Abstract
Background
Bupropion, an antidepressant and smoking cessation medication, is metabolized to hydroxybupropion (HB), an active metabolite, primarily by CYP2B6.
Objectives
To compare plasma concentrations of bupropion and metabolites at steady state in healthy volunteers with and without CYP2B6 genetic variants.
Methods
In a genotype-guided study of 42 healthy subjects we measured plasma and urine concentrations of bupropion and its metabolites, HB, threohydrobupropion (TB) and erythrohydrobupropion (EB) after 7 days of sustained release bupropion dosing.
Results
CYP2B6*6 and *18 gene variants were associated with approximately 33% reduced concentrations of HB, with no effects on concentrations of bupropion or other metabolites. We could account for 50% of the variation in HB concentrations in a model including genotype and sex.
Conclusions
Since HB is active and steady state concentrations of HB are more than 10 times higher than bupropion, CYP2B6 variants are likely to affect pharmacological activity. Due to the large individual variation within genotype group, the use of therapeutic drug monitoring for dose optimization may be necessary.
Introduction
Bupropion, a norepinephrine and dopamine uptake inhibitor and nicotinic receptor antagonist, is widely used in the treatment of depression and for smoking cessation. Bupropion is extensively metabolized, with conversion to three main plasma metabolites: hydroxybupropion (HB), threohydrobupropion (TB) and erythrohydrobupropion (EB).[1, 2] The metabolites of bupropion are pharmacologically active: HB is 50%-100% as active and TB and EB 20% as active as bupropion in animal models of inhibition of neurotransmitter uptake or models of depression.[3-6] Bupropion has a half-life ranging from 10 to 20 hrs, and HB has a half-life of about 20 hr, while TB and EB have half-lives of 37 and 33 hours, respectively.[1, 7]
The primary enzyme involved in metabolism of bupropion to hydroxybupropion is the liver enzyme CYP2B6.[8-11] The metabolism of bupropion to TB and EB is primarily by carbonyl reductase, a nonmicrosomal enzyme in the liver and gut. CYP2C19 appears to contribute to oxidation of bupropion to metabolites other than HB.[11] CYP2B6 is polymorphic, with CYP2B6*6 the most common variant allele [http://www.pharmgkb.org/]. A genotype containing at least one CYP2B6*6 allele is found in about 25%, 45%, 50% and 70% of Asians, Caucasians, African Americans and Yupik Alaskan Native Peoples, respectively.[12-14] The other common variants include *4 (2-4% allele frequency in Caucasians and Asians, but rare in African Americans) and *5 (10%, 5% and 1% allele frequency in Caucasians, African Americans and Asians, respectively). Relevant to the present study, the CYP2B6*18 allele is found essentially exclusively in people of African descent, with an allele frequency of 4-7%.
Pharmacokinetic studies in individuals with different genotypes have been performed using single doses of bupropion. Kirchheiner showed that CYP2B6*4 is associated with faster clearance of bupropion, but that CYP2B6*6 did not affect bupropion clearance [15]. Loboz [16] reported that one individual who was a homozygote for CYP2B6 *6 had slower bupropion metabolism than individuals homozygous for the CYP2B6*1 reference allele. In vitro studies support the idea that the CYP2B6*6 variants are associated with reduced concentrations of functional mRNA, protein and enzymatic activity, thought to be related to aberrant splicing with loss of exons 4 to 6.[17-20] Recombinant expression of CYP2B6.18 in COS-1 cells resulted in marked reduction of expression and reduced bupropion hydroxylase activity. [21] We are aware of no human data on the effect of CYP2B6*18 on bupropion metabolism.
While single dose studies indicate genetic variation in bupropion metabolism, usual usage for depression or smoking cessation involves prolonged dosing regiments. Thus steady state analyses of the impact of genotype on concentrations of bupropion and metabolites are needed. We conducted a genotype-stratified pharmacokinetic study to compare plasma concentrations of bupropion, HB, TB and EB throughout the day after 7 days of daily dosing with sustained release bupropion in healthy volunteers with and without CYP2B6*6 variants. We hypothesized that the presence of CYP2B6*6 alleles would be associated with significant higher steady state plasma concentrations of bupropion and lower plasma concentrations of HB compared to people with reference alleles. We were also able to include a few subjects with the CYP2B6*18 allele. We hypothesized that CYP2B6*18 allele would be associated with significant higher steady state plasma concentrations of bupropion and lower plasma concentrations of HB compared to people with reference alleles.
Methods
Subjects
Forty-two healthy subjects were studied, of which 62% were men. Self-declared race included 21 white (14 non-Hispanic, 7 Hispanic), 14 African Americans and 7 Asians. The subjects were selected from a pool of people who had given DNA for genotyping for potential future participation in pharmacokinetic studies. We enriched the subject pool for those with CYP2B6*6 alleles. Subjects could not be taking regular medications (including oral contraceptives) and could not be current alcohol or illicit drug abusers. Subjects were screened by medical history, physical examination, complete blood count, liver and kidney function and EKG. Individuals with a history of seizures, significant prior head trauma or eating disorders were excluded, as these are risk factors for bupropion-induced seizures. Subjects were compensated for participation. The study was approved the Institutional Review Boards at the University of California San Francisco and University of Toronto.
Procedures
After screening, subjects signed informed consents and were asked to take one bupropion 150 mg XL (sustained release) tablet in the morning before breakfast daily for 7 days. Seven days was taken as adequate time to reach steady state concentrations of bupropion and metabolites.[1] Subjects were asked to call the research associate daily to confirm that he/she had taken the medication and to give the exact time. On Day 7, subjects were admitted to Clinical Study Unit at San Francisco Hospital Medical Center at about 8 am for a 24 hour admission. Blood samples were collected every 4 hours during the day until 12 midnight then again at 8 am the next day. Bupropion degrades in blood, a process that is temperature and pH dependent.[22] Therefore, blood samples were collected in chilled heparinized tubes and centrifuged in a cold centrifuge. Plasma was then separated into a citric acid tube and frozen until analysis. We found that this procedure for handling blood specimens prevented loss of bupropion that has been seen with other sampling procedures.
Genotyping
Common CYP2B6 alleles (*4 K262R, *5 R487C, *6 K262R & Q172H, and *9 Q172H) were genotyped using two-step allele-specific PCR assay as previously described.[13, 23]. In addition, new genotyping assays were developed for CYP2B6*18. In order to avoid the amplification of CYP2B7 pseudogene, each assay included a gene-specific amplification followed by an allele specific amplification. The gene-specific amplification primers were forward 5’CCTCTCGGTCTGCCCATCTATAAAC3’ and reverse 5’AGGAACCCTGTCTCTGTGTGACT3’. The allele specific amplification primers were forward wild type 5’GGACCCCAGCGCCCCCAA3’, forward variant 5’GGACCCCAGCGCCCCCAG3’, reverse wild type 5’CGATGTGGGCCAATCACCTGTTCAA3’, and reverse variant 5’CGATGTGGGCCAATCACCTGTTCAG3’. The assays were verified by sequencing (ACGT Corporation, Toronto ON, Canada).
Analytical Chemistry
Plasma samples were analyzed for concentrations of bupropion and its metabolites, HB, TB and EB using liquid chromatography – tandem mass spectroscopy, as previously described [24, 25]. The lower limit of quantitation for bupropion was 1 ng/ml. Data on within-run and between-run accuracy and precision are provided in supplemental table 1. Urine samples were assayed with and without glucuronide deconjugation. The deconjugation procedure followed the method of Petsalo, modified with overnight instead of 6 hour incubation.[26] Concentrations of conjugated bupropion or metabolites were determined as the difference between concentrations measured with compared to without deconjugation. Concentrations of bupropion and metabolite standards were unaffected by the deconjugation procedure.
Data Analysis
The main measure of exposure to bupropion and its major metabolites was the area under the plasma concentration-time curve for 24 hours on day 7 (AUC24). Oral clearance of bupropion was also computed as Dose/AUC24, normalized for body weight. Clearance via different metabolic pathways was estimated using the ratio of AUCs for each of the metabolites over bupropion. We also analyzed the maximal concentration (Cmax) for bupropion and metabolites. Urine data were examined both as absolute concentrations and the molar percentage of each analyte in relation to the sum of bupropion and its metabolites.
Pairwise statistical comparisons were performed by Wilcoxon’s tests. Linear regression analysis was used to examine the effects of having zero, one or two copies of CYP2B6*6 or CYP2B6*18 allele, race/ethnicity, sex, and BMI on HB AUC and the HB/BUP ratio. To enable better interpretation of the regression results, the PK parameters were not normalized. Normalization by log transformation did not change the significances of the finding. All analyses were performed using Stata 11 (StataCorp, College Station, TX). P values below 0.05 were considered to be statistically significant.
Results
Demographic data and genotype frequencies for the subjects are shown in Table 1. Figure 1 shows the average plasma concentrations of bupropion and metabolites over 24 hours. Steady state HB concentrations were on average more than 10-fold higher than bupropion or EB concentrations, while TB concentrations were intermediate. Pharmacokinetic data for subjects with and without reduced function CYP2B6 alleles are shown in Table 2. The Cmax, Css (mean steady state concentration) and AUC of HB were significantly lower in subjects with at least one reduced function alleles (i.e either CYP2B6*6 or *18) compared to those without, which could also be seen within the CYP2B6*6 group alone. Consistent with this finding, the ratio of HB/BUP was significantly lower in those with CYP2B6*6 variants. Other comparisons were not statistically significantly different. Figure 2 shows HB concentrations (AUC) according to CYP2B6 genotype, sex and race. On average, homozygous and heterozygous CYP2B6 *6 subjects had lower HB concentrations compared to those without reduced function alleles. Urine bupropion and metabolite excretion data are shown in Table 3. Less than 1% of the daily bupropion dose was excreted unchanged as bupropion. The greatest percentage of the bupropion dose was recovered by TB followed by HB. Bupropion was not appreciably glucuronidated, while glucuronide metabolites accounted for about 75%, 25% and 10% of total excreted HB, EB and TB respectively. Subjects with CYP2B6 reduced function variants, in particular CYP2B6*6, excreted significantly less HB compared to those without reduced function variants. Renal clearances averaged 0.17, 0.03, 0.37 and 0.50 ml/min/kg for bupropion, HB, EB and TB, respectively.
Table 1.
Demographics and Genotype Distribution
| White (N=21) |
African American (N=14) |
Asians (N=7) |
|
|---|---|---|---|
| Sex (% male) | 61.9 | 50 | 85.7 |
| Agea (y) | 29.2 (19,51) | 33.1 (22,64) | 37.0 (22,60) |
| Weight (kg) | 72.0 (65.9,78.1) | 75.6 (66.8,84.4) | 80.8 (56.2,105.4) |
| BMI (kg/m2) | 24.1 (22.3,25.9) | 25.6 (23.2,27.9) | 29.1 (18.7,39.5) |
| CYP2B6 Genotype | |||
| *1/*1 | 6 (29%) | 3 (21%) | 2 (29%) |
| *1/*4 | - | - | 2 (29%) |
| *1/*5 | 2 (10%) | 1 (7%) | - |
| *1/*6 | 6 (28%) | 5 (36%) | 3 (43%) |
| *5/*6 or *1/*7 | 3 (14%) | - | - |
| *6/*6 | 4 (19%) | 2 (14%) | - |
| *1/*18 | - | 2 (14%) | - |
| *6/*18 | - | 1 (7%) | - |
mean and range
men (95% C.I.)
Figure 1.
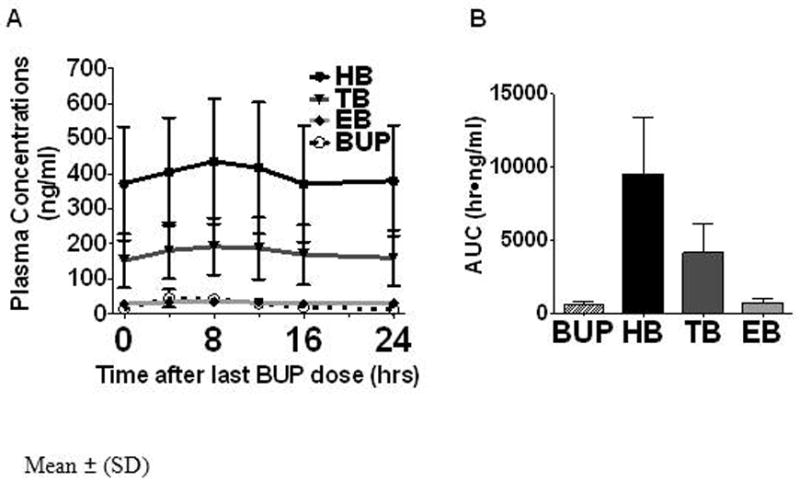
Panel A: Plasma concentrations of bupropion (BUP) and its metabolites hydroxybupropion (HB), threohydrobupropion (TB) and erythrohydrobupropion (EB) over 24 hours. Mean ± SD
Panel B: Area under the 24 hour plasma concentration-time curve for bupropion and metabolites.
Table 2.
Pharmacokinetic Parameters for Bupropion and Metabolites (mean, 95% CI)
| All (N=42) | No reduced function allele (N=16) | With reduced function allele(s) (N=26) | *6 allele (N= 23) | *18 allele (N= 2) | *6/18 allele (N= 1) | |
|---|---|---|---|---|---|---|
| BUP | ||||||
| Cmax (ng/mL) | 58 (52,63) | 58 (51,65) | 57 (48, 66) | 58 (50,67) | 59 (-2,121) | 33 |
|
|
||||||
| Css (ng/mL) | 28.5 (26.3,30.8) | 29.4 (26.6,32.1) | 28.0 (24.7,31.4) | 28.6 (25.6,31.7) | 25.9 (25.6,226.5) | 18.8 |
|
|
||||||
| AUC (hr•ng/mL) | 685 (631,739) | 705 (639,770) | 673 (593, 753) | 687 (614,761) | 621 (-614,5437) | 450 |
|
| ||||||
| HB | ||||||
| Cmax (ng/mL) | 464 (406,522) | 592 (493,690) | 386a (330, 442) | 405a (352,459) | 291 (130,452) | 137 |
|
|
||||||
| Css (ng/mL) | 397 (347,447) | 501 (410,591) | 333b (285,381) | 349b (301,397) | 258 (-992,1509) | 115 |
|
|
||||||
| AUC (hr•ng/mL) | 9524 (8319,10729) | 12015 (9852,14177) | 7996a (6833,9149) | 8375b (7219,9531) | 6200 (-23812,36212) | 2753 |
|
|
||||||
| HB/BUP | 14.4 (12.6,16.2) | 17.4 (14.1,20.8) | 12.5c (10.6, 14.4) | 12.8c (10.8,14.8) | 12.2 (-34.2,58.6) | 6.1 |
|
| ||||||
| EB | ||||||
| Cmax (ng/mL) | 38 (35,42) | 43 (37,49) | 35 (31,41) | 36 (31,41) | 38 (-1,77) | 22 |
|
|
||||||
| Css (ng/mL) | 32 (29,36) | 36 (30,42) | 30 (25,34) | 30 (26,34) | 31 (-183,246) | 19 |
|
|
||||||
| AUC (hr·ng/mL) | 772 (689,856) | 868 (727,1008) | 713 (609,817) | 721 (618,825) | 753 (-4393,5899) | 444 |
|
|
||||||
| EB/BUP | 1.15 (1.05,1.25) | 1.24 (1.07,1.42) | 1.09 (0.96,1.22) | 1.08 (0.93,1.23) | 1.30 (-0.48,3.08) | 0.99 |
|
| ||||||
| TB | ||||||
| Cmax (ng/mL) | 208 (181,236) | 241 (190,293) | 188 (157, 218) | 188 (159,216) | 222 (-9,453) | 122 |
|
|
||||||
| Css (ng/mL) | 175 (150,200) | 204 (153,255) | 158 (132,182) | 157 (132,182) | 190 (-1220,1599) | 108 |
|
|
||||||
| AUC (hr•ng/mL) | 4209 (3612,4807) | 4893 (3661,6126) | 3789 (3170, 4407) | 3775 (3178,4371) | 4549 (-29275,38373) | 2582 |
|
|
||||||
| TB/BUP | 6.21 (5.53,6.90) | 6.93 (5.53,8.32) | 5.77 (5.04, 6.50) | 5.62 (4.82,6.42) | 7.50 (3.78,11.23) | 5.73 |
Significant differences compared to no reduced function allele group
P = 0.005
P < 0.001
P = 0.01
Figure 2.
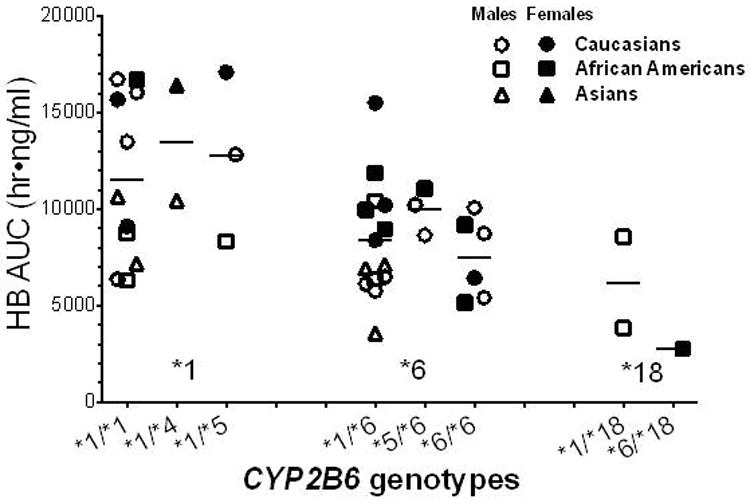
Area under the 24 hour hydroxybupropion (HB) concentration-time curve in individuals with different CYP2B6 genotypes, shown by sex and race.
Table 3.
24-hour Urine Bupropion and Metabolite Excretion
| 24-hour recovery (nmol, mean = 95%) | ||||||
|---|---|---|---|---|---|---|
| All (N=42) | No reduced function allele (N=16) | With reduced function allele(s) (N=26) | *6 allele (N= 23) | *18 allele (N= 2) | *6/18 allele (N= 1) | |
| BUP | 2087 (1512,2662) | 2568 (1169,3968) | 1790 (1346,2234) | 1800 (1305,2295) | 1301 (-3755,6356) | 2543 |
| HB – free | 3672 (2782,4561) | 4698 (2776,6619) | 3041 (2187,3895) | 3207 (2257,4157) | 1736 (884,2587) | 1824 |
| HB – gluc | 12283 (10393,14173) | 14902 (11602,18201) | 12283 (8450, 12892) | 11068 (8612,13525) | 9039 (-4153,22232) | 4787 |
| EB–free | 4872 (3753,5991) | 6186 (3634,8738) | 4063 (3107,5020) | 4067 (2996,5138) | 3711 (-15768,23191) | 4692 |
| EB – gluc | 1718 (1438,1998) | 1905 (1354,2456) | 1603 (1278,1928) | 1602 (1235,1969) | 1763 (-2118,5643) | 1304 |
| TB–free | 38376 (27180,49572) | 53034 (25558,80501) | 29356 (22366,36344) | 28990 (21286,36694) | 32492 (-163420,228404) | 31495 |
| TB – gluc | 4778 (2539,7017) | 7731 (2047,13415) | 2961 (1862,4060) | 2877 (1788,3966) | 5402 (-39287,50083) | 0 |
| % Daily Bupropion dose | ||||||
| All (N=42) | No reduced function allele (N=16) | With reduced function allele(s) (N=26) | *6 allele (N= 23) | *18 allele (N= 2) | *6/18 allele (N= 1) | |
| BUP | 0.33% (0.24%,0.43%) | 0.41% (0.19%,0.63%) | 0.29% (0.22%,0.36%) | 0.29% (0.21%,0.37%) | 0.21% (-0.60%,1.02%) | 0.41% |
| HB – free | 0.59% (0.45%,0.73%) | 0.75% (0.44%,1.06%) | 0.49% (0.35%,0.62%) | 0.51% (0.36%,0.67%) | 0.28% (0.14%,0.41%) | 0.29% |
| HB – gluc | 1.97% (1.66%,2.27%) | 2.38% (1.86%,2.91%) | 1.70% (1.35%,2.06%) | 1.77% (1.38%,2.16%) | 1.45% (-0.66%,3.56%) | 0.77% |
| EB–free | 0.78% (0.60%,0.96%) | 0.99% (0.58%,1.40%) | 0.65% (0.50%,0.80%) | 0.65% (0.48%,0.82%) | 0.59% (-2.52%,3.71%) | 0.75% |
| EB – gluc | 0.27% (0.23%,0.32%) | 0.30% (0.22%,0.39%) | 0.26% (0.20%,0.31%) | 0.26% (0.20%,0.32%) | 0.28% (-0.34%,0.90%) | 0.21% |
| TB–free | 6.14% (4.35%,7.93%) | 8.49% (4.09%,12.88%) | 4.70% (3.58%,5.82%) | 4.64% (3.41%,5.87%) | 5.20% (-26.18%,36.54%) | 5.04% |
| TB – gluc | 0.76% (0.41%,1.12%) | 1.24% (0.33%,2.15%) | 0.47% (0.30%,0.65%) | 0.46% (0.29%,0.64%) | 0.86% (6.29%,8.01%) | 0.00% |
| Renal clearance (ml/min/kg) | ||||||
| All (N=42) | No reduced function allele (N=16) | With reduced function allele(s) (N=23) | *6 allele (N= 23) | *18 allele (N= 2) | *6/18 allele (N= 1) | |
| BUP | 0.17 (0.12,0.21) | 0.18 (0.10,0.26) | 0.16 (0.10,0.21) | 0.16 (0.09,0.22) | 0.10 (-0.33,0.53) | 0.28 |
| HB | 0.02 (0.02,0.03) | 0.03 (0.02,0.04) | 0.03 (0.02,0.03) | 0.03 (0.02,0.03) | 0.01 (-0.04,0.06) | 0.04 |
| EB | 0.36 (0.28,0.45) | 0.41 (0.26,0.56) | 0.34 (0.24,0.43) | 0.34 (0.23,0.45) | 0.20 (-0.12,0.52) | 0.53 |
| TB | 0.50 (0.39,0.61) | 0.58 (0.36,0.81) | 0.44 (0.33,0.56) | 0.45 (0.32,0.58) | 0.29 (-0.14,0.73) | 0.62 |
Bold – significant difference compared to no reduced function allele group, p = 0.03.
Table 4 presents results of a regression analysis of factors that influence HB AUC and the HB/BUP ratio. The presence of the CYP2B6*6 allele was associated with significantly reduced HB concentrations and HB/BUP ratio. The CYP2B6*18 allele was associated with significantly reduced HB concentrations and borderline (p=0.056) lower HB/BUP ratio. Women had a significantly higher HB AUC than men, although this difference was not observed with HB/BUP ratios. Once CYP2B6*6 and CYP2B6*18 genotype and sex were accounted for in the regression model, there was no effect of African American race. Asians had a borderline lower level of HB AUC and HB/BUP ratio (P=0.05 and 0.058) which may reflect the presence of additional untested variant alleles.
Table 4.
Regression Analysis of Predictors
| Hydroxybupropion AUC‡ R2 = 0.50 (p < 0.001) | ||||
|---|---|---|---|---|
| Predictor | B | β | 95% CI | P |
| CYP2B6*6 (per # alleles) | -3049 | -0.56 | -4411, 1-687 | 0.001 |
| CYP2B6*18 (per # alleles) | -5095 | -0.34 | -9018, -1171 | 0.012 |
| Sex (female) | 2287 | 0.37 | 917, 4856 | 0.005 |
| Race (Asian) | -2071 | -0.2 | -4734, 592 | 0.123 |
| Race (African American) | -1667 | -0.21 | -3893, 559 | 0.138 |
| Hydroxybupropion/Bupropion R2 = 0.32 (p = 0.01) | ||||
| Predictor | B | β | 95% CI | P |
| CYP2B6*6 (per # alleles) | -4.22 | -0.51 | -6.60, -1.83 | 0.001 |
| CYP2B6*18 (per # alleles) | -6.69 | -0.30 | -13.56, 0.18 | 0.056 |
| Sex (female) | 1.88 | 0.16 | -1.57, 5.33 | 0.277 |
| Race (Asian) | -4.00 | -0.26 | -8.66, 0.66 | 0.090 |
| Race (African American) | -0.25 | -0.02 | -4.15, 3.65 | 0.897 |
AUC in hr*ng/mL
The B and β provided refers to the variables listed in parentheses beside each categorical predictor.
Discussion
We report several novel findings with respect to the disposition of bupropion in relation to CYP2B6 genotype. We present the first analysis of steady state concentrations of bupropion and its metabolites in relationship to genotype and race. We confirm findings from previous studies that CYP2B6*6 does not affect bupropion clearance, but does affect the generation of HB. [15] We present novel data on the functional significance of the CYP2B6*18 allele in people. We present novel data on renal excretion and clearance of bupropion and metabolites at steady state.
Bupropion is metabolized to HB primarily by CYP2B6, with possible contributions from CYP2E1 and CYP3A4 at high substrate concentrations.[8-11] Bupropion is also metabolized to hydroxylated metabolites other than HB by CYP2C19.[11] Bupropion metabolism to EB and TB are thought to be catalyzed by carbonyl reductase enzymes.[11]
The CYP2B6*6 results in a disruption of an exonic splicing enhancer, resulting in a lower amount of functional CYP2B6 mRNA and less protein expression. [17, 18, 20, 23] In addition amino acid changes may contribute to altered function of the protein[27]. In human liver samples, the CYP2B6*6 variant has been associated with reduced bupropion hydroxylation activities [19, 20], but in human studies this variant did not affect bupropion clearance. [15] Our findings on bupropion blood concentrations during steady state confirm the prior observations with single doses that while it substantially alters HB formation from bupropion, the CYP2B6*6 variant does not affect bupropion clearance. Presumably this is explained by the multiple metabolic pathways, other than via CYP2B6, through which bupropion is metabolized.
Our results extend the in vitro and in vivo results of other studies that the concentrations of HB are substantially reduced among those with at least on CYP2B6*6 allele.[15, 16, 19, 25] We found that the HB/BUP plasma AUC ratio decreases in a gene-dose related manner. The magnitude of the effect is substantial with the AUC of HB being decreased by 50% in CYP2B6*6 homozygotes.
While both bupropion and HB are active, the blood concentrations of HB at steady state are 10-30 times higher than that of bupropion. The pharmacologic effect of a drug is determined by the free drug concentration, which is the product of total blood concentration and % unbound. The % unbound for bupropion is 16% and for HB is 23%[1]. Given the much higher steady state concentration and the greater % unbound, HB is likely to contribute most of the pharmacologic activity of bupropion. The reduction of 50% in HB activity in the CYP2B6*6 homozygotes is expected to have a substantial impact on the pharmacologic activity and therapeutic efficacy of bupropion; this may alter drug dosing and/or therapeutic response more substantially in some populations compared to others as CYP2B6*6 homozygotes range from 3% in Asians to over 25% in Yupik peoples.[14]
A limitation of our kinetic study is that we did not measure enantiomers of bupropion or its metabolites. Bupropion is marketed as a racemic mixture. [28] After dosing with a racemic mixture, the blood concentrations of the R- are 3-fold higher than those of S-bupropion[29]. Similarly, HB is enantiomeric, with 2S,3S-HB having greater potency than the 2R,3R- HB at nicotine receptors, while blood concentrations of the 2R,3R are higher than those of 2S,3S after a single dose.[3-5]
Racial differences in the prevalence of the CYP2B6*6 allele are known: approximately 25% of Asians, 45% of Caucasians, 50% of African Americans have at least one CYP2B6*6 allele.[12, 13, 25] We are aware of no prior research examining the impact of the CYP2B6*6 variant on bupropion kinetics in people of different races. In a multiple regression analysis, race had no effect on the HB/BUP ratio when genotype was included in the model, suggesting that the impact of the variant genotypes is similar across races.
Three of our African American subjects had the CYP2B6*18 variant. While the number of subjects is small, in multiple regression analysis this variant was significantly associated with reduced HB AUC and borderline significantly associated with reduced HB/BUP ratio. The single nucleotide polymorphism that characterizes the CYP2B6*18 variant causes a change in amino acid sequence which leads to very low CYP2B6 protein expression in mammalian and yeast expression systems[21, 30]. This suggests that CYP2B6*18 is the causal variant for pharmacokinetic effects, although we cannot exclude the possibility that CYP2B6*18 has other additional variants in linkage disequilibrium specific to African ancestry. The impact of this allele appears to be greater than the reduction of function found with CYP2B6*6, although with observations in just three subjects this conclusion remains tentative. Based on the allele frequencies of *6 and *18 (6% and 35%, respectively)[25], approximately 16% of African Americans have two copies of either the *6 and/or *18 allele and 49% of African Americans have one copy of either the *6 or *18 allele. This suggests that a large proportion of African Americans have reduced metabolism for CYP2B6 substrates.
We measured bupropion and its major metabolites including glucuronide conjugates in a 24 hour urine at steady state. This allowed us to determine the percent recovery of the daily bupropion dose. Of the daily bupropion dose we could account for only 0.3% as unchanged bupropion and about 10% as the sum of all of measured metabolites in urine. These findings are similar to those seen with administration of C14 labeled bupropion.[1] Petsalo has identified 20 metabolites of bupropion in human urine, with the most abundant being m-chlorohippuric acid. [26] Thus, most of the metabolites of bupropion excreted in urine are not the main metabolites measured in plasma.
The most prevalent urine metabolite was TB, followed by HB and EB, with relatively low concentrations of bupropion. As expected, subjects with the CYP2B6*6 variant excreted significantly less HB compared with those without any reduced function variants. Petsalo identified 8 glucuronide conjugates of bupropion metabolites, including conjugates of HB, TB and EB. We provide the first human data indicating the glucuronidation is greatest for HB, less for TB and EB, and minimal or none for bupropion.
The renal clearances of bupropion and its metabolites were all low, much lower than glomerular filtration rate. The lowest clearances were for bupropion and HB, most likely related at least in part to a high degree of protein binding (84% and 77%, respectively). [1] The renal clearance of TB and EB were higher, consistent with a lower extent of plasma protein binding. Our data on renal clearance of bupropion may have forensic utility, allowing extrapolation from urine concentrations to plasma concentrations at steady state.
With respect to using genotyping to personalize treatment, our data indicate that people who have CYP2B6*6 and CYP2B6*18 variants have reduced concentrations of HB and would be expected to have reduced pharmacologic activity from a given dose of bupropion. Consistent with this idea, Zhu et al recently reported that when bupropion is used for smoking cessation in African American smokers, the quit rates were associated with plasma HB but not bupropion concentrations.[25] As expected the HB concentrations were lower in smokers who have reduced function CYP2B6 variants. It was speculated that smokers with reduced HB formation might need higher doses of bupropion to achieve the same smoking cessation effect, and possibly for its antidepressant effects, compared to those who are normal metabolizers.
Our paper focuses on the effects of the CYP2B6 genotype on bupropion metabolism. CYP2B6 is also involved in the metabolism of other drugs, such as efavirenz, cyclophosphamide, artemisinin, ketamine, propofol, methadone and selegeline. Studies with efavirenz have shown that the CYP2B6*6 variant is associated with higher concentrations of parent drug and increased toxicity.[31] This finding of reduced enzymatic activity is consistent with our finding that the CYP2B6*6 variant is associated with reduced generation of the bupropion metabolite, HB. Whether a person with a CYP2B6*6 allele requires dose reduction (efavirenz) or dose escalation (possibly bupropion) depends on the extent to which the parent drug vs metabolite mediates pharmacologic activity.
In conclusion we demonstrate for CYP2B6*6 and provide strong evidence for CYP2B6*18 that these gene variants are associated with reduced concentrations of the active metabolite HB at steady state, with no effect of concentrations of bupropion or other metabolites. The effect of the gene variants appears to be independent of race. Our data provide quantitative estimates of gene variant effects, with our model including sex accounting for 50% of the variation in HB concentrations. There is wide variability in HB concentrations within any particular genotype, likely due to additional untested alleles, potential impact of CYP2B6 inducers, and the known impact of sex. Therapeutic drug monitoring should be evaluated as a way to optimize therapeutic dosing.
Supplementary Material
Acknowledgments
We thank Dr Faith Allen for data management, Dr Delia Dempsey for medical supervision, Cotys Winston for clinical research coordination, Olivia Yturralde, and Lisa Yu for analytical chemistry, Ewa Hoffmann for laboratory research coordination, and Marc Olmsted for editorial assistance.
FUNDING
Studies described in this report were funded by National Institutes of Health grants DA U01 020830, CA 78603, DA 12393, NCRR UCSF CTSI UL1 RR 024131 and by Canadian Institute of Health (CIHR) operating grants MOP#86471 and TMH-109787 and the Canada Foundation for Innovation (#20289 and #16014), the CAMH Foundation and the Ontario Ministry of Research and Innovation. Dr. Tyndale was supported in part by CAMH.
Footnotes
Conflict of interest statements
Dr Benowitz consults with Pfizer, McNeil and GlaxoSmithKline on smoking cessation medications and has provided paid expert testimony concerning nicotine addiction in litigation against tobacco companies.
Dr. Tyndale has participated in one day advisory meetings for Novartis and McNeil.
References
- 1.Johnston AJ, Ascher J, Leadbetter R, Schmith VD, Patel DK, Durcan M, et al. Pharmacokinetic optimisation of sustained-release bupropion for smoking cessation. Drugs. 2002;62(Suppl 2):11–24. doi: 10.2165/00003495-200262002-00002. [DOI] [PubMed] [Google Scholar]
- 2.Jefferson JW, Pradko JF, Muir KT. Bupropion for major depressive disorder: Pharmacokinetic and formulation considerations. Clin Ther. 2005;27:1685–95. doi: 10.1016/j.clinthera.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 3.Damaj MI, Carroll FI, Eaton JB, Navarro HA, Blough BE, Mirza S, et al. Enantioselective effects of hydroxy metabolites of bupropion on behavior and on function of monoamine transporters and nicotinic receptors. Mol Pharmacol. 2004;66:675–82. doi: 10.1124/mol.104.001313. [DOI] [PubMed] [Google Scholar]
- 4.Damaj MI, Grabus SD, Navarro HA, Vann RE, Warner JA, King LS, et al. Effects of hydroxymetabolites of bupropion on nicotine dependence behavior in mice. J Pharmacol Exp Ther. 2010;334:1087–95. doi: 10.1124/jpet.110.166850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bondarev ML, Bondareva TS, Young R, Glennon RA. Behavioral and biochemical investigations of bupropion metabolites. Eur J Pharmacol. 2003;474:85–93. doi: 10.1016/s0014-2999(03)02010-7. [DOI] [PubMed] [Google Scholar]
- 6.Grabus SD, Carroll FI, Damaj MI. Bupropion and its Main Metabolite Reverse Nicotine Chronic Tolerance in the Mouse. Nicotine Tob Res. 2012 doi: 10.1093/ntr/nts088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Findlay JW, Van Wyck Fleet J, Smith PG, Butz RF, Hinton ML, Blum MR, et al. Pharmacokinetics of bupropion, a novel antidepressant agent, following oral administration to healthy subjects. Eur J Clin Pharmacol. 1981;21:127–35. doi: 10.1007/BF00637513. [DOI] [PubMed] [Google Scholar]
- 8.Hesse LM, Venkatakrishnan K, Court MH, von Moltke LL, Duan SX, Shader RI, et al. CYP2B6 mediates the in vitro hydroxylation of bupropion: potential drug interactions with other antidepressants. Drug Metab Dispos. 2000;28:1176–83. [PubMed] [Google Scholar]
- 9.Faucette SR, Hawke RL, Lecluyse EL, Shord SS, Yan B, Laethem RM, et al. Validation of bupropion hydroxylation as a selective marker of human cytochrome P450 2B6 catalytic activity. Drug Metab Dispos. 2000;28:1222–30. [PubMed] [Google Scholar]
- 10.Faucette SR, Hawke RL, Shord SS, Lecluyse EL, Lindley CM. Evaluation of the contribution of cytochrome P450 3A4 to human liver microsomal bupropion hydroxylation. Drug Metab Dispos. 2001;29:1123–9. [PubMed] [Google Scholar]
- 11.Chen Y, Liu HF, Liu L, Nguyen K, Jones EB, Fretland AJ. The in vitro metabolism of bupropion revisited: concentration dependent involvement of cytochrome P450 2C19. Xenobiotica. 2010;40:536–46. doi: 10.3109/00498254.2010.492880. [DOI] [PubMed] [Google Scholar]
- 12.Zanger UM, Klein K, Saussele T, Blievernicht J, Hofmann MH, Schwab M. Polymorphic CYP2B6: molecular mechanisms and emerging clinical significance. Pharmacogenomics. 2007;8:743–59. doi: 10.2217/14622416.8.7.743. [DOI] [PubMed] [Google Scholar]
- 13.Lee AM, Jepson C, Hoffmann E, Epstein L, Hawk LW, Lerman C, et al. CYP2B6 genotype alters abstinence rates in a bupropion smoking cessation trial. Biol Psychiatry. 2007;62:635–41. doi: 10.1016/j.biopsych.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Binnington MJ, Zhu AZ, Renner CC, Lanier AP, Hatsukami DK, Benowitz NL, et al. CYP2A6 and CYP2B6 genetic variation and its association with nicotine metabolism in South Western Alaska Native people. Pharmacogenet Genomics. 2012;22:429–40. doi: 10.1097/FPC.0b013e3283527c1c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirchheiner J, Klein C, Meineke I, Sasse J, Zanger UM, Murdter TE, et al. Bupropion and 4-OH-bupropion pharmacokinetics in relation to genetic polymorphisms in CYP2B6. Pharmacogenetics. 2003;13:619–26. doi: 10.1097/00008571-200310000-00005. [DOI] [PubMed] [Google Scholar]
- 16.Loboz KK, Gross AS, Williams KM, Liauw WS, Day RO, Blievernicht JK, et al. Cytochrome P450 2B6 activity as measured by bupropion hydroxylation: effect of induction by rifampin and ethnicity. Clin Pharmacol Ther. 2006;80:75–84. doi: 10.1016/j.clpt.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 17.Lamba V, Lamba J, Yasuda K, Strom S, Davila J, Hancock ML, et al. Hepatic CYP2B6 expression: gender and ethnic differences and relationship to CYP2B6 genotype and CAR (constitutive androstane receptor) expression. J Pharmacol Exp Ther. 2003;307:906–22. doi: 10.1124/jpet.103.054866. [DOI] [PubMed] [Google Scholar]
- 18.Hofmann MH, Blievernicht JK, Klein K, Saussele T, Schaeffeler E, Schwab M, et al. Aberrant splicing caused by single nucleotide polymorphism c.516G>T [Q172H], a marker of CYP2B6*6, is responsible for decreased expression and activity of CYP2B6 in liver. J Pharmacol Exp Ther. 2008;325:284–92. doi: 10.1124/jpet.107.133306. [DOI] [PubMed] [Google Scholar]
- 19.Hesse LM, He P, Krishnaswamy S, Hao Q, Hogan K, von Moltke LL, et al. Pharmacogenetic determinants of interindividual variability in bupropion hydroxylation by cytochrome P450 2B6 in human liver microsomes. Pharmacogenetics. 2004;14:225–38. doi: 10.1097/00008571-200404000-00002. [DOI] [PubMed] [Google Scholar]
- 20.Desta Z, Saussele T, Ward B, Blievernicht J, Li L, Klein K, et al. Impact of CYP2B6 polymorphism on hepatic efavirenz metabolism in vitro. Pharmacogenomics. 2007;8:547–58. doi: 10.2217/14622416.8.6.547. [DOI] [PubMed] [Google Scholar]
- 21.Klein K, Lang T, Saussele T, Barbosa-Sicard E, Schunck WH, Eichelbaum M, et al. Genetic variability of CYP2B6 in populations of African and Asian origin: allele frequencies, novel functional variants, and possible implications for anti-HIV therapy with efavirenz. Pharmacogenet Genomics. 2005;15:861–73. doi: 10.1097/01213011-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Laizure SC, DeVane CL. Stability of bupropion and its major metabolites in human plasma. Ther Drug Monit. 1985;7:447–50. doi: 10.1097/00007691-198512000-00015. [DOI] [PubMed] [Google Scholar]
- 23.Al Koudsi N, Tyndale RF. Hepatic CYP2B6 is altered by genetic, physiologic, and environmental factors but plays little role in nicotine metabolism. Xenobiotica. 2010;40:381–92. doi: 10.3109/00498251003713958. [DOI] [PubMed] [Google Scholar]
- 24.Haas JS, Kaplan CP, Barenboim D, Jacob P, 3rd, Benowitz NL. Bupropion in breast milk: an exposure assessment for potential treatment to prevent post-partum tobacco use. Tob Control. 2004;13:52–6. doi: 10.1136/tc.2003.004093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu AZ, Cox LS, Nollen N, Mayo M, Faseru B, Okuyemi KS, et al. CYP2B6 and bupropion’s smoking cessation pharmacology: the role of hydroxybupropion. Clin Pharmacol Ther. 2012;96:771–7. doi: 10.1038/clpt.2012.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Petsalo A, Turpeinen M, Tolonen A. Identification of bupropion urinary metabolites by liquid chromatography/mass spectrometry. Rapid Commun Mass Spectrom. 2007;21:2547–54. doi: 10.1002/rcm.3117. [DOI] [PubMed] [Google Scholar]
- 27.Zhang H, Sridar C, Kenaan C, Amunugama H, Ballou DP, Hollenberg PF. Polymorphic variants of cytochrome P450 2B6 (CYP2B6.4-CYP2B6.9) exhibit altered rates of metabolism for bupropion and efavirenz: a charge-reversal mutation in the K139E variant (CYP2B6.8) impairs formation of a functional cytochrome p450-reductase complex. J Pharmacol Exp Ther. 2011;338:803–9. doi: 10.1124/jpet.111.183111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu H, Loboz KK, Gross AS, McLachlan AJ. Stereoselective analysis of hydroxybupropion and application to drug interaction studies. Chirality. 2007;19:163–70. doi: 10.1002/chir.20356. [DOI] [PubMed] [Google Scholar]
- 29.Kharasch ED, Mitchell D, Coles R. Stereoselective bupropion hydroxylation as an in vivo phenotypic probe for cytochrome P4502B6 (CYP2B6) activity. J Clin Pharmacol. 2008;48:464–74. doi: 10.1177/0091270008314254. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, Sonnerborg A, Rane A, Josephson F, Lundgren S, Stahle L, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genomics. 2006;16:191–8. doi: 10.1097/01.fpc.0000189797.03845.90. [DOI] [PubMed] [Google Scholar]
- 31.Ngaimisi E, Mugusi S, Minzi O, Sasi P, Riedel KD, Suda A, et al. Effect of rifampicin and CYP2B6 genotype on long-term efavirenz autoinduction and plasma exposure in HIV patients with or without tuberculosis. Clin Pharmacol Ther. 2011;90:406–13. doi: 10.1038/clpt.2011.129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


