: TIMING ~ 5 MIN PER EMBRYO
1 At the required gestational age, kill the pregnant dam in a humane manner (for example by cervical dislocation). !CAUTION. Animal procedures must be carried out in accordance with relevant institutional and government ethics guidelines.
2 Dissect the uterus by cutting a V-shape into the abdominal skin and membrane. Pull the uterine horns out of the body cavity with forceps and cut off the birth canal, fatty tissue and both oviducts. Transfer the uterine horns into a clean plastic dish containing ice cold PBS.
3 Under a dissecting stereomicroscope, use forceps to remove each single embryo sac from the uterine horns by rupturing the surrounding muscular layers. This can be done after cutting the uterus into pieces containing single embryos or by removing the embryos one after another from the intact uterine horns (supplementary videos 1 and 2).
4 Rupture each embryo sac until the embryo emerges, sever the umbilical cord, collect the embryos with a plastic Pasteur pipette into a dish with clean PBS and place the dish on ice (supplementary videos 1 and 2). CRITICAL STEP. Perform this step with the embryos submerged in PBS to prevent the surface tension from rupturing the embryo. This is particularly relevant for dissection of embryos at 10.5-11.5 dpc.
5 Transfer one embryo at a time with a plastic Pasteur pipette to a fresh dish containing ice-cold PBS for hindbrain dissection. If the experiment requires genotyping of the litter, keep a piece of embryonic tail or yolk sac tissue for genomic DNA isolation.
HINDBRAIN DISSECTION: TIMING ~ 5 MIN PER HINDBRAIN
6 By squeezing with forceps, sever the head above the forelimbs (Fig. 4a-d, line 1; Fig. S2a) and remove the front of the head and face (Fig. 4e-h, line 2; Fig. S2b).
7 Rupture the thin and translucent roof-plate overlying the hindbrain at the level of the fourth ventricle by pulling gently with sharp forceps (Fig. 4i,j, Fig. S2c and supplementary videos 1 and 2). Continue to tear the membrane towards the midbrain (Fig. 4i,k, small arrows; Fig. S2d,e), then towards the spinal cord to open up the hindbrain (Fig. 4i.l, curved arrows; Fig. S2f; supplementary videos 1 and 2). The hindbrain tissue can now be seen clearly under the dissecting stereomicroscope. CRITICAL STEP. Do not insert the forceps too deep, as this may damage the hindbrain tissue.
8 Tease off the pial membrane from underneath the hindbrain (Fig. 4m,n; Fig. S2g,h and supplementary videos 1 and 2); from E12.5 onwards it comes off very easily after opening the roof-plate, at earlier stages it is a matter of tugging a bit here and there, like peeling an orange - some regions give way easier than others.
9 Remove the midbrain and spinal cord to allow the hindbrain to unfurl (Fig. 4m, lines 6 and 7, respectively; Fig. S2i,j). Transfer the isolated hindbrains (Fig. 4o, Fig. S2k,l) with a plastic Pasteur pipette flat onto the bottom of a well in a 24-well plate that has been placed on ice. CRITICAL STEP. Do not allow embryos to dehydrate. Keep embryos in PBS until adding the fixative in step 10, or omit steps 10-13C and proceed to step 13D for AP fusion protein binding assay.
FIXATION: TIMING 2 HOURS
10 Remove PBS and add cold 4% formaldehyde to the hindbrain. CRITICAL STEP. For all antibodies mentioned in this protocol (Tables 1,2), freshly prepared or freshly thawed formaldehyde fixative is a suitable fixative. Other antibodies may require an alternative fixation protocol.
11 Fix for 2 h on ice with gentle agitation, rinse three times in PBS and proceed to staining protocol. CRITICAL STEP: For PECAM staining, it is best to stain freshly fixed tissues and avoid fixation longer than 2 h or methanol storage. If staining cannot be initiated immediately after fixation, samples can be stored over night at 4 °C in PBS before staining the following day. PAUSE POINT. Alternatively, transfer through a rising methanol gradient to store at −20 °C for up to 3 months (gradient: 25%, 50% and 75% absolute methanol in PBS, then absolute methanol).
STAINING PROCEDURE: TIMING 3 DAYS
12 For tissue stored in methanol, transfer through a decreasing methanol gradient (75%, 50% and 25% absolute methanol in PBT, then PBT alone).
13 Place hindbrains in 2.0 ml round-bottomed safe-lock tubes ready for fluorescent or histochemical labelling. Follow option A for PECAM and/or IB4 fluorescent staining. Follow option B for PECAM or IB4 histochemical staining. Guidance on using alternative antibodies is given in Box 1. Follow option C for vibratome sectioning and option D for whole-mount AP fusion protein binding. CRITICAL STEP. To save antibody and to standardise antibody exposure between different samples for comparison of staining intensity, individual hindbrains can be tagged to distinguish them from each other morphologically and then pooled into one tube (for example, midbrain or spinal cord or both can be left attached; a maximum number of 4 tagged hindbrains per tube is recommended).
- PECAM and/or IB4 fluorescent staining TIMING 3d
- Incubate in blocking solution for 30 minutes with gentle rolling at room temperature. For PECAM labelling, use 10% (vol/vol) normal goat serum in PBT as blocking solution. For biotinylated IB4 labelling, use 10% (vol/vol) normal goat or rabbit serum in PBT as blocking solution.
- Incubate overnight at 4 °C with gentle rolling in blocking solution containing primary antibody (rat anti-mouse PECAM, diluted 1:200 or biotinylated IB4, diluted 1:200).
- Thoroughly wash the hindbrains five times for 15-30 minutes each in PBT at room temperature.
- Incubate overnight at 4 °C with gentle rolling in blocking solution containing secondary antibody diluted 1:200. For PECAM detection, use AlexaFluor488-tagged goat anti-rat antibodies. For biotinylated IB4 detection, use AlexaFluor488-tagged streptavidin.
- Wash the hindbrains as in step 13 A iii.
- Post-fix in 4% (wt/vol) formaldehyde solution for 30 minutes at room temperature. PAUSE POINT. At this stage samples can either be processed for imaging or can be stored at 4 °C in PBS for up to one week before imaging (see below).
- PECAM or IB4 histochemical staining: TIMING 3d
- Eliminate endogenous peroxidase activity by incubating in 1% (vol/vol) hydrogen peroxide in PBS for 30 minutes at room temperature with gentle rolling.
- Wash samples in PBT twice for 5 minutes each.
- Incubate in blocking solution for 30 minutes at room temperature with gentle rolling. For PECAM, use 10% (vol/vol) normal rabbit serum in PBT as blocking solution. For biotinylated IB4, use 10% (vol/vol) normal goat or rabbit serum in PBT as blocking solution.
- Incubate overnight at 4 °C in blocking solution containing primary antibody (rat anti-mouse PECAM, diluted 1:200 or biotinylated IB4, diluted 1:200).
- Thoroughly wash the hindbrains five times for 15-30 minutes in PBT at room temperature.
- Incubate overnight at 4 °C with gentle rolling in blocking solution containing secondary antibodies, diluted 1:200. For PECAM detection, use HRP-tagged rabbit anti-rat antibody. For biotinylated IB4 detection, use HRP-tagged streptavidin.
- Wash the hindbrains as in step 13 B v.
- Incubate the hindbrains at room temperature (with gentle rolling) in a solution of diaminobenzidine for 10 minutes, and then in a solution of diaminobenzidine and urea/hydrogen peroxide for 5 minutes, or until colour develops. CAUTION. Samples need to be protected from light, as diaminobenzidine and urea/hydrogen peroxide solutions are photosensitive. CRITICAL STEP. To avoid overdeveloping the reaction, periodically observe the samples under a stereomicroscope. ?TROUBLESHOOTING
- To stop the reaction, wash the hindbrains briefly in ddH2O and then PBS and post-fix in 4% (wt/vol) formaldehyde for 30 minutes at room temperature. PAUSE POINT. At this stage, samples can either be processed for imaging or can be stored at 4 °C in PBS until imaging (see below).
- VIBRATOME SECTIONING AFTER STAINING: TIMING 30 MIN PER HINDBRAIN
- Place 3 g agarose in 100 ml ddH20 in a glass bottle and melt in the microwave or a boiling water bath. Leave to cool, for example in a 60°C oven
- Place the hindbrain in a square plastic mould and remove excess liquid attached to the exterior of the hindbrain with a pipette, followed by brief, gentle blotting with paper tissue.
- When the molten agarose has cooled sufficiently to comfortably touch the glass, pour it into the mould and quickly position the hindbrain before the agarose begins to set.
- After the agarose has set, remove the agarose block containing the hindbrain from the mould (which can be re-used) and cut 80-100 μm transverse sections with a vibratome. ?TROUBLESHOOTING
- AP FUSION PROTEIN BINDING ASSAY: TIMING 2 DAYS
- Incubate the hindbrains in 10% (vol/vol) FBS in PBS for 30 minutes with gentle rolling at 4°C. CRITICAL STEP. Magnesium chloride at a final concentration of 4 mM must be added to PBS when assessing protein-protein interactions involving semaphorins. ?TROUBLESHOOTING
- Incubate the hindbrains in conditioned medium containing AP-fusion protein overnight at 4°C. CRITICAL STEP. Incubate one hindbrain with conditioned medium containing AP protein not fused to any other protein as a negative control.
- Thoroughly wash the hindbrains three times for 15 minutes in PBS at room temperature. ?TROUBLESHOOTING
- Fix in 4% (wt/vol) formaldehyde for 1 h at 4°C with gentle agitation. CRITICAL STEP. Do not overfix. Longer fixation may decrease AP enzymatic activity.
- Thoroughly wash the hindbrains two times for 15 minutes in PBT at room temperature.
- Incubate the hindbrains in PBS at 65°C for 3 h to inactivate endogenous alkaline phosphatases.
- Incubate the hindbrains 10 minutes in developing buffer.
- Detect tissue-bound heat-stable recombinant AP activity by incubating the hindbrains in developing solution at room temperature for 15 minutes, or until colour develops. CAUTION! Samples need to be protected from light, as NBT and BCIP contained in the developing solution are photosensitive. CRITICAL STEP. To avoid overdeveloping the reaction, periodically observe the samples under a stereomicroscope. ?TROUBLESHOOTING
- To stop the reaction, wash the hindbrains briefly in ddH2O and then PBS and post-fix in 4% formaldehyde for 30 minutes at room temperature. PAUSE POINT. At this stage, samples can either be processed for imaging (see below) or can be stored at 4 °C in PBS until imaging.
IMAGING: TIMING VARIABLE, 0.5-1 HOUR PER HINDBRAIN
14 Image wholemount fluorescently labelled hindbrains as described in option A, for wholemount HRP- or AP-labelled hindbrains follow option B. To image vibratome sections follow option C.
- Fluorescently labelled hindbrains.
- stick two layers of black electrical tape onto a glass slide and cut rectangular pockets to create a spacer. Flatmount each hindbrain with SlowFade reagent into one pocket and cover with a glass coverslip.
- Image the hindbrains with a fluorescence stereomicroscope or by confocal scanner laser microscopy.
- HRP- or AP-labelled hindbrains
- Place each hindbrain individually into an empty plastic dish (60 mm diameter), remove excess liquid and cover with a glass coverslip. Alternatively, hindbrains can be flattened more gently if placed into a well on an agarose dish (the well is created by scooping a little agarose out with a pipette tip). This prevents tearing of the tissue when the glass coverslip is pushed down gently to flatten the hindbrain.
- Image the hindbrains with a standard stereomicroscope equipped with a suitable camera. Note that it may be necessary to wet the coverslip before taking the image, to remove dust and allow the hindbrain to flat properly.
- To image vibratome sections.
- Mount sections on a glass slide in 90% (vol/vol) glycerol in PBS for HRP-labelled samples or SlowFade reagent for fluorescently labelled samples and cover with a glass coverslip. ?TROUBLESHOOTING
BOX 1: ALTERNATIVE IMMUNOSTAINING PROTOCOLS: TIMING SAME AS SINGLE STAINING PROCEDURE (3 DAYS)
Multi-labelling of vessels and other cell types can be achieved using a combination of primary antibodies from different species, or antibodies together with IB4 (e.g. IB4 and F4/80 double staining, shown in Fig. 1c,d). For such experiments, apply the primary antibodies, or IB4 and the primary antibodies, together in the blocking solution; after washing, apply the secondary antibodies (and streptavidin in the case of IB4) together in the blocking solution. The antibodies used above and some further examples of monoclonal antibodies suitable to label macrophages, smooth muscle cells or capillary and venous endothelia are described in Table 1. A separate list of polyclonal antibodies useful for labelling vascular basement membrane, pericytes, mitotic cells and the VEGF-A receptors NRP1 and VEGFR2 can be found in Table 2. However, polyclonal antibodies can show batch-specific variations and need to be tested for their suitability by the users.
To block unspecific antibody binding, staining protocols usually include serum that is compatible with the primary and secondary antibodies, i.e. from the same species as the one in which the secondary antibodies are raised. In contrast, for staining protocols that include primary antibodies raised in goats, we recommend using serum-free serum block (e.g. from DAKO, cat. no. X0909); it is not necessary to include this block in the antibody dilutions. Moreover, we recommend using only fluorophore-conjugated Fab fragments as secondary antibodies in experiments that include primary antibodies raised in goat.
IB4 binds α-D-galactosyl residues on glycoproteins, and such lectin epitopes are generally more stable than protein epitopes; IB4 staining is therefore compatible with immunolabelling after acid hydrolysis for BrdU immunolabelling or in situ hybridisation.
SUMMARY OF TIMING
Expression of AP fusion protein, optional (Reagent Setup): 2 d
Steps 1-5, embryo isolation: ~ 5 min per embryo
Steps 6-9, hindbrain dissection: ~ 5 min per hindbrain
Steps 10-11, fixation: 2 h
Steps 12-13, staining procedure: ~ 3 d
Step 14, imaging: variable, 0.5-1 h per hindbrain
Box 1, alternative staining protocols: same as single staining procedure (3 d)
Data analysis (discussed in Anticipated Results): variable, depending on number of samples, 0.5-2 h
TROUBLESHOOTING
Troubleshooting advice can be found in Table 4.
ANTICIPATED RESULTS
To quantify the angiogenic parameters described in Table 3, image several randomly chosen fields from each side of the hindbrain and average the numeric values obtained for each field to minimise technical error. For high power bright-field images obtained with a stereomicroscope, e.g. 11.5× magnification (depending on the specifications of the microscope), acquire a minimum of three fields of 0.25 mm2 (Fig. 2c,f; Fig. S1c,f) and count vascular intersections in each field (e.g. Fig. 2h and Fig. S1h). To quantify angiogenic parameters in low power confocal images use a minimum of two fields of approximately 0.85 mm2 (with 10× objectives) or 0.2 mm2 (with 20× objectives), one from each side of the hindbrain (Fig. 5c,d). To quantify angiogenic parameters in higher magnification confocal images, e.g. 40× or 63x, use a minimum of 4 fields such as those shown in Fig. 5e-g. In all cases, the averaged numeric value for all fields from one hindbrain will yield the final value for that hindbrain.
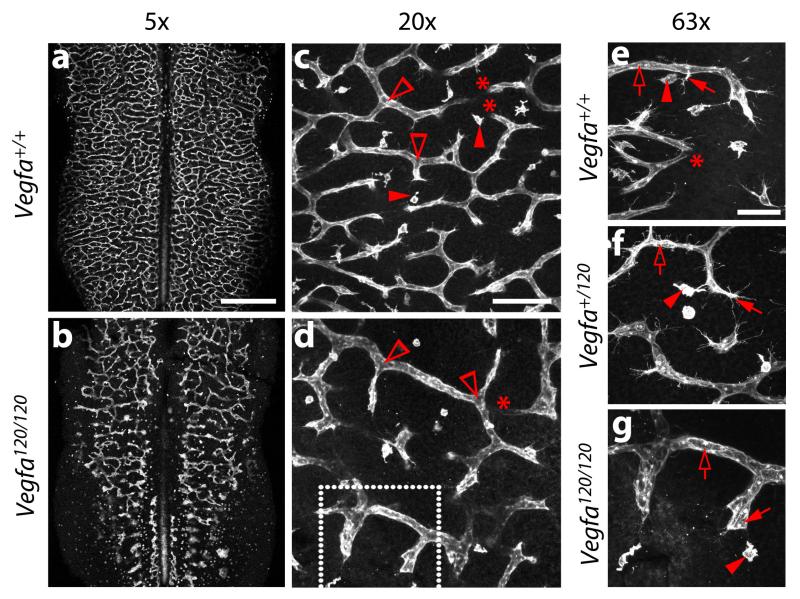
Fig. 5. Examples of vascular defects in the hindbrain of mice from Vegfa+/120 intercrosses.
E11.5 littermate hindbrains from timed matings of Vegfa+/120 parents were fluorescently labelled with IB4 and imaged at the indicated magnifications. Examples of vascular intersections (clear arrowheads) for branch point quantification and tissue macrophages (solid arrowheads) are indicated in (c,d). Examples of tissue macrophages (solid arrowheads), endothelial tip cells (solid arrows) and endothelial stalks cells (clear arrows) are indicated in (e-g). Areas indicated with asterisks show vessels that dive into deeper layers of the hindbrain and appear ‘cut-off’, because the confocal z-stack does not span the entire SVP. The area shown in (g) is a higher magnification of the area indicated with a white box in (d). Scale bars: (a,b) 500 μm; (c,d) 100 μm, (e-g) 50 μm. All animal procedures were performed in accordance with institutional and UK Home Office guidelines.
For manual counting of angiogenic features such as endothelial tip cells, vascular branchpoints, intersections or tissue macrophages, it is advisable to track all counted structures by marking them with a symbol in the digital image or by using the ‘Count Tool’ in Adobe Photoshop (Adobe Inc). Length measurements, for example of vessel segments or vascular diameters, can be achieved with the ‘Ruler Tool’ function in Adobe Photoshop or appropriate imaging software, such as Openlab (Improvision). After careful validation, automated protocols may also be used to quantify vascular intersections and other angiogenic parameters, for example by using the freely available AngioTool 25, which is based on the free software ImageJ (http://rsb.info.nih.gov/ij; National Institute of Health, Bethesda,MD). ImageJ may also be modified to develop alternative quantification methods that suit specific needs. Alternatively, commercial software such as that provided by Imaris (Bitplane) or Volocity (Improvision) are useful to quantitate angiogenic parameters.
In our experience, the analysis of a minimum of 3 hindbrains of each genotype from at least 2 litters will yield statistical significance when phenotypic differences between genotypes are large (e.g. hindbrains expressing all VEGF-A isoforms compared to hindbrains expressing only VEGF120, Fig. 5a-e,g). For more subtle phenotypes, the analysis of 5-10 hindbrains is usually appropriate to obtain sufficient statistical power (e.g. hindbrains expressing all VEGF-A isoforms from both alleles compared to hindbrains expressing VEGF120 only from one allele; Fig. 5e, f). When representing data obtained from biological experiments consider published advice on the use of error bars 26.
Figure 1 shows examples of typical fluorescent staining results for labelling of endothelial tip and stalks cells as well as tissue macrophages. Figure 2 shows typical histochemical staining for PECAM and an example of a quantitation analysis for radial vessels on the pial side of the hindbrain and vascular intersections in the subventricular zone; the fields shown contain 44 radial vessels per 0.25 mm2 and 95 intersections per 0.25 mm2 in the subventricular vascular plexus. Some typical values for vessel numbers between E10.5 and E12.5 have previously been published 1 and may be used as guidelines for expected values at different developmental stages; however, the precise number may vary depending on the precise developmental stage or mouse genetic background, and littermate controls should therefore always be used to assess possible mutant phenotypes.
Figure 3 shows examples of freshly dissected whole-mount hindbrains reacted with AP-tagged VEGF165 or SEMA3A proteins. This assay visualises AP-ligand binding to both radial vessels on the pial side (Fig, 3a-c) and SVP vessels on the ventricular side (Fig. 3d-f) 6. VEGF165 binds hindbrain vessels with high affinity (Fig. 3a,b and d,e) 6, due to the expression by endothelial cells of several receptors, including VEGFR2 and the VEGF165-specific receptor NRP1 1, Because NRP1 can additionally bind semaphorins, SEMA3A fusion protein also binds hindbrain vessels (Fig. 3c,f) 6.
Figure 5 shows the vascular phenotype of E11.5 hindbrains containing a knockin mutation that allows production of the VEGF120 isoform, but disrupts the production of the heparin/neuropilin binding VEGF-A isoforms. Compared to wild type littermates, homozygous Vegfa120/120 show a dramatic decrease in vessel branching and an increase in vessel calibre (compare Fig. 5a,c,e with 5b,d,g), as previously published 2. The phenotype of Vegfa+/120 mutants is more subtle, with an intermediate amount of vessel branching and vessel calibre compared to wild type and homozygous littermates (compare Fig. 5f with 5e,g). A common problem in endothelial tip cell quantitation is highlighted in Fig. 5c-e, where areas indicated with asterisks show vessels that dive into deeper layers of the hindbrain and appear ‘cut-off’; care should be taken to not confuse such vessel fragments with new vessel sprouts, which are headed by filopodia-studded tip cells.
Supplementary video 2 shows the dissection of a hindbrain in which NRP1 was deleted in endothelial cells using Cre-Lox technology to yield Tie2-Cre;Nrp1fl/− hindbrains 27. This approach delays the embryonic lethality caused by two null alleles of Nrp1 from E12.5 to the time of birth 14,27, but preserved the formation of hindbrain vascular malformations, which are already visible during the dissection procedure (compare dissected hindbrains in supplementary videos 1 and 2).
Supplementary Material
ACKNOWLEDGMENTS
We thank the staff of the Biological Resources Unit at the UCL Institute of Ophthalmology for help with mouse husbandry and the Imaging Facility of the UCL Institute of Ophthalmology for maintenance of the confocal microscopes. The protocol described in this manuscript was developed with funding from the Wellcome Turst [095623/Z/11/Z] and MRC [project grant G0601093] to C. R. and PhD studentships from the British Heart Foundation to A. F. [ref. 44626] and A. P. [ref. FS/10/54/28680] and a PhD studentship from the MRC to C.H.M. [G0700020].
List of abbreviations
- AP
alkaline phosphatase
- BCIP
5-bromo-4-chloro-3-indolyl-phosphate
- BrdU
5-bromo-2′-deoxyuridine
- ddH2O
double-distilled H2O
- dpc
days post coitum
- EC
endothelial cell
- FBS
fetal bovine serum
- HRP
horseradish peroxidase
- IB4
isolectin B4
- NBT
4-nitro blue tetrazolium
- NGS
normal goat serum
- NRP1
neuropilin 1
- NRS
normal rabbit serum
- PBS
phosphate buffered saline
- PBT
PBS + 0.1% Triton×100
- PDGF-BB
platelet-derived growth factor B
- PECAM
platelet endothelial cell adhesion molecule
- PFA
paraformaldehyde
- pH3
phosphorylated histone 3
- PI3K
phosphatidylinositol 3-kinase
- SMA
smooth muscle alpha actin
- SVP
subventricular vascular plexus
- VEGF
vascular endothelial growth factor
- VEGFR
vascular endothelial growth factor receptor
- Vol
volume
- Wt
weight
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
AUTHOR CONTRIBUTIONS STATEMENTS
A.F., A. P., C.H.M. and C.R. prepared the figures and supplementary videos. All authors contributed to the development of the protocol and wrote the manuscript.
REFERENCES
- 1.Fantin A, et al. Tissue macrophages act as cellular chaperones for vascular anastomosis downstream of VEGF-mediated endothelial tip cell induction. Blood. 2010;116:829–840. doi: 10.1182/blood-2009-12-257832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ruhrberg C, et al. Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes & Development. 2002;16:2684–2698. doi: 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerhardt H, et al. VEGF guides angiogenic sprouting utilizing endothelial tip cell filopodia. The Journal of Cell Biology. 2003;161:1163–1177. doi: 10.1083/jcb.200302047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Raab S, et al. Impaired brain angiogenesis and neuronal apoptosis induced by conditional homozygous inactivation of vascular endothelial growth factor. Thromb Haemost. 2004;91:595–605. doi: 10.1160/TH03-09-0582. [DOI] [PubMed] [Google Scholar]
- 5.Gerhardt H, et al. Neuropilin-1 is required for endothelial tip cell guidance in the developing central nervous system. Developmental Dynamics. 2004;231:503–509. doi: 10.1002/dvdy.20148. [DOI] [PubMed] [Google Scholar]
- 6.Vieira JM, Schwarz Q, Ruhrberg C. Selective requirements for NRP1 ligands during neurovascular patterning. Development. 2007;134:1833–1843. doi: 10.1242/dev.002402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Graupera M, et al. Angiogenesis selectively requires the p110alpha isoform of PI3K to control endothelial cell migration. Nature. 2008;453:662–666. doi: 10.1038/nature06892. [DOI] [PubMed] [Google Scholar]
- 8.Lu X, et al. The netrin receptor UNC5B mediates guidance events controlling morphogenesis of the vascular system. Nature. 2004;432:179–186. doi: 10.1038/nature03080. [DOI] [PubMed] [Google Scholar]
- 9.Abramsson A, et al. Defective N-sulfation of heparan sulfate proteoglycans limits PDGF-BB binding and pericyte recruitment in vascular development. Genes & Development. 2007;21:316–331. doi: 10.1101/gad.398207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tammela T, et al. VEGFR-3 controls tip to stalk conversion at vessel fusion sites by reinforcing Notch signalling. Nature Cell Biology. 2011;13:1202–1213. doi: 10.1038/ncb2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pitulescu ME, Schmidt I, Benedito R, Adams RH. Inducible gene targeting in the neonatal vasculature and analysis of retinal angiogenesis in mice. Nat Protoc. 2010;5:1518–1534. doi: 10.1038/nprot.2010.113. [DOI] [PubMed] [Google Scholar]
- 12.Fruttiger M. Development of the retinal vasculature. Angiogenesis. 2007;10:77–88. doi: 10.1007/s10456-007-9065-1. [DOI] [PubMed] [Google Scholar]
- 13.Carmeliet P, et al. Impaired myocardial angiogenesis and ischemic cardiomyopathy in mice lacking the vascular endothelial growth factor isoforms VEGF164 and VEGF188. Nat Med. 1999;5:495–502. doi: 10.1038/8379. [DOI] [PubMed] [Google Scholar]
- 14.Kawasaki T, et al. A requirement for neuropilin-1 in embryonic vessel formation. Development. 1999;126:4895–4902. doi: 10.1242/dev.126.21.4895. [DOI] [PubMed] [Google Scholar]
- 15.Ruhrberg C. Growing and shaping the vascular tree: multiple roles for VEGF. BioEssays. 2003;25:1052–1060. doi: 10.1002/bies.10351. [DOI] [PubMed] [Google Scholar]
- 16.Nagy A. Cre recombinase: the universal reagent for genome tailoring. Genesis. 2000;26:99–109. [PubMed] [Google Scholar]
- 17.Ferrara N, et al. Heterozygous embryonic lethality induced by targeted inactivation of the VEGF gene. Nature. 1996;380:439–442. doi: 10.1038/380439a0. [DOI] [PubMed] [Google Scholar]
- 18.Carmeliet P, et al. Abnormal blood vessel development and lethality in embryos lacking a single VEGF allele. Nature. 1996;380:435–439. doi: 10.1038/380435a0. [DOI] [PubMed] [Google Scholar]
- 19.Schwarz Q, et al. Vascular endothelial growth factor controls neuronal migration and cooperates with Sema3A to pattern distinct compartments of the facial nerve. Genes & Development. 2004;18:2822–2834. doi: 10.1101/gad.322904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sawamiphak S, Ritter M, Acker-Palmer A. Preparation of retinal explant cultures to study ex vivo tip endothelial cell responses. Nat Protoc. 2010;5:1659–1665. doi: 10.1038/nprot.2010.130. [DOI] [PubMed] [Google Scholar]
- 21.Laitinen L. Griffonia simplicifolia lectins bind specifically to endothelial cells and some epithelial cells in mouse tissues. The Histochemical Journal. 1987;19:225–234. doi: 10.1007/BF01680633. [DOI] [PubMed] [Google Scholar]
- 22.Sorokin SP, Hoyt RF., Jr. Macrophage development: I. Rationale for using Griffonia simplicifolia isolectin B4 as a marker for the line. Anat Rec. 1992;232:520–526. doi: 10.1002/ar.1092320409. [DOI] [PubMed] [Google Scholar]
- 23.Albelda SM, Muller WA, Buck CA, Newman PJ. Molecular and cellular properties of PECAM-1 (endoCAM/CD31): a novel vascular cell-cell adhesion molecule. J Cell Biol. 1991;114:1059–1068. doi: 10.1083/jcb.114.5.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brennan C, Fabes J. Alkaline phosphatase fusion proteins as affinity probes for protein localization studies. Science’s STKE : signal transduction knowledge environment. 2003;2003:PL2. doi: 10.1126/stke.2003.168.pl2. [DOI] [PubMed] [Google Scholar]
- 25.Zudaire E, Gambardella L, Kurcz C, Vermeren S. A computational tool for quantitative analysis of vascular networks. PLoS ONE. 2011;6:e27385. doi: 10.1371/journal.pone.0027385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cumming G, Fidler F, Vaux DL. Error bars in experimental biology. The Journal of Cell Biology. 2007;177:7–11. doi: 10.1083/jcb.200611141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gu C, et al. Neuropilin-1 conveys semaphorin and VEGF signaling during neural and cardiovascular development. Developmental Cell. 2003;5:45–57. doi: 10.1016/s1534-5807(03)00169-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maden CH, Ruhrberg C. The murine hindbrain as a model to study the molecular and cellular mechanisms of angiogenesis in intact tissues. In: Zudaire E, Cuttita F, editors. The Textbook of Angiogenesis and Lymphangiogenesis. 2012. Springer pp. 205–216. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.