Abstract
Objective
To understand the HIV-hepatitis B virus (HBV) epidemic from a global perspective by clinically and virologically characterizing these viruses at the time of antiretroviral therapy (ART) initiation in a multi-national cohort.
Methods and design
HIV-infected subjects enrolled in two international studies were classified as HIV-HBV co-infected or HIV monoinfected prior to ART. HIV-HBV co-infected subjects were tested for HBV characteristics, hepatitis D virus (HDV), a novel non-invasive marker of liver disease, and drug-resistant HBV. Comparisons between discrete covariates used chi-square or Fisher’s exact tests (and Jonchkheere-Terpstra for trend tests) while continuous covariates were compared using Wilcoxon Rank-Sum Test.
Results
Of the 2105 HIV-infected subjects from 11 countries, the median age was 34 years and 63% were Black. The 115 HIV-HBV co-infected subjects had significantly higher ALT and AST values, lower body mass index, and lower CD4+ T-cell counts than HIV monoinfected subjects (median 159 cells/mL and 137 cells/mL, respectively, P=0.04). In the co-infected subjects, 49.6% had HBeAg-negative HBV, 60.2% had genotype A HBV, and 13% were HDV positive. Of the HBeAg-negative subjects, 66% had HBV DNA ≤2000 IU/ml compared to 5.2% of the HBeAg-positive subjects. Drug-resistant HBV was not detected.
Conclusions
Screening for HBV in HIV-infected patients in resource-limited settings is important since it is associated with lower CD4+ T-cell counts. In settings where HBV DNA is not available, HBeAg may be useful to assess the need for HBV treatment. Screening for drug-resistant HBV is not needed prior to starting ART in settings where this study was conducted.
Keywords: HIV, HBV, coinfection, global
Introduction
Chronic hepatitis B (CH-B), which is the leading cause of end stage liver disease worldwide, is common in human immunodeficiency virus type-1 (HIV)-infected individuals with a prevalence ranging from 5–20% in various studies of HIV-infected subjects [1–5]. Although co-infection with HIV and hepatitis B virus (HBV) is recognized as being common, there are limited data to provide an international perspective on this epidemic. Such a perspective is especially important as highly active antiretroviral therapy (HAART) is being introduced worldwide including in resource-limited settings with high HBV endemicity such as Asia and Africa. Since liver disease is a leading cause of non-AIDS death in HIV-infected patients receiving HAART, characterizing HBV prior to HAART is important in order to better understand the scope of the disease and to prioritize treatment needs in resource-limited settings.
Most studies of HIV-HBV co-infected subjects have been conducted in countries with low HBV endemicity and have shown a negative effect of HIV on CH-B with decreased hepatitis B e antigen (HBeAg) clearance, high HBV DNA levels, and increased risk for cirrhosis and liver-related death [6, 7]. Data from countries with low HBV endemicity are informative, but they represent HBV disease that occurs primarily in adulthood. In contrast, in countries with high HBV endemicity, transmission primarily occurs in infancy or early childhood; thus, in most of these cases, HBV has been present for years prior to HIV infection. It is not known if the worldwide epidemic is similar to what has been reported from countries with low HBV endemicity. One study from Nigeria suggests that this may not be the case since HBV DNA levels were lower than what has been seen in studies from North America and Europe [8]. Thus, data on HIV and HBV characteristics with a more global perspective in these co-infected individuals at the time HAART is initiated are needed.
Two randomized clinical trials of antiretroviral therapy were conducted by the Adult AIDS Clinical Trials Group (ACTG) in diverse resource-limited settings. The first, ACTG A5175, enrolled HIV-infected men and women worldwide to compare different initial HAART regimens [9]. The second, A5208, compared the response to first-line HAART in African women who had received single-dose nevirapine during a prior pregnancy to those who had not received nevirapine [10]. Both of these trials tested subjects for HBV at HAART initiation; thus, they are useful to characterize untreated HIV and HBV co-infection from a multi-national perspective.
Methods
Study subjects
The subjects participated in one of the following parent ACTG studies: 1) A5175, which was a randomized, open-label comparison of once-daily protease inhibitor or non-nucleoside reverse transcriptase inhibitor-containing regimens for HAART-naïve HIV-infected men and women (CD4 + T-cell count <300 cells/mm3 and ALT/AST <5x upper limit of normal (ULN)) from Brazil, Haiti, India, Malawi, Peru, South Africa, Thailand, United States, and Zimbabwe, as previously described (n=1571) [9] or 2) A5208, which studied whether peripartum exposure to single-dose nevirapine diminished the response to a nevirapine-based initial HAART regimen (n=745), enrolled women with CD4+ T-cell count <200 cells/mm3 and baseline AST/ALT <2.5x the ULN from the following African countries: South Africa, Kenya, Zimbabwe, Botswana, Zambia, Malawi, and Uganda, as previously described [10]. All A5175 sites and seven of the ten A5208 sites had local IRB approval for participation in this study. Informed consent was obtained from the study subjects in their native language, and the study was approved by the Johns Hopkins University IRB. Of the sites that participated, Brazil excluded people who had CH-B from enrolling in the parent study; thus, Brazilian participants were not included in prevalence estimates but are included in the HIV monoinfected study group. In addition, four of the 25 US sites, which enrolled 13% of the subjects from the US, excluded people with CH-B.
Participants were classified as HIV-HBV co-infected if they were hepatitis B surface antigen (HBsAg) positive at the visit prior to randomization and positive for HBeAg, HBV DNA, or hepatitis B e antibody (anti-HBe) at the study entry visit. All others were classified as HIV monoinfected. Data abstracted from the parent study database included the following: age, sex, race, body-mass index (BMI), history of liver disease, hemoglobin, HIV disease stage, HIV RNA, CD4+ T-cell count, ALT, and AST. The HIV-HBV co-infected participants also had the following performed using study entry visit serum: HBeAg, anti-HBe, HBV genotype, HBV DNA, hepatitis D antibody (anti-HDV), hepatitis C virus (HCV) antibody (anti-HCV), HCV RNA if anti-HCV positive, HBV Pol sequencing, and a non-invasive marker of liver disease that correlates with liver disease [11]. This non-invasive marker measures the change in glycosylation of an immunoglobulin G reactive to specific alpha-galactose epitope, which is measured by reactivity to the fucose-binding lectin Aleuria aurantia lectin (AAL-reactivity). This marker was selected since other readily available markers use parameters such as platelets that are affected by HIV infection.
Laboratory testing
All specimens were stored at −80°C. Serological testing for HBeAg (ETI-EBK Plus, Diasorin, Stillwater MN or Abbott ARCHITECT® HBeAg v17.0, Abbott Park, IL), anti-HBe (ETI-AB-EBK Plus or Abbott ARCHITECT® Anti HBe v13.0), anti-HDV (ETI-AB-DELTAK-2, Diasorin, Stillwater, MN), and anti-HCV (Ortho HCV v3.0, Ortho Diagnostics, Raritan NJ) were performed according to manufacturer’s instructions. HBV DNA testing was done with real-time PCR using either RealARTTM HBV LC PCR v 3.0 (Qiagen, Valencia, CA) or Abbott RealTime HBV DNA (Abbott Molecular, Des Plains, IL). The highest common lower limit of detection of these assays was 200 IU/ml, which is the value used in the analyses. HBV genotype and drug-resistance mutations were determined by HBV Pol sequencing, performed as previously described [12]. HCV RNA was determined with the Abbott RealTime HCV RNA (Abbott Molecular, Des Plaines, IL), performed according to manufacturer’s instructions. AAL-reactivity was tested as previously described [11] with values >5 representing cirrhosis, values 3–5 representing moderate liver disease, and values of 1–2 representing no or mild liver disease.
Statistical analysis
ALT and AST levels were graded according to standard ACTG definitions [13]. Comparisons between groups were made using chi-square or Fisher’s exact test for discrete covariates and Wilcoxon Rank-Sum Test for continuous covariates. For 3-level ordinal group comparisons, the Jonchkheere-Terpstra trend test was performed. To test for associations of pre-treatment covariates with baseline HBV DNA level, linear regression modeling on the log 10 transformed HBV DNA was performed. Pre-treatment covariates tested include screening CD4 T-cell count category (< 50, 50–199, and 200–299), log10 HIV-1 RNA copies/mL, sex, age, BMI, anti-HDV, HBeAg, anti-HBe, HBV genotype, hemoglobin per 5 g/dL increase, creatinine clearance (Crockoft-Gault equation), ALT, and AST. P-values <0.05 were considered statistically significant and were not adjusted for multiplicity.
Results
A5175 and A5208 recruited 2316 participants of whom 2105 (90.9%) were enrolled in sites that obtained IRB approval for this substudy. Of these 2105 subjects, 115 (5.5%) were HIV-HBV coinfected. There was no significant difference in the prevalence of a positive HBsAg between the sites that did (6.7%) and did not (5.6%) have IRB approval for this substudy. The HIV-HBV co-infection prevalence varied with the highest prevalence in Zimbabwe (11%) and the lowest in Kenya (2.2%) (Table 1). The prevalence of HIV-HBV co-infection across continents was similar with Asia at 5.9%, Africa at 6.7%, Central/South America at 5.1% (excluding Brazil), and North America (US) at 4.8%. Anti-HCV was positive in 6 subjects who all had undetectable HCV RNA.
Table 1.
Distribution of study subjects by country
| Country | HIV monoinfected Number (% of group) | HIV-HBV co-infected Number (% of group) | HIV-HBV co-infected Estimated Prevalence (%) (95% CI) |
|---|---|---|---|
| Africa | |||
| Botswana | 86 (4) | 4 (3) | 4.4 (1.2–11.0) |
| Kenya | 135 (7) | 3 (3) | 2.2 (0.5–6.2) |
| Malawi | 265 (13) | 24 (21) | 8.3 (5.4–12.0) |
| South Africa | 314 (16) | 17 (15) | 5.1 (3.0–8.1) |
| Zimbabwe | 203 (10) | 24 (21) | 11.0 (6.9–15) |
| South/Central America | |||
| Brazil | 231 (12) | 0* | N/A |
| Haiti | 94 (5) | 6 (5) | 6 (2.2–13.0) |
| Peru | 128 (6) | 6 (5) | 4.5 (1.7–9.5) |
| North America | |||
| United States | 200 (10) | 10 (9) | 4.8 (2.3–8.6) |
| Asia | |||
| India | 242 (12) | 13 (11) | 5.1 (2.7–8.6) |
| Thailand | 92 (5) | 8 (6) | 8.0 (3.5–15.0) |
excluded HIV-HBV co-infected subjects at enrollment
The median age of the 2105 subjects was 34 years and 39% were male (Table 2). The racial make-up was majority Black (62.7%) followed by Asian (17%), White (11.9%), and Other (7.4%). The HIV-HBV co-infected subjects had more Blacks and fewer Whites than the HIV monoinfected subjects (P=0.07). The BMIs were lower among the HIV-HBV co-infected (median 21.7) compared to the HIV monoinfected participants (median 22.4, P=0.02); however, the distribution of numbers in each BMI category (underweight, normal, overweight, and obese) was similar.
Table 2.
Characteristics of study subjects
| HIV monoinfected N=1990 | HIV-HBV co-infected N=115 | P | |
|---|---|---|---|
| Age (IQR) (years)* | 34 (29–40) | 34 (30–40) | 0.53 |
| Male sex (%) | 39 | 47 | 0.09 |
| Race | 0.07 | ||
| Black (%) | 62.1 | 73.9 | |
| White (%) | 12.5 | 2.6 | |
| Asian (%) | 16.9 | 18.2 | |
| Other (%) | 7.5 | 5.2 | |
| BMI (IQR)* | 22.4 (20.3–25.3) | 21.7 (19.8–24.0) | 0.02 |
| ALT (IQR)* (U/L) | 23 (16.5–35.0) | 25.5 (19.0–38.0) | 0.03 |
| Normal | 1804 (91.0%) | 100 (87.0%) | 0.14 |
| Mild | 154 (7.8%) | 11 (9.6%) | |
| Moderate | 20 (1.0%) | 4 (3.5%) | |
| Severe | 3 (0.2%) | 0 (0.0%) | |
| Life Threatening | 1 (0.01%) | 0 (0.0%) | |
| AST (IQR)* (U/L) | 28.8 (23.0–38.0) | 34 (27.0–44.0) | <0.001 |
| Normal | 1703 (85.9%) | 86 (74.8%) | 0.001 |
| Mild | 242 (12.2%) | 28 (24.3%) | |
| Moderate | 35 (1.8%) | 1 (0.9%) | |
| Severe | 2 (0.1%) | 0 (0.0%) | |
| Life Threatening | 1 (0.1%) | 0 (0.0%) | |
| HIV characteristics | |||
| CD4 T-cell count (IQR) (cells/mm3)* | 159 (90,218) | 137 (68,210) | 0.04 |
| HIV RNA (IQR) (log cp/ml)* | 5.1 (4.6–5.5) | 5.1 (4.6–5.6) | 0.23 |
| WHO Stage IV or clinical AIDS (%) | 8.6 | 12.2 | 0.19 |
median values
Liver function and HIV parameters
Given the entry criteria for the parent studies, only seven subjects had grade 3 to 4 elevations of ALT or AST, all of whom were HIV monoinfected. Although ALT and AST values were typically low (median 23 and 29 U/L, respectively), the distributions of these values were both higher among the HIV-HBV co-infected (median ALT 25.5 U/L, median AST 34 U/L) compared to the HIV monoinfected subjects (median ALT 23 U/L (P=0.03), median AST 28.8 U/L (P<0.001)) (Table 2).
The median CD4 T-cell count was 157 cells/mm3 (interquartile range (IQR) 88–218 cells/mm3) and was significantly lower in the HIV-HBV co-infected (137 cells/mm3, IQR 68–210 cells/mm3) than in the HIV monoinfected subjects (159 cells/mm3 (IQR 90–218 cells/mm3, P = 0.04) (Table 2). However, the two groups did not differ in HIV RNA level (median 5.10 and 5.05 log10 cp/ml, respectively P=0.23). Also, the CD4 T-cell differences between the groups could not be accounted for by differences in HBV prevalence by country (Figure 1). Consistent with the difference in CD4 T-cell counts, the HIV-HBV co-infected subjects had a higher prevalence of clinical AIDS, which was defined as WHO stage IV or history of AIDS, but the difference was not statistically significant (12.2% versus 8.6% in HIV-HBV co-infected and HIV monoinfected, respectively, P=0.19).
Figure 1.
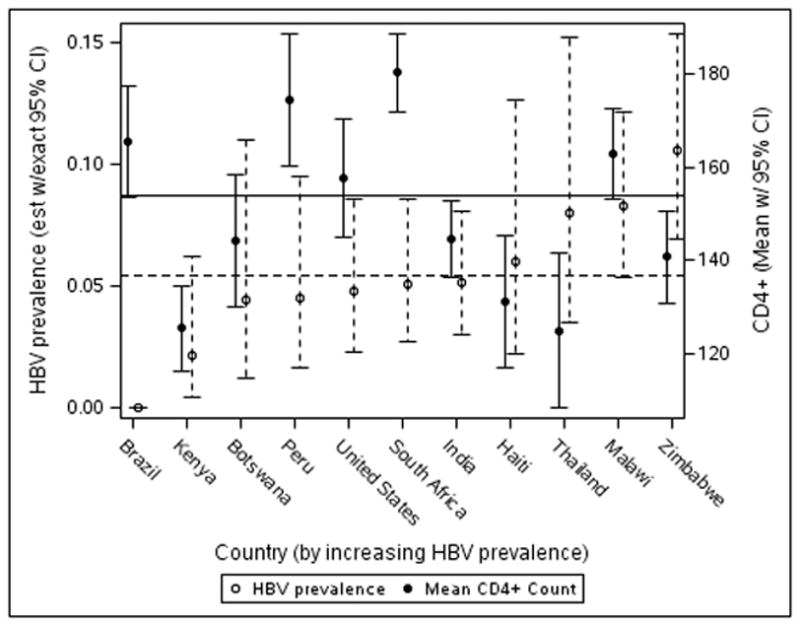
Distribution of HBV prevalence and mean CD4+ T-cell count by country. There is no clear relationship between HBV prevalence and mean CD4+ T-cell count.
As a sensitivity analysis, we repeated the above analyses excluding participants from Brazil since they were all HIV monoinfected by that site’s inclusion criteria. Comparisons between HIV monoinfected and HIV/HBV co-infected participants were similar with the following exceptions. The proportion of men was significantly higher in the HIV/HBV co-infected (46%) compared to the HIV monoinfected group (36%, P=0.02). The distribution of races between groups was attenuated (P=0.7). However, the difference in the ALT distribution was strengthened (P=0.002).
HBV characteristics in HIV-HBV co-infected subjects
From the study entry visit, serum was available for additional HBV testing in the 113 of the 115 HIV-HBV co-infected subjects. Approximately half, 57/113 (50.4%), were HBeAg-positive, and this proportion was similar in A5175 (51.8%) and A5208 (46.7%). The median age was 34 years in both the HBeAg-negative and –positive subjects and the proportion of females was similar between the HBeAg groups (48% and 52%). By country, subjects from Haiti (83%), India (69%), Kenya (67%), and Peru (83%) were predominantly HBeAg-positive, while those from Malawi (65%), Thailand (63%), and the United States (80%) were mainly HBeAg-negative.
HBV genotype could not be determined for 18 (16%) subjects due to low HBV DNA. Among observed genotypes, the most common were A (71.5%) and D (15.8%), but the distribution varied geographically with Africa having 95% genotype A, US with 78% genotype A, Thailand and India with 100% genotype C and D, respectively, and South and Central America with 55% genotype A, 27% genotype F, and 18% genotype E. Genotypes A and C were evenly divided between HBeAg-positive and −negative disease whereas D, E, and F were predominantly in HBeAg-positive subjects (Table 3).
Table 3.
HBV Characteristics in HIV-HBV co-infected subjects by HBeAg status
| Overall N=113 | HBeAg-negative N=56 | HBeAg-positive N=57 | |
|---|---|---|---|
| Median ALT (IQR)(U/L)* | 25.5 (19.0–38.0) | 23 (17.5–33.0) | 28 (21.9–49.0) |
| Median AST (IQR)(U/L)* | 34.0 (27.0–44.0) | 31.5 (24.0–40.0) | 36 (29.0–55.0) |
| Detectable HBV DNA (>200 IU/ml (%))* | 91.2 | 83.9 | 98.2 |
| Median HBV DNA (IQR)(log IU/ml)* | 5.1 (2.7–8.1) | 2.7 (2.3–4.4) | 8.0 (5.6–8.6) |
| HBV genotype (number)† | |||
| A | 68 | 33 | 35 |
| C | 5 | 2 | 3 |
| D | 15 | 4 | 11 |
| E | 4 | 1 | 3 |
| F | 3 | 0 | 3 |
| AAL reactivity (number)‡ | |||
| 1–2 | 39 | 18 | 21 |
| 3–5 | 48 | 26 | 22 |
| >5 | 16 | 8 | 8 |
P< 0.05 comparing HBeAg-negative to HBeAg-positive
HBV genotype not determined in 15 due to inability to amplify
Missing in 12 subjects
Anti-HDV was detected in 15 of 113 (13.3%) HBV co-infected subjects, which is within the range of the prevalence reported in HBV monoinfection [14]. Anti-HDV was detected only among subjects from Botswana, Kenya, Peru, South Africa, Thailand, or Zimbabwe with Botswana having the highest relative number of subjects (three of four). When stratified by HBeAg status, a similar proportion of the HBeAg-negative and –positive subjects (14 and 12%, respectively) were anti-HDV positive.
HBV DNA was detected in 91.2% of the 113 co-infected subjects and was significantly more common in the HBeAg-positive (98.2%) compared to the HBeAg-negative subjects (83.9%, P=0.008) (Table 3). The median HBV DNA was significantly higher in the HBeAg-positive (median 7.96 log IU/ml) compared to the HBeAg-negative subjects (median 2.74 log IU/ml, P <0.001). In fact, 66.1% of HBeAg-negative subjects had an HBV DNA ≤2000 IU/ml, which is the level above which treatment should be considered in HBeAg-negative patients [15]. In contrast, only 5.2% of HBeAg-positive subjects had HBV DNA ≤2000 IU/ml. Only 12% of the HBeAg-positive subjects had values below the cut-off recommended by some treatment guidelines for HBeAg-positive monoinfected patients (20,000 IU/ml) [16]. Genotypes C, D, and F had a higher proportion of subjects with HBV DNA >20,000 IU/ml compared to the other genotypes (P =0.049).
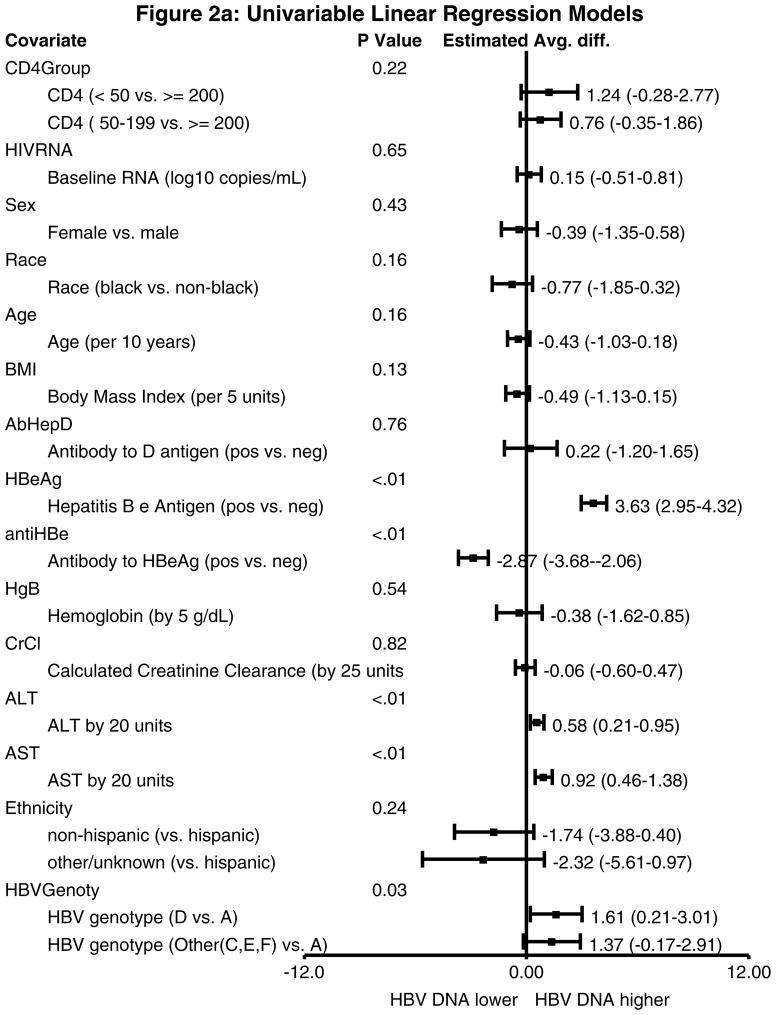
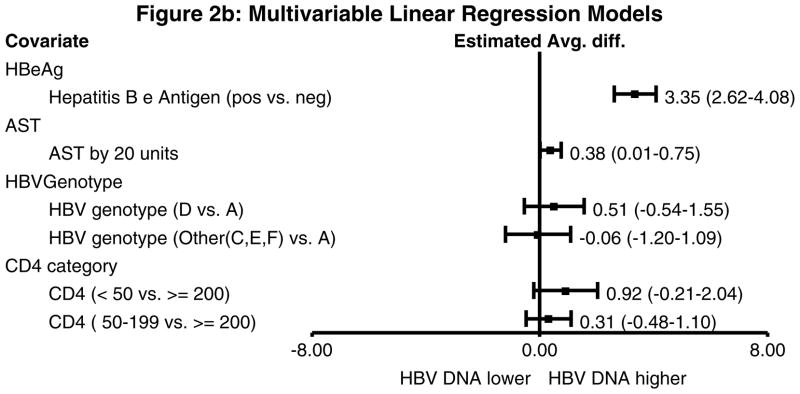
In univariable analysis with log HBV DNA as the outcome, HBeAg-positive subjects had a 3.63 log IU/ml (95% confidence interval (CI) 2.95–4.32 log IU/mL) higher HBV DNA than those who were HBeAg-negative (P<0.01) (Figure 2a). Those with higher ALT and AST also had higher log HBV DNA values. When compared to subjects infected with genotype A, those infected with genotype D had a 1.61 log IU/ml higher HBV DNA (95% CI 0.21–3.01 log IU/mL) and those infected with genotypes C, E, or F had a 1.37 log IU/mL higher HBV DNA (95% CI −0.17–2.91 log IU/mL). In a multivariable model, only HBeAg status and AST remained significantly associated with higher HBV DNA (Figure 2b).
Figure 2.
Figure 2a. Univariable linear regression models with log HBV DNA as the outcome. HBeAg positive status was associated with a 3.63 log IU/ml higher HBV DNA, anti-HBe positive status was associated with a 2.87 log IU/ml lower HBV DNA, both ALT and AST were associated with higher HBV DNA levels, and non-A genotype HBV was associated with higher HBV DNA levels.
Figure 2b. Multivariable linear regression models with log HBV DNA as the outcome. The associated variables from the univariable models were included in this model and CD4 count was forced into the model. Anti-HBe was not included since it is collinear with HBeAg status. HBeAg-positive status was associated with a significantly higher HBV DNA level (3.35 log IU/ml higher compared to HBeAg-negative subjects). AST was also associated with higher HBV DNA level.
Of the 95 subjects with HBV Pol sequencing, none (95% CI 0–3.7%) had mutations in HBV Pol that are associated with known resistance to lamivudine, adefovir dipivoxil, or entecavir [17].
Liver disease characteristics in HIV-HBV co-infected subjects
AST values were higher in the HBeAg-positive (median 36.0 U/L) compared to the HBeAg-negative subjects (median 31.5 U/L, P=0.008) with a similar relationship in ALT (median 28.0 and 23.0 U/L, respectively, P=0.03) (Table 3). Liver disease was measured by AAL-reactivity in 103 subjects since the samples from India were not available for this assay. No or mild liver disease was present in 39 (37.8%), moderate liver disease in 48 (46.6%), and cirrhosis in 16 subjects (15.5%). All 16 with cirrhosis were from A5175, 15 of whom were Black. The 16 with AAL values consistent with cirrhosis (> 5) trended towards a lower albumin (3.29 g/dL) compared to the 87 without cirrhosis (3.62 g/dL)(P =0.06); however, none of these 16 subjects had clinical signs or symptoms of liver disease.
HIV disease characteristics in HIV-HBV co-infected subjects
Since the HIV-HBV co-infected subjects had lower CD4+ T-cell counts than the HIV monoinfected subjects, we further explored the relationship between HBV DNA and CD4+ T-cell counts amongst the co-infected subjects. The median CD4+ T-cell count did not differ by HBeAg status. To determine if CD4+ T-cell count was associated with HBV DNA, the CD4+ T-cell count was stratified into 3 groups of <50, 51–199, and >200 cells/mm3. The proportion of subjects with HBV DNA >200, 000 IU/ml decreased significantly as CD4+ T-cell count increased (P=0.04) (Figure 3).
Figure 3.
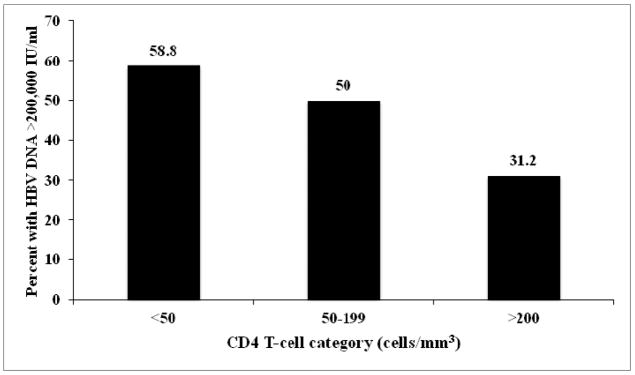
Frequency (in percent) of subjects with HBV DNA >200,000 IU/ml by CD4+ T-cell category. Subjects with CD4 T-cells <50 cells/mm3 had the highest proportion with HBV DNA >200, 000 IU/ml.
Discussion
This is the first multi-national study of HIV-HBV co-infected individuals characterizing both HBV and HIV parameters at the time of HAART initiation. Several notable findings include an association between HBV co-infection and lower CD4+ T-cell count especially those with high HBV DNA levels, the low HBV DNA levels in the majority of HBeAg-negative subjects, and the absence of drug-resistant HBV. Such characterization of HBV in HIV-infected patients at the time of HAART initiation advances our understanding of HBV in this setting in order to focus treatment efforts.
The association of lower CD4+ T-cell counts with HIV-HBV co-infection was first noted in a Nigerian study, but it was not clear if that finding was universal or specific to Nigeria [8]. This is the first multi-national study demonstrating that HIV-HBV co-infected subjects have lower CD4+ T-cell counts than HIV monoinfected subjects. If the finding were region-specific, then the association would have been diminished with the diverse countries included in this study. This finding is also robust because it was detected despite the fact that study entry criteria for the parent studies required a CD4+ T-cell count <300 cells/ mm3 (for A5175) or 200 cells/ mm3 (for A5208). Furthermore, this is the first study to demonstrate that the HIV-HBV co-infected subjects with CD4+ T-cell counts <50 cells/mm3 were more likely to have high levels of HBV replication (HBV DNA >200, 000 IU/ml) than were those with higher CD4+ T-cell counts. One possible mechanism for this association is that CH-B may lead to immune activation, which increases CD4+ T-cell apoptosis. This association is not due to differences in geographical distribution of CD4+ T-cell counts among countries since those with higher HBV prevalence did not recruit subjects with lower CD4+ T-cell counts.
The low levels of HBV DNA in a large proportion of individuals was unexpected since HIV-HBV co-infection is associated with higher HBV DNA in subjects from areas of the world with low HBV endemicity [6]. In our study, 16% of the HBeAg negative subjects had an undetectable HBV DNA, and an additional 50% had a HBV DNA <2000 IU/ml, which is the level above which treatment is considered. In contrast, the majority of HBeAg-positive subjects (75%) had detectable HBV DNA levels that were >200,000 IU/ml. These findings are supported by the multivariable analysis demonstrating that HBeAg-positive status was the only factor associated with higher HBV DNA levels. The low HBV DNA in this cohort is not simply due to exclusion of subjects with higher ALT or AST levels since studies from Nigeria and South Africa that did not exclude such patients also show low HBV DNA in a substantial proportion of HIV-HBV co-infected subjects [8, 18]. In the Nigerian study, these low levels were also found primarily in HBeAg-negative subjects, but HBeAg was not determined in the other study. Taken together, these studies suggest that determining HBeAg may provide a strategy to prioritize the need for HBV treatment in HIV-infected individuals when HBV DNA assays are not available in resource-limited settings.
In our study, no subject had pre-existing drug-resistant mutations in the majority population of the quasispecies. In two prior Japanese studies of therapy naïve subjects, the prevalence of drug-resistant mutants using population sequencing was 0 and 1.6% [19, 20]. One of these studies used a second assay that could detect as little as 0.001% of mutant virus and found that 11% of subjects had a drug-resistant virus in the minority population [19]. However, whether such low levels of virus lead to HBV treatment failure is not known. One study found lamivudine-resistant strains in 10 of 20 HIV-HBV co-infected patients prior to therapy, but the pattern of mutations was identical in the majority of the tested samples suggesting that contamination may explain this high prevalence [21]. Our study does not support the need for HBV drug-resistance testing prior to starting anti-HBV therapy.
Our study had several limitations. First, the inclusion criteria of the parent antiretroviral treatment trials may have skewed our study population. Even though both parent studies were focused on participants with CD4 T-cell counts < 300 cells/mm3, we were still able to see an association between CD4 T-cell count and HBV infection. The exclusion of subjects with very high AST and ALT, may have underestimated the prevalence of CH-B especially those with HBeAg-positive disease. However, we believe that this exclusion did not substantially alter the HBV DNA findings or the association with lower CD4+ T-cell counts, as discussed above. In addition, HIV-HBV co-infected subjects are recognized to have lower ALT levels than are HBV monoinfected subjects despite higher HBV DNA levels [6]. Second, we were not able to characterize liver disease using a liver biopsy since biopsies were not feasible and are not readily available in resource-limited countries. However, we did use a non-invasive marker of liver disease (AAL reactivity) that correlates with the level of fibrosis [11] and does not use platelet count, which can be affected by HIV. About 15% of the cohort had a level of AAL reactivity that was consistent with cirrhosis, which was almost exclusively found in subjects of African descent; however, the only sign of liver disease in these subjects was a trend towards lower albumin. This prevalence is similar to that described in HBV monoinfected cohorts with ALT cut-offs that were lower at <2x the ULN [22]. A Ugandan study found a 17% prevalence of significant fibrosis in HIV mono-infected subjects [23]; thus, the cirrhosis prevalence in our study of HIV-HBV co-infected individuals is possible. Further studies are needed to characterize liver disease in HIV-HBV co-infected subjects from resource-limited settings including those with high ALT/AST levels. Third, we did not find an association in the multivariable analysis with HBV genotype and HBV DNA, but these data are limited by the inability to genotype 16% of the subjects. This lack of association was also seen in a study from Denmark of 784 HBV monoinfected patients [24]. However, in another study HBV DNA levels were the highest in HBeAg-positive subjects with HBV genotype D [25].
In summary, we found a variable prevalence (2–11%) of hepatitis B in HIV-infected subjects from diverse settings prior to starting HAART. Thus, it is important for providers to know the prevalence of HBV in their country. HBV DNA levels were relatively low in HBeAg negative subjects; thus, this serologic marker may be useful in prioritizing patients on their need for HBV treatment in settings where HBV DNA is not available. HIV-HBV co-infection, especially those with high HBV DNA, was associated with lower CD4+ T-cell counts prior to HAART initiation, which emphasizes the need for HBV testing in HIV-infected patients. Further work is needed to study HIV-HBV co-infection in subjects with high ALT/AST levels, to understand the mechanism for CD4 + T-cell loss in HIV-HBV co-infection, and to optimize treatment for HIV-HBV co-infection in resource-limited settings.
Acknowledgments
Funding support:
This work was supported by the National Institute of Allergy and Infectious Diseases at the National Institutes of Health [R01 AI071820 (CT), K24 AI56933 (JC), AI068636 (AIDS Clinical Trials Group), AI69450 (TC), U01 AI068634 (Statistical and Data Management Center of the AIDS Clinical Trials Group), R01CA120206 (AM), R01CA136607 (AM)]. It was also supported by Award Number U01AI068636 from the National Institute of Allergy and Infectious Diseases, the National Institute of Mental Health (NIMH), and the National Institute of Dental and Craniofacial Research (NIDCR). Study drugs were provided by Abbott Laboratories, Boehringer Ingelheim Pharmaceuticals, Gilead Sciences, Bristol-Myers Squibb, GlaxoSmithKline, and Merck & Co. Inc.
Additional funding for each site includes:
UZ-Parirenyatwa CRS (Site 30313) CTU Grant: 1U01AI069436-01
Wits HIV CRS (Site 11101) CTU Grant: 1U01AI069463-01
University of North Carolina Project, Kamuzu Central Hospital (Site 12001) CTU Grant: 1U01AI069518-01
Durban Adult HIV CRS (Site 11201) CTU Grant: 1U01AI069426-01
YRG CARE Medical Ctr., VHS CRS (Site 11701) CTU Grant: 1U01AI069432-01
Instituto de Pesquisa Clinica Evandro Chagas CRS (Site 12101) CTU Grant: 1U01AI069476-01
College of Med. JHU CRS (Site 30301) CTU Grant: 1U01AI069518-01
Chiang Mai Univ. ACTG CRS (Site 11501) CTU Grant: 1U01AI069399-01
Hospital Nossa Senhora da Conceicao CRS (Site 12201) CTU Grant: 1U01AI069401-01
Les Centres GHESKIO CRS (Site 30022) CTU Grant: 1U01AI069421-01
NARI Pune CRS (Site 11601) CTU Grant: 1U01AI069417-01
Asociacion Civil Impacta Salud y Educacion, Sede Barranco(Site 11301) CTU Grant: 5U01 AI069438
Walter Reed Project - Kenya Med. Research Institute (Site 12501) CTU Grant: IAA#Y1-AI-8374-01
AMPATH at MOI Univ. Teaching Hosp. Eldoret CRS (Site 12601) CTU Grant: IAA#Y1-AI-8374-01
Kalingalinga Clinic CRS (Site 12801) CTU Grant: 1U01AI069518-01
Gaborone Prevention/Treatment Trials CRS (Site 12701) CTU Grant: 1U01AI069456-01 (Site 12401) CTU Grant: 1U01AI069456-01
San Miguel CRS (Site 11302) CTU Grant: 5U01AI069438
Molepolole Prevention/Treatment Trials CRS (Site 12702) CTU Grant: 1U01AI069456-01
University of Texas Southwestern Medical Center (Site 3751) Grant: AI 046376
NARI Clinic at NIV CRS (Site 11603) CTU Grant: 1U01AI069417-01
University of Cincinnati (Site 2401) CTU Grant: AI-069513
Univ. of California Davis Med. Ctr., ACTU (Site 3851) Grant: AI38858-09S1
NARI Clinic at Gadikhana Dr. Kotnis Municipal Disp (Site 11602) CTU Grant: 1U01AI069417-01
University of Colorado Hospital CRS (Site 6101) CTU Grant: AI69450
The Ohio State University (Site 2301) CTU Grant: AI069474
Northwestern University CRS (Site 2701) CTU Grant: AI069471
University of Minnesota (Site 1501) CTU Grant: AI27661
Washington U CRS CTU (Site 2101) Grant: 1U01AI069495
Beth Israel Med. Ctr., ACTU (Site 2851) CTU Grant: AI46370
The Miriam Hospital (Site 2951) CTU Grant: AI069472-01
Duke University Medical Center CRS (Site 1601) CTU Grant: AI069484-06
University of Southern California CRS (Site 1201) CTU Grant: AI069428
Harbor-UCLA Medical Center (Site 603) CTU Grant: U01-A1 069424
UNC AIDS CRS (Site 3201) CTU Grant: AI069423; RR 025747; AI050410
Hospital of the Univ. of Pennsylvania CRS (Site 6201) CTU Grant: U01-AI- AI69467-05; CFAR Grant: P30-AI-045008-12
HIV Prevention & Treatment CRS (Site 30329) CTU Grant: 1U01AI069470
Vanderbilt Therapeutics CRS (Site 3652) CTU Grant: U01-AI069439
Rush Univ. Med. Ctr. ACTG CRS (Site 2702) CTU Grant: 1U01AI069471-01
University of Texas, Galveston (Site 6301) Grant: AI32782
New York University/NYC HHC at Bellevue Hospital (Site 401) CTU Grant: Al-27665; Al069532
Christine Hurley, RN and Roberto Corales, DO- AIDS Care CRS (Site 1108) CTU Grant: U01AI069511-02 (as of 2/12/08); CTSI: UL1 RR 024160
UCLA CARE Center CRS (Site 0601) CTU Grant: 1U01AI069424-01
University of Rochester (Site 1101) CTU Grant: U01AI069511-02 (as of 2/12/08); CRC: UL1 RR 024160
Cook County Hosp. CORE Ctr. (Site 2705) CTU Grant:1U01AI069471-01
SSTAR, Family Healthcare Ctr. (Site 2954) Grant: AI46381
Wake County Health and Human Services Clinical Research Site (Site 3206) CTU Grant: AI25868
University of Hawaii at Manoa, Leahi Hosp. (Site 5201) Grant: AI34853
Todd Stroberg, R.N., and Christina Megill, PA-C. -Cornell CTU (Site 7804) CTU Grant # AI069419 CTSC# RR024996
Footnotes
Author contributions: Study design (CT, JH, MN, UL, SL, TC, JC), data analysis (CT, LS, KH, JC), data collection (MS, HS, SS, SK, HI, AM), drafting of manuscript (CT, LS, KH, JC), critical reading of manuscript (all authors). The authors would like to acknowledge the efforts of the investigators at each of the sites participating in this study, Ken Braun and James Tutko for managing the database, and the A5175 and A5208 study participants.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
None of the authors have a potential conflict of interest.
References
- 1.Alter MJ. Epidemiology of viral hepatitis and HIV co-infection. J Hepatol. 2006;44(1 Suppl):S6–S9. doi: 10.1016/j.jhep.2005.11.004. [DOI] [PubMed] [Google Scholar]
- 2.Zhou J, Dore GJ, Zhang F, Lim PL, Chen YM. Hepatitis B and C virus coinfection in The TREAT Asia HIV Observational Database. J Gastroenterol Hepatol. 2007 Sep;22(9):1510–8. doi: 10.1111/j.1440-1746.2007.05062.x. [DOI] [PubMed] [Google Scholar]
- 3.Nyirenda M, Beadsworth MB, Stephany P, et al. Prevalence of infection with hepatitis B and C virus and coinfection with HIV in medical inpatients in Malawi. J Infect. 2008 Jul;57(1):72–7. doi: 10.1016/j.jinf.2008.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Diop-Ndiaye H, Toure-Kane C, Etard JF, et al. Hepatitis B, C seroprevalence and delta viruses in HIV-1 Senegalese patients at HAART initiation (retrospective study) J Med Virol. 2008 Aug;80(8):1332–6. doi: 10.1002/jmv.21236. [DOI] [PubMed] [Google Scholar]
- 5.Lee HC, Ko NY, Lee NY, Chang CM, Ko WC. Seroprevalence of viral hepatitis and sexually transmitted disease among adults with recently diagnosed HIV infection in Southern Taiwan, 2000–2005: upsurge in hepatitis C virus infections among injection drug users. J Formos Med Assoc. 2008 May;107(5):404–11. doi: 10.1016/S0929-6646(08)60106-0. [DOI] [PubMed] [Google Scholar]
- 6.Colin JF, Cazals-Hatem D, Loriot MA, et al. Influence of human immunodeficiency virus infection on chronic hepatitis B in homosexual men. Hepatology. 1999 Apr;29(4):1306–10. doi: 10.1002/hep.510290447. [DOI] [PubMed] [Google Scholar]
- 7.Thio CL, Seaberg EC, Skolasky RL, et al. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter AIDS Cohort Study (MACS) Lancet. 2002 Dec 14;360:1921–6. doi: 10.1016/s0140-6736(02)11913-1. [DOI] [PubMed] [Google Scholar]
- 8.Idoko J, Meloni S, Muazu M, et al. Nigerian Cohort. 49. 2007. Hepatitis B Virus Co-Infection Impacts Baseline HIV parameters and HAART-related Hepatotoxicity Risk in an HIV-infected; pp. 1268–73. [Google Scholar]
- 9.Campbell TC, Smeaton L, Kumarasamy N, et al. Efficacy and safety of EFV with either co-formulated 3TC/ZDV or FTC/TDF for initial treatment of HIV-1 infected men and women in diverse multinational settings: ACTG PEARLS Study. 18th Conference on Retroviruses and Opportunistic Infections; 2011 Feb. [Google Scholar]
- 10.Lockman S, Hughes MD, McIntyre J, et al. Antiretroviral therapies in women after single-dose nevirapine exposure. N Engl J Med. 2010 Oct 14;363(16):1499–509. doi: 10.1056/NEJMoa0906626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehta AS, Long RE, Communale MA, et al. Increased levels of galactose-deficient anti-gal IgG in the sera of hepatitis C virus-infected individuals with fibrosis and cirrhosis. J Virol. 2008 Feb;82(3):1259–70. doi: 10.1128/JVI.01600-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews GV, Bartholomeusz A, Locarnini S, et al. Characteristics of drug resistant HBV in an international collaborative study of HIV-HBV-infected individuals on extended lamivudine therapy. AIDS. 2006 Apr 4;20(6):863–70. doi: 10.1097/01.aids.0000218550.85081.59. [DOI] [PubMed] [Google Scholar]
- 13.Division of AIDS. Table for grading the severity of adult and pediatric adverse events 2009. 2009 Available at http://rsc.tech-res.com/Document/safetyandpharmacovigilance/Table_for_Grading_Severity_of_Adult_Pediatric_Adverse_Events.pdf.
- 14.Hughes SA, Wedemeyer H, Harrison PM. Hepatitis delta virus. Lancet. 2011 Jul 2;378(9785):73–85. doi: 10.1016/S0140-6736(10)61931-9. [DOI] [PubMed] [Google Scholar]
- 15.Neuschwander-Tetri BA, Caldwell SH. Nonalcoholic steatohepatitis: summary of an AASLD Single Topic Conference. Hepatology. 2003 May;37(5):1202–19. doi: 10.1053/jhep.2003.50193. [DOI] [PubMed] [Google Scholar]
- 16.Lok AS, McMahon BJ. Chronic hepatitis B: update 2009. Hepatology. 2009 Sep;50(3):661–2. doi: 10.1002/hep.23190. [DOI] [PubMed] [Google Scholar]
- 17.Deng L, Tang H. Hepatitis B virus drug resistance to current nucleos(t)ide analogs: Mechanisms and mutation sites. Hepatol Res. 2011 Nov;41(11):1017–24. doi: 10.1111/j.1872-034X.2011.00873.x. [DOI] [PubMed] [Google Scholar]
- 18.Hoffmann CJ, Charalambous S, Martin DJ, et al. Hepatitis B virus infection and response to antiretroviral therapy (ART) in a South African ART program. Clin Infect Dis. 2008 Dec 1;47(11):1479–85. doi: 10.1086/593104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirishima T, Okanoue T, Daimon Y, et al. Detection of YMDD mutant using a novel sensitive method in chronic liver disease type B patients before and during lamivudine treatment. J Hepatol. 2002 Aug;37(2):259–65. doi: 10.1016/s0168-8278(02)00145-9. [DOI] [PubMed] [Google Scholar]
- 20.Ohishi W, Shirakawa H, Kawakami Y, et al. Identification of rare polymerase variants of hepatitis B virus using a two-stage PCR with peptide nucleic acid clamping. J Med Virol. 2004 Apr;72(4):558–65. doi: 10.1002/jmv.20026. [DOI] [PubMed] [Google Scholar]
- 21.Selabe SG, Lukhwareni A, Song E, Leeuw YG, Burnett RJ, Mphahlele MJ. Mutations associated with lamivudine-resistance in therapy-naive hepatitis B virus (HBV) infected patients with and without HIV co-infection: implications for antiretroviral therapy in HBV and HIV co-infected South African patients. J Med Virol. 2007 Nov;79(11):1650–4. doi: 10.1002/jmv.20974. [DOI] [PubMed] [Google Scholar]
- 22.Gobel T, Erhardt A, Herwig M, et al. High prevalence of significant liver fibrosis and cirrhosis in chronic hepatitis B patients with normal ALT in central Europe. J Med Virol. 2011 Jun;83(6):968–73. doi: 10.1002/jmv.22048. [DOI] [PubMed] [Google Scholar]
- 23.Stabinski L, Reynolds SJ, Ocama P, et al. High prevalence of liver fibrosis associated with HIV infection: a study in rural Rakai, Uganda. Antivir Ther. 2011;16(3):405–11. doi: 10.3851/IMP1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krarup H, Andersen S, Madsen PH, et al. HBeAg and not genotypes predicts viral load in patients with hepatitis B in Denmark: a nationwide cohort study. Scand J Gastroenterol. 2011 Dec;46(12):1484–91. doi: 10.3109/00365521.2011.619273. [DOI] [PubMed] [Google Scholar]
- 25.Lindh M, Horal P, Dhillon AP, Norkrans G. Hepatitis B virus DNA levels, precore mutations, genotypes and histological activity in chronic hepatitis B. J Viral Hepat. 2000 Jul;7(4):258–67. doi: 10.1046/j.1365-2893.2000.00236.x. [DOI] [PubMed] [Google Scholar]