Abstract
A large fraction of human tumors carry p53 mutations, which allow tumor initiation and progression; furthermore, it is now clear that restoration or reactivation of wild-type p53 function prompts rapid elimination of tumors. The discovery and design of compounds that reactivate or enhance the p53 pathway has resulted in the identification of promising drug candidates that have now entered clinical trials for anti-cancer strategies. However, some of these agents appear to elicit undesirable toxic effects on normal cells and tissues and therefore are restricted in the dose that can be applied in tumors. In this Review, we discuss the concerns about and promise of these p53 activators and propose ways to expand and optimize screening strategies to identify such molecules.
INTRODUCTION
The p53 tumor suppressor protein serves as a genome guardian and has been intensively studied for nearly 30 years (1, 2). It functions mainly as a transcription factor by binding to specific DNA sequences and by transactivating or repressing a large group of target genes (3–5). These downstream targets of p53 regulate the pathways of cell cycle arrest, apoptosis, and DNA repair to maintain a dynamic equilibrium between cell growth and arrest in response to factors including DNA damage, hypoxia (oxygen deprivation), and a deficiency of growth factors or nutrients (4, 6, 7).
The gene encoding p53, TP53, is the most frequently inactivated tumor suppressor gene in human malignancies; its inactivation is beneficial for tumor survival (8, 9). On this basis, restoration of wild-type p53 activity seems to be one of the most attractive goals for a successful tumor therapy. In the clinic, the functional status of p53 has been associated with the prognosis, progression, and therapeutic response of tumors (10). Tumor cells containing wild-type p53 are usually more sensitive to radiotherapy or chemotherapy than those bearing mutant p53. However, recent findings have yielded unexpected insight about the incidence of selective pro-survival effects of wild-type p53 in cancer cells, in which it might confer a selective advantage that antagonizes cancer therapy (11–14). Moreover, there is a major concern about developing p53 activators as cancer therapeutics, because activating p53 might not only impose on tumor growth but would also lead to harmful effects in normal cells and tissues. Nevertheless, all these characteristics make p53 an important molecular target for tumor suppression and reconstitution of p53 function in tumors may provide a new form of cancer therapy. In view of the importance of of cell death/apoptosis on therapy outcome, strategies to restore apoptotic p53 pathways in tumor cells have been thoroughly pursued in recent years.
THE BRIGHT SIDE OF p53 IN CANCER THERAPEUTICS
Cells are continuously subjected to conditions and stimuli that can result in genotoxic stress, including oxidative metabolism and irradiation. Thus, the ability to detect and repair consequent DNA damage is a critical function of normal cells. Failure to efficiently repair DNA damage can ultimately result in malignant transformation. Induction of a cell cycle arrest (a transient or permanent block of cell proliferation) or the activation of cell death pathways in response to genotoxic stress comprises the major arms of the survival-death axis governed by p53. Because of these biological properties, inactivation of wild-type p53 is a crucial step in tumor development and progression, reflected by the high incidence of TP53 mutations in a variety of human cancer types.
p53 is normally expressed at low levels so that it does not disrupt the cell cycle or cause the cell to undergo untimely death. Such low concentrations are achieved through the operation of a negative feedback loop consisting of wild-type p53 and the MDM2 gene and its product. MDM2 is a p53 transcriptional target whose product ubiquitinates p53, thus marking it for proteosome-mediated degradation (15–17). However, p53 is stabilized and accumulates upon stresses such as DNA damage or oncogene activation, resulting in cell cycle arrest, senescence, and/or cell death through transactivation of its target genes, including those encoding p21 (which promotes cell cycle arrest) and the pro-apoptotic proteins Bax (Bcl-2-associated protein), PUMA (p53 up-regulated modulator of apoptosis), and Noxa (phorbol-12-myristate-13-acetate-induced protein 1) (4, 6) (Fig. 1). Because the downstream targets that have been identified play a critical role in the p53 tumor suppression response, these targets should be a major avenue for therapeutic intervention in p53 activation in cancer cells.
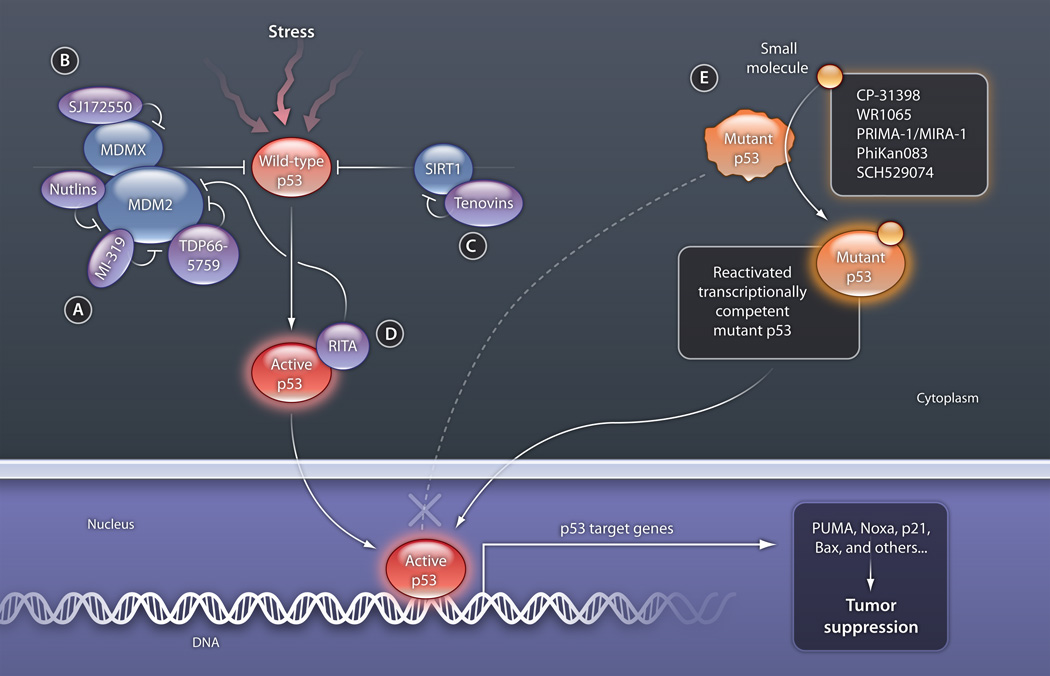
Fig 1. Modulating the p53 pathway with small molecules.
Wild-type p53 is activated by a variety of stressors, including DNA damage, oncogene expression, nutrient starvation, oxidative stress, and depletion of ribonucleotide triphosphates (used in RNA synthesis). (A) Small molecules that target MDM2 and block p53 binding stabilize p53. (B) SJ172550 targets the p53 binding pocket of MDMX, also promoting p53 stabilization. (C) Tenovin-6 inhibits the protein deacetylase activity of SIRT. Acetylation results in the stabilization of p53 and interferes with MDM2-mediated degradation. (D) RITA binds to p53 and interferes with the interaction of MDM2 and p53, activating p53 function. (E) Small molecules designed to bind transcriptionally inert mutant p53 proteins stabilize the core domain, restore the native state, and eventually enable binding to DNA.
CREDIT: C. BICKEL/SCIENCE TRANSLATIONAL MEDICINE
It has also been demonstrated that in a substantial percentage of human tumors, p53 is inactivated by MDM2 overexpression and/or amplification (18). Furthermore, it is well established that MDMX/MDM4—a nonredundant homolog of MDM2—also regulates p53 and is overexpressed in many cancers (15, 19). However, unlike MDM2, MDMX expression is not regulated by p53 and its product lacks intrinsic ubiquitin ligase activity; thus, it is not an essential part of the negative feedback loop described above. However, MDMX forms heterodimers with MDM2, which enhances the ability of MDM2 to induce p53 degradation (16, 17). Interrupting the interactions between p53 and its negative regulators such as MDM2 to activate or stabilize p53 is a promising therapeutic strategy for the treatment of cancers retaining wild-type p53.
Tumors that carry mutations in TP53 often overexpress mutant p53, resulting in increased resistance to conventional chemotherapy and radiotherapy as compared with cells that do not overexpress mutant p53. This finding indicates that such mutant p53 provides a certain selective advantage for tumor development—an oncogenic gain-of-function phenotype. Tumor cells containing mutant p53 should become sensitive to chemotherapy upon restoration of the wild-type p53 pathway (Fig. 1). This makes mutant p53 an attractive target for selective cancer therapy that would not affect normal cells, because normal cells do not contain mutant p53.
THE DARK SIDE OF p53 IN CANCER THERAPEUTICS
p53 in normal tissue
DNA damage-induced cell death through the action of chemotherapeutic drugs is the most widely used strategy in cancer therapy. However, selectivity remains a great concern because most such drugs kill both cancer cells and the surrounding normal cells, which is an important cause of the side effects of cancer chemotherapy that severely limit current treatment regimes. Although the key to successful anti-cancer therapies is to target critical nodes that are required for the survival of cancer cells, such therapies should not be harmful to normal cells.
The idea of restoring wild-type p53 pathways (apoptosis and cell cycle arrest) by inhibiting proteosomal degradation of p53 (for example, via MDM2 inhibition) is a promising therapeutic strategy (20). So far, the identification of small molecules that either (i) inhibit the E3 ligase activity of MDM2 or (ii) can occupy the hydrophobic p53-binding pocket/cleft in MDM2 is quite feasible (20, 21). These MDM2 inhibitors all cause tumor regression through cell death in xenograft models, although it is unclear how such molecules would affect healthy cells and tissues in humans.
p53 as a guardian of cancer cells
Wild-type p53 can commit cancer cells to a survival path in response to stress such as DNA damage (11, 12, 22). Survival-promoting activities of wild-type p53 include the induction of a broad range of cellular responses, including cell cycle regulation, facilitation of DNA repair pathways, the maintenance of the cellular redox state, and activation of genes with anti-apoptotic activities (13, 14). The major role of p53 is to guard normal cells against malignant transformation and to protect the genome under stress conditions. For example, p53 can directly participate in repair processes by binding to DNA and resolving abnormal DNA structures or by activating p53 target genes that are involved in the regulation of DNA repair (23). Because of its functions in DNA repair and cell cycle regulation, wild-type p53 might also function as a “guardian” in cancer cells to promote cancer cell survival in the presence of stress, thereby raising the view that wild-type p53 activity may not always be helpful in preventing tumorigenesis. Indeed, some cases, the retention of wt-p53 has been shown to protect cancers from some forms of cytotoxic chemotherapy and so can be associated with a poor response to treatment (24). Moreover, the use of p53 inhibitors has been proposed for chemoprotection of normal cells since much of the toxicity seen in response to conventional genotoxic chemotherapies is due to the activation of p53, thus inhibition of p53 in normal cells may protect them from cell death (25, 26). More recently, accumulating evidence indicated that p53 engages potent pro-survival pathways which are critical for maintenance of a normal, healthy cell/tissue but may contribute to tumor cell survival against cytotoxic damage (2, 13, 14, 27). A growing number of p53-induced survival genes have been identified, which function through several diverse mechanisms (2, 12, 22, 28, 29). Numerous p53 downstream or interacting target proteins function to inhibit apoptosis, including genes involved in DNA repair, cell cycle control, metabolism, oxidative stress response, transcription, growth factors and receptors etc (2, 13, 14). It is clear that wt-p53 is not only a powerful proapoptotic inducer in response to cytotoxic stress but also a potent inducer of cell cycle arrest, protecting tumor cells from further excessive damages. While the responses of cancer cells to p53 seem more complicated than we previously thought, such genes or pathways toward p53-induced pro-survival in cancer cells may be used as new targets for the development of cancer therapeutics.
DEVELOPMENT OF p53 ACTIVATORS AS THERAPEUTIC DRUGS
Challenges ahead
Recent genetic studies in mouse models [involving three tumor types (lymphoma, sarcoma, and hepatocellularcarcinoma)] have shown that reactivation of the p53 pathway in tumors with inhibited p53 activity is an exceptionally effective intervention (30–32). Such a reactivation process appears to be feasible because the p53 protein is usually highly expressed in tumors, although it may be functionally inert. However, from a conventional drug discovery point of view, p53 is a challenging target: it doesn’t offer the accessibility of a receptor-ligand interaction or an enzyme active site. Instead, it is a homotetrameric transcription factor with complicated protein-protein interactions, rendering it as an “undruggable” target. Researchers must therefore overcome this challenge, as well as identify the means to manipulate this target appropriately in its environment to achieve the desired therapeutic efficacy with minimal toxicity. Nevertheless, recent advances in drug discovery and progress in technology development are complementing the last 30 years of p53-based research and are creating a realistic expectation that tumor specific p53 restoration therapies are approaching a level at which they could be used for clinical applications.
Targeting the wild-type p53 pathway
Several approaches have been undertaken to restore wild-type p53 function. Initially, tumor therapy based on exogenous wild-type p53 expression via gene therapy was exploited. Adenovirus-based gene therapy, involving introduction of a functional copy of TP53 into tumors by local injection at the tumor site, killed a high percentage of cancer cells but also resulted in high toxicity, partially because of strong bystander effects (such that neighboring cells were killed because of signals originating from the tumor cells) (33). In addition, obvious problems with this approach are the inability to infect every cell in the tumor with virus as well as a lack of stability for the newly synthesized functional p53 protein.
More recently, the use of small molecules for endogenous p53 activation, in tumors retaining a wild-type p53 gene, was intensively investigated (Fig. 1). The majority of these efforts were based on the assumption that subtle variation of p53 protein levels could affect the threshold of the tumor cell tolerance for apoptotic signals. Therefore, compounds modulating the stability of wild-type p53 through inhibition of its negative regulators would be expected to induce cancer cell death effectively. The best-known naturally occurring p53 inhibitor is its own downstream target MDM2 (17, 34, 35). In past years, several strategies for tumor therapy have been developed to increase p53 activity by neutralizing MDM2 function at different levels. Vassilev and colleagues (20) identified a group of small molecules that target the MDM2-p53 interaction. These imidazoline derivatives, designated Nutlins, specifically bind and dissociate MDM2 from p53, thereby rescuing p53 from degradation and inducing cell cycle and apoptosis (20). In vivo experiments also showed a significant antitumor effect of Nutlins (36).
The benzodiazepenes and the spiro-oxidole based compounds are another class of small molecules that have been found to target the p53-MDM2 interaction (37, 38). These experiments led to the preclinical development of TDP665759 and MI-319, which disrupt the binding of MDM2 to p53 in vitro and suppress the growth of tumor cells both in vitro and in vivo. Both compounds limit tumor growth without causing major toxicity in the surrounding tissue, although they induce low levels of p53 by increasing its stability in normal cells, which doesn’t seem to be sufficient for the activation of the apoptotic cascade (39).
Considering the speed of current drug discovery efforts, the development of optimized analogs that target the p53-MDM2 interaction will likely occur in the very near future. However, this strategy is also associated with several potential problems. Because MDM2 is a classical p53 target gene and is part of an inducible feedback loop that inhibits the p53 pathway, MDM2 targeting compounds will eventually cause the induction of their own target (that is, MDM2) and will thus limit their own efficacy. In addition, the use of MDM2 targeting compounds may be problematic in tumors expressing high levels of mutant p53. Given the pro-oncogenic impact of several mutations in the p53 molecule, releasing these mutants from the effects of MDM2-mediated degradation may actually promote further tumorogenesis (40).
One of the key regulators of the p53 pathway is another member of the MDM2 family of proteins, MDMX. Although MDM2 and MDMX share homology in their p53 binding domain, Nutlin-like compounds fail to efficiently affect MDMX, which will form a heterodimer with MDM2 and thereby stimulate the degradation of p53. Therefore, a general consensus is emerging that an effective induction of wild type p53 in tumors could be achieved successfully by the simultaneous inhibition of MDM2 and MDMX. Recently, the identification of the first MDMX inhibitor, SJ172550, was published (41) and although its molecular mechanism of action is not completely clear yet, initial data indicate moderate success in combination with the MDM2 inhibitors in mouse cancer models.
SIRT1 and 2 are two members of the nicotinamide adenine dinucleotide-dependent class III histone deacetylases (42). It is well established that acetylation of p53 at Lys382 enhances DNA binding of p53. Therefore, SIRT1 deacetylation of Lys382 in the p53 protein has a negative effect on p53 activity, establishing SIRT1 as yet another possible target for modulation of the p53 pathway (43). Indeed, a p53-based phenotypic screen indentified a class of small molecules called tenovins, which are potent inhibitors of SIRT1 and 2 as shown later through a yeast genetic screen and subsequent enzymatic assays (44). Tenovins rapidly increased accumulation of acetylated p53 both in vitro and in vivo and represent a previously unknown class of p53-activating agents that can be further developed for clinical use.
In another attempt to activate the p53 pathway through stabilization of the wild-type p53 protein, a small molecule referred to as RITA (reactivation of p53 and induction of tumor cell apoptosis) was discovered and found to directly bind p53 at the N terminus and prevent degradation (45). Whether this is the only mechanism by which RITA increases p53 activity in cells is still controversial, as there is evidence that RITA can bind to multiple proteins and activate the DNA damage response pathway independently of p53 (46).
The rapid advances in the understanding of the p53 pathway in recent years have allowed the identification of number of targets suitable for drug discovery (as described above) that should function as modulators of the intact wild-type p53 pathway (Fig. 1). However, a number of identified non-genotoxic compounds might be expected to induce p53 in normal tissues as well, and their therapeutic index (a value that compares the amount of drug that causes a therapeutic effect to the amount that is toxic, therefore, will depend strictly on the difference in p53 concentrations between normal and cancer cells as well as on the level of p53 induction (47). Thus, new approaches for the modulation of wild-type p53 signaling are vitally needed. Furthermore, as we gain a better understanding of p53 signaling pathways, additional p53-linked targets can be identified and validated for future therapeutic studies. An intense fresh look at the p53 pathway should result in both (i) new cell-based screens for the identification of small molecule modulators of selected p53 targets that will control p53-mediated cell fate decisions such as reversible growth arrest, apoptosis, senescence, or survival and (ii) target-based biochemical screens for the development of small molecule protein-protein interaction inhibitors (Fig. 2). In many of these cases, the p53 system, as challenging as it is, might become the unprecedented model for the invention and development of a variety of new drug discovery approaches.
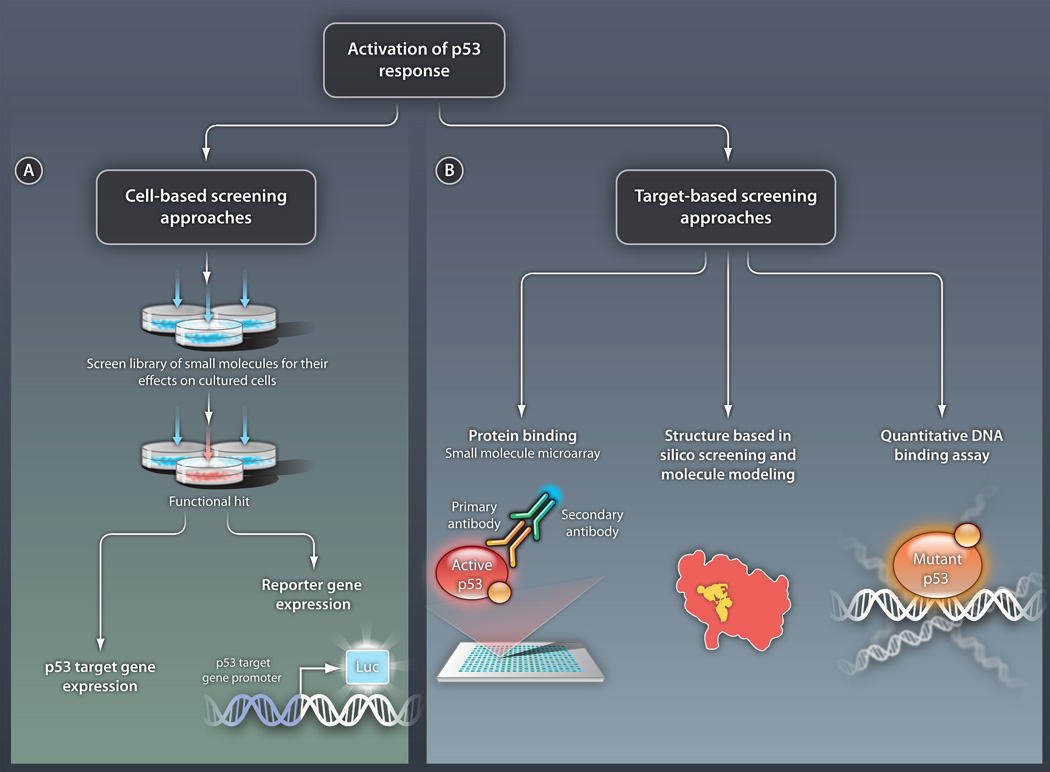
Fig 2. Strategies for identifying activators of the p53 response.
(A) Cell-based screening. In this approach, small molecules are tested for their ability to activate p53 pathways in cultured cells. Activation can be detected by measuring endogenous levels of transcripts from p53 target genes or by using various reporter systems. In this second method, the promoter of a p53 target gene is placed upstream of a gene encoding a product that can be easily monitored, such as the enzyme luciferase (luc). Cell-based screening will identify compounds with certain biological activity but with an unknown direct target molecule. (B) Target-based screening. In this approach, p53 protein activators are sought in vitro. Specific methods include the following: (i) Direct protein binding assays, such as the SMM assay. In this example, a library of small molecules is assembled on a microarray and the ability of p53 to bind the molecules is tested. Here, p53 binding is detected by a set of antibodies. (ii) Forward structural design, in which the structures of mutant versions of p53 are analyzed and molecules are designed to interact with relevant regions of these structures. (iii) Direct DNA binding assays, in which molecules that allow mutant p53 to bind DNA are sought. Such methods will enable the identification of compounds with a defined molecular mechanism.
CREDIT: C. BICKEL/SCIENCE TRANSLATIONAL MEDICINE
Targeting mutant p53: reactivation by small molecules
Around half of all human tumors overexpress a nonfunctional mutant p53 that accumulates to high concentrations in tumor cells (8, 10, 21). Because of these high p53 concentrations, restoration of the function of such mutant p53 proteins would be an extremely efficient strategy for selective elimination of tumor cells. A small molecule targeting mutant p53 should not affect wild-type p53 in normal cells because it is already properly folded and because of its low concentrations, owing to continuous degradation by MDM2 in the absence of stress. However, activating mutant p53 with small molecules appears to be an even greater challenge than activating wild-type p53, mainly because of the wide range of mutant proteins that are expressed in tumors, which might exhibit different structural alterations. Notably, about 95% of the cancer-associated mutations occur in the thermally-unstable central core DNA binding domain of the protein and result in further destabilization of the structure, abrogation of DNA binding, and impairment of the p53 response (48). Therefore, it is reasonable to postulate that small molecules that bind specifically to the mutant DNA binding domain and stabilize p53 in its active biological conformation—in effect, functioning as chaperones could potentially rescue wild-type p53 function independently of the exact mutation present (49).
Early progress has demonstrated the feasibility of this approach: antibodies that bind the p53 C terminus (pAb421) and a synthetic peptide (p53C) derived from the C-terminal domain showed a stimulatory effect on the DNA binding of several proteins with mutations in the central DNA binding domain (at residues His175, Ala143, and Ser249) as well as induction of apoptosis in cancer cells expressing such contact mutants (50). However, the challenges that are associated with peptide or protein stability and transport into cancer cells are preventing this approach from being tested clinically. Therefore, in recent years several screening attempts for mutant p53 reactivating compounds have been carried out using either protein binding assays or cell–based assays involving mutant p53-containing tumor cells (Fig. 2). New compounds targeting mutant p53 have been identified using both types of screening assays (Fig. 1). CP-31398, identified by Pfizer through an in vitro p53 stabilization assay, showed promising initial pre-clinical results in p53 mutant and null cells (51). Subsequent analysis, however, revealed that the mechanism of action of this compound is not based on direct binding with p53 but instead is mediated by an interaction with DNA (52).
Another compound that was identified, WR1065, is the active metabolite of amifostine (a drug that controls some side effects of radiation therapy and chemotherapy and has been reported to restore completely or partially the transcriptional activity of p53 mutant proteins by a yet unknown mechanism (53). Similarly, a cell-based screening assay—involving a Saos-2 cell line (derived from a human osteosarcoma that was engineered to express arg273his mutant p53—yielded PRIMA-1 and MIRA-1 (a more potent second generation analog), two small molecules capable of selective induction of apoptosis in cells expressing mutant p53. These compounds were shown to induce expression of p53 target genes such as those encoding p21, MDM2, and/or PUMA and inhibit tumor growth in vivo in mice (54). Extensive characterization of these compounds revealed that they form adducts with thiols in the mutant p53 core domain (55). How this covalent modification drives the equilibrium between mutant and wild-type conformations toward the native conformation, however, is an open question that needs a clear answer before full clinical development is possible.
Another approach used structural data and took advantage of computational techniques. In this case, the crystal structure of a version of the p53 protein that contained a Tyr220 → Cys220 mutation was determined and found to include a binding pocket. This finding allowed an in silico screening of interacting molecules and led to the identification of a small molecule, PhiKan083, that fits into the pocket and stabilizes the mutant core domain in the wild-type conformation (56).
Very recently, a p53 DNA binding assay identified the small molecule SCH529074, which promotes the DNA binding activity of mutant p53 in cell-free systems, induces apoptosis in tumor cells, and reduces tumor growth in a xenograft model (57). The very early preclinical data with this compound are encouraging and are paving the way for the pharmacological optimization of this compound.
Thus, mutant p53 reactivation by small molecules is a rapidly evolving approach with great potential for the development of new anti-cancer drugs. As described above, several drug discovery strategies have been undertaken; some, including biochemical screening, crystal structure-guided molecular design, and in silico screening, are based on target engagement by p53 (Fig. 2). A recently developed methodology, based on the direct binding of a target protein to small molecules chemically immobilized on a glass slide, called a small molecule microarray (SMM), may offer an advantage for future efforts. This method offers high-throughput screening at a very low cost (58). The remaining p53-associated drug discovery strategies, as described in this Review, were cell-based assays. These approaches offer the advantage of selecting for in vivo activity; identification of the drug target, however, can be challenging (Fig. 2). Considering the complicated and problematic nature of the p53 pathway as a drug target, a combination of screening approaches—for example, target-based biochemical assays (such as SMM) and cell-based phenotypic approaches—may be the optimal way to identify small molecules with clear biological functions and known mechanisms of action that are poised for optimization via comprehensive medicinal chemistry. All of these approaches may provide the basis for the rational design of more efficient and selective anti-cancer drugs that restore and enhance the p53 pathway.
Acknowledgement
We thank to Kiki Chu, Xinbin Chen and CBRC colleagues for the critical readings of manuscript and their helpful discussion.
Funding: The academic research of the authors in the field of cancer therapeutics is supported by NIH grants 5RO1CA127247; 1RO1CA142805; 1RO1CA149477; 5P01 CA80058; 2R01CA085681 (S.W.L.) and 5R01CA140615 (A.M.)
Footnotes
Competing interests: The authors don’t have any competing interests.
Contributor Information
Anna Mandinova, Cutaneous Biology Research Center, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA 02129 USA; Broad Institute of MIT and Harvard, Cambridge, MA 02139, USA.
Sam W. Lee, Cutaneous Biology Research Center, Massachusetts General Hospital and Harvard Medical School, Charlestown, MA 02129 USA Broad Institute of MIT and Harvard, Cambridge, MA 02139, USA, swlee@partners.org.
REFERENCES AND NOTES
- 1.Levine AJ, Oren M. The first 30 years of p53: growing ever more complex. Nat Rev Cancer. 2009;9:749. doi: 10.1038/nrc2723. published online EpubOct (nrc2723 [pii] 10.1038/nrc2723) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vousden KH, Prives C. Blinded by the Light: The Growing Complexity of p53. Cell. 2009;137:413. doi: 10.1016/j.cell.2009.04.037. published online EpubMay 1 (S0092-8674(09)00459-0 [pii] 10.1016/j.cell.2009.04.037) [DOI] [PubMed] [Google Scholar]
- 3.el-Deiry WS, Kern SE, Pietenpol JA, Kinzler KW, Vogelstein B. Definition of a consensus binding site for p53. Nat Genet. 1992;1:45. doi: 10.1038/ng0492-45. published online EpubApr (10.1038/ng0492-45) [DOI] [PubMed] [Google Scholar]
- 4.Laptenko O, Prives C. Transcriptional regulation by p53: one protein, many possibilities. Cell Death Differ. 2006;13:951. doi: 10.1038/sj.cdd.4401916. published online EpubJun (4401916 [pii] 10.1038/sj.cdd.4401916) [DOI] [PubMed] [Google Scholar]
- 5.Raycroft L, Wu HY, Lozano G. Transcriptional activation by wild-type but not transforming mutants of the p53 anti-oncogene. Science. 1990;249:1049. doi: 10.1126/science.2144364. published online EpubAug 31 ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruse JP, Gu W. Modes of p53 regulation. Cell. 2009;137:609. doi: 10.1016/j.cell.2009.04.050. published online EpubMay 15 (S0092-8674(09)00511-X [pii] 10.1016/j.cell.2009.04.050) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275. doi: 10.1038/nrm2147. published online EpubApr (nrm2147 [pii] 10.1038/nrm2147) [DOI] [PubMed] [Google Scholar]
- 8.Nigro JM, Baker SJ, Preisinger AC, Jessup JM, Hostetter R, Cleary K, Bigner SH, Davidson N, Baylin S, Devilee P, et al. Mutations in the p53 gene occur in diverse human tumour types. Nature. 1989;342:705. doi: 10.1038/342705a0. published online EpubDec 7 (10.1038/342705a0) [DOI] [PubMed] [Google Scholar]
- 9.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789. doi: 10.1038/nm1087. published online EpubAug (10.1038/nm1087 nm1087 [pii]) [DOI] [PubMed] [Google Scholar]
- 10.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9:701. doi: 10.1038/nrc2693. published online EpubOct (nrc2693 [pii] 10.1038/nrc2693) [DOI] [PubMed] [Google Scholar]
- 11.Bar J, Feniger-Barish R, Lukashchuk N, Shaham H, Moskovits N, Goldfinger N, Simansky D, Perlman M, Papa M, Yosepovich A, Rechavi G, Rotter V, Oren M. Cancer cells suppress p53 in adjacent fibroblasts. Oncogene. 2009;28:933. doi: 10.1038/onc.2008.445. published online EpubFeb 12 (onc2008445 [pii] 10.1038/onc.2008.445) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Das S, Raj L, Zhao B, Kimura Y, Bernstein A, Aaronson SA, Lee SW. Hzf Determines cell survival upon genotoxic stress by modulating p53 transactivation. Cell. 2007;130:624. doi: 10.1016/j.cell.2007.06.013. published online EpubAug 24 (S0092-8674(07)00779-9 [pii] 10.1016/j.cell.2007.06.013) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Janicke RU, Sohn D, Schulze-Osthoff K. The dark side of a tumor suppressor: anti-apoptotic p53. Cell Death Differ. 2008;15:959. doi: 10.1038/cdd.2008.33. published online EpubJun (cdd200833 [pii] 10.1038/cdd.2008.33) [DOI] [PubMed] [Google Scholar]
- 14.Vousden KH. Outcomes of p53 activation--spoilt for choice. J Cell Sci. 2006;119:5015. doi: 10.1242/jcs.03293. published online EpubDec 15 (119/24/5015 [pii] 10.1242/jcs.03293) [DOI] [PubMed] [Google Scholar]
- 15.Barak Y, Juven T, Haffner R, Oren M. mdm2 expression is induced by wild type p53 activity. EMBO J. 1993;12:461. doi: 10.1002/j.1460-2075.1993.tb05678.x. published online EpubFeb ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manfredi JJ. The Mdm2-p53 relationship evolves: Mdm2 swings both ways as an oncogene and a tumor suppressor. Genes Dev. 2010;24:1580. doi: 10.1101/gad.1941710. published online EpubAug 1 (24/15/1580 [pii] 10.1101/gad.1941710) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wade M, Wang YV, Wahl GM. The p53 orchestra: Mdm2 and Mdmx set the tone. Trends Cell Biol. 2010;20:299. doi: 10.1016/j.tcb.2010.01.009. published online EpubMay (S0962-8924(10)00033-4 [pii] 10.1016/j.tcb.2010.01.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Momand J, Jung D, Wilczynski S, Niland J. The MDM2 gene amplification database. Nucleic Acids Res. 1998;26:3453. doi: 10.1093/nar/26.15.3453. published online EpubAug 1 (gkb548 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fakharzadeh SS, Trusko SP, George DL. Tumorigenic potential associated with enhanced expression of a gene that is amplified in a mouse tumor cell line. EMBO J. 1991;10:1565. doi: 10.1002/j.1460-2075.1991.tb07676.x. published online EpubJun ( [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844. doi: 10.1126/science.1092472. published online EpubFeb 6 (10.1126/science.1092472 1092472 [pii]) [DOI] [PubMed] [Google Scholar]
- 21.Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862. doi: 10.1038/nrc2763. published online EpubDec (nrc2763 [pii] 10.1038/nrc2763) [DOI] [PubMed] [Google Scholar]
- 22.Fang L, Li G, Liu G, Lee SW, Aaronson SA. p53 induction of heparin-binding EGF-like growth factor counteracts p53 growth suppression through activation of MAPK and PI3K/Akt signaling cascades. EMBO J. 2001;20:1931. doi: 10.1093/emboj/20.8.1931. published online EpubApr 17 (10.1093/emboj/20.8.1931) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat Rev Mol Cell Biol. 2005;6:44. doi: 10.1038/nrm1546. published online EpubJan (nrm1546 [pii] 10.1038/nrm1546) [DOI] [PubMed] [Google Scholar]
- 24.Bertheau P, Espie M, Turpin E, Lehmann J, Plassa LF, Varna M, Janin A, de H. The, TP53 status and response to chemotherapy in breast cancer. Pathobiology. 2008;75:132. doi: 10.1159/000123851. 000123851 [pii] 10.1159/000123851) [DOI] [PubMed] [Google Scholar]
- 25.Gudkov AV, Komarova EA. Prospective therapeutic applications of p53 inhibitors. Biochem Biophys Res Commun. 2005;331:726. doi: 10.1016/j.bbrc.2005.03.153. published online EpubJun 10 (S0006-291×(05)00659-5 [pii] 10.1016/j.bbrc.2005.03.153) [DOI] [PubMed] [Google Scholar]
- 26.Strom E, Sathe S, Komarov PG, Chernova OB, Pavlovska I, Shyshynova I, Bosykh DA, Burdelya LG, Macklis RM, Skaliter R, Komarova EA, Gudkov AV. Small-molecule inhibitor of p53 binding to mitochondria protects mice from gamma radiation. Nat Chem Biol. 2006;2:474. doi: 10.1038/nchembio809. published online EpubSep (nchembio809 [pii] 10.1038/nchembio809) [DOI] [PubMed] [Google Scholar]
- 27.Aylon Y, Oren M. Living with p53, dying of p53. Cell. 2007;130:597. doi: 10.1016/j.cell.2007.08.005. published online EpubAug 24 (S0092-8674(07)01027-6 [pii] 10.1016/j.cell.2007.08.005) [DOI] [PubMed] [Google Scholar]
- 28.Brooks CL, Gu W. p53 Activation: a case against Sir. Cancer Cell. 2008;13:377. doi: 10.1016/j.ccr.2008.04.009. published online EpubMay (S1535-6108(08)00126-8 [pii] 10.1016/j.ccr.2008.04.009) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, Li KC, Hong TM, Yang PC. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11:694. doi: 10.1038/ncb1875. published online EpubJun (ncb1875 [pii] 10.1038/ncb1875) [DOI] [PubMed] [Google Scholar]
- 30.Martins CP, Brown-Swigart L, Evan GI. Modeling the therapeutic efficacy of p53 restoration in tumors. Cell. 2006;127:1323. doi: 10.1016/j.cell.2006.12.007. published online EpubDec 29 (S0092-8674(06)01597-2 [pii] 10.1016/j.cell.2006.12.007) [DOI] [PubMed] [Google Scholar]
- 31.Ventura A, Kirsch DG, McLaughlin ME, Tuveson DA, Grimm J, Lintault L, Newman J, Reczek EE, Weissleder R, Jacks T. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661. doi: 10.1038/nature05541. published online EpubFeb 8 (nature05541 [pii] 10.1038/nature05541) [DOI] [PubMed] [Google Scholar]
- 32.Xue W, Zender L, Miething C, Dickins RA, Hernando E, Krizhanovsky V, Cordon-Cardo C, Lowe SW. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656. doi: 10.1038/nature05529. published online EpubFeb 8 (nature05529 [pii] 10.1038/nature05529) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Roth JA. Adenovirus p53 gene therapy. Expert Opin Biol Ther. 2006;6:55. doi: 10.1517/14712598.6.1.55. published online EpubJan (10.1517/14712598.6.1.55) [DOI] [PubMed] [Google Scholar]
- 34.Brooks CL, Gu W. p53 ubiquitination: Mdm2 and beyond. Mol Cell. 2006;21:307. doi: 10.1016/j.molcel.2006.01.020. published online EpubFeb 3 (S1097-2765(06)00040-2 [pii] 10.1016/j.molcel.2006.01.020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shan J, Brooks C, Kon N, Li M, Gu W. Dissecting roles of ubiquitination in the p53 pathway. Ernst Schering Found Symp Proc. 2008:127. doi: 10.1007/2789_2008_105. [DOI] [PubMed] [Google Scholar]
- 36.Tovar C, Rosinski J, Filipovic Z, Higgins B, Kolinsky K, Hilton H, Zhao X, Vu BT, Qing W, Packman K, Myklebost O, Heimbrook DC, Vassilev LT. Small-molecule MDM2 antagonists reveal aberrant p53 signaling in cancer: implications for therapy. Proc Natl Acad Sci U S A. 2006;103:1888. doi: 10.1073/pnas.0507493103. published online EpubFeb 7 (0507493103 [pii] 10.1073/pnas.0507493103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ding K, Lu Y, Nikolovska-Coleska Z, Wang G, Qiu S, Shangary S, Gao W, Qin D, Stuckey J, Krajewski K, Roller PP, Wang S. Structure-based design of spiro-oxindoles as potent, specific small-molecule inhibitors of the MDM2-p53 interaction. J Med Chem. 2006;49:3432. doi: 10.1021/jm051122a. published online EpubJun 15 (10.1021/jm051122a) [DOI] [PubMed] [Google Scholar]
- 38.Grasberger BL, Lu T, Schubert C, Parks DJ, Carver TE, Koblish HK, Cummings MD, LaFrance LV, Milkiewicz KL, Calvo RR, Maguire D, Lattanze J, Franks CF, Zhao S, Ramachandren K, Bylebyl GR, Zhang M, Manthey CL, Petrella EC, Pantoliano MW, Deckman IC, Spurlino JC, Maroney AC, Tomczuk BE, Molloy CJ, Bone RF. Discovery and cocrystal structure of benzodiazepinedione HDM2 antagonists that activate p53 in cells. J Med Chem. 2005;48:909. doi: 10.1021/jm049137g. published online EpubFeb 24 (10.1021/jm049137g) [DOI] [PubMed] [Google Scholar]
- 39.Mohammad RM, Wu J, Azmi AS, Aboukameel A, Sosin A, Wu S, Yang D, Wang S, Al-Katib AM. An MDM2 antagonist (MI-319) restores p53 functions and increases the life span of orally treated follicular lymphoma bearing animals. Mol Cancer. 2009;8:115. doi: 10.1186/1476-4598-8-115. 1476-4598-8-115 [pii] 10.1186/1476-4598-8-115) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prives C, White E. Does control of mutant p53 by Mdm2 complicate cancer therapy? Genes Dev. 2008;22:1259. doi: 10.1101/gad.1680508. published online EpubMay 15 (22/10/1259 [pii] 10.1101/gad.1680508) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reed D, Shen Y, Shelat AA, Arnold LA, Ferreira AM, Zhu F, Mills N, Smithson DC, Regni CA, Bashford D, Cicero SA, Schulman BA, Jochemsen AG, Guy RK, Dyer MA. Identification and characterization of the first small molecule inhibitor of MDMX. J Biol Chem. 2010;285:10786. doi: 10.1074/jbc.M109.056747. published online EpubApr 2 (M109.056747 [pii] 10.1074/jbc.M109.056747) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Haigis MC, Guarente LP. Mammalian sirtuins--emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913. doi: 10.1101/gad.1467506. published online EpubNov 1 (20/21/2913 [pii] 10.1101/gad.1467506) [DOI] [PubMed] [Google Scholar]
- 43.Luo J, Li M, Tang Y, Laszkowska M, Roeder RG, Gu W. Acetylation of p53 augments its site-specific DNA binding both in vitro and in vivo. Proc Natl Acad Sci U S A. 2004;101:2259. doi: 10.1073/pnas.0308762101. published online EpubFeb 24 (101/8/2259 [pii]) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lain S, Hollick JJ, Campbell J, Staples OD, Higgins M, Aoubala M, McCarthy A, Appleyard V, Murray KE, Baker L, Thompson A, Mathers J, Holland SJ, Stark MJ, Pass G, Woods J, Lane DP, Westwood NJ. Discovery, in vivo activity, and mechanism of action of a small-molecule p53 activator. Cancer Cell. 2008;13:454. doi: 10.1016/j.ccr.2008.03.004. published online EpubMay (S1535-6108(08)00089-5 [pii] 10.1016/j.ccr.2008.03.004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Issaeva N, Bozko P, Enge M, Protopopova M, Verhoef LG, Masucci M, Pramanik A, Selivanova G. Small molecule RITA binds to p53, blocks p53-HDM-2 interaction and activates p53 function in tumors. Nat Med. 2004;10:1321. doi: 10.1038/nm1146. published online EpubDec (nm1146 [pii] 10.1038/nm1146) [DOI] [PubMed] [Google Scholar]
- 46.Krajewski M, Ozdowy P, D'Silva L, Rothweiler U, Holak TA. NMR indicates that the small molecule RITA does not block p53-MDM2 binding in vitro. Nat Med. 2005;11:1135. doi: 10.1038/nm1105-1135. published online EpubNov (nm1105-1135 [pii] 10.1038/nm1105-1135) [DOI] [PubMed] [Google Scholar]
- 47.Willis AC, Chen X. The promise and obstacle of p53 as a cancer therapeutic agent. Curr Mol Med. 2002;2:329. doi: 10.2174/1566524023362474. published online EpubJun ( [DOI] [PubMed] [Google Scholar]
- 48.Terzian T, Suh YA, Iwakuma T, Post SM, Neumann M, Lang GA, Van Pelt CS, Lozano G. The inherent instability of mutant p53 is alleviated by Mdm2 or p16INK4a loss. Genes Dev. 2008;22:1337. doi: 10.1101/gad.1662908. published online EpubMay 15 (22/10/1337 [pii] 10.1101/gad.1662908) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brachmann RK, Yu K, Eby Y, Pavletich NP, Boeke JD. Genetic selection of intragenic suppressor mutations that reverse the effect of common p53 cancer mutations. EMBO J. 1998;17:1847. doi: 10.1093/emboj/17.7.1847. published online EpubApr 1 (10.1093/emboj/17.7.1847) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875. doi: 10.1016/0092-8674(92)90562-q. published online EpubNov 27 (0092-8674(92)90562-Q [pii]) [DOI] [PubMed] [Google Scholar]
- 51.Foster BA, Coffey HA, Morin MJ, Rastinejad F. Pharmacological rescue of mutant p53 conformation and function. Science. 1999;286:2507. doi: 10.1126/science.286.5449.2507. published online EpubDec 24 (8097 [pii]) [DOI] [PubMed] [Google Scholar]
- 52.Rippin TM, Bykov VJ, Freund SM, Selivanova G, Wiman KG, Fersht AR. Characterization of the p53-rescue drug CP-31398 in vitro and in living cells. Oncogene. 2002;21:2119. doi: 10.1038/sj.onc.1205362. published online EpubMar 28 (10.1038/sj.onc.1205362) [DOI] [PubMed] [Google Scholar]
- 53.Grdina DJ, Shigematsu N, Dale P, Newton GL, Aguilera JA, Fahey RC. Thiol and disulfide metabolites of the radiation protector and potential chemopreventive agent WR-2721 are linked to both its anti-cytotoxic and anti-mutagenic mechanisms of action. Carcinogenesis. 1995;16:767. doi: 10.1093/carcin/16.4.767. published online EpubApr ( [DOI] [PubMed] [Google Scholar]
- 54.Bykov VJ, Issaeva N, Shilov A, Hultcrantz M, Pugacheva E, Chumakov P, Bergman J, Wiman KG, Selivanova G. Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat Med. 2002;8:282. doi: 10.1038/nm0302-282. published online EpubMar (10.1038/nm0302-282 nm0302-282 [pii]) [DOI] [PubMed] [Google Scholar]
- 55.Lambert JM, Gorzov P, Veprintsev DB, Soderqvist M, Segerback D, Bergman J, Fersht AR, Hainaut P, Wiman KG, Bykov VJ. PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell. 2009;15:376. doi: 10.1016/j.ccr.2009.03.003. published online EpubMay 5 (S1535-6108(09)00078-6 [pii] 10.1016/j.ccr.2009.03.003) [DOI] [PubMed] [Google Scholar]
- 56.Boeckler FM, Joerger AC, Jaggi G, Rutherford TJ, Veprintsev DB, Fersht AR. Targeted rescue of a destabilized mutant of p53 by an in silico screened drug. Proc Natl Acad Sci U S A. 2008;105:10360. doi: 10.1073/pnas.0805326105. published online EpubJul 29 (0805326105 [pii] 10.1073/pnas.0805326105) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Demma M, Maxwell E, Ramos R, Liang L, Li C, Hesk D, Rossman R, Mallams A, Doll R, Liu M, Seidel-Dugan C, Bishop WR, Dasmahapatra B. SCH529074, a small molecule activator of mutant p53, which binds p53 DNA binding domain (DBD), restores growth-suppressive function to mutant p53 and interrupts HDM2-mediated ubiquitination of wild type p53. J Biol Chem. 2010;285:10198. doi: 10.1074/jbc.M109.083469. published online EpubApr 2 (M109.083469 [pii] 10.1074/jbc.M109.083469) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J Am Chem Soc. 2003;125:8420. doi: 10.1021/ja0352698. published online EpubJul 16 (10.1021/ja0352698) [DOI] [PubMed] [Google Scholar]