Abstract
Genomic sequencing is becoming accurate, fast, and inexpensive, and is rapidly being incorporated into clinical practice. Incidental findings, which result in large numbers from genomic sequencing, are a potential barrier to the utility of this new technology due to their high prevalence and the lack of evidence or guidelines available to guide their clinical interpretation. This unit reviews the definition, classification, and management of incidental findings from genomic sequencing. The unit focuses on the clinical aspects of handling incidental findings, with an emphasis on the key role of clinical context in defining incidental findings and determining their clinical relevance and utility.
Introduction
Since the completion of the Human Genome Project in 2001, there has been a rapid expansion of knowledge about human DNA structural and sequence variation. Even more exciting for the practice of medicine, there is an increasing understanding of the connections between genetic variation and human health signifying that the long anticipated genomic revolution is underway (Feero et al., 2010; Guttmacher et al., 2010). However, genomic technologies remain expensive, the interpretation of large-scale genomic data is difficult, and most practicing physicians have no experience in applying genomic knowledge and data in medical practice.
Examples of the medical use of wholeexome and whole-genome sequencing (GS) are becoming increasingly common, and many innovative applications of sequencing technologies have been undertaken. Hundreds of pathogenic mutations identified through genomic sequencing have been reported in recent years (Gilissen et al., 2011; Gonzaga-Jauregui et al., 2012). Laboratories providing GS for a variety of diagnostic indications have estimated that they find a causative mutation in 27% of cases (Eng et al., 2012) and a preliminary report of GS for developmental delay claimed a 15% to 35% diagnostic rate in identifying the genetic cause (Kogelenberg et al., 2012). Evaluation of sick neonates may benefit from point of care diagnostics using rapid turnaround sequencing and interpretation (Saunders et al., 2012). In some cases, identification of a causative mutation through genome sequencing has helped to formulate a treatment plan and in other cases offered new opportunities for reproductive planning. One of the earliest, and most publicized, cases involved identification of an unexpected mutation in the X-linked inhibitor of apoptosis gene and subsequent life-saving treatment plan in a young boy with inflammatory bowel disease (Worthey et al., 2011).
In addition to diagnostic uses, there will be many other applications of GS. One area of great excitement is the sequencing of tumors to individualize cancer treatments (Macconaill and Garraway, 2010; McDermott et al., 2011). Sequencing can also rapidly and comprehensively identify recessive conditions for use in family planning. While carrier screening is already used in clinical practice to a limited extent (e.g., focused carrier screening in individuals of Ashkenazi Jewish descent), genomic technologies will allow broader, more comprehensive screening to those who seek it. Additionally, genomic sequencing is a powerful tool to screen for multiple pharmacogenomic variants simultaneously, creating the opportunity for personalized medication selection and dosing regimens based on an individual’s genotype (Wiita and Schrijver, 2011). Patients will look to their physicians for insight and advice on how to utilize genomic technologies for all of these applications (McGuire et al., 2009; Gollust et al., 2012), but neither doctors nor the health care infrastructure are well prepared. Among the problems to be solved are the integration of genomic data into electronic medical records, generating more detailed and useful phenotypic data, sharing genomic and phenotypic data across institutions, refining automated translational tools, and integrating new discoveries quickly into clinical practice (Cordero and Ashley, 2012).
In all of these clinical applications of genomic technologies, one of the most pressing concerns is the challenge posed by incidental (or secondary) findings. Incidental findings in genomics may be defined as genomic variants of potential medical relevance unrelated to the medical reason for ordering the test (Green et al., 2013a). This unit explores the definitions of incidental findings in medical practice, the scope of the issue in clinical GS, the molecular characterization of incidental findings, and approaches to classifying and communicating incidental findings. Additionally, we will consider the role of clinical context in handling incidental findings and address the issues facing clinicians given the paucity of existing guidelines and recommendations on when and how to report incidental findings.
What are Genomic Incidental Findings?
Deciding what is, and what is not, an incidental finding is not always straightforward and may be specific to a given clinical situation. The use of the term “incidental” implies that GS is being ordered for a specific diagnostic indication, such as a particular disease phenotype in a patient or a family history of a suspected genetic disease. While genetic variants considered relevant to the diagnosis would be reported as a primary result, any finding unrelated to the specific diagnostic indication would be incidental. If, on the other hand, GS is obtained for a healthy individual without any personal or family history of disease, then all clinically meaningful findings in the sequence could be considered to be incidental. In addition to these two extremes, when sequencing becomes more common in clinical medicine, one can imagine patients with vague family histories, or ambiguous clinical findings, undergoing GS. The clinically relevant discoveries made under these circumstances could be considered “quasi-incidental.” Such findings would lack the typical prior probabilities that are used in conventional genetic testing, but would nevertheless be the result of more directed attention to a particular disease domain than in entirely healthy individuals without a relevant family history.
Characterizing incidental findings is also complicated because not all clinicians would agree on whether a given variant is clinically meaningful (Green et al., 2012b). Some practitioners might argue that a variant with an uncertain association to a tiny fraction of increased risk for a given condition should nonetheless be considered as medically important and communicated to the patient. Others might argue that only clinically validated findings of medically actionable results would be worthy of note in a clinical context. There are no data available on the downstream risks and benefits of disclosing incidental genomic findings at all, much less with varying degrees of supporting evidence. Thus, there is no standard for the degree of evidence necessary to consider an incidental finding worthwhile to be reported.
Incidental findings are not new to medicine and parallel medical situations may offer useful insights. For example, there is an extensive literature of incidental findings in radiology (Ahmed et al., 2010; Alpert and Naidich, 2011; van Vugt et al., 2012). One study reviewing whole-body CT scans found an incidental finding rate of 86% (Furtado et al., 2005), and review and analysis of 44 publications on incidental findings suggested that the overall frequency of incidental findings in radiological studies was 23.6% (Lumbreras et al., 2010). The field of radiology has incorporated the broad examination of radiological films and the documentation of any incidental findings into day-to-day practice. When examining a particular radiographic study, a radiologist is obligated by current standards of care to examine the entire film and report abnormalities whether or not they are linked to the reason for which the film was ordered. Similar standards have not yet been agreed upon for the evaluation of GS. In performing GS, procedures have been piloted that would allow sequencing and interpretation to be focused upon the specific phenotype of the patient (Saunders et al., 2012) and to ignore other parts of the genome. Nonetheless, most molecular laboratories will soon be using interpretive pipelines to analyze their clinical sequence data that make it relatively easy to examine all disease-associated genes in the exome or genome, and depending upon the filtration parameters of the pipeline, one could observe dozens to hundreds of disease-associated variants in each sequence (Ashley et al., 2010; Xue et al., 2012). Under these circumstances, there is increasing concern that a deluge of incidental findings will overwhelm a clinician’s efforts to use sequencing to answer specific clinical questions and negate some of the benefits of GS.
The problem of incidental findings is one manifestation of the rapidly evolving knowledge base in genetics (Feero et al., 2010; Varmus, 2010). For some, this is a reason to reject the integration of incidental findings in clinical practice until evidence about such information is better established. Yet, it is important to remember that physicians are accustomed to managing patients in spite of an incomplete evidence base. A wealth of literature has explored the gradual development of reliable evidence in clinical medicine (Downing, 2009; Petitti et al., 2009), and there are many examples of the necessity of utilizing tests and tools in medicine with insufficient evidence (Travis, 2006; Brauer and Bozic, 2009; Moussa, 2011). Thus, genomic incidental findings represent a new type of clinical information for which clinicians will decipher with their existing expertise.
Genetic Variation and the Limitations of Genomic Sequencing
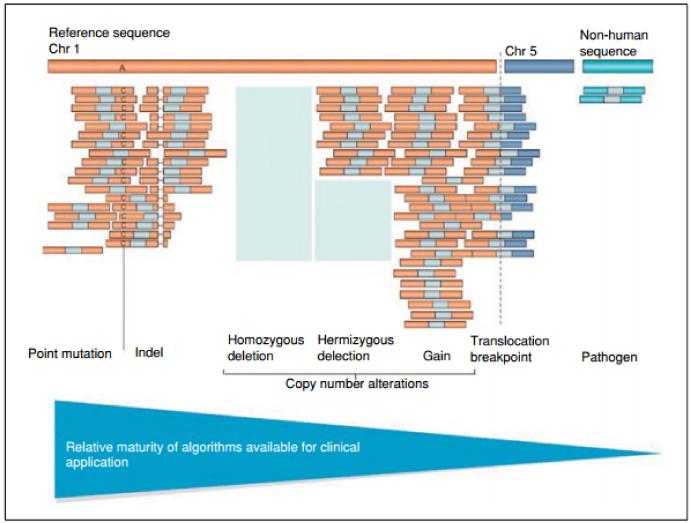
In considering the issues surrounding the interpretation and management of incidental findings in clinical GS, it is important to understand the types of variants that can be detected effectively through GS. The most common type of variant that can be accurately detected by GS today is a single nucleotide substitution, and these will be the primary focus of this unit. Other common forms of variation include “indels” or small insertions and deletions, from 1 to 100 base pairs in length. Indels can only be detected in GS if the programs for computational alignment of short DNA sequences are tolerant of the missing or added bases. The detection of even larger structural changes or of repeat elements in the genome is even more challenging, though gains or losses of large segments of genomic material in an individual can potentially be inferred from regional coverage differences compared to a reference. Bioinformatics tools to more sensitively detect small and large structural change are under active development and improved sequencing coverage, and refined analytical algorithms are expected. Figure 9.23.1 depicts the different types of variation detectable by GS and the relative maturity of the detection technologies being applied to these classes (Meyerson et al.,2010).
Figure 9.23.1.
Detecting types of genitic variation by whole genome sequencing (from Meyerson et al., 2010).
Mendelian and Common Variants
Another way to think about disease associated genetic variation is the degree to which the disease phenotype is, or is not, strongly associated with that variant. Early understanding of genetic disorders and gene discovery relied heavily on diseases with Mendelian inheritance patterns, i.e., classic dominant, recessive, and X-linked disorders. Whether or not the variant was always associated with signs or symptoms defined the “penetrance” of the variant, while the extent or severity of the clinical features defined the “expressivity” of the variant. Rare variants that are highly penetrant and highly expressive in clinical practice were among the earliest to be discovered and managed by practitioners in clinical genetics, and these precedents have colored both internal and external perceptions about the entire field of genetics with the aura of determinism. Mendelian variants were discovered and understood largely in the context of patients with a particular phenotype. Thus, the search for these variants in unaffected individuals usually took place where there was a known family history that implicated a particular pattern of inheritance. As discussed further below, evaluating Mendelian variants discovered as incidental findings is challenging because penetrance estimates derived from affected families may be much higher than for unaffected families.
In addition to highly penetrant Mendelian variants, extensive research using genomewide association studies has accelerated the discovery of genetic variants that typically confer very modest risk of diseases like diabetes or heart disease. Unlike Mendelian variants, neither the diseases nor the variants under study are rare. A subset of these common variants is associated with drug effectiveness, sensitivity, and risk of side effects. These pharmacogenomic variants could allow for more accurate initial drug dosing, ongoing management, and risk profiling for adverse events (Relling et al., 2010; Relling and Klein, 2011). Both common disease risk variants and pharmacogenomic variants are typically associated with very modest effect sizes, making it unclear at times whether they alter risk profiles in any medically meaningful way. Moreover, they are usually derived from studies that may be restricted to a single population or ethnic group, and no information may exist on persons outside that group. Thus, incidental findings associated with common variants may also be difficult to interpret.
Filtering Variants in Genome Sequencing
As described in the latest release of 1000 Genomes data, an individual genome contains an average of 3.6 million single nucleotide variations, 344,000 indels, and 717 large deletions (Abecasis et al., 2012). In a recent study that evaluated nine whole genome sequences on healthy patients, 3.6 to 3.9 million genomic variants were found, the majority of which were single-nucleotide polymorphisms and more than 30,000 (less than 1%) fell in coding regions in each patient (Kohane et al., 2012). Additionally, 12% of the variations consisted of insertions, deletions, or multiple-base substitutions. Yet if this many variations occur in every genome, how can we decide which ones to be concerned about? The answer is that variants in GS are typically filtered by comparison against common variation in the population and in comparison to disease-associated databases.
The clinical context in which GS is ordered is a key factor in determining how such a large number of variants are filtered and what subset of variants might be included in both the primary and incidental portion of a clinical report. In the most common current scenario, GS is ordered to search for a cause of a rare disease in a symptomatic patient; therefore, one of the first steps is to filter out variants that are common (usually >1% frequency) and known to be benign. Using databases of known, common variation in generally healthy controls, such as 1000Genomes and dbSNP, a large variant list from an individual genome can be reduced by ~90% (Bainbridge et al., 2011; Worthey et al., 2011). A further step in the filtering of such a large number of variants involves comparing the results of GS in a patient with databases of disease-associated mutations (Kohane et al., 2006; Stenson et al., 2009; Fokkema et al., 2011; Samuels and Rouleau, 2011). This step is problematic at the present time because such databases are generally acknowledged to be replete with errors.
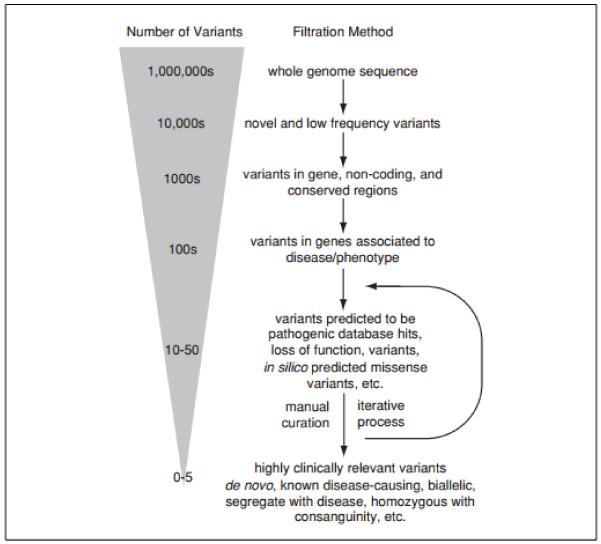
Additional filtering can help further reduce the number of variants, depending on the clinical context and availability of parental genomes, including selecting for novel variants, de novo variants, protein truncating variants, and missense variants, homozygous variants in the context of consanguineous families, biallelic variants where recessive inheritance is suspected, and variants in genes with expression patterns that match the involved organ sites. Additional in silico tools can be employed to filter variants for those that would be predicted to be deleterious to protein structure (Adzhubei et al., 2010; Kumar et al., 2009; Ng and Henikoff, 2003). See Figure 9.23.2 for a visual representation of the whole genome sequence analytical process and estimated number of variants at each stage.
Figure 9.23.2.
Variant frequency and filtering layers in whole genome sequencing (from Green, et al., 2013a)
Significant limitations exist, however, in these strategies and in the many filtering tools and databases used for variant analysis. When frequency filters are used, there is an assumption that affected individuals will not be included in the populations used for comparison. This may be true if the genetic disease in question is a fatal disease of childhood, but might not be at all true if the disease in question has more benign or late-onset presentations. Thus, single-nucleotide polymorphism (SNP) databases intended to document benign variation, such as dbSNP, undoubtedly include pathogenic variants due to the inclusion of data from persons with symptomatic disease, or due to the inclusion of data from individuals who have not yet manifested disease. Conversely, databases of pathogenic mutations, such as Human Genetic Mutation Database, have been shown to include a large proportion of common, benign variants that were incorrectly classified as pathogenic based on small, biased studies in disease cohorts (Tong et al., 2011). In addition, computational tools that predict protein structure and function are not fully validated, and results from these tools require caution in using them to independently define variant pathogenicity (Jordan et al., 2011). Finally, there is a paucity of data and tools to aid in the interpretation of rare intronic variants. The recently published ENCODE project findings advanced knowledge of intronic function significantly, but clinical application of these data have yet to be realized (ENCODE Project Consortium, 2012).
The Incidentalome
Given the enormous amount of variation in the human genome and the limitations described above regarding the interpretation of variants possibly associated with human disease, there is a large potential for overinterpretation of incidental findings. Kohane and co-authors outlined multiple mechanisms by which genomic sequencing can lead to falsepositive incidental findings, including: measurement error rate in genomescale sequencing, inherent errors in testing multiple hypotheses across millions of variants, mutations incorrectly documented as pathogenic, and incorrect assignment of prior probabilities for the interpretation of variants (Kohane et al., 2012). To demonstrate this point, they analyzed the genomes of nine reportedly healthy individuals, yielding large numbers of apparently pathogenic variants (see Table 9.23.1 below). The large numbers of “pathogenic” findings of questionable validity and utility has been labeled the “incidentalome” (Kohane et al., 2006).
Table 9.23,1.
Results of Annolation Filler Application lo Nine Reportedly Healthy Individuals of European Descent (from Kohane el al_, 5012)
| Filtering | Number of genes/individual |
Number of variants/individual |
|---|---|---|
| Genes with rare/novel, nonsynonymous mutations as highly conserved loci |
400±10 | 455±9 |
| Genes with rare/novel, nonsynonymous mutations at highly conserved loci and predicted to be deleterious |
I36±6 | I47±6 |
| Genes with mutations implicated in disease | 199±3 | 22fi±4 |
| Genes with homozygous mutations implicated in disease |
55±2 | 59± 2 |
| Genes with rare/novel mutations implicated in disease | 3±0 | 4±0 |
| Genes with rare/novel mutations implicated in disease and predicted to be deleterious |
2 ±0 | 2±0 |
As new pathogenic variants are discovered, the challenge of handling incidental findings becomes even larger in scope. We have estimated that based upon currently published recommendations for the return of genomic research results, there are an estimated 4,000 to 17,000 variants that might be considered for return and that this number could grow by 36% over the next four years based on the rate of new variant identification and the inclusion of variants associated with variable disease expression (Cassa et al., 2012).
A Clinical Approach to Incidental Findings
Should Incidental Findings be reported at all?
Given the limitations described above, it is reasonable to ask whether incidental findings should ever be reported in clinical GS. One intriguing argument against reporting of incidental findings was raised in a set of recommendations on the future of GS in the United Kingdom’s National Health Service (NHS) (Wright et al., 2011). These writers suggested a definitional distinction between an “assay,” i.e., the sequencing of an individual’s genome, and a “test,” i.e., analyzing a particular portion of the genome for clinical purposes. In their view, the assay contains no medical information per se, whereas the test is an interpretative step requiring purposeful analysis that can only be directed by clinical symptoms or by prior knowledge of the likely pathogenicity of variants. The report maintains that analyzing genomes beyond what is strictly called for by the clinical context represents “opportunistic screening” that has no scientific basis, and that the NHS does not have an obligation to support this activity in the absence of demonstrated clinical utility.
The issue of whether or not to report incidental findings is further complicated by the availability of genetic information from personal genomics (or direct-to-consumer genetic testing) companies that operate outside of the medical establishment, and the controversies surrounding medical utility, the role of medical expertise, and individual autonomy that these services have provoked (Evans and Green, 2009; Frueh et al., 2011). A thorough discussion of this issue is outside the scope of this unit, but it is worth noting that most, if not all, of the health-related information provided by personal genomics companies is incidental. While some customers of personal genomics companies have family histories of relevant conditions that spark their interest, most surveys of customers reveal that curiosity, along with a general desire to improve their health, are the most powerful motivators (Gollust et al., 2012; Green et al., 2012a). The phenomenon of personal genomics companies has highlighted the fact that many individuals are interested in learning genetic information about themselves and their children, whether or not such information leads to prevention. In a society that is trending towards self-empowerment through Internet mediated information-seeking, the growing desire for genetic information is sure to influence the question of what patients will request from their doctors when GS is performed.
Opinions on whether it is appropriate to seek and report incidental findings in clinical GS often depend upon whether one believes that such findings have clinical utility and whether such utility is cost-effective. Clinical utility refers to the likelihood that the information could or would be used to make a meaningful medical intervention with an appropriate balance of risks, benefits, and costs. Clinical utility is best defined by empirical research, and such research has not been conducted to determine the downstream benefits, risks or costs of disclosing unexpected results from GS. This is a difficult area to study, since the penetrance and expressivity of variants discovered by screening have not been ascertained for most variants and are almost universally unknown. In the absence of empirical data, policies about seeking and returning incidental findings in GS may be guided by best practices in the clinical environment. But since GS technologies are so new, and because there is such limited clinical experience in discovering and reporting primary or incidental findings in GS, the clinical environment offers little help. To add to the uncertainty about incidental findings, there are prevailing clinical traditions and ethical norms, as well as strongly held opinions from a variety of experts, which are sometimes in conflict. Genetic testing for adultonset genetic diagnoses in children has traditionally been avoided, and this position has been reinforced by several policy recommendations (Boards of Directors, American Society of Human Genetics and American College of Medical Genetics, 1995; Committee on Bioethics [American Academy of Pediatrics], 2001). Genetic testing for recessive traits has the potential to influence reproductive choices, including termination of pregnancies, and this is an area of extreme sensitivity. In addition, as we have shown in a series of clinical trials carried out over the last decade, implications of primary or incidental genetic information may vary greatly depending upon the age, the family history or the psychology of the individual (Roberts et al., 2011; Lautenbach et al., 2013). The downstream medical procedures, costs, and iatrogenic consequences of predictive genetic information revealed by incidental findings are only recently being explored using empirical studies and remain largely unknown.
Incidental findings in GS are different from incidental findings from other medical tests due to their potential significance for family members. For example, the implications of incidentally discovering a highly penetrant cancer risk variant in a middle-aged individual with no children, no siblings, no living parents, and small extended family are different from identifying the same variant in a middle-aged patient with young adult children and siblings with their own children. In many cases, the adult patient’s primary reason for pursuing genetic testing relates to concern over genetic risks that might be passed to their own children or to future generations. While broad standards and practices in returning incidental findings should not necessarily be different in these two scenarios, the physician and patient’s application of the incidental findings will be different in each case and will drive the clinical utility of potential incidental findings.
Conceptual frameworks for the return of incidental genomic findings
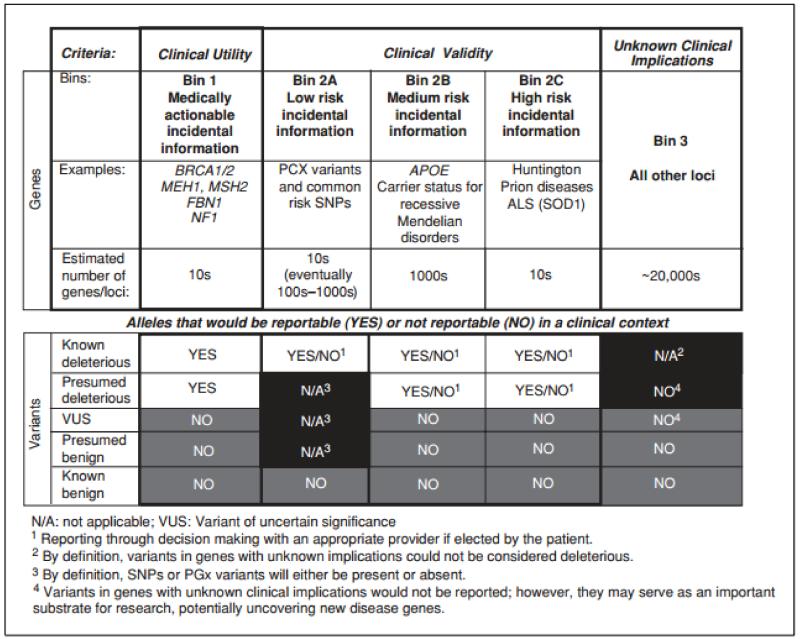
As noted above, in the clinical domain where GS is now being introduced, there are few sources of empirical data and there is not yet any consensus on best practices to guide clinicians. But there have been several attempts to apply reasonable clinical principles to address this issue. One proposed schema is a “binning” approach (Berg et al., 2011). These authors created bins representing categories of incidental findings that may be distinguished by varying degrees of clinical utility and validity. Clinical validity is determined by the degree of evidence supporting associations between genotype and clinical features, such as well-documented pharmacogenetic associations or disease risk established in GWAS studies. The authors’ first bin includes those variants that are clinically actionable, such as pathogenic variants in cancer risk genes. The second bin includes variants that are considered to be clinically valid but not actionable. Clinically valid variants are further divided into subcategories based on effect size of the association between the variant and disease (e.g., large effect size for Huntington disease, low effect size for common risk variants). The authors argue that such a categorical approach not only enables more efficient interpretation and reporting of incidental findings, but also facilitates a meaningful and relatively efficient informed consent process prior to a patient undergoing genomic sequencing.
A preliminary report by some of the same authors offered an example of the binning approach in practice (Berg et al., 2013). The group categorized 2,016 Mendelian genes into the bins described (see Fig. 9.23.3), and subsequently evaluated 80 whole genome sequences to identify variants that would fall in each category. After filtering for disease-causing mutations and population frequency (excluded variants with frequency of more than 5%), they found an average of 17.4 variants per person among bins 1 and 2 that warranted manual analysis, with the vast majority (15.7) falling into bins 2b and bin R (a separate bin for heterozygous variants identified in genes associated with recessive inheritance). Manual analysis further removed ~50% of the variants identified from the above filtering.
Figure 9.23.3.
Binning approach to incidental findings (from Berg et al., 2011)
The template of bins could be a valuable tool as data become available on clinical utility, but in the absence of such data, it is hard to know how useful the framework of bins will be. This is because the definitions of the bins themselves are somewhat subjective (one person’s sense of “high risk” may be another person’s “medium risk”). Perhaps the greatest subjectivity around this framework of binning lies in the definition of what is or is not “actionable.” The concept of medically actionable information seems logical and appealing on the surface, but it is not easy to reach consensus on what this means. Medical action could mean regular surveillance for the appearance of cancer, reproductive screening coupled with pre-natal diagnosis and pregnancy termination or even psychological preparation for an untreatable diagnosis. It turns out that both patients and clinicians have divergent views of medical actionability. For example, if a genetic test could identify children destined to develop untreatable intellectual disability, some would maintain that this is not actionable, while others would argue that such knowledge would allow for additional enrichment of the infant’s early environment that could mitigate the degree of disability.
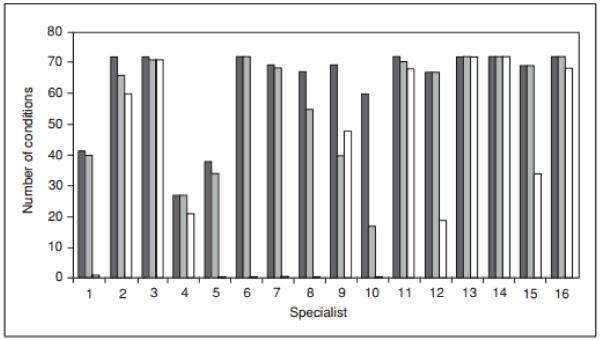
The authors of the binning approach suggest an “iterative, centralized, evidence-based, and consensus-driven process” (Berg et al., 2011); however, clinical variant classification is not an area that easily lends itself to consensus. In a recently published study, only partial concordance was observed across 16 genetics experts who were asked to independently evaluate 99 common genetic conditions and individual genes and select those they would recommend reporting back to the patient’s physician as incidental findings after GS (Green et al., 2012b). The study found that concordance was higher for reporting incidental information upon which clinicians could act (such as cancer predisposition syndromes) than for those which lacked effective medical interventions. Concordance was also higher when reporting incidental findings about adults rather than about children, and for reporting known pathogenic mutations and presumed pathogenic truncating variants rather than missense variants predicted to be pathogenic. One striking observation in this study was the degree to which the individual experts differed in the logic of their decision-making when presented with different types of variants in disease-associated genes. Figure 9.23.4 shows the number of conditions chosen by each expert when the question was asked separately about three different types of variants. Several experts (such as experts 9 and 10) selected many more variants when presented with known pathogenic mutations, while others (such as experts 11, 13, 14 and 16) put nearly equal weight on all three variant types. This exercise illustrates the problem with any ad hoc framework. In the absence of data, even highly skilled practitioners can have very different notions about appropriate clinical actions.
Figure 9.23.4.
Number of conditions/genes out of a total of 72 that each of 16 specialists chose for incidental return in an adult patient (from Green et al., 2012b). Black lines indicate choices for “known pathogenic mutations,” dark gray lines indicate choices for “truncating mutations” and white lines indicate “missense mutation predicted in silico to be pathogenic.
Despite the absence of data, GS is being so rapidly integrated into the practice of medicine that a set of recommendations was recently released by the American College of Genetics and Genomics (ACMG) encouraging the evaluation and reporting of a small number of variants based upon a presumption of clinical utility (Levenson, 2012; Green et al., 2013b). The rationale for this recommendation is twofold. First, the ACMG Working Group that developed these guidelines believes that there is sufficient clinical consensus that a small number of highly penetrant variants should be sought and reported whenever clinical sequencing is performed. Secondly, there is a proliferation of interpretative pipelines in genomics emerging from both established molecular laboratories and from vendors, and it is easy to imagine an “arms race” in which these services compete to present as much “clinical information” as possible when interpreting the genome for clinicians. The ACMG Working Group believes it is important to emphasize that only a small number of genomic findings rise to the level of being recommended for return when discovered incidentally. In creating these recommendations, the ACMG recognizes a distinction between opportunistic screening or the balance of benefitrisk-cost after a genome has already been ordered and performed, and population screening where the evidence supporting a favorable benefit-risk-cost would need to be quite high and certain.
Reporting of incidental findings to clinicians
Clinical molecular testing for specific candidate genes or gene panels is well established, and the format for these reports is relatively standardized among molecular laboratories. But there are few precedents for generating reports for patients that are undergoing GS. In particular, there are no specific guidelines to suggest how molecular laboratories should seek or report incidental findings. This is more problematic than is sounds because the interpretation of candidate gene sequencing is usually anchored by the clinical presentation of the patient, or by the family history, often with prior knowledge of specific variants being sought. In these circumstances, it often makes sense for the laboratory to report variants that are deemed pathogenic, along with variants of unknown significance (VUS), and sometimes even describe variants thought to be benign. When reports are provided in this manner, they offer very complete information for the informed geneticist and genetic counselor to interpret to the family.
GS reports that incorporate a larger portion of the genome than gene panel studies will force molecular laboratories to modify these reports for several reasons. First, they will have to decide whether and how extensively to seek and report any incidental findings. Next, if even a small proportion of incidental findings were reported, the number of VUS and benign variants would be excessive. Third, the interpretation of variants discovered as incidental findings, particularly VUS, cannot be provided with the same confidence as in a more directed gene sequencing study based upon patient phenotype or family history. Thus, a more arbitrary cut-off for reporting variants of clinical interest may need to be selected, such one that only reports certain categories of variants as defined by ACMG guidelines (Richards et al.,2008).
Early programs in GS have used different approaches. Some may analyze and report GS for clinical purposes in ways specifically designed to avoid incidental findings. For example, the issue of incidental findings in children may be avoided by preselecting likely candidate genes for exploration using infant phenotype as a guide (Saunders et al., 2012). Other laboratories have devised a broader approach to report findings of their GS service. The University of Wisconsin laboratory creates two categories: findings that are mandatory and findings that are optional for disclosure. Findings considered mandatory for reporting include diagnostic findings and unrelated findings associated with treatable, childhood-onset conditions. They further describe three optional subcategories, including findings related to non-actionable childhood conditions, non-actionable adult conditions, and actionable adult conditions. After an extensive period of genetic counseling, parents of pediatric patients may choose which of the above categories they wish to be disclosed. Baylor College of Medicine’s molecular laboratory takes yet another approach, in which an expanded report of incidental and uncertain findings is available on request and is separate from a primary, diagnosis-centered report.
At the present time, most GS is still ordered by clinicians with specialized knowledge of genetics, but as the price of GS continues to fall and the applications of GS expand, the form and content of the GS report will become increasingly important. In the near future, it is easy to imagine clinicians who are not geneticists ordering GS in the same manner that clinicians who are not radiologists might order X-ray films today. In this circumstance, the GS report will need to be understandable, somehow communicating the complex nuances of uncertainty around incidental findings in ways that are understandable to nongenetic clinicians, and even to primary care physicians. With the ongoing rapid expansion of genomic knowledge, GS results, including incidental findings, present the opportunity for an innovative, iterative clinical reporting process.
Examples of efforts to communicate and update variant interpretations and notifying clinicians of changes are being implemented and evaluated (Aronson et al., 2011, 2012). In order to meet the demand for return and ongoing evaluation of incidental findings, it will become essential to integrate sequence information into a medical record system and incorporate clinical decision support systems to interact, on an as-needed basis, with the patient’s genome over the course of a lifetime.
Conclusions and Future Directions
GS is a disruptive technology that, as exemplified by the issue of incidental findings, will challenge the ability of patients and physicians to incorporate a new type and quantity of information into everyday clinical practice. Given our ever-expanding yet still limited capacity to interpret most genetic variation, the abilities of clinicians to communicate and manage uncertainty with patients will be tested. Incomplete or ambiguous information has always been part of the practice of medicine, so this is not an unfamiliar dilemma. Sequencing is only the beginning of comprehensive molecular applications to medicine. Epigenetics, proteomics, and metabolomics will all be layered onto the interpretation genomic data, adding new insights and even greater ambiguities. Using advanced technologies before their clinical utility has been fully vetted may become the norm rather than the exception. If so, this will require a more sophisticated clinical workforce, along with a greater emphasis on clinician numeracy and probabilistic thinking, as well as realignment of current incentives. The management of incidental findings in clinical genomic sequencing today may well point the way towards new models for transforming our medical care system tomorrow.
Acknowledgements
This work was supported by NIH grants HG006500, HG005092, HG003178, HG00603, HG006615, HG003170,GM007748, AG027841, CA154517. We thank Dr. Heidi Rehm for her contributions to many of the ideas and insights described in this article.
Literature Cited
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I, Schmidt S, Peshkin L, Ramensky V, Gerasimova A, Bork P, Kondrashov A, Sunyaev S. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed S, Horton KM, Jeffrey RB, Jr., Sheth S, Fishman EK. Incidental thyroid nodules on chest CT: Review of the literature and management suggestions. AJR Am. J. Roentgenol. 2010;195:1066–1071. doi: 10.2214/AJR.10.4506. [DOI] [PubMed] [Google Scholar]
- Alpert JB, Naidich DP. Imaging of incidental findings on thoracic computed tomography. Radiol. Clin. North Am. 2011;49:267–289. doi: 10.1016/j.rcl.2010.10.005. [DOI] [PubMed] [Google Scholar]
- Aronson SJ, Clark EH, Babb LJ, Baxter S, Farwell LM, Funke BH, Hernandez AL, Joshi VA, Lyon E, Parthum AR, Russell FJ, Varugheese M, Venman TC, Rehm HL. The GeneInsight Suite: A platform to support laboratory and provider use of DNAbased genetic testing. Hum. Mutat. 2011;32:532–536. doi: 10.1002/humu.21470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronson SJ, Clark EH, Varugheese M, Baxter S, Babb LJ, Rehm HL. Communicating new knowledge on previously reported genetic variants. Genet. Med. 2012 doi: 10.1038/gim.2012.19. doi:10.1038/gim.2012.19 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashley EA, Butte AJ, Wheeler MT, Chen R, Klein TE, Dewey FE, Dudley JT, Ormond KE, Pavlovic A, Morgan AA, Pushkarev D, Neff NF, Hudgins L, Gong L, Hodges LM, Berlin DS, Thorn CF, Sangkuhl K, Hebert JM, Woon M, Sagreiya H, Whaley R, Knowles JW, Chou MF, Thakuria JV, Rosenbaum AM, Zaranek AW, Church GM, Greely HT, Quake SR, Altman RB. Clinical assessment incorporating a personal genome. Lancet. 2010;375:1525–1535. doi: 10.1016/S0140-6736(10)60452-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bainbridge MN, Wiszniewski W, Murdock DR, Friedman J, Gonzaga-Jauregui C, Newsham I, Reid JG, Fink JK, Morgan MB, Gingras MC, Muzny DM, Hoang LD, Yousaf S, Lupski JR, Gibbs RA. Whole-genome sequencing for optimized patient management. Sci. Transl. Med. 2011;3:87re83. doi: 10.1126/scitranslmed.3002243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg JS, Khoury MJ, Evans JP. Deploying whole genome sequencing in clinical practice and public health: Meeting the challenge one bin at a time. Genet. Med. 2011;13:499–504. doi: 10.1097/GIM.0b013e318220aaba. [DOI] [PubMed] [Google Scholar]
- Berg JS, Adams M, Nassar N, Bizon C, Lee K, Schmitt CP, Wilhelmsen KC, Evans JP. An informatics approach to analyzing the incidentalome. Genet. Med. 2013;1:36–44. doi: 10.1038/gim.2012.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boards of Directors, American Society of Human Genetics and American College of Medical Genetics Points to consider: Ethical, legal, and psychosocial implications of genetic testing in children and adolescents. Am. J. Hum. Genet. 1995;57:1233–1241. [PMC free article] [PubMed] [Google Scholar]
- Brauer CA, Bozic KJ. Using observational data for decision analysis and economic analysis. J. Bone Joint Surg. Am. 2009;91:73–79. doi: 10.2106/JBJS.H.01537. [DOI] [PubMed] [Google Scholar]
- Cassa CA, Savage SK, Taylor PL, Green RC, McGuire AL, Mandl KD. Disclosing pathogenic genetic variants to research participants: Quantifying an emerging ethical responsibility. Genome Res. 2012;22:421–428. doi: 10.1101/gr.127845.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Committee on Bioethics (American Academy of Pediatrics) Ethical issues with genetic testing in pediatrics. Pediatrics. 2001;107:1451–1455. doi: 10.1542/peds.107.6.1451. [DOI] [PubMed] [Google Scholar]
- Cordero P, Ashley EA. Whole-genome sequencing in personalized therapeutics. Clin. Pharmacol. Therapeut. 2012;91:1001–1009. doi: 10.1038/clpt.2012.51. [DOI] [PubMed] [Google Scholar]
- Downing GJ. Key aspects of health system change on the path to personalized medicine. Transl. Res. 2009;154:272–276. doi: 10.1016/j.trsl.2009.09.003. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng CM, Muzny D, Reid J, Bainbridge M, Pham P, Bekheirnia MR, Beuten J, Hardison M, Niu Z, Person R, Vatta M, Xia F, Hawes A, Wang M, Ding Y, Sun H, Scheel M, Saada N, Liu W, Braxton A, Ward P, Willis A, Wiszniewska J, Plon SE, Lupski JR, Beaudet AL, Gibbs RA, Yang Y. American Society of Human Genetics Annual Meeting. San Francisco, CA: Nov 7, 2012. 2012. Clinical whole exome sequencing for the diagnosis of Mendelian disorders: Program design, implementation, and first year reporting experience. [Google Scholar]
- Evans JP, Green RC. Direct to consumer genetic testing: Avoiding a culture war. Genet. Med. 2009;11:568–569. doi: 10.1097/GIM.0b013e3181afbaed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feero WG, Guttmacher AE, Collins FS. Genomic medicine-an updated primer. N. Engl. J. Med. 2010;362:2001–2011. doi: 10.1056/NEJMra0907175. [DOI] [PubMed] [Google Scholar]
- Fokkema IF, Taschner PE, Schaafsma GC, Celli J, Laros JF, den Dunnen JT. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011;32:557–563. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- Frueh FW, Greely HT, Green RC, Hogarth S, Siegel S. The future of direct-toconsumer clinical genetic tests. Nat. Rev. Genet. 2011;12:511–515. doi: 10.1038/nrg3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furtado CD, Aguirre DA, Sirlin CB, Dang D, Stamato SK, Lee P, Sani F, Brown MA, Levin DL, Casola G. Wholebody CT screening: spectrum of findings and recommendations in 1192 patients. Radiology. 2005;237:385–394. doi: 10.1148/radiol.2372041741. [DOI] [PubMed] [Google Scholar]
- Gilissen C, Hoischen A, Brunner HG, Veltman JA. Unlocking Mendelian disease using exome sequencing. Genome Biol. 2011;12:228. doi: 10.1186/gb-2011-12-9-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollust SE, Gordon ES, Zayac C, Griffin G, Christman MF, Pyeritz RE, Wawak L, Bernhardt BA. Motivations and perceptions of early adopters of personalized genomics: Perspectives from research participants. Public Health Genomics. 2012;15:22–30. doi: 10.1159/000327296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzaga-Jauregui C, Lupski JR, Gibbs RA. Human genome sequencing in health and disease. Annu. Rev. Med. 2012;63:35–61. doi: 10.1146/annurev-med-051010-162644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Mountain J, Kiefer A, Moreno T, MacBean E, Kalia SS, Roberts JS, PGen Study Group . American Society of Human Genetics, Annual Meeting. San Francisco, CA: Nov 7, 2012a. 2012. Consumer Genomics: Motivations and Intentions. [Google Scholar]
- Green RC, Berg JS, Berry GT, Biesecker LG, Dimmock DP, Evans JP, Grody WW, Hegde MR, Kalia S, Korf BR, Krantz I, McGuire AL, Miller DT, Murray MF, Nussbaum RL, Plon SE, Rehm HL, Jacob HJ. Exploring concordance and discordance for return of incidental findings from clinical sequencing. Genet. Med. 2012b;14:405–410. doi: 10.1038/gim.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RC, Rehm HL, Kohane I. Challenges in the Clinical Use of Genome Sequencing. In: Willard HFG, G.S., editors. Genomic and Personalized Medicine. 2nd ed Academic Press; New York: 2013a. [Google Scholar]
- Green RC, Berg JS, Grody WW, Kalia SS, Korf BR, Martin CL, McGuire A, Nussbaum RL, O’Daniel JM, Ormond KE, Rehm HL, Watson MS, Williams MS, Biesecker LG. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet. Med. 2013b doi: 10.1038/gim.2013.73. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttmacher AE, McGuire AL, Ponder B, Stefansson K. Personalized genomic information: Preparing for the future of genetic medicine. Nat. Rev. Genet. 2010;11:161–165. doi: 10.1038/nrg2735. [DOI] [PubMed] [Google Scholar]
- Jordan DM, Kiezum A, Baxter SM, Agarwala V, Green RC, Murray MF, Pugh T, Lebo MS, Rehm HL, Funke BH, Sunyaev S. Development and validation of a computational method for assessment of missense variants in hypertrophic cardiomyopathy. Am. J. Hum. Genet. 2011;88:183–192. doi: 10.1016/j.ajhg.2011.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kogelenberg MV, Morley K, Fitzgerald T, Gerety S, Tivey A, Al-Turki K, Clayton S, Wright C, Barrett J, Firth H, FitzPatrick D, Carter N, Hurles N, On behalf of the DDD project . American Society of Human Genetics Annual Meeting. San Francisco, CA: 2012. Exome sequencing results in 23 patients with severe developmental disorders in the DDD project. [Google Scholar]
- Kohane IS, Masys DR, Altman RB. The incidentalome: A threat to genomic medicine. JAMA. 2006;296:212–215. doi: 10.1001/jama.296.2.212. [DOI] [PubMed] [Google Scholar]
- Kohane IS, Hsing M, Kong SW. Taxonomizing, sizing, and overcoming the incidentalome. Genet. Med. 2012;14:399–404. doi: 10.1038/gim.2011.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lautenbach D, Christensen K, Sparks J, Green RC. Communicating Genetic Risk Information for Common Disorders in the Era of Genomic Medicine. In: Chakraverti A, Green E, editors. Annual Reviews in Genetics and Human Genomics. Annual Reviews. Vol. 14. Calif; Palo Alto: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenson D. The tricky matter of secondary genomic findings: ACMG plans to issue recommendations. Am. J. Med. Genet. 2012;A 158A:ix–x. doi: 10.1002/ajmg.a.35521. [DOI] [PubMed] [Google Scholar]
- Lumbreras B, Donat L, Hernandez-Aguado I. Incidental findings in imaging diagnostic tests: A systematic review. Br. J. Radiol. 2010;83:276–289. doi: 10.1259/bjr/98067945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macconaill LE, Garraway LA. Clinical implications of the cancer genome. J. Clin. Oncol. 2010;28:5219–5228. doi: 10.1200/JCO.2009.27.4944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDermott U, Downing JR, Stratton MR. Genomics and the continuum of cancer care. N. Engl. J. Med. 2011;364:340–350. doi: 10.1056/NEJMra0907178. [DOI] [PubMed] [Google Scholar]
- McGuire AL, Diaz CM, Wang T, Hilsenbeck SG. Social networkers’ attitudes toward direct-to-consumer personal genome testing. Am. J. Bioethics. 2009;9:3–10. doi: 10.1080/15265160902928209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G. Advances in understanding cancer genomes through second-generation sequencing. Nat. Rev. Genet. 2010;11:685–696. doi: 10.1038/nrg2841. [DOI] [PubMed] [Google Scholar]
- Moussa ID. Coronary artery bifurcation interventions: The disconnect between randomized clinical trials and patient centered decisionmaking. Catheter Cardiovasc. Interv. 2011;77:537–545. doi: 10.1002/ccd.22865. [DOI] [PubMed] [Google Scholar]
- Ng PC, Henikoff S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003;31:3812–3814. doi: 10.1093/nar/gkg509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petitti DB, Teutsch SM, Barton MB, Sawaya GF, Ockene JK, DeWitt T, Force USPST. Update on the methods of the U.S. Preventive Services Task Force: Insufficient evidence. Ann. Intern. Med. 2009;150:199–205. doi: 10.7326/0003-4819-150-3-200902030-00010. [DOI] [PubMed] [Google Scholar]
- Relling MV, Altman RB, Goetz MP, Evans WE. Clinical implementation of pharmacogenomics: Overcoming genetic exceptionalism. Lancet Oncol. 2010;11:507–509. doi: 10.1016/S1470-2045(10)70097-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relling MV, Klein TE. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin.Pharmacol.Ther. 2011;89:464–467. doi: 10.1038/clpt.2010.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards CS, Bale S, Bellissimo DB, Das S, Grody WW, Hegde MR, Lyon E, Ward BE. ACMG recommendations for standards for interpretation and reporting of sequence variations: Revisions 2007. Genet. Med. 2008;10:294–300. doi: 10.1097/GIM.0b013e31816b5cae. [DOI] [PubMed] [Google Scholar]
- Roberts JS, Christensen KD, Green RC. Using Alzheimer’s disease as a model for genetic risk disclosure: Implications for personal genomics. Clin. Genet. 2011;80:407–414. doi: 10.1111/j.1399-0004.2011.01739.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuels ME, Rouleau GA. The case for locus-specific databases. Nat. Rev. Genet. 2011;12:378–379. doi: 10.1038/nrg3011. [DOI] [PubMed] [Google Scholar]
- Saunders CJ, Miller NA, Soden SE, Dinwiddie DL, Noll A, Alnadi NA, Andraws N, Patterson ML, Krivohlavek LA, Fellis J, Humphray S, Saffrey P, Kingsbury Z, Weir JC, Betley J, Grocock RJ, Margulies EH, Farrow EG, Artman M, Safina NP, Petrikin JE, Hall KP, Kingsmore SF. Rapid whole-genome sequencing for genetic disease diagnosis in neonatal intensive care units. Sci. Transl. Med. 2012;4:154ra135. doi: 10.1126/scitranslmed.3004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenson P, Mort M, Ball E, Howells K, Phillips A, Thomas N, Cooper D. The human gene mutation database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong MY, Cassa CA, Kohane IS. Automated validation of genetic variants from large databases: Ensuring that variant references refer to the same genomic locations. Bioinformatics. 2011;27:891–893. doi: 10.1093/bioinformatics/btr029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travis SP. New thinking: Theory vs. practice. A case study illustrating evidence-based therapeutic decision making. Colorectal Dis. 2006;8:25–29. doi: 10.1111/j.1463-1318.2006.00989.x. [DOI] [PubMed] [Google Scholar]
- van Vugt S, Broekhuizen L, Zuithoff N, de Jong P, Butler C, Hood K, Coenen S, Goossens H, Little P, Almirall J, Blasi F, Chlabicz S, Davies M, Godycki-Cwirko M, Hupkova H, Kersnik J, Mierzecki A, Molstad S, Moore M, Schaberg T, De Sutter A, Torres A, Touboul P, Verheij T, On behalf of the, G.P.G. Incidental chest radiographic findings in adult patients with acute cough. Ann. Fam. Med. 2012;10:510–515. doi: 10.1370/afm.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varmus H. Ten years on-the human genome and medicine. N. Engl. J. Med. 2010;362:2028–2029. doi: 10.1056/NEJMe0911933. [DOI] [PubMed] [Google Scholar]
- Wiita AP, Schrijver I. Clinical application of high throughput molecular screening techniques for pharmacogenomics. Pharmgenomics Pers. Med. 2011;4:109–121. doi: 10.2147/PGPM.S15302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worthey EA, Mayer AN, Syverson GD, Helbling D, Bonacci BB, Decker B, Serpe JM, Dasu T, Tschannen MR, Veith RL, Basehore MJ, Broeckel U, Tomita-Mitchell A, Arca MJ, Casper JT, Margolis DA, Bick DP, Hessner MJ, Routes JM, Verbsky JW, Jacob HJ, Dimmock DP. Making a definitive diagnosis: Successful clinical application of whole exome sequencing in a child with intractable inflammatory bowel disease. Genet. Med. 2011;13:255–262. doi: 10.1097/GIM.0b013e3182088158. [DOI] [PubMed] [Google Scholar]
- Wright C, Burton H, Hall A, Moorthie S, Pokorska-Bocci A, Sagoo G, Sanderson S, Skinner R. Next steps in the sequence. The implications of whole genome sequencing for health in the UK. PHG Foundation; 2011. http://www.phgfoundation.org. [Google Scholar]
- Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN, Tyler-Smith C. Deleterious- and disease-allele prevalence in healthy individuals: Insights from current predictions, mutation databases, and population-scale resequencing. Am. J. Hum. Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]