Abstract
Duchenne muscular dystrophy (DMD) is a devastating disease that dramatically decreases the lifespan and abilities of affected young people. The primary molecular cause of the disease is the absence of functional dystrophin protein, which is critical to proper muscle function. Those with DMD vary in disease presentation and dystrophin mutation; the same causal mutation may be associated with drastically different levels of disease severity. Also contributing to this variation are the influences of additional modifying genes and/or changes in functional elements governing such modifiers. This genetic heterogeneity complicates the efficacy of treatment methods and to date medical interventions are limited to treating symptoms. Animal models of DMD have been instrumental in teasing out the intricacies of DMD disease and hold great promise for advancing knowledge of its variable presentation and treatment. This review addresses the utility of comparative genomics in elucidating the complex background behind phenotypic variation in a canine model of DMD, Golden Retriever muscular dystrophy (GRMD). This knowledge can be exploited in the development of improved, more personalized treatments for DMD patients, such as therapies that can be tailor-matched to the disease course and genomic background of individual patients.
Keywords: Genomics, Comparative genomics, Golden retriever muscular dystrophy, Duchenne muscular dystrophy, Animal models of DMD, Canine dystrophin
INTRODUCTION
Duchenne muscular dystrophy (DMD) is a lethal X-linked disease in humans characterized by the absence of dystrophin protein, which leads to progressive muscle weakness, respiratory insufficiency, and cardiomyopathy [1]. The disease results from mutations in the DMD gene and occurs in approximately 1 in 3,500 live male human births. DMD patients are often wheelchair bound by age 14 [2] and typically succumb to cardiomyopathies and/or breathing complications well before age 30. A similar condition, Becker muscular dystrophy (BMD), is also caused by mutation of the DMD gene; however, unlike DMD, the reading frame remains intact in BMD patients. This truncated but still-functional transcript results in a milder clinical phenotype, with ambulation preserved well past the teenage years. Currently, there is no cure for DMD, and available therapies are restricted in their utility. An urgent need exists for novel therapeutic measures that are tailored to the individual.
The DMD gene, at 2.2Mb in size, is the largest one identified to date in the human genome. It is also one of the most complex genes yet identified. DMD contains at least 8 promoters and 2 polyadenylation sites and is differentially spliced, producing several tissue-specific isoforms. The gene encodes dystrophin, a cytoskeletal protein, part of the dystrophin-glyoprotein complex located between the extracellular matrix and inner cytoskeleton of muscle fibers [3]. It stiffens muscle fibers, acting as a type of shock absorber by providing resistance against deformation [4]. A deficiency of dystrophin leaves the fibers susceptible to contraction-induced microfissures, which disrupt calcium homeostasis, ultimately resulting in cellular necrosis [5, 6].
The DMD gene is also present in the genomes of at least 48 non-human species (Ensembl release 71; [7]). One tenet of comparative genomics is that much can be learned about the human genome – and, by extension, human disease – via comparison with the genomes of other species. Mutations in DMD homologs of mice, dogs, and cats have been linked to analogous but variable diseases. As an example, the mdx mouse has a relatively mild phenotype, while dystrophin-deficient dogs have clinical disease more in keeping with that of DMD. Comparing genomic features across species enables the identification of common mechanisms contributing to their DMD-like phenotypes. Differences can reveal a separate evolutionary path and/or a novel function or relationship for some genomic element. Importantly, these differences may also hold the key to phenotypic differences between and within DMD animal models.
This review summarizes present knowledge about the genomic variations underlying the phenotypic variation seen in DMD and its animal models – particularly golden retriever muscular dystrophy (GRMD). We also discuss the utility of comparative genomics in identifying molecular targets for improved, personalized treatments.
OVERVIEW OF GENETIC VARIATION IN DMD
Genetic Variation in Dystrophin
The human DMD gene contains 79 exons, separated by introns that vary greatly in size from 107bp to over 248kb. The enormous size of some of the introns appears to be correlated with the high mutation rate in two regions of the gene: the major mutational hotspot located at exons 45-55 (intron 44-45 is the largest of the gene), and the minor hotspot located around exons 2-20 (introns 1-2 and 2-3 are the second and third largest, respectively) [8-11].
Mutations in DMD Gene
Mutations within the DMD gene are responsible for the loss of fully-functional dystrophin protein at the muscle plasma membrane [1]. In-frame mutations resulting in a premature stop codon and truncated protein product cause Becker muscular dystrophy, while insertion/deletion mutations resulting in a disrupted reading frame can cause premature truncation of protein synthesis – and the more-severe Duchenne muscular dystrophy phenotype [12]. Mutations in the DMD gene have been catalogued extensively in humans (e.g., [13-16]).
Databases developed in recent years serve as repositories of information about genetic variations identified within the DMD gene. The UMD-DMD France national database catalogs mutations of the DMD gene found in (primarily) French patients with dystrophinopathies [14]. This site currently lists 2,898 mutations, over 77% of which are duplications or deletions that affect ≥1 exon. UMD-DMD further classifies patient phenotype based on age of wheelchair dependency (as described in [17]) and includes symptomatic female carriers, asymptomatic affected males, and “pending” (patients with unknown phenotype). The Leiden Open Variation Database (LOVD; [15]) has segregated small mutations (<1 exon in size) from larger mutations involving whole-exon changes [16]. LOVD currently lists nearly 26,000 “small” mutations in the DMD gene, which frequently result in frame shifts, nonsense codons, or disruption of normal splicing mechanisms. The majority (78.5%) of these smaller changes are reported as “substitutions”, such as single-nucleotide polymorphisms. Over 9,000 whole-exon changes in the DMD gene were reported by LOVD as of January 8, 2013; 83.1% of these are deletions. Duplications account for nearly 12.2% of this category, and other mutations (insertions, insertion/deletions, inversions, and others) make up the remaining fraction of these very large changes.
The Leiden database describes most of the mutations identified in 1,111 patients included a 2009 study by Flanigan et al. [13]. Deletions accounted for an uncharacteristically low proportion of mutations described in this study (43%) but this low figure probably reflects selection bias, described by the authors. Exon duplications made up 11%, and the rest identified (46%) were point mutations. Exon 2 was identified as a duplication hotspot.
Magri et al. [18] identified several forms of genetic variation (deletions, duplications, nucleotide substitutions and other microrearrangements) in the DMD genes of a cohort of 320 patients (205 DMD and 115 BMD). Deletions and duplications accounted for 65.8% and 13.6% of these mutations, respectively, and localized within exons 1-60. Point mutations (20.6% of mutations identified) were found throughout the gene and were significantly correlated with lower intelligence quotient (IQ) levels, particularly those located distally to exon 45. Regardless of the type of mutation, patients bearing a mutation in the proximal part of the gene (defined here as exons 1-45) displayed earlier cardiac symptoms. This study found that the type of variation itself was not correlated with any particular clinical phenotype in DMD patients. This is not surprising, as by definition DMD diagnosis is predicated on a lack of dystrophin, no matter the precipitating genetic variation. Instead, phenotype was primarily influenced by the size and location of the mutation. Deletion of any of the first 20 exons (which encode actin-binding sites) and/or deletions affecting more than 25 exons had the most severe consequences.
A population-based survey in Canada performed over a ten-year period [19] sought to catalog mutations in the DMD genes of 529 DMD and 137 BMD patients. This study also identified a mutational hotspot around exons 45-55, as well as a lesser hotspot around exons 2-20. Deletions accounted for 64% of the mutations identified; 11% and 25% of individuals surveyed had duplications and point mutations, respectively.
While this is by no means meant to be an exhaustive listing of studies cataloging mutations in the DMD gene, commonalities in the data sets described above provide insight into the phenotypic variation seen in dystrophinopathies. First, deletions were the most common mutations identified in all but one study. In some cases, massive deletions were described, including multiple exons, though phenotype was not always proportionately affected. Next, these studies concurred regarding the existence and locations of two mutational hotspots in the gene, encompassing exons 2-20 and 45-55. It is not clear why these hotspots exist, or whether mutations in other parts of the gene result in a loss of viability that would preclude their identification. Lastly, these studies acknowledged the complexity of the DMD gene and the unclear connection between DMD mutation and pathogenesis of dystrophinopathies.
Exceptions to the Reading-Frame Rule
In the studies described above, exceptions to the reading-frame rule [12] were paradoxical but not uncommon. Ambulation is lost by 14 years of age for DMD boys, while BMD patients maintain the ability to walk beyond age 16 [2]. However, this is not always the case. These studies support the reading frame exception rate, originally postulated to include up to 10% of patients [15]. Flanigan et al. [13] found out-of-frame mutations in 79 patients with BMD or an intermediate phenotype (IMD), and in-frame mutations were identified in 37 DMD patients. This study did not report any genotype-phenotype correlations, suggesting instead that unknown influences such as changes in the sequence or function of regulatory elements may account for the phenotypic variation not sufficiently explained by the reading-frame rule. In the study done by Magri et al. [18], 11 patients classified as DMD based on phenotype and lack of dystrophin were found to bear in-frame mutations. These patients harbored large deletions of 4 to 45 exons, all located within exons 3-51. Finally, in the aforementioned survey of Canadian patients [19] 7 identical mutations (6 deletions and 1 duplication) were associated with both severely-affected DMD and mildly-affected BMD patients. These mutations included exceptions to the reading-frame rule, involving 13 DMD patients harboring in-frame deletions and 6 BMD patients with an out-of-frame deletion or duplication.
Reading-frame-rule exceptions in BMD patients may be attributed to alternate start codons or alternate splicing in the 5’ (proximal) end of the DMD gene that “rescue” the dystrophin transcript [2, 20-22]. This region includes intron 7, which is particularly vulnerable to deletions and insertions of various mobile elements (such as LINEs and LTR sequences) [23, 24].
In conclusion, while the reading-frame rule is a very good indicator for disease severity in terms of progression to wheelchair, in reality a spectrum of disease severity exists which is not necessarily attributed to mutations in the DMD gene alone.
Symptomatic Female Carriers
A dystrophic phenotype is present in up to 22% of female carriers [25, 26]. Clinical presentation ranges in severity from a mild, BMD-like appearance to DMD-like disease, and may include symptoms such as muscle weakness and cardiomyopathy [26-29]. One simple explanation for these occurrences is the presence of only one X chromosome in manifesting carriers, as seen in Turners Syndrome patients [30-32] and in at least one XY male pseudohermaphrodite with female secondary sex characteristics [33]. Uniparental disomy of the X chromosome harboring the defective DMD locus has also been found in symptomatic female carriers [34]. Another possible cause is X:autosome translocations that disrupt the DMD gene [35]. These may furthermore be associated with non-random X-chromosome inactivation (XCI). Skewed XCI is a relatively common finding in manifesting carriers [29, 36]. The amount of skewing is not directly correlated with phenotypic severity [36, 37], though XCI patterns can differ between tissues [38]. Indeed, asymmetric muscle weakness in symptomatic females may be attributed to XCI pattern variation between muscles. This asymmetry has also been associated with somatic and germline mosaicism [39, 40].
Non-DMD Genetic Influences
Even though the DMD gene has been identified as the mutated locus causal for Duchenne muscular dystrophy [1], multiple other loci are suspected to be involved in the phenotypic variability in DMD patients, perhaps via epistatic interactions [41]. Indeed, as the list of mutations found in the DMD genes of affected patients grows longer, so does the list of non-DMD loci with apparent involvement in dystrophinopathic phenotypes. An excellent example is that of osteopontin. The osteopontin gene has been identified as a disease modifier [42]; the genotype of a lone single nucleotide polymorphism (SNP) within the gene has been associated with significant differences in DMD phenotype [43, 44].
Gene expression profiling in DMD patients has helped to elucidate the complex network of molecular pathways involved in DMD pathogenesis. One such study of skeletal muscle (quadriceps) from DMD patients identified 105 genes that are differentially regulated relative to controls; many are up-regulated and play a role in muscle regeneration and structure [45]. Another study showed that even before clinical symptoms of the disease are visible, the muscles of children with DMD exhibit a gene expression profile that is distinctly different from those of healthy children the same age [46]. The advent of microarray technology has facilitated the comparison of gene expression states in whatever tissue, age group, or phenotypic status is pertinent to the question being addressed. These data can support and even augment the findings of the traditional histological methods used for identifying significant dissimilarities between groups.
The development of novel therapeutics for DMD depends on studying all aspects of the molecular background of the disease. While mutation of the DMD gene itself is the primary genetic lesion, the variation observed in phenotype and gene mutation limits the possibility of a single drug abrogating the disease for all patients. Modifier genes and related molecular pathways offer innovative options for drug development. Regulatory elements responsible for the up- or down-regulation of modifier gene expression are additional candidates for consideration [47]. For ameliorating specific facets of the disease, drug targets outside of dystrophin itself must be considered.
CURRENT TREATMENT METHODS
At the present time, the standard protocol for treating DMD involves steroids (e.g. prednisone and deflazacort) to reduce inflammation, slow the disease process, and prolong ambulation. Unfortunately long-term steroid treatment has its own negative side effects, such as weight gain, immunosuppression, and increased risk of bone fractures [48].
Other treatment methods also seek to delay disease progression and prolong functionality – for example, pharmacologic methods [49, 50], therapies utilizing “substitute” proteins to compensate for the lack of dystrophin [51-56], and cell-based therapies in which stem cells are used to stimulate muscle regeneration and replacement [57-64]. The treatment most studied today involves replacing the missing normal dystrophin protein, either by repairing the defective gene or introducing exogenous dystrophin, e.g. exon skipping [65-69], stop codon read-through [13, 15, 70, 71], and other methods that introduce the DMD gene directly [72-76].
Many promising treatment methods have been developed with the use of animal models, which provide invaluable analogs of the disease without risking the welfare of DMD boys.
ANIMAL MODELS
Animal models of DMD provide insight into phenotypic and genotypic variation in the disease and the molecular basis for such diversity, suggesting therapeutic targets for the development of personalized treatments. Several animal models of the disease exist, including the dystrophin-deficient mdx mouse [77, 78], and feline [79] and canine X-linked models of muscular dystrophy [80-90]. Dystrophin deficiency affecting cardiac and skeletal muscle has also been described in swine [91] but little information has been published regarding the causal mutation and the associated phenotype.
Mouse
The mdx mouse model of DMD is caused by a naturally-occurring point mutation that causes a frame shift, resulting in a premature stop codon in the dystrophin gene [77, 92]. The mdx mouse shows a relatively mild phenotype and minimal shortening of the lifespan [93, 94].
Studies using the mdx model have uncovered a substantial amount of the current body of knowledge regarding DMD. This model has had a significant role in elucidating the effects of non-DMD loci on disease pathogenesis and/or modification. For example, gene expression profiles of mdx mouse hindlimb muscle have revealed important contributors to disease pathogenesis, such as pathways involved in inflammation and muscle maintenance [95, 96]. The mdx model has also been invaluable for exploring potential new therapeutic measures. Many of the pharmacologic, cell-based, and gene repair treatment methods for DMD described above were originally developed and/or tested using mdx mice.
Despite the clear utility of this model, the pathology of the mdx model is somewhat different from human DMD, in part due to the increased telomere length and greater muscle stem cell reserve in mice compared with humans or dogs [97, 98]. Muscle degeneration is relatively milder in the mouse than in humans [93, 99]. Muscle necrosis and regeneration occur in phases in mdx mice as in DMD [100-102], though regenerative capacity is ultimately overwhelmed by muscle degeneration in DMD [94]. Results from murine models do not always translate reliably to humans [103] so results obtained from the mdx mouse should be interpreted cautiously. In the case of DMD, large animal models – particularly the dog – offer an intermediate species in which to further elucidate therapeutic efficacy.
Cat
A feline model of DMD has been described [79, 104] but is not as well studied as mouse and dog models. The causative mutation in the cat is a deletion of promoters specific to the muscle and Purkinje cell isoforms of dystrophin [105]. This model is sometimes called hypertrophic feline muscular dystrophy (HFMD) because of the marked muscle hypertrophy observed in these cats. The relevancy of this model toward understanding disease pathogenesis and developing novel treatments for DMD is not yet clear: the model shares less similarity with human DMD than dog models and has not been utilized in treatment development.
Dog
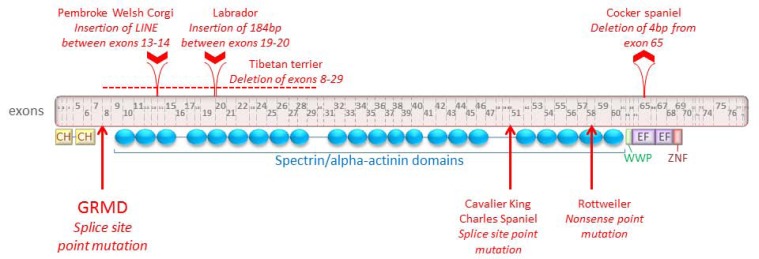
Several canine models of DMD have been identified, all of which carry mutations that result in a lack of functional dystrophin (Fig. 1). In the German shorthaired pointer, cocker spaniel and Tibetan terrier, the causative mutation is a deletion. The entire dystrophin gene is deleted in the pointer [85], though there appears to be some variation in phenotype even between littermates [106]. In the cocker spaniel, there is a 4 nucleotide deletion in exon 65 and in the Tibetan terrier, exons 8-29 are deleted [84]. The culpable mutations in the Pembroke Welsh corgi and Labrador retriever models are both insertions: the insertion of a long interspersed nuclear element (LINE-1) in intron 13 for the corgi [88], and of 184 nucleotides (a “pseudoexon”) in intron 19 for the Labrador [84, 87]. Point mutations account for the dystrophic phenotype in three additional dog breeds: the golden retriever (splice site mutation which causes exon 7 to be skipped [86]), Rottweiler (nonsense mutation in exon 58 [90]), and Cavalier King Charles spaniel (splice site mutation which causes exon 50 to be skipped [89]).
Fig. (1).
The canine dystrophin protein (Ensembl protein ID ENSCAFP00000031637), along with mutation information for seven dog breeds known to exhibit DMD-linked muscular dystrophy. “CH” indicates calponin homology domains, which are actin-binding domains. “WWP” indicates the WW domain, which binds proline-rich polypeptides and is the primary interaction site for dystrophin and dystroglycan. “EF” indicates members of the EF-hand family; this domain stabilizes the dystrophin-dystroglycan complex. “ZNF” represents a putative zincbinding domain, ZnF_ZZ, which is present in dystrophin-like proteins and may bind to calmodulin. All 79 exons are represented. Exons and protein domains are approximately shown to scale. Insertion and deletion mutations are shown above the exons. Point mutations are indicated by arrows at the bottom of the figure.
Compared to mice, the canine models of DMD share a greater degree of similarity with the human disease, and clinical features of muscular dystrophy are more severe in dogs than in mice. This makes the dog arguably more relevant for studying the clinical and molecular aspects of DMD. While mice have several obvious advantages for treatment development and testing – shorter generation time, larger litters, easier handling and husbandry, etc. – preclinical studies in dog may better predict the potential success of a treatment in humans. In particular, organ size and the immune response in dogs are more similar to those of humans [107].
GOLDEN RETRIEVER MUSCULAR DYSTROPHY (GRMD)
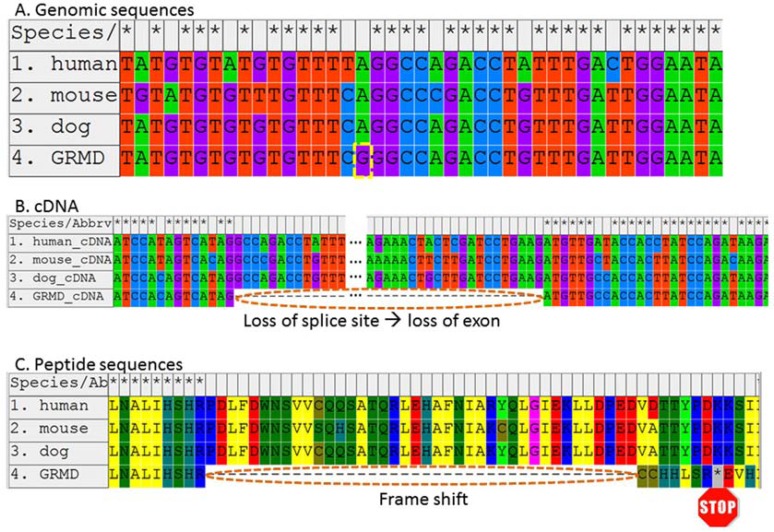
The best-studied canine ortholog of DMD is Golden Retriever muscular dystrophy (GMRD) [83, 108]. Our group described the causative mutation for GMRD as a splice site mutation resulting in the elimination of exon 7 and creation of a premature stop codon in exon 8 of the dystrophin mRNA (Fig. 2; [86]). Since that discovery, all affected descendants of the single original founder/proband of GRMD are presumed to possess the same mutation [109].
Fig. (2).
The causative mutation and flanking sequence for GRMD, compared with normal DMD sequences of humans, mice, and dogs. GRMD is caused by a point mutation (A; the site of the mutation is shown with a dashed yellow border). This mutation is located within a splice site and causes improper splicing and a frame shift, as shown within the contexts of the cDNA (B) and resulting peptide (C) sequences. This, in turn, creates a premature stop codon (indicated here by an asterisk with a “STOP” sign beneath). This figure is an updated version of those found in the original paper describing the GRMD causal mutation [86].
GRMD bears a striking similarity to human DMD, making it a strong model for studies directed towards development of treatments for the human disease. Dogs with GRMD are afflicted with a progressive, fatal disease with skeletal and cardiac muscle phenotypes and selective muscle involvement [110].
Phenotypic variability is frequently observed in GRMD, as in humans. Our group has observed differences in phenotypic severity for various biological markers of the disease, such as those described in Table 1 (e.g., [111-114]) with some being analogous to DMD phenotypes in humans. Severity/age-of-onset of a particular metric is often predictive of the severity of other traits associated with DMD [115]; disease severity tends to show variable progression in GRMD, as well. Variable cognitive dysfunction, cardiac involvement, and respiratory complications are observed among DMD patients (e.g., [115, 116]); similar variation in cardiac and skeletal muscle phenotypes is found in GRMD [84]. Rare cases have been documented wherein a child completely lacking the dystrophin protein – thus fitting the molecular diagnostic criteria of DMD – displays a phenotype so mild that clinical diagnosis is ambiguous (for example, see [117]). Likewise, all GRMD dogs feature a total absence of dystrophin except for rare revertant fibers, but do not share a similar disease course. Some severely-affected pups survive only a few days, while other dogs that survive for years with mild clinical involvement have also been documented [118, 119].
Table 1.
Objective Biomarkers to Evaluate Disease Progression and to Establish Phenotype-Genotype Associations in GRMD Dogs
| Age 6 Months | Age 12 Months | |||
|---|---|---|---|---|
| Affected | Normal | Affected | Normal | |
| range ave ± std dev |
range ave ± std dev |
range ave ± std dev |
range ave ± std dev |
|
| Tetanic flexion (Newton/kg) |
0.438-0.794 0.595 ± 0.141 |
1.23-1.48 1.35 ± 0.107 |
0.402-1.08 0.781 ± 0.347 |
1.26-1.65 1.42 ± 0.167 |
| Tetanic extension (Newton/kg) |
0.366-2.07 1.15 ± 0.649 |
2.00-3.20 2.63 ± 0.508 |
1.34-2.54 2.00 ± 0.610 |
1.98-2.65 2.37 ± 0.315 |
| Tibiotarsal joint angle (degrees) |
140-160 151 ± 9.00 |
158-162 161 ± 1.91 |
130-155 145 ± 13.1 |
149-158 154 ± 3.74 |
| % Eccentric contraction decrement (@ 10 stims). |
8.10-40.7 23.8 ± 13.6 |
8.22-11.9 9.91 ± 1.61 |
29.5-59.0 44.1 ± 14.8 |
3.47-12.3 7.53 ± 3.83 |
| % Eccentric contraction decrement (@ 30 stims). |
29.7-74.8 50.4 ± 18.3 |
17.2-24.7 20.1 ± 3.24 |
64.1-72.6 67.7 ± 4.37 |
7.59-20.1 13.2 ± 5.21 |
| Maximum hip flexion angle |
45-105 67.4 ± 23.0 |
56-80 63.0 ± 11.5 |
42-100 76.0 ± 30.3 |
52-70 58.5 ± 7.90 |
| Pelvic angle |
36-57 47.8 ± 7.56 |
25-40 35.0 ± 6.88 |
44-54 49.3 ± 5.03 |
37-50 42.5 ± 5.45 |
| Cranial sartorius circumference (mm/kg) |
3.00-5.10 4.30 ± 0.782 |
2.16-2.74 2.38 ± 0.251 |
4.62-4.62 4.62 ± n/a |
2.17-2.34 2.26 ± 0.120 |
| Quadriceps femoris weight (g) |
79.5-110 96.2 ± 14.7 |
172-223 192 ± 22.9 |
98.3-120.2 110 ± 11.0 |
203-258 227 ± 22.6 |
| Quadriceps femoris weight (g/kg body weight) |
6.65-8.27 7.31 ± 0.786 |
9.20-12.7 10.8 ± 1.51 |
8.19-8.86 8.59 ± 0.354 |
8.99-10.8 9.97 ± 0.744 |
Data were collected from 8 each GRMD and normal dogs. Functional measurements were made at both 6 and 12 months in half of the dogs, and at only 6 months in the others. Half of the dogs (4 of each group) were euthanized at 6 months; the others were euthanized at 12 months.
GRMD shows a remarkable likeness to DMD and, as such a strong analog, provides important comparative data for DMD research. Currently this model has been used primarily for therapeutic discovery and testing. Regarding genetic variation in GRMD, studies have largely focused on the DMD gene (e.g., [120]). Most recently, a gene expression microarray has been used to interrogate expression profiles for variably involved muscles of age-matched GRMD dogs (P. Nghiem, submitted for publication; [84]). Association studies seeking to connect genomic variation with phenotypic variation in GRMD are also currently underway (C. Brinkmeyer-Langford, in preparation).
COMPARATIVE GENOMICS OF GRMD
Many inherited canine diseases bear remarkable similarities to human diseases [121-124], and it is easier to identify the genetic contributions in the dog. Artificial selection has been used in many dog breeds for generations (for example, using a prize-winning sire to breed multiple dams, resulting in a large number of half-sibling offspring). The consequences of this include relatively small effective population sizes and long (>1Mb) runs of homozygosity [125, 126]. This has resulted in extensive linkage disequilibrium (LD) within many dog breeds, often extending to 1Mb or greater [125, 127, 128]. Long-range LD like this facilitates the identification of regions that may be associated with some trait or disease of interest, as fewer markers (typically SNPs) are needed. In addition, the multi-generational pedigree information available for many dog breeds provides a resource that is not easily accessible for most human studies [122, 128, 129]. Pedigrees facilitate linkage studies that can connect hereditary conditions, such as diseases, with their causative gene(s). Consequently, genetic effects are more easily distinguished and susceptibility genes are more readily identified. Finally, dogs and humans share a gene repertoire that is very much alike, making the dog a powerful model for investigating disease pathogenesis in human conditions, as well [125, 130].
Genome-wide association studies (GWAS) in dogs are very useful for correlating genetic variants with phenotypes of interest. The GRMD model presents a powerful GWAS opportunity for several reasons. All GRMD colonies in the world today are descended from a single founding sire [86, 112]. The artificial breeding strategy used in these colonies has resulted in substantial inbreeding [112]. Also, even with the occasional outcrossing necessary to reduce litter mortality rates [112], it is highly likely that genetic drift has resulted from the isolation of GRMD populations. The high degree of relatedness between GRMD dogs within the same colony eliminates a lot of extraneous data “clutter” and adds substantial power to association studies by streamlining the identification of genomic regions correlated with phenotypic variation, such as biomarker values in GRMD.
Phenotypic variation in GMRD, observed even between littermates, likely reflects the influence of factors in addition to the causal splice site mutation in the DMD gene. Such factors may include additional DMD gene mutations, copy number variations (CNVs), epigenetic changes, or modifying genes that influence the clinical course of the disease.
Comparison of DNA and Protein Sequences
Although the full-length DMD gene and protein sequences of GRMD dogs have not been completely characterized, we can compare these sequences from the published genomes of human and animal models of DMD, including the dog. This comparative information can help demonstrate the relevancy of an animal model by illustrating its similarity to the human at the DNA/transcript level.
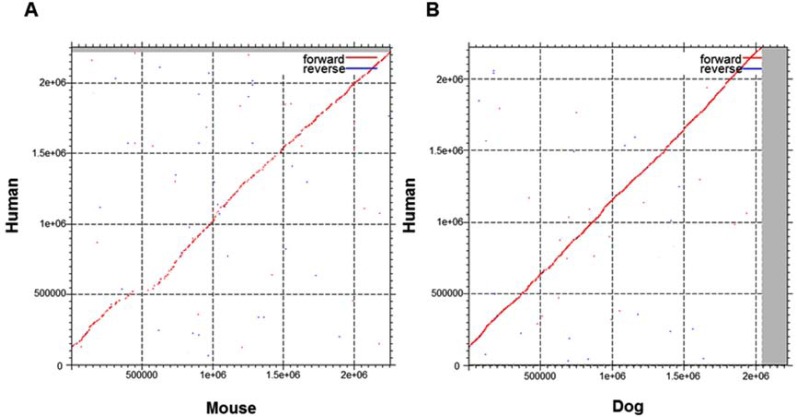
The DMD gene is well-conserved and present in at least 48 species (Ensembl version 71; [7]). The human nucleotide sequence is approximately 2.22Mb in length (GRCh37.p10); the homologous sequence in dog is 2.04Mb long (CanFam3.1). Approximate gene lengths in cats (Felis_catus-6.2) and mice (GRCm38.p1) are 0.148Mb and 2.26Mb, respectively. Aside from differences in gene lengths, species-specific variation exists in the DMD gene sequences of humans, dogs and mice, as observed by aligning mouse and dog sequences with the human sequence (Fig. 3). Humans, dogs, and mice have 79 exons each within their DMD genes; this is not the case for cats and many other species. This may be attributed to the incomplete status of the genome assemblies for this region.
Fig. (3).
Dot plots showing sequence conservation between human, mouse and dog DMD genomic sequences. Plots were created using MAFFT multiple sequence alignment online software [155, 156]. The human sequence used was 2220382bp in length (sequence ID: gi224589822); lengths of the mouse and dog sequences used were 2256181bp (gi372099090) and 2042811bp (gi357579592), respectively.
The dystrophin protein itself also varies in size and constitution. Currently 18 dystrophin isoforms have been identified in the human; 4 have been found in the cat and only 1 each for dogs and mice. Additional isoforms, not yet identified, may also exist in these species. The sequences of the longest dystrophin isoforms of humans, dogs, and mice share ~92% sequence similarity (Supplementary Fig. 1 (583.7KB, pdf) ). The available dystrophin protein sequences for the cat are considerably shorter and therefore not included in this figure.
It is interesting to note that regions with the most diversity in the protein sequence correspond to mutational hotspots in the human DMD gene. The longest dystrophin isoform in humans, Dp427m, contains a string of 7 amino acids (EIYNQPN) that is unique to humans and located within repeat region 19 of the central rod domain, just upstream from hinge region 3. The surrounding sequence lies within spectrin repeat 19 of dogs and mice and is less conserved compared to the rest of the protein sequence. This divergent region is encoded within exons 48-51 of the human dystrophin transcript, encompassing ~10kb inside the major mutational “hotspot” of this gene in humans. The lack of conservation here supports that this may indeed be a hotspot for variation: not only within humans, but also between humans and other mammalian models of DMD. Functionally, it is less clear what sort of effect these 7 amino acids have on differences between human and dog/mouse/cat DMD models – though their location in humans, coupled with the frequency of deleterious mutations in this region, strongly suggests they may play a critical role in the normal function of dystrophin, such as that of a “shock absorber” or force transducer. In addition, a second poorly-conserved segment corresponds to part of exon 8 in humans, which is located within the minor mutational hotspot in humans. Similar hotspots are suspected in animal models of DMD [89].
Copy Number Variation (CNV)
Copy number variation (CNV) has garnered a great deal of interest in recent years, and has been linked to a number of complex diseases and phenotypes [131-133]. Any segment of DNA, regardless of size, which exists in a copy number different from some reference genome, can be called a CNV [134, 135]. CNVs feature a mutation rate considerably higher – 1000 to 10000 times more frequent – than that of single nucleotide changes [136]. Because of this, disease susceptibility may be more strongly influenced by CNVs than SNPs, which are older [137].
Many DMD-causing mutations in the DMD gene, such as exonic deletions, can be defined as CNVs. Genomic instability of the so-called “mutational hotspots” within the human DMD gene may render these regions more susceptible to mutation/CNV. CNVs have been identified in the dog genome [138, 139], but these studies examined the genome from a broad perspective which did not account for smaller variations (1kb or smaller in size). To date, the canine DMD gene itself has not been subject to an in-depth search for CNVs. This will be a priority for future studies seeking to better define the causal mutations behind canine models of DMD. Investigating CNVs will also provide an improved understanding of the evolutionary dynamics behind the inception of these mutations. This knowledge can help us determine why and how different dog breeds have developed breed-specific versions of DMD-like disease.
Epigenetics
In muscular dystrophies, including DMD, the decision for injured muscles to regenerate or degenerate is directed by epigenetic cues. Fibroadipocyte progenitor cells promote regeneration in normal muscle that has been injured [140]. Dystrophic muscle, however, may ultimately signal these progenitors to become fibroadipocytes that promote fibrosis and fat deposition rather than muscle regeneration [141, 142]. These signals, which determine the fate of the fibroadipocytes, are driven by epigenetic marks – specifically, chromatin modifications such as histone acetylation – that regulate gene expression [143, 144]. Even in genetically-identical cells, gene expression may be regulated differently via epigenetic mechanisms, including methylation of DNA and acetylation of histone proteins [145-147].
Pharmacologic agents targeting epigenetic regulation, such as histone deacetylase inhibitors (HDACi), currently show great potential for the treatment of DMD and other diseases [148, 149]. HDACi have been tested in the mdx model with promising results [150], though to date these have not been used with any of the dog models. However, HDACi have been tested in the treatment of other conditions shared by both humans and dogs (for example, osteosarcoma [151] and hemangiosarcoma [152], and renal transplant rejection [153]). Because the pathogenesis of the GRMD model is highly similar to that of DMD, HDACi represent a potential (though currently unexplored) treatment avenue in GRMD dogs.
Modifier Genes
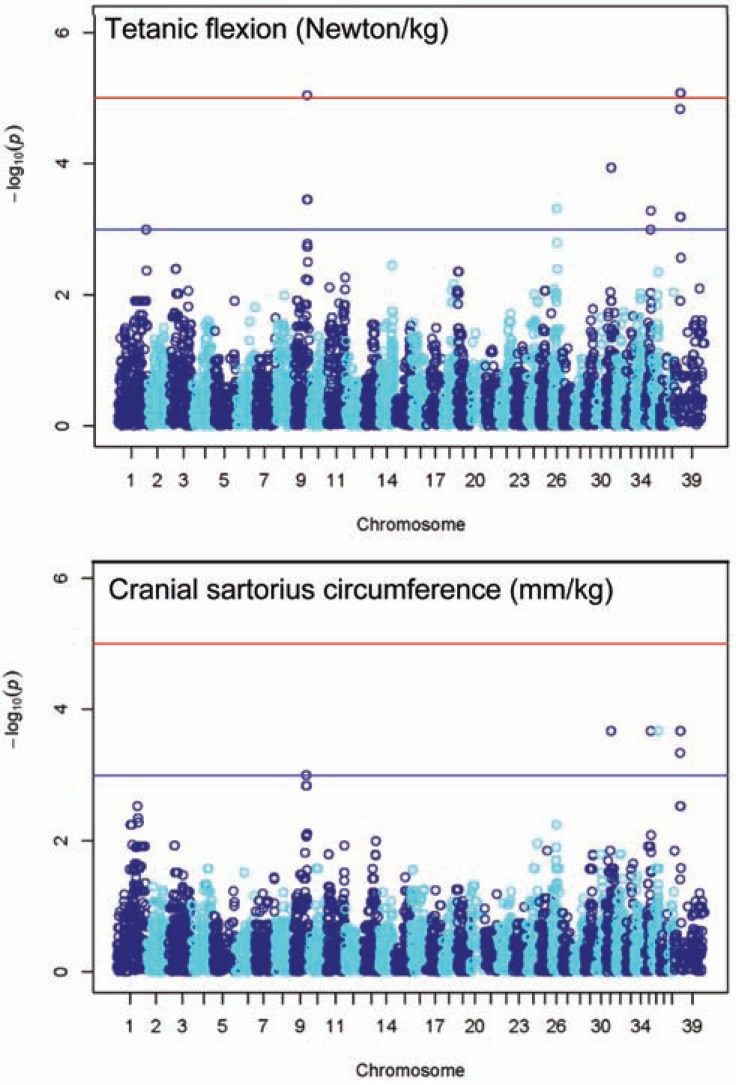
The phenotypic variation seen in dogs may be attributable to non-DMD modifier genes such as those identified in humans and mice. In some cases, outbreeding may have also contributed to the phenotypic heterogeneity [154]. We have performed a GWAS in our lab using 8 GRMD-affected and 8 age-matched unaffected dogs (including sibling pairs when available). This study has revealed at least 3 chromosomal regions, in addition to the DMD gene itself, that harbor statistically-significant associations with specific quantitative biomarkers (C. Brinkmeyer-Langford in preparation; Table 1; see two examples in Fig. 4). Candidate genes in these regions have been subjected to quantitative PCR using RNA isolated from muscle samples taken from these 16 dogs. Many of these genes encode proteins that work in association with dystrophin and/or are connected to muscle regeneration, and several others are affiliated with cardiomyopathy such as that observed in conjunction with DMD and GRMD. Variations within these candidate genes, such as differences in expression level and/or sequence variation, may explain some of the phenotypic heterogeneity observed in our highly inbred colony.
Fig. (4).
Manhattan plots for 2 GRMD biomarkers, showing –log10 P values for SNP associations. Note that Chromosome 39 is, in fact, the X chromosome. The blue (lower) line represents significance threshold -log10(1e-3); red (upper) line represents -log10(1e-5). Figure from C. Brinkmeyer-Langford, in preparation.
CONCLUSIONS
Humans and dogs have been companions for thousands of years. Today, dogs are providing valuable information toward understanding and treating human diseases, making the phrase “man’s best friend” more appropriate than ever. The GRMD model of DMD shares a superior level of pathologic similarity to the human disease, making it a powerful model for the development of novel therapeutics. This similarity goes even deeper than the macroscopic level: the sequences and structures of the DMD genes and proteins of humans and dogs bear a strong likeness to each other. In this era of One Health medicine and personalized genomics, the future holds great promise for the GRMD model and the human application of the data it reveals.
ACKNOWLEDGEMENTS
Declared none.
CONFLICT OF INTEREST
The author(s) confirm that this article content has no conflicts of interest.
SUPPLEMENTARY MATERIAL
Supplementary material is available on the publisher’s web site along with the published article.
REFERENCES
- 1.Hoffman E, Brown RJ, Kunkel L. Dystrophin: the protein product of the Duchenne muscular dystrophy locus. Cell. 1987;51:919–928. doi: 10.1016/0092-8674(87)90579-4. [DOI] [PubMed] [Google Scholar]
- 2.Malhotra SB, Hart KA, Klamut HJ, Thomas NS, Bodrug SE, Burghes AH, Bobrow M, Harper PS, Thompson MW, Ray PN, Worton RG. Frame-shift deletions in patients with Duchenne and Becker muscular dystrophy. Science. 1988;242:755–759. doi: 10.1126/science.3055295. [DOI] [PubMed] [Google Scholar]
- 3.Ervasti JM, Campbell KP. A role for the dystrophin-glycoprotein complex as a transmembrane linker between laminin and actin. J Cell Biol. 1993;122:809–823. doi: 10.1083/jcb.122.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pasternak C, Wong S, Elson EL. Mechanical function of dystrophin in muscle cells. J Cell Biol. 1995;128:355–361. doi: 10.1083/jcb.128.3.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deconinck N, Dan B. Pathophysiology of duchenne muscular dystrophy current hypotheses. Pediatr Neurol. 2007;36:1–7. doi: 10.1016/j.pediatrneurol.2006.09.016. [DOI] [PubMed] [Google Scholar]
- 6.Turner PR, Westwood T, Regen CM, Steinhardt RA. Increased protein degradation results from elevated free calcium levels found in muscle from mdx mice. Nature. 1988;335:735–738. doi: 10.1038/335735a0. [DOI] [PubMed] [Google Scholar]
- 7.Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, Fitzgerald S, Gil L, Garcia-Giron C, Gordon L, Hourlier T, Hunt S, Juettemann T, Kahari AK, Keenan S, Komorowska M, Kulesha E, Longden I, Maurel T, McLaren WM, Muffato M, Nag R, Overduin B, Pignatelli M, Pritchard B, Pritchard E, Riat HS, Ritchie GR, Ruffier M, Schuster M, Sheppard D, Sobral D, Taylor K, Thormann A, Trevanion S, White S, Wilder SP, Aken BL, Birney E, Cunningham F, Dunham I, Har-row J, Herrero J, Hubbard TJ, Johnson N, Kinsella R, Parker A, Spudich G, Yates A, Zadissa A, Searle SM. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.denDunnen JT, Bakker E, Breteler EG, Pearson PL, vanOmmen GJ. Direct detection of more than 50% of the Duchenne muscular dystrophy mutations by field inversion gels. Nature. 1987;329:640–642. doi: 10.1038/329640a0. [DOI] [PubMed] [Google Scholar]
- 9.DenDunnen JT, Grootscholten PM, Bakker E, Blonden LA, Ginjaar HB, Wapenaar MC, vanPaassen HM, vanBroeckhoven C, Pearson PL, vanOmmen GJ. Topography of the Duchenne muscular dystrophy (DMD) gene.FIGE and cDNA analysis of 194 cases reveals 115 deletions and 13 duplications. Am J Hum Genet. 1989;45:835–847. [PMC free article] [PubMed] [Google Scholar]
- 10.Forrest SM, Cross GS, Speer A, Gardner-Medwin D, Burn J, Davies KE. Preferential deletion of exons in Duchenne and Becker muscular dystrophies. Nature. 1987;329:638–640. doi: 10.1038/329638a0. [DOI] [PubMed] [Google Scholar]
- 11.Koenig M, Hoffman EP, Bertelson CJ, Monaco AP, Feener C, Kunkel LM. Complete cloning of the Duchenne muscular dystrophy (DMD) cDNA and preliminary genomic organization of the DMD gene in normal and affected individuals. Cell. 1987;50:509–517. doi: 10.1016/0092-8674(87)90504-6. [DOI] [PubMed] [Google Scholar]
- 12.Monaco AP, Bertelson CJ, Liechti-Gallati S, Moser H, Kunkel LM. An explanation for the phenotypic differences between patients bearing partial deletions of the DMD locus. Genomics. 1988;2:90–95. doi: 10.1016/0888-7543(88)90113-9. [DOI] [PubMed] [Google Scholar]
- 13.Flanigan KM, Dunn DM, vonNiederhausern A, Soltanzadeh P, Gappmaier E, Howard MT, Sampson JB, Mendell JR, Wall C, King WM, Pestronk A, Florence JM, Connolly AM, Mathews KD, Stephan CM, Laubenthal KS, Wong BL, Morehart PJ, Meyer A, Finkel RS, Bonnemann CG, Medne L, Day JW, Dalton JC, Margolis MK, Hinton VJ, Weiss RB. Mutational spectrum of DMD mutations in dystrophinopathy patients application of modern diagnostic techniques to a large cohort. Hum Mutat. 2009;30:1657–1666. doi: 10.1002/humu.21114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tuffery-Giraud S, Beroud C, Leturcq F, Yaou RB, Hamroun D, Michel-Calemard L, Moizard MP, Bernard R, Cossee M, Boisseau P, Blayau M, Creveaux I, Guiochon-Mantel A, deMartinville B, Philippe C, Monnier N, Bieth E, Khau Van, Kien P, Desmet FO, Humbertclaude V, Kaplan JC, Chelly J, Claustres M. Genotype-phenotype analysis in 2,405 patients with a dystrophinopathy using the UMD-DMD database a model of nationwide knowledgebase. Hum Mutat. 2009;30:934–945. doi: 10.1002/humu.20976. [DOI] [PubMed] [Google Scholar]
- 15.Aartsma-Rus A, VanDeutekom JC, Fokkema IF, VanOmmen GJ, DenDunnen JT. Entries in the Leiden Duchenne muscular dystrophy mutation database an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve. 2006;34:135–144. doi: 10.1002/mus.20586. [DOI] [PubMed] [Google Scholar]
- 16.White SJ, denDunnen JT. Copy number variation in the genome, the human DMD gene as an example. Cytogenet Genome Res. 2006;115:240–246. doi: 10.1159/000095920. [DOI] [PubMed] [Google Scholar]
- 17.Emery AEH. Diagnostic criteria for neuromuscular disorders.European Neuromuscular Center Baarn. The Netherlands. 1994 [Google Scholar]
- 18.Magri F, Govoni A, D'Angelo MG, DelBo R, Ghezzi S, Sandra G, Turconi AC, Sciacco M, Ciscato P, Bordoni A, Tedeschi S, Fortunato F, Lucchini V, Bonato S, Lamperti C, Coviello D, Torrente Y, Corti S, Moggio M, Bresolin N, Comi GP. Genotype and phenotype charac-terization in a large dystrophinopathic cohort with extended follow-up. J Neurol. 2011;258:1610–1623. doi: 10.1007/s00415-011-5979-z. [DOI] [PubMed] [Google Scholar]
- 19.Mah JK, Selby K, Campbell C, Nadeau A, Tarnopolsky M, McCormick A, Dooley JM, Kolski H, Skalsky AJ, Smith RG, Buckley D, Ray PN, Yoon G. A population-based study of dystrophin mutations in Canada. Can J Neurol Sci. 2011;38:465–474. doi: 10.1017/s0317167100011896. [DOI] [PubMed] [Google Scholar]
- 20.Gualandi F, Rimessi P, Trabanelli C, Spitali P, Neri M, Patarnello T, Angelini C, Yau SC, Abbs S, Muntoni F, Calzolari E, Ferlini A. Intronic breakpoint definition and transcription analysis in DMD/BMD patients with deletion/duplication at the 5' mutation hot spot of the dystro-phin gene. Gene. 2006;370:26–33. doi: 10.1016/j.gene.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 21.Kesari A, Pirra LN, Bremadesam L, McIntyre O, Gordon E, Dubrovsky AL, Viswanathan V, Hoffman EP. Integrated DNA cDNA and protein studies in Becker muscular dystrophy show high exception to the reading frame rule. Hum Mutat. 2008;29:728–737. doi: 10.1002/humu.20722. [DOI] [PubMed] [Google Scholar]
- 22.Gurvich OL, Maiti B, Weiss RB, Aggarwal G, Howard MT, Flanigan KM. DMD exon 1 truncating point mutations amelioration of phenotype by alternative translation initiation in exon 6. Hum Mutat. 2009;30:633–640. doi: 10.1002/humu.20913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McNaughton JC, Hughes G, Jones WA, Stockwell PA, Klamut HJ, Petersen GB. The evolution of an intron analysis of a long, deletion-prone intron in the human dystrophin gene. Genomics. 1997;40:294–304. doi: 10.1006/geno.1996.4543. [DOI] [PubMed] [Google Scholar]
- 24.McNaughton JC, Broom JE, Hill DF, Jones WA, Marshall CJ, Renwick NM, Stockwell PA, Petersen GB. A cluster of transposon-like repetitive sequences in intron 7 of the human dystrophin gene. J Mol Biol. 1993;232:314–321. doi: 10.1006/jmbi.1993.1389. [DOI] [PubMed] [Google Scholar]
- 25.Moser H, Emery AE. The manifesting carrier in Duchenne muscular dystrophy. Clin Genet. 1974;5:271–284. doi: 10.1111/j.1399-0004.1974.tb01694.x. [DOI] [PubMed] [Google Scholar]
- 26.Hoogerwaard EM, Bakker E, Ippel PF, Oosterwijk JC, Majoor-Krakauer DF, Leschot NJ, VanEssen AJ, Brunner HG, vanderWouw PA, Wilde AA, deVisser M. Signs and symptoms of Duchenne muscular dystrophy and Becker muscular dystrophy among carriers in The Netherlands: a cohort study. Lancet. 1999;353:2116–2119. doi: 10.1016/s0140-6736(98)10028-4. [DOI] [PubMed] [Google Scholar]
- 27.Soltanzadeh P, Friez MJ, Dunn D, vonNiederhausern A, Gurvich OL, Swoboda KJ, Sampson JB, Pestronk A, Connolly AM, Florence JM, Finkel RS, Bonnemann CG, Medne L, Mendell JR, Mathews KD, Wong BL, Sussman MD, Zonana J, Kovak K, Gospe SMJr, Gappmaier E, Taylor LE, Howard MT, Weiss RB, Flanigan KM. Clinical and genetic characterization of manifesting carriers of DMD mutations. Neuromuscular disorders. NMD. 2010;20:499–504. doi: 10.1016/j.nmd.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Norman A, Harper P. A survey of manifesting carriers of Duchenne and Becker muscular dystrophy in Wales. Clin Genet. 1989;36:31–37. doi: 10.1111/j.1399-0004.1989.tb03363.x. [DOI] [PubMed] [Google Scholar]
- 29.Bushby KM, Goodship JA, Nicholson LV, Johnson MA, Haggerty ID, Gardner-Medwin D. Variability in clinical genetic and protein abnormalities in manifesting carriers of Duchenne and Becker muscular dystrophy. Neuromuscular disorders. NMD. 1993;3:57–64. doi: 10.1016/0960-8966(93)90042-i. [DOI] [PubMed] [Google Scholar]
- 30.BjerglundNielsen L, Nielsen IM. Turner's syndrome and Duchenne muscular dystrophy in a girl with an X autosome translocation. Ann Genet. 1984;27:173–177. [PubMed] [Google Scholar]
- 31.Chelly J, Marlhens F, LeMarec B, Jeanpierre M, Lambert M, Hamard G, Dutrillaux B, Kaplan JC. De novo DNA microdeletion in a girl with Turner syndrome and Duchenne muscular dystrophy. Hum Genet. 1986;74:193–196. doi: 10.1007/BF00282093. [DOI] [PubMed] [Google Scholar]
- 32.Ferrier P, Bamatter F, Klein D. Muscular Dystrophy (Duchenne) in a Girl with Turner's Syndrome. J Med Genet. 1965;2:38–46. doi: 10.1136/jmg.2.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katayama Y, Tran VK, Hoan NT, Zhang Z, Goji K, Yagi M, Takeshima Y, Saiki K, Nhan NT, Matsuo M. Co-occurrence of mutations in both dystrophin- and androgen-receptor genes is a novel cause of female Duchenne muscular dystrophy. Hum Genet. 2006;119:516–519. doi: 10.1007/s00439-006-0159-4. [DOI] [PubMed] [Google Scholar]
- 34.Quan F, Janas J, Toth-Fejel S, Johnson DB, Wolford JK, Popovich BW. Uniparental disomy of the entire X chromosome in a female with Duchenne muscular dystrophy. Am J Hum Genet. 1997;60:160–165. [PMC free article] [PubMed] [Google Scholar]
- 35.Boyd Y, Buckle V, Holt S, Munro E, Hunter D, Craig I. Muscular dystrophy in girls with X autosome translocations. J Med Genet. 1986;23:484–490. doi: 10.1136/jmg.23.6.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Seemann N, Selby K, McAdam L, Biggar D, Kolski H, Goobie S, Yoon G, Campbell C. Symptomatic dystrophinopathies in female children. Neuromuscular disorders. NMD. 2011;21:172–177. doi: 10.1016/j.nmd.2010.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Sewry CA, Sansome A, Clerk A, Sherratt TG, Hasson N, Rodillo E, Heckmatt JZ, Strong PN, Dubowitz V. Manifesting carriers of Xp21 muscular dystrophy lack of correlation between dystrophin expression and clinical weakness. Neuromuscular disorders. NMD. 1993;3:141–148. doi: 10.1016/0960-8966(93)90006-6. [DOI] [PubMed] [Google Scholar]
- 38.Matthews PM, Benjamin D, VanBakel I, Squier MV, Nicholson LV, Sewry C, Barnes PR, Hopkin J, Brown R, Hilton-Jones D, Boyd Y, Karpati G, Brown GK, Craig IW. Muscle X-inactivation patterns and dystrophin expression in Duchenne muscular dystrophy carriers. Neuro-muscular disorders. NMD. 1995;5:209–220. doi: 10.1016/0960-8966(94)00057-g. [DOI] [PubMed] [Google Scholar]
- 39.vanEssen AJ, Mulder IM, vanderVlies P, vanderHout AH, Buys CH, Hofstra RM, denDunnen JT. Detection of point mutation in dystrophin gene reveals somatic and germline mosaicism in the mother of a patient with Duchenne muscular dystrophy. Am J Med Genet A. 2003;118A:296–298. doi: 10.1002/ajmg.a.10056. [DOI] [PubMed] [Google Scholar]
- 40.Bakker E, Veenema H, DenDunnen JT, vanBroeckhoven C, Grootscholten PM, Bonten EJ, vanOmmen GJ, Pearson PL. Germinal mosaicism increases the recurrence risk for 'new' Duchenne muscular dystrophy mutations. J Med Genet. 1989;26:553–559. doi: 10.1136/jmg.26.9.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Infante JP, Huszagh VA. On the nature of the Duchenne muscular dystrophy locus a portion of a complex of related gene clusters of recent pseudoautosomal origin. Mol Cell Biochem. 1988;81:103–119. doi: 10.1007/BF00219313. [DOI] [PubMed] [Google Scholar]
- 42.Pegoraro E, Hoffman EP, Piva L, Gavassini BF, Cagnin S, Ermani M, Bello L, Soraru G, Pacchioni B, Bonifati MD, Lanfranchi G, Angelini C, Kesari A, Lee I, Gordish-Dressman H, Devaney JM, McDonald CM. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy. Neurology. 2011;76:219–226. doi: 10.1212/WNL.0b013e318207afeb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bello L, Piva L, Barp A, Taglia A, Picillo E, Vasco G, Pane M, Previtali SC, Torrente Y, Gazzerro E, Motta MC, Grieco GS, Napolitano S, Magri F, D'Amico A, Astrea G, Messina S, Sframeli M, Vita GL, Boffi P, Mongini T, Ferlini A, Gualandi F, Soraru G, Ermani M, Vita G, Battini R, Bertini E, Comi GP, Berardinelli A, Minetti C, Bruno C, Mercuri E, Politano L, Angelini C, Hoffman EP, Pegoraro E. Importance of SPP1 genotype as a covariate in clinical trials in Duchenne muscular dystrophy. Neurology. 2012;79:159–162. doi: 10.1212/WNL.0b013e31825f04ea. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kyriakides T, Pegoraro E, Hoffman EP, Piva L, Cagnin S, Lanfranchi G, Griggs RC, Nelson SF. SPP1 genotype is a determinant of disease severity in Duchenne muscular dystrophy predicting the severity of Duchenne muscular dystrophy: implications for treatment. Neurology. 2011;77:1858–1859. doi: 10.1212/WNL.0b013e318239b9ae. [DOI] [PubMed] [Google Scholar]
- 45.Haslett JN, Sanoudou D, Kho AT, Han M, Bennett RR, Kohane IS, Beggs AH, Kunkel LM. Gene expression profiling of Duchenne muscular dystrophy skeletal muscle. Neurogenetics. 2003;4:163–171. doi: 10.1007/s10048-003-0148-x. [DOI] [PubMed] [Google Scholar]
- 46.Pescatori M, Broccolini A, Minetti C, Bertini E, Bruno C, D'Amico A, Bernardini C, Mirabella M, Silvestri G, Giglio V, Modoni A, Pedemonte M, Tasca G, Galluzzi G, Mercuri E, Tonali PA, Ricci E. Gene expression profiling in the early phases of DMD a constant molecular signature characterizes DMD muscle from early postnatal life throughout disease progression. FASEB J. 2007;21:1210–1226. doi: 10.1096/fj.06-7285com. [DOI] [PubMed] [Google Scholar]
- 47.Kotelnikova E, Shkrob MA, Pyatnitskiy MA, Ferlini A, Daraselia N. Novel approach to meta-analysis of microarray datasets reveals muscle remodeling-related drug targets and biomarkers in Duchenne muscular dystrophy. PLoS Comput Biol. 2012;8:e1002365. doi: 10.1371/journal.pcbi.1002365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.King WM, Ruttencutter R, Nagaraja HN, Matkovic V, Landoll J, Hoyle C, Mendell JR, Kissel JT. Orthopedic outcomes of long-term daily corticosteroid treatment in Duchenne muscular dystrophy. Neurology. 2007;68:1607–1613. doi: 10.1212/01.wnl.0000260974.41514.83. [DOI] [PubMed] [Google Scholar]
- 49.Abdel-Hamid H, Clemens PR. Pharmacological therapies for muscular dystrophies. Curr Opin Neurol. 2012;25:604–608. doi: 10.1097/WCO.0b013e328357f44c. [DOI] [PubMed] [Google Scholar]
- 50.Partridge TA. Impending therapies for Duchenne muscular dystrophy. Curr Opin Neurol. 2011;24:415–422. doi: 10.1097/WCO.0b013e32834aa3f1. [DOI] [PubMed] [Google Scholar]
- 51.Tinsley JM, Fairclough RJ, Storer R, Wilkes FJ, Potter AC, Squire SE, Powell DS, Cozzoli A, Capogrosso RF, Lambert A, Wilson FX, Wren SP, DeLuca A, Davies KE. Daily treatment with SMTC1100 a novel small molecule utrophin upregulator dramatically reduces the dystrophic symptoms in the mdx mouse. PLoS One. 2011;6:e19189. doi: 10.1371/journal.pone.0019189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tinsley J, Deconinck N, Fisher R, Kahn D, Phelps S, Gillis JM, Davies K. Expression of full-length utrophin prevents muscular dystrophy in mdx mice. Nat Med. 1998;4:1441–1444. doi: 10.1038/4033. [DOI] [PubMed] [Google Scholar]
- 53.Selsby JT, Morine KJ, Pendrak K, Barton ER, Sweeney HL. Rescue of dystrophic skeletal muscle by PGC-1alpha involves a fast to slow fiber type shift in the mdx mouse. PLoS One. 2012;7:e30063. doi: 10.1371/journal.pone.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amenta AR, Yilmaz A, Bogdanovich S, McKechnie BA, Abedi M, Khurana TS, Fallon JR. Biglycan recruits utrophin to the sarcolemma and counters dystrophic pathology in mdx mice. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:762–767. doi: 10.1073/pnas.1013067108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burkin DJ, Wallace GQ, Nicol KJ, Kaufman DJ, Kaufman SJ. Enhanced expression of the alpha 7 beta 1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J Cell Biol. 2001;152:1207–1218. doi: 10.1083/jcb.152.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wehling M, Spencer MJ, Tidball JG. A nitric oxide synthase transgene ameliorates muscular dystrophy in mdx mice. J Cell Biol. 2001;155:123–131. doi: 10.1083/jcb.200105110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Partridge TA, Morgan JE, Coulton GR, Hoffman EP, Kunkel LM. Conversion of mdx myofibres from dystrophin-negative to -positive by injection of normal myoblasts. Nature. 1989;337:176–179. doi: 10.1038/337176a0. [DOI] [PubMed] [Google Scholar]
- 58.Partridge TA, Grounds M, Sloper JC. Evidence of fusion between host and donor myoblasts in skeletal muscle grafts. Nature. 1978;273:306–308. doi: 10.1038/273306a0. [DOI] [PubMed] [Google Scholar]
- 59.Sampaolesi M, Blot S, D'Antona G, Granger N, Tonlorenzi R, Innocenzi A, Mognol P, Thibaud JL, Galvez BG, Barthelemy I, Perani L, Mantero S, Guttinger M, Pansarasa O, Rinaldi C, Cusella De, Angelis MG, Torrente Y, Bordignon C, Bottinelli R, Cossu G. Mesoan-gioblast stem cells ameliorate muscle function in dystrophic dogs. Nature. 2006;444:574–579. doi: 10.1038/nature05282. [DOI] [PubMed] [Google Scholar]
- 60.Gussoni E, Soneoka Y, Strickland CD, Buzney EA, Khan MK, Flint AF, Kunkel LM, Mulligan RC. Dystrophin expression in the mdx mouse restored by stem cell transplantation. Nature. 1999;401:390–394. doi: 10.1038/43919. [DOI] [PubMed] [Google Scholar]
- 61.Fan Y, Maley M, Beilharz M, Grounds M. Rapid death of injected myoblasts in myoblast transfer therapy. Muscle Nerve. 1996;19:853–860. doi: 10.1002/(SICI)1097-4598(199607)19:7<853::AID-MUS7>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 62.Law PK, Goodwin TG, Fang Q, Hall TL, Quinley T, Vastagh G, Duggirala V, Larkin C, Florendo JA, Li L, Jackson T, Yoo TJ, Chase N, Neel M, Krahn T, Holcomb R. First human myoblast transfer therapy continues to show dystrophin after 6 years. Cell Transplant. 1997;6:95–100. doi: 10.1177/096368979700600114. [DOI] [PubMed] [Google Scholar]
- 63.Skuk D, Goulet M, Roy B, Piette V, Cote CH, Chapdelaine P, Hogrel JY, Paradis M, Bouchard JP, Sylvain M, Lachance JG, Tremblay JP. First test of a "high-density injection" protocol for myogenic cell transplantation throughout large volumes of muscles in a Duchenne muscular dystrophy patient eighteen months follow-up. Neuromuscular disorders. NMD. 2007;17:38–46. doi: 10.1016/j.nmd.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 64.Tremblay JP, Malouin F, Roy R, Huard J, Bouchard JP, Satoh A, Richards CL. Results of a triple blind clinical study of myoblast transplantations without immunosuppressive treatment in young boys with Duchenne muscular dystrophy. Cell Transplant. 1993;2:99–112. doi: 10.1177/096368979300200203. [DOI] [PubMed] [Google Scholar]
- 65.vanDeutekom JC, Janson AA, Ginjaar IB, Frankhuizen WS, Aartsma-Rus A, Bremmer-Bout M, denDunnen JT, Koop K, vanderKooi AJ, Goemans NM, deKimpe SJ, Ekhart PF, Venneker EH, Platenburg GJ, Verschuuren JJ, vanOmmen GJ. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. [DOI] [PubMed] [Google Scholar]
- 66.Kinali M, Arechavala-Gomeza V, Feng L, Cirak S, Hunt D, Adkin C, Guglieri M, Ashton E, Abbs S, Nihoyannopoulos P, Garralda ME, Rutherford M, McCulley C, Popplewell L, Graham IR, Dickson G, Wood MJ, Wells DJ, Wilton SD, Kole R, Straub V, Bushby K, Sewry C, Morgan JE, Muntoni F. Local restoration of dystrophin expression with the morpholino oligomer AVI-4658 in Duchenne muscular dystrophy a single-blind placebo-controlled dose-escalation proof-of-concept study. Lancet Neurol. 2009;8:918–928. doi: 10.1016/S1474-4422(09)70211-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cirak S, Arechavala-Gomeza V, Guglieri M, Feng L, Torelli S, Anthony K, Abbs S, Garralda ME, Bourke J, Wells DJ, Dickson G, Wood MJ, Wilton SD, Straub V, Kole R, Shrewsbury SB, Sewry C, Morgan JE, Bushby K, Muntoni F. Exon skipping and dystrophin restoration in patients with Duchenne muscular dystrophy after systemic phosphorodiamidate morpholino oligomer treatment an open-label phase 2 dose-escalation study. Lancet. 2011;378:595–605. doi: 10.1016/S0140-6736(11)60756-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mann CJ, Honeyman K, Cheng AJ, Ly T, Lloyd F, Fletcher S, Morgan JE, Partridge TA, Wilton SD. Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:42–47. doi: 10.1073/pnas.011408598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yokota T, Lu QL, Partridge T, Kobayashi M, Nakamura A, Takeda S, Hoffman E. Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol. 2009;65:667–676. doi: 10.1002/ana.21627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Barton-Davis ER, Cordier L, Shoturma DI, Leland SE, Sweeney HL. Aminoglycoside antibiotics restore dystrophin function to skeletal muscles of mdx mice. J Clin Invest. 1999;104:375–381. doi: 10.1172/JCI7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wagner KR, Hamed S, Hadley DW, Gropman AL, Burstein AH, Escolar DM, Hoffman EP, Fischbeck KH. Gentamicin treatment of Duchenne and Becker muscular dystrophy due to nonsense mutations. Ann Neurol. 2001;49:706–711. [PubMed] [Google Scholar]
- 72.Wang B, Li J, Xiao X. Adeno-associated virus vector carrying human minidystrophin genes effectively ameliorates muscular dystrophy in mdx mouse model. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:13714–13719. doi: 10.1073/pnas.240335297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kornegay JN, Li J, Bogan JR, Bogan DJ, Chen C, Zheng H, Wang B, Qiao C, Howard JFJr, Xiao X. Widespread muscle expression of an AAV9 human mini-dystrophin vector after intravenous injection in neonatal dystrophin-deficient dogs. Mol Ther. 2010;18:1501–1508. doi: 10.1038/mt.2010.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Mendell JR, Campbell K, Rodino-Klapac L, Sahenk Z, Shilling C, Lewis S, Bowles D, Gray S, Li C, Galloway G, Malik V, Coley B, Clark KR, Li J, Xiao X, Samulski J, McPhee SW, Samulski RJ, Walker CM. Dystrophin immunity in Duchenne's muscular dystrophy. N Engl J Med. 2010;363:1429–1437. doi: 10.1056/NEJMoa1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Pichavant C, Chapdelaine P, Cerri DG, Bizario JC, Tremblay JP. Electrotransfer of the full-length dog dys-trophin into mouse and dystrophic dog muscles. Hum Gene Ther. 2010;21:1591–1601. doi: 10.1089/hum.2010.024. [DOI] [PubMed] [Google Scholar]
- 76.Zhang G, Wooddell CI, Hegge JO, Griffin JB, Huss T, Braun S, Wolff JA. Functional efficacy of dystrophin expression from plasmids delivered to mdx mice by hydrodynamic limb vein injection. Hum Gene Ther. 2010;21:221–237. doi: 10.1089/hum.2009.133. [DOI] [PubMed] [Google Scholar]
- 77.Bulfield G, Siller WG, Wight PA, Moore KJ. X chromosome-linked muscular dystrophy (mdx) in the mouse. Proceedings of the National Academy of Sciences of the United States of America. 1984;81:1189–1192. doi: 10.1073/pnas.81.4.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gillis JM. Understanding dystrophinopathies an inventory of the structural and functional consequences of the absence of dystrophin in muscles of the mdx mouse. J Muscle Res Cell Motil. 1999;20:605–625. doi: 10.1023/a:1005545325254. [DOI] [PubMed] [Google Scholar]
- 79.Carpenter JL, Hoffman EP, Romanul FC, Kunkel LM, Rosales RK, Ma NS, Dasbach JJ, Rae JF, Moore FM, McAfee MB, Pearce LK. Feline muscular dystrophy with dystrophin deficiency. Am J Pathol. 1989;135:909–919. [PMC free article] [PubMed] [Google Scholar]
- 80.Baltzer WI, Calise DV, Levine JM, Shelton GD, Edwards JF, Steiner JM. Dystrophin-deficient muscular dystrophy in a Weimaraner. J Am Anim Hosp Assoc. 2007;43:227–232. doi: 10.5326/0430227. [DOI] [PubMed] [Google Scholar]
- 81.Cooper BJ, Winand NJ, Stedman H, Valentine BA, Hoffman EP, Kunkel LM, Scott MO, Fischbeck KH, Kornegay JN, Avery RJ, Williams JR, Schmickel RD, Sylvester JE. The homologue of the Duchenne locus is defective in X-linked muscular dystrophy of dogs. Nature. 1988;334:154–156. doi: 10.1038/334154a0. [DOI] [PubMed] [Google Scholar]
- 82.Jones BR, Brennan S, Mooney CT, Callanan JJ, McAllister H, Guo LT, Martin PT, Engvall E, Shelton GD. Muscular dystrophy with truncated dystrophin in a family of Japanese Spitz dogs. J Neurol Sci. 2004;217:143–149. doi: 10.1016/j.jns.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 83.Kornegay JN, Tuler SM, Miller DM, Levesque DC. Muscular dystrophy in a litter of golden retriever dogs. Muscle Nerve. 1988;11:1056–1064. doi: 10.1002/mus.880111008. [DOI] [PubMed] [Google Scholar]
- 84.Kornegay JN, Bogan JR, Bogan DJ, Childers MK, Li J, Nghiem P, Detwiler DA, Larsen CA, Grange RW, Bhavaraju-Sanka RK, Tou S, Keene BP, Howard JFJr, Wang J, Fan Z, Schatzberg SJ, Styner MA, Flanigan KM, Xiao X, Hoffman EP. Canine models of Duchenne muscular dystrophy and their use in therapeutic strategies. Mamm Genome. 2012;23:85–108. doi: 10.1007/s00335-011-9382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Schatzberg SJ, Olby NJ, Breen M, Anderson LV, Langford CF, Dickens HF, Wilton SD, Zeiss CJ, Binns MM, Kornegay JN, Morris GE, Sharp NJ. Molecular analysis of a spontaneous dystrophin 'knockout' dog. Neuromuscular disorders. NMD. 1999;9:289–295. doi: 10.1016/s0960-8966(99)00011-5. [DOI] [PubMed] [Google Scholar]
- 86.Sharp NJ, Kornegay JN, VanCamp SD, Herbstreith MH, Secore SL, Kettle S, Hung WY, Constantinou CD, Dykstra MJ, Roses AD, Bartlett RJ. An error in dystrophin mRNA processing in golden retriever muscular dystrophy an animal homologue of Duchenne muscular dystrophy. Genomics. 1992;13:115–121. doi: 10.1016/0888-7543(92)90210-j. [DOI] [PubMed] [Google Scholar]
- 87.Smith BF, Kornegay JN, Duan D. Independent canine models of Duchenne muscular dystrophy due to intronic insertions of repetitive DNA. Mol Ther. 2007;15:S51. [Google Scholar]
- 88.Smith BF, Yue Y, Woods PR, Kornegay JN, Shin JH, Williams RR, Duan D. An intronic LINE-1 element insertion in the dystrophin gene aborts dystrophin expression and results in Duchenne-like muscular dystrophy in the corgi breed. Lab Invest. 2011;91:216–231. doi: 10.1038/labinvest.2010.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Walmsley GL, Arechavala-Gomeza V, Fernandez-Fuente M, Burke MM, Nagel N, Holder A, Stanley R, Chandler K, Marks SL, Muntoni F, Shelton GD, Piercy RJ. A duchenne muscular dystrophy gene hot spot mutation in dystrophin-deficient cavalier king charles spaniels is ame-nable to exon 51 skipping. PLoS One. 2010;5:e8647. doi: 10.1371/journal.pone.0008647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Winand NJ, Pradham D, Cooper BJ. Molecular characterization of severe Duchenne-type muscular dystrophy in a family of Rottweiler dogs. In Molecular mechanism of neuromuscular disease Muscular Dystrophy Association. Tucson. 1994 [Google Scholar]
- 91.Nonneman DJ, Brown-Brandl T, Jones SA, Wiedmann RT, Rohrer GA. A defect in dystrophin causes a novel porcine stress syndrome. BMC Genomics. 2012;13:233. doi: 10.1186/1471-2164-13-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sicinski P, Geng Y, Ryder-Cook AS, Barnard EA, Darlison MG, Barnard PJ. The molecular basis of muscular dystrophy in the mdx mouse a point mutation. Science. 1989;244:1578–1580. doi: 10.1126/science.2662404. [DOI] [PubMed] [Google Scholar]
- 93.Pastoret C, Sebille A. mdx mice show progressive weakness and muscle deterioration with age. J Neurol Sci. 1995;129:97–105. doi: 10.1016/0022-510x(94)00276-t. [DOI] [PubMed] [Google Scholar]
- 94.Chamberlain JS, Metzger J, Reyes M, Townsend D, Faulkner JA. Dystrophin-deficient mdx mice display a reduced life span and are susceptible to spontaneous rhabdomyosarcoma. FASEB J. 2007;21:2195–2204. doi: 10.1096/fj.06-7353com. [DOI] [PubMed] [Google Scholar]
- 95.Porter JD, Khanna S, Kaminski HJ, Rao JS, Merriam AP, Richmonds CR, Leahy P, Li J, Guo W, Andrade FH. A chronic inflammatory response dominates the skeletal muscle molecular signature in dystrophin-deficient mdx mice. Hum Mol Genet. 2002;11:263–272. doi: 10.1093/hmg/11.3.263. [DOI] [PubMed] [Google Scholar]
- 96.Tseng BS, Zhao P, Pattison JS, Gordon SE, Granchelli JA, Madsen RW, Folk LC, Hoffman EP, Booth FW. Regenerated mdx mouse skeletal muscle shows differential mRNA expression. J Appl Physiol. 2002;93:537–545. doi: 10.1152/japplphysiol.00202.2002. [DOI] [PubMed] [Google Scholar]
- 97.Sacco A, Mourkioti F, Tran R, Choi J, Llewellyn M, Kraft P, Shkreli M, Delp S, Pomerantz JH, Artandi SE, Blau HM. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 2010;143:1059–1071. doi: 10.1016/j.cell.2010.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mourkioti F, Kustan J, Kraft P, Day JW, Zhao MM, Kost-Alimova M, Protopopov A, Depinho RA, Bernstein D, Meeker AK, Blau HM. Role of telomere dysfunction in cardiac failure in Duchenne muscular dystrophy. Nature cell biology. 2013 doi: 10.1038/ncb2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Banks GB, Chamberlain JS. The value of mammalian models for duchenne muscular dystrophy in developing therapeutic strategies. Curr Top Dev Biol. 2008;84:431–453. doi: 10.1016/S0070-2153(08)00609-1. [DOI] [PubMed] [Google Scholar]
- 100.McGeachie JK, Grounds MD, Partridge TA, Morgan JE. Age-related changes in replication of myogenic cells in mdx mice quantitative autoradiographic studies. J Neurol Sci. 1993;119:169–179. doi: 10.1016/0022-510x(93)90130-q. [DOI] [PubMed] [Google Scholar]
- 101.Tanabe Y, Esaki K, Nomura T. Skeletal muscle pathology in X chromosome-linked muscular dystrophy (mdx) mouse. Acta Neuropathol. 1986;69:91–95. doi: 10.1007/BF00687043. [DOI] [PubMed] [Google Scholar]
- 102.Dangain J, Vrbova G. Muscle development in mdx mutant mice. Muscle Nerve. 1984;7:700–704. doi: 10.1002/mus.880070903. [DOI] [PubMed] [Google Scholar]
- 103.Seok J, Warren HS, Cuenca AG, Mindrinos MN, Baker HV, Xu W, Richards DR, McDonald-Smith GP, Gao H, Hennessy L, Finnerty CC, Lopez CM, Honari S, Moore EE, Minei JP, Cuschieri J, Bankey PE, Johnson JL, Sperry J, Nathens AB, Billiar TR, West MA, Jeschke MG, Klein MB, Gamelli RL, Gibran NS, Brownstein BH, Miller-Graziano C, Calvano SE, Mason PH, Cobb JP, Rahme LG, Lowry SF, Maier RV, Moldawer LL, Herndon DN, Davis RW, Xiao W, Tompkins RG. Genomic responses in mouse models poorly mimic human inflammatory diseases. Proceedings of the National Academy of Sciences of the United States of America. 2013 doi: 10.1073/pnas.1222878110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Gaschen FP, Hoffman EP, Gorospe JR, Uhl EW, Senior DF, Cardinet GH3rd, Pearce LK. Dystrophin deficiency causes lethal muscle hypertrophy in cats. J Neurol Sci. 1992;110:149–159. doi: 10.1016/0022-510x(92)90022-d. [DOI] [PubMed] [Google Scholar]
- 105.Winand NJ, Edwards M, Pradhan D, Berian CA, Cooper BJ. Deletion of the dystrophin muscle promoter in feline muscular dystrophy. Neuromuscular disorders. NMD. 1994;4:433–445. doi: 10.1016/0960-8966(94)90082-5. [DOI] [PubMed] [Google Scholar]
- 106.Olby NJ, Sharp NJ, Nghiem PE, Keene BW, DeFrancesco TC, Sidley JA, Kornegay JN, Schatzberg SJ. Clinical progression of X-linked muscular dystrophy in two German Shorthaired Pointers. J Am Vet Med Assoc. 2011;238:207–212. doi: 10.2460/javma.238.2.207. [DOI] [PubMed] [Google Scholar]
- 107.Nowend KL, Starr-Moss AN, Murphy KE. The function of dog models in developing gene therapy strategies for human health. Mamm Genome. 2011;22:476–485. doi: 10.1007/s00335-011-9348-0. [DOI] [PubMed] [Google Scholar]
- 108.Valentine BA, Cooper BJ, deLahunta A, O'Quinn R, Blue JT. Canine X-linked muscular dystrophy.An animal model of Duchenne muscular dystrophy clinical studies. J Neurol Sci. 1988;88:69–81. doi: 10.1016/0022-510x(88)90206-7. [DOI] [PubMed] [Google Scholar]
- 109.Bartlett RJ, Winand NJ, Secore SL, Singer JT, Fletcher S, Wilton S, Bogan DJ, Metcalf-Bogan JR, Bartlett WT, Howell JM, Cooper BJ, Kornegay JN. Mutation segregation and rapid carrier detection of X-linked muscular dystrophy in dogs. Am J Vet Res. 1996;57:650–654. [PubMed] [Google Scholar]
- 110.Kornegay JN, Cundiff DD, Bogan DJ, Bogan JR, Okamura CS. The cranial sartorius muscle undergoes true hypertrophy in dogs with golden retriever muscular dystrophy. Neuromuscular disorders. NMD. 2003;13:493–500. doi: 10.1016/s0960-8966(03)00025-7. [DOI] [PubMed] [Google Scholar]
- 111.Kornegay JN, Bogan DJ, Bogan JR, Childers MK, Cundiff DD, Petroski GF, Schueler RO. Contraction force generated by tarsal joint flexion and extension in dogs with golden retriever muscular dystrophy. J Neurol Sci. 1999;166:115–121. doi: 10.1016/s0022-510x(99)00118-5. [DOI] [PubMed] [Google Scholar]
- 112.Kornegay JN, Bogan JR, Bogan DJ, Childers MK, Grange RW. Golden retriever muscular dystrophy (GRMD).Developing and maintaining a colony and physiological functional measurements. Methods Mol Biol. 2011;709:105–123. doi: 10.1007/978-1-61737-982-6_7. [DOI] [PubMed] [Google Scholar]
- 113.Kornegay JN, Sharp NJ, Bogan DJ, VanCamp SD, Metcalf JR, Schueler RO. Contraction tension and kinetics of the peroneus longus muscle in golden retriever muscular dystrophy. J Neurol Sci. 1994;123:100–107. doi: 10.1016/0022-510x(94)90210-0. [DOI] [PubMed] [Google Scholar]
- 114.Kornegay JN, Sharp NJ, Schueler RO, Betts CW. Tarsal joint contracture in dogs with golden retriever muscular dystrophy. Lab Anim Sci. 1994;44:331–333. [PubMed] [Google Scholar]
- 115.Desguerre I, Christov C, Mayer M, Zeller R, Becane HM, Bastuji-Garin S, Leturcq F, Chiron C, Chelly J, Gherardi RK. Clinical heterogeneity of duchenne muscular dystrophy (DMD) definition of sub-phenotypes and predictive criteria by long-term follow-up. PLoS One. 2009;4:e4347. doi: 10.1371/journal.pone.0004347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Brooke MH, Fenichel GM, Griggs RC, Mendell JR, Moxley R, Miller JP, Province MA. Clinical investigation in Duchenne dystrophy 2.Determination of the "power" of therapeutic trials based on the natural history. Muscle Nerve. 1983;6:91–103. doi: 10.1002/mus.880060204. [DOI] [PubMed] [Google Scholar]
- 117.Dubowitz V. Enigmatic conflict of clinical and molecular diagnosis in Duchenne/Becker muscular dystrophy. Neuromuscular disorders. NMD. 2006;16:865–866. doi: 10.1016/j.nmd.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 118.Ambrosio CE, Valadares MC, Zucconi E, Cabral R, Pearson PL, Gaiad TP, Canovas M, Vainzof M, Miglino MA, Zatz M. Ringo a Golden Retriever Muscular Dystrophy (GRMD) dog with absent dystrophin but normal strength. Neuromuscular disorders. NMD. 2008;18:892–893. doi: 10.1016/j.nmd.2008.06.385. [DOI] [PubMed] [Google Scholar]
- 119.Zucconi E, Valadares MC, Vieira NM, Bueno CRJr, Secco M, Jazedje T, daSilva HC, Vainzof M, Zatz M. Ringo discordance between the molecular and clinical manifestation in a golden retriever muscular dystrophy dog. Neuromuscular disorders NMD. 2010;20:64–70. doi: 10.1016/j.nmd.2009.10.011. [DOI] [PubMed] [Google Scholar]
- 120.Schatzberg SJ, Anderson LV, Wilton SD, Kornegay JN, Mann CJ, Solomon GG, Sharp NJ. Alternative dystrophin gene transcripts in golden retriever muscular dystrophy. Muscle Nerve. 1998;21:991–998. doi: 10.1002/(sici)1097-4598(199808)21:8<991::aid-mus2>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 121.Tsai KL, Clark LA, Murphy KE. Understanding hereditary diseases using the dog and human as companion model systems. Mamm Genome. 2007;18:444–451. doi: 10.1007/s00335-007-9037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Ostrander EA, Franklin H. Epstein Lecture.Both ends of the leash--the human links to good dogs with bad genes. N Engl J Med. 2012;367:636–646. doi: 10.1056/NEJMra1204453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Karlsson EK, Lindblad-Toh K. Leader of the pack gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9:713–725. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- 124.Ostrander EA, Giniger E. Semper fidelis what man's best friend can teach us about human biology and disease. Am J Hum Genet. 1997;61:475–480. doi: 10.1086/515522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, Alvarez P, Biagi T, Brockman W, Butler J, Chin CW, Cook A, Cuff J, Daly MJ, DeCaprio D, Gnerre S, Grabherr M, Kellis M, Kleber M, Bardeleben C, Goodstadt L, Heger A, Hitte C, Kim L, Koepfli KP, Parker HG, Pollinger JP, Searle SM, Sutter NB, Thomas R, Webber C, Baldwin J, Abebe A, Abouelleil A, Aftuck L, Ait-Zahra M, Aldredge T, Allen N, An P, Anderson S, Antoine C, Arachchi H, Aslam A, Ayotte L, Bachantsang P, Barry A, Bayul T, Benamara M, Berlin A, Bessette D, Blitshteyn B, Bloom T, Blye J, Boguslavskiy L, Bonnet C, Boukhgalter B, Brown A, Cahill P, Calixte N, Camarata J, Cheshatsang , Chu J, Citroen M, Collymore A, Cooke P, Dawoe T, Daza R, Decktor K, DeGray S, Dhargay N, Dooley K, Dorje P, Dorjee K, Dorris L, Duffey N, Dupes A, Egbiremolen O, Elong R, Falk J, Farina A, Faro S, Ferguson D, Ferreira P, Fisher S, FitzGerald M, Foley K, Foley C, Franke A, Friedrich D, Gage D, Garber M, Gearin G, Giannoukos G, Goode T, Goyette A, Graham J, Grandbois E, Gyaltsen K, Hafez N, Hagopian D, Hagos B, Hall J, Healy C, Hegarty R, Honan T, Horn A, Houde N, Hughes L, Hunnicutt L, Husby M, Jester B, Jones C, Kamat A, Kanga B, Kells C, Khazanovich D, Kieu AC, Kisner P, Kumar M, Lance K, Landers T, Lara M, Lee W, Leger JP, Lennon N, Leuper L, LeVine S, Liu J, Liu X, Lokyitsang Y, Lokyitsang T, Lui A, Macdonald J, Major J, Marabella R, Maru K, Matthews C, McDonough S, Mehta T, Meldrim J, Melnikov A, Me-neus L, Mihalev A, Mihova T, Miller K, Mittelman R, Mlenga V, Mulrain L, Munson G, Navidi A, Naylor J, Nguyen T, Nguyen N, Nguyen C, Nicol R, Norbu N, Norbu C, Novod N, Nyima T, Olandt P, O'Neill B, O'Neill K, Osman S, Oyono L, Patti C, Perrin D, Phunkhang P, Pierre F, Priest M, Rachupka A, Raghuraman S, Rameau R, Ray V, Raymond C, Rege F, Rise C, Rogers J, Rogov P, Sahalie J, Settipalli S, Sharpe T, Shea T, Sheehan M, Sherpa N, Shi J, Shih D, Sloan J, Smith C, Sparrow T, Stalker J, Stange-Thomann N, Stavropoulos S, Stone C, Stone S, Sykes S, Tchuinga P, Tenzing P, Tesfaye S, Thoulutsang D, Thoulutsang Y, Topham K, Topping I, Tsamla T, Vassiliev H, Venkataraman V, Vo A, Wangchuk T, Wangdi T, Weiand M, Wilkinson J, Wilson A, Yadav S, Yang S, Yang X, Young G, Yu Q, Zainoun J, Zembek L, Zimmer A, Lander ES. Genome sequence comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 126.Vonholdt BM, Pollinger JP, Lohmueller KE, Han E, Parker HG, Quignon P, Degenhardt JD, Boyko AR, Earl DA, Auton A, Reynolds A, Bryc K, Brisbin A, Knowles JC, Mosher DS, Spady TC, Elkahloun A, Geffen E, Pilot M, Jedrzejewski W, Greco C, Randi E, Bannasch D, Wilton A, Shearman J, Musiani M, Cargill M, Jones PG, Qian Z, Huang W, Ding ZL, Zhang YP, Bustamante CD, Ostrander EA, Novembre J, Wayne RK. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature. 2010;464:898–902. doi: 10.1038/nature08837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Boyko AR, Quignon P, Li L, Schoenebeck JJ, Degenhardt JD, Lohmueller KE, Zhao K, Brisbin A, Parker HG, vonHoldt BM, Cargill M, Auton A, Reynolds A, Elkahloun AG, Castelhano M, Mosher DS, Sutter NB, Johnson GS, Novembre J, Hubisz MJ, Siepel A, Wayne RK, Bustamante CD, Ostrander EA. A simple genetic architecture underlies morphological variation in dogs. PLoS Biol. 2010;8:e1000451. doi: 10.1371/journal.pbio.1000451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sutter NB, Eberle MA, Parker HG, Pullar BJ, Kirkness EF, Kruglyak L, Ostrander EA. Extensive and breed-specific linkage disequilibrium in Canis familiaris. Genome Res. 2004;14:2388–2396. doi: 10.1101/gr.3147604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Karlsson EK, Baranowska I, Wade CM, SalmonHillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ3rd, Comstock KE, Keller ET, Mesirov JP, vonEuler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39:1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- 130.Derrien T, Theze J, Vaysse A, Andre C, Ostrander EA, Galibert F, Hitte C. Revisiting the missing protein-coding gene catalog of the domestic dog. BMC Genomics. 2009;10:62. doi: 10.1186/1471-2164-10-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.McCarroll SA, Altshuler DM. Copy-number variation and association studies of human disease. Nat Genet. 2007;39:S37–42. doi: 10.1038/ng2080. [DOI] [PubMed] [Google Scholar]
- 132.Feuk L, Marshall CR, Wintle RF, Scherer SW. Structural variants: changing the landscape of chromosomes and design of disease studies. Hum Mol Genet. 2006;15 Spec No 1:R57–66. doi: 10.1093/hmg/ddl057. [DOI] [PubMed] [Google Scholar]
- 133.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu Rev Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 134.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 135.Zhang F, Carvalho CM, Lupski JR. Complex human chromosomal and genomic rearrangements. Trends Genet. 2009;25:298–307. doi: 10.1016/j.tig.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- 137.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen WK, Swartz JD, Rush LJ, Alvarez CE. Mapping DNA structural variation in dogs. Genome Res. 2009;19:500–509. doi: 10.1101/gr.083741.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nicholas TJ, Baker C, Eichler EE, Akey JM. A high-resolution integrated map of copy number polymorphisms within and between breeds of the modern domesticated dog. BMC Genomics. 2011;12:414. doi: 10.1186/1471-2164-12-414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Joe AW, Yi L, Natarajan A, LeGrand F, So L, Wang J, Rudnicki MA, Rossi FM. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nature cell biology. 2010;12:153–163. doi: 10.1038/ncb2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Uezumi A, Fukada S, Yamamoto N, Takeda S, Tsuchida K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nature cell biology. 2010;12:143–152. doi: 10.1038/ncb2014. [DOI] [PubMed] [Google Scholar]
- 142.Uezumi A, Ito T, Morikawa D, Shimizu N, Yoneda T, Segawa M, Yamaguchi M, Ogawa R, Matev MM, Miyagoe-Suzuki Y, Takeda S, Tsujikawa K, Tsuchida K, Yamamoto H, Fukada S. Fibrosis and adipogenesis originate from a common mesenchymal progenitor in skeletal muscle. J Cell Sci. 2011;124:3654–3664. doi: 10.1242/jcs.086629. [DOI] [PubMed] [Google Scholar]
- 143.Guasconi V, Puri PL. Chromatin the interface between extrinsic cues and the epigenetic regulation of muscle regeneration. Trends Cell Biol. 2009;19:286–294. doi: 10.1016/j.tcb.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Guasconi V, Puri PL. Epigenetic drugs in the treatment of skeletal muscle atrophy. Curr Opin Clin Nutr Metab Care. 2008;11:233–241. doi: 10.1097/MCO.0b013e3282fa1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Lanzuolo C, Orlando V. The function of the epigenome in cell reprogramming. Cell Mol Life Sci. 2007;64:1043–1062. doi: 10.1007/s00018-007-6420-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 147.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 148.Consalvi S, Saccone V, Giordani L, Minetti G, Mozzetta C, Puri PL. Histone deacetylase inhibitors in the treatment of muscular dystrophies epigenetic drugs for genetic diseases. Mol Med. 2011;17:457–465. doi: 10.2119/molmed.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mozzetta C, Minetti G, Puri PL. Regenerative pharmacology in the treatment of genetic diseases the paradigm of muscular dystrophy. Int J Biochem Cell Biol. 2009;41:701–710. doi: 10.1016/j.biocel.2008.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Minetti GC, Colussi C, Adami R, Serra C, Mozzetta C, Parente V, Fortuni S, Straino S, Sampaolesi M, DiPadova M, Illi B, Gallinari P, Steinkuhler C, Capogrossi MC, Sartorelli V, Bottinelli R, Gaetano C, Puri PL. Functional and morphological recovery of dystrophic muscles in mice treated with deacetylase inhibi-tors. Nat Med. 2006;12:1147–1150. doi: 10.1038/nm1479. [DOI] [PubMed] [Google Scholar]
- 151.Wittenburg LA, Ptitsyn AA, Thamm DH. A systems biology approach to identify molecular pathways altered by HDAC inhibition in osteosarcoma. J Cell Biochem. 2012;113:773–783. doi: 10.1002/jcb.23403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cohen LA, Powers B, Amin S, Desai D. Treatment of canine haemangiosarcoma with suberoylanilide hydroxamic acid a histone deacetylase inhibitor. Vet Comp Oncol. 2004;2:243–248. doi: 10.1111/j.1476-5810.2004.00057.x. [DOI] [PubMed] [Google Scholar]
- 153.Kinugasa F, Nagatomi I, Nakanishi T, Noto T, Mori H, Matsuoka H, Sudo Y, Mutoh S. Effect of the immunosuppressant histone deacetylase inhibitor FR276457 in a canine renal transplant model. Transpl Immunol. 2009;21:198–202. doi: 10.1016/j.trim.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 154.Miyazato LG, Moraes JR, Beretta DC, Kornegay JN. Muscular dystrophy in dogs does the crossing of breeds influence disease phenotype. Vet Pathol. 2011;48:655–662. doi: 10.1177/0300985810387070. [DOI] [PubMed] [Google Scholar]
- 155.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7 improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Heringa J. Two strategies for sequence comparison profile-preprocessed and secondary structure-induced multiple alignment. Comput Chem. 1999;23:341–364. doi: 10.1016/s0097-8485(99)00012-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material is available on the publisher’s web site along with the published article.