Abstract
Objectives
Genome-wide association studies have recently identified genetic polymorphisms associated with common, etiologically complex diseases, for which direct-to-consumer genetic testing with provision of absolute genetic risk estimates is marketed by commercial companies. Polymorphisms associated with atrial fibrillation (AF) have shown relatively large risk estimates but the robustness of such estimates across populations and study designs has not been studied.
Design
A systematic literature review with meta-analysis and assessment of between-study heterogeneity was performed for single nucleotide polymorphisms (SNPs) in the six genetic regions associated with AF in genome-wide or candidate gene studies.
Results
Data from 18 samples of European ancestry (n=12,100 cases; 115,702 controls) were identified for the SNP on chromosome 4q25 (rs220733), 16 samples (n=12,694 cases; 132,602 controls) for the SNP on 16q22 (rs2106261) and 4 samples (n=5,272 cases; 59,725 controls) for the SNP in KCNH2 (rs1805123). Only the discovery studies were identified for SNPs on 1q21 and in GJA5 and IL6R, why no meta-analyses were performed for those SNPs. In overall random-effects meta-analyses, association with AF was observed for both SNPs from genome-wide studies on 4q25 (OR 1.67, 95% CI=1.50–1.86, p=2×10−21) and 16q22 (OR 1.21, 95% CI=1.13–1.29, p=1×10−8), but not the SNP in KCNH2 from candidate gene studies (p=0.15). There was substantial effect heterogeneity across case-control and cross-sectional studies for both polymorphisms (I2=0.50–0.78, p<0.05), but not across prospective cohort studies (I2=0.39, p=0.15). Both polymorphisms were robustly associated with AF for each study design individually (p<0.05).
Conclusions
In meta-analyses including up to 150,000 individuals, polymorphisms in two genetic regions were robustly associated with AF across all study designs but with substantial context-dependency of risk estimates.
Keywords: atrial fibrillation, genetics, genome-wide, prediction, SNP, meta-analysis
Introduction
Since genetic association studies became feasible on a genome-wide scale in 2006, reproducible associations of common variants (polymorphisms) with many complex diseases have been established. Whereas rare genetic variants (mutations), exclusive to individual families, can have a nearly deterministic impact on disease, polymorphisms confer a probabilistic increment in risk, set against other clinical and environmental risk factors in causing disease. Direct-to-consumer genetic testing for such disease-associated polymorphisms is currently marketed by commercial companies, providing the customer with an estimate of absolute genetic risk, derived from average population risk and genotypic risk estimates in published studies. However, absolute risk estimates have been reported to vary widely between companies and research on the clinical utility of genetic testing is only in its infancy with many questions still to be answered [1].
First, limited data have been published on the consistency of genotypic risk estimates across studies and populations. As genome-wide association (GWA) studies are tools for discovery of novel genetic susceptibility regions, narrow sample selection criteria are often utilized to improve statistical power, including the use of cases with early disease onset or with a family history of disease or the use of “super-controls”, who have not developed disease despite a high age. Such strategies can potentially inflate risk estimates and the results of GWA studies may therefore not be directly generalizable to genetic prediction on the population level. Second, the clinical validity of polymorphisms for prediction has for most diseases been shown to be limited due to relatively small conferred risks. For example, for coronary artery disease, one of the most well-studied diseases, risk estimates for associated polymorphisms have been shown to be similar across studies [2,3] but modest – odds ratio ~1.2 per risk allele, corresponding to an approximate risk increase of 20% compared to individuals without the risk allele – and contribute minimally to conventional risk factors in prediction, as quantified by the area under the receiver-operating characteristic (ROC) curve [3,4] which scale with the magnitude of risk estimates [5].
Relatively higher risk estimates with odds ratios up to 2.46, corresponding to a risk increase of 146% per risk allele, have been reported for polymorphisms associated with atrial fibrillation (AF)[6–10], another common cardiac disease and major risk factor for stroke, heart failure and death, which is known to have a heritable component [11–13]. To date, polymorphisms at six genomic regions have been reproducibly associated with AF in the community; chromosome 4q25, located 150 kb from the closest gene - a transcription factor (PITX2) involved in cardiac development; chromosome 16q22, located in an intron of another transcription factor of unknown function, expressed in cardiac tissue (ZFHX3); an amino acid-altering variant in KCNH2, the gene encoding one of the major cardiac voltage-gated potassium channels; and a variant located in an intron of the IL6R gene, encoding the receptor for the cytokine interleukin-6 [6–8,10,14]. Furthermore, a polymorphism on chromosome 1q21, located in an intron of a cardiac potassium channel (KCNN3) [9], and another polymorphism in the promoter region upstream of the GJA5 gene, encoding a component of cardiac gap junctions [15], have been reproducibly associated with lone AF, i e AF presenting at a young age in the absence of precipitating factors such as heart failure, valvular disease and hyperthyroidism. However, the consistency of risk estimates has not been evaluated for these polymorphisms.
Thus, we performed a systematic literature review of studies testing the association of these polymorphisms with AF and evaluated the robustness of risk estimates across studies of various design.
Materials and Methods
Systematic literature review
A systematic literature search was performed in PubMed for studies testing the association of AF with single nucleotide polymorphisms (SNPs) in the six genetic regions for which reproducible association has been reported: chromosome 4q25 (rs2200733), chromosome 16q22 (rs2106261) and chromosome 1q21 (rs13376333) identified in genome-wide association studies [6–9], and a missense SNP (rs1805123) in the KCNH2 gene and SNPs in GJA5 (rs10465885) and IL6R (rs4845625) identified in candidate gene studies [10,14,15]. The following search criterion was used: “atrial fibrillation” AND (4q25 OR 16q22 OR 1q21 OR PITX2 OR ZFHX3 OR KCNN3 OR KCNH2 OR GJA5 OR IL6R OR rs2200733 OR rs2106261 OR rs1337633 OR rs1805123 OR rs10465885 OR rs4845625). Reference lists of included articles were hand searched. We excluded studies reported in other languages than English or reporting only on interventional subgroups, such as patients undergoing coronary artery bypass graft surgery or cardiac catheter ablation procedures. Several published studies reported consortium results with multiple independent populations. These samples were listed and analyzed as separate datasets. Samples reporting results in more than one study were only included once, but population-based cohort samples could contribute to both cross-sectional and prospective analyses.
Meta-analyses
Meta-analyses were performed separately for samples of different primary continental ancestries (European, Asian, African). Heterogeneity was assessed using Cochran’s Q test for heterogeneity, computed as the sum of the squared deviations of each study’s effect from the weighted mean over the study variance, and the I2 test, the percentage of total variation across studies that is due to heterogeneity rather than chance (I2 = [Q − df]/Q) [16,17]. For each genetic polymorphism, effect estimates were combined across all studies and per study design (case-control, cross-sectional, prospective cohort) using random-effects meta-analysis as described by DerSimonian and Laird [18] or fixed-effects meta-analysis with inverse variance weights. To assess the potential role of study design or sample age as the source of any observed heterogeneity, additional heterogeneity analyses were performed by study design and sample mean age was meta-regressed on point estimates for cross-sectional and prospective cohort studies. Meta-regression was performed separately for cross-sectional and prospective cohort studies and was not performed for case-control studies as most studies had substantial differences in age distribution between cases and controls. Sample inclusion bias was evaluated for each SNP both overall and for each study design from funnel plots of individual study samples and Egger’s linear regression asymmetry test, where the standard normal deviate of the effect estimate is regressed against the inverse of its standard error and the hypothesis is tested that the intercept of the regression line differs from the origin [19]. Several genetic inheritance models were explored (additive, recessive and dominant models) in the largest published sample to study the relation of these genetic polymorphisms with AF – the prospective Malmö Diet and Cancer study (Smith JG et al, Arch Intern Med 2012: in press). Genetic models were compared using two measures of global fit: the Akaike information criterion (AIC) [20] and the Schwarz bayesian information criterion (BIC) [21]. Statistical analyses were performed in SAS 9.2 (SAS Institute, Cary, North Carolina, USA) or Stata version 11.1 (StataCorp, College Station, Texas, USA).
Results
Systematic literature review
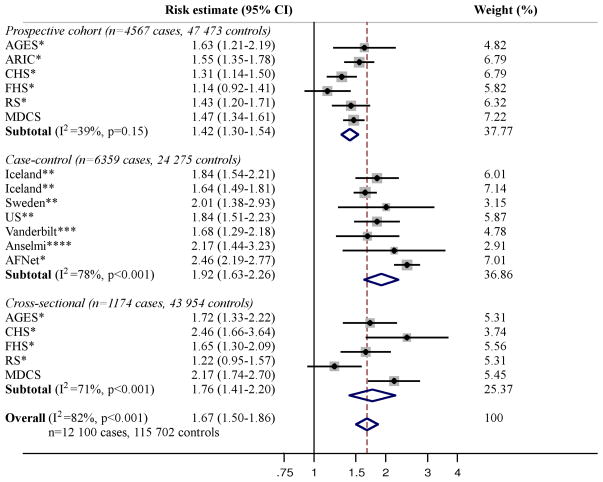
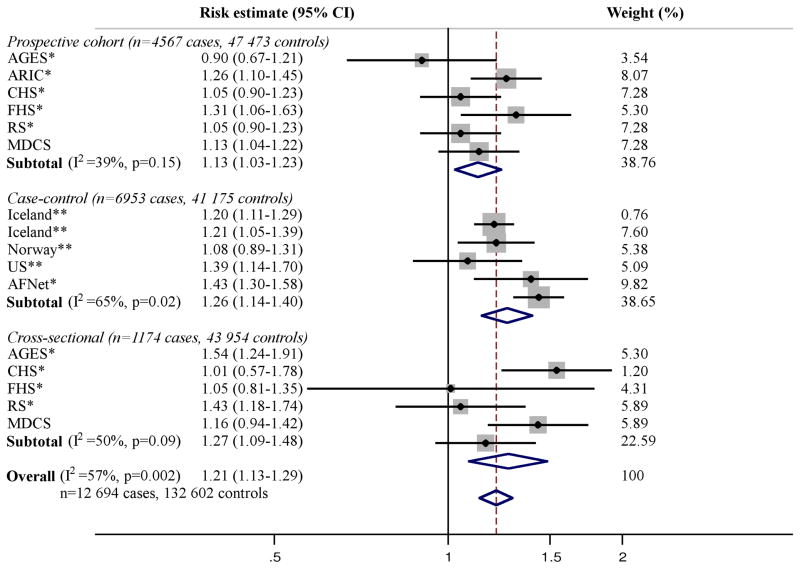
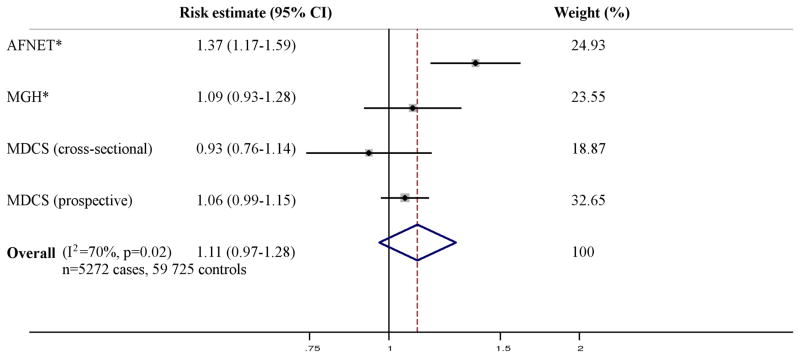
The literature review identified five publications including five cross-sectional, seven case-control, and six prospective European samples with a total of 12,100 cases and 115,702 controls, testing the association of the SNP on chromosome 4q25 and AF (Figure 1) [Smith JG et al, Arch Intern Med 2012: in press] [6,7,22,23]. For the SNP on chromosome 16q22, three publications were identified, including five cross-sectional, five case-control, and six prospective samples with a total of 12,694 cases and 132,602 controls (Figure 2) [Smith JG et al, Arch Intern Med 2012: in press] [7,8]. For the SNP in KCNH2, three publications including one cross-sectional, two case-control, and one prospective European samples with 5,272 cases and 59,725 controls were identified (Figure 3) [Smith JG et al, Arch Intern Med 2012: in press] [10,24]. For the SNPs on chromosome 1q21 and in IL6R and GJA5 we only identified the publications in which the associations were initially described [9,14,15], why no meta-analyses were performed for these SNPs.
Figure 1. Meta-analysis of the SNP on chromosome 4q25 and AF.
All results are presented as relative risk estimates (95% confidence intervals) from unadjusted, additive genetic models (adjusted for age and sex in the study by Kääb et al[22] and MDCS [Smith JG et al, Arch Intern Med 2012: in press]). Heterogeneity analysis and random-effects meta-analysis was performed per study design and across all studies. P-values in heterogeneity analyses refer to Cochrans Q test. Weights refer to DerSimonian-Laird weights. * Benjamin et al 2009 [7]. A proxy SNP (rs17042171) was used in this study (r2=1.0 in HapMap CEU). ** Gudbjartsson et al 2007 [6]. *** Kääb et al 2009 [22]. **** Viviani Anselmi et al 2009 [23].
Figure 2. Meta-analysis of the SNP on chromosome 16q22 and AF.
All results are presented as relative risk estimates (95% confidence intervals) from unadjusted, additive genetic models (adjusted for age and sex in MDCS [Smith JG et al, Arch Intern Med 2012: in press]). Heterogeneity analysis and random-effects meta-analysis was performed per study design and across all studies. P-values in heterogeneity analyses refer to Cochrans Q test. Weights refer to DerSimonian-Laird weights. * Benjamin et al 2009 [7]. ** Gudbjartsson et al 2009 [8]. A proxy SNP (rs7193343) was used in this study (r2=0.78 in HapMap CEU).
Figure 3. Meta-analysis of a missense SNP in KCNH2 and AF.
All results are presented as relative risk estimates (95% confidence intervals) from additive genetic models adjusted for age and sex (in MDCS [Smith JG et al, Arch Intern Med 2012: in press]), and additionally for hypertension in AFNET and MGH. Heterogeneity analysis and random-effects meta-analysis was performed per study design and across all studies. P-values in heterogeneity analyses refer to Cochrans Q test. P-values in heterogeneity analyses refer to Cochrans Q test. Weights refer to DerSimonian-Laird weights. * Sinner et al 2010 [24].
Few studies in samples of primarily non-European ancestry were identified: 3 samples of Asian ancestry from 3 publications [6,25,26] tested the association of the SNP on chromosome 4q25 and AF, with a total of 916 cases and 3845 controls. One study reporting association of the SNP on 16q22 with AF in a Chinese sample of 650 cases and 1447 controls (p=0.001) was also identified [27]. Two studies [14,28] reported association of 4q25 with AF and one study [28] tested the association of 16q22 with AF in African Americans.
Meta-analysis in samples of European ancestry
Characteristics for samples of European ancestry in previous studies included in the meta-analysis are shown in Table 1. All six population-based samples reported distribution of sex and age at baseline. Population-based samples had a similar proportion of male participants but varied widely in distribution of baseline age, ranging from a sample mean of 58.1 (SD 7.6) in the Malmö Diet and Cancer study (MDCS) to 76.5 (SD 5.5) in the Age, Gene/Environment Susceptibility study (AGES). Of the twelve case-control samples, two did not report distribution of sex and age at diagnosis for participants, and four additional samples did not provide characteristics for both cases and controls. The distribution of age and sex varied widely between cases and controls in most case-control studies. The age at diagnosis in cases ranged from 54.2 (13.7) in a case-control study from Nashville to 74.4 (8.7) in a study from Sweden. In individuals of European ancestry, the minor allele was the same for all cohorts (4q25: T. 16q22: A. KCNH2: C) and allele frequencies were similar across cohorts as shown in Supplementary Table 1 with weighted average minor allele frequencies of 0.11 for 4q25, 0.18 for 16q22 and 0.22 for KCNH2.
Table 1.
Baseline characteristics of samples of European ancestry included in meta-analyses
| Study | Sample size (cases/controls) | Case age (mean, SD) | Control age (mean, SD) | Case sex (% male) | Control sex (% male) | Genomic region | Reference |
|---|---|---|---|---|---|---|---|
| Case-control studies | |||||||
| Iceland | 550/4,476 | 72.5 (11.0) | 61.5 (15.8) | 67.3 | 49.2 | 4q25 | Gudbjartsson 2007 [6] |
| Iceland | 2,251/13,238 | 70.5 (13.0) | 61.9 (18.4) | 59.8 | 42.7 | 4q25 | Gudbjartsson 2007 [6] |
| Sweden | 143/738 | 74.4 (8.7) | 43.1 (12.3) | 46.2 | 59.7 | 4q25 | Gudbjartsson 2007 [6] |
| USA | 636/804 | NA | 67.4 (12.3) | NA | 50.9 | 4q25 | Gudbjartsson 2007 [6] |
| Germany* | 2,145/4,073 | 61.0 (11.6) | 49.2 (13.9) | 72.9 | 49.2 | 4q25, 16q22, KCNH2 | Benjamin 2009 [7] Sinner 2011 [24] |
| Iceland | 2381/33,723 | 72.9 (12.0) | NA | 59.2 | 41.4 | 16q22 | Gudbjartsson 2009 [8] |
| Iceland | 970/1,939 | 67.0 (13.5) | NA | 66.8 | 56.1 | 16q22 | Gudbjartsson 2009 [8] |
| Norway | 722/711 | NA | NA | NA | NA | 16q22 | Gudbjartsson 2009 [8] |
| USA | 735/729 | NA | NA | NA | NA | 16q22 | Gudbjartsson 2009 [8] |
| Nashville | 556/598 | 54.2 (13.7) | 56.6 (14.2) | 67.8 | 66.6 | 4q25 | Kääb 2009 [22] |
| Italy | 78/348 | 64.0 (12.3) | 35 | NA | NA | 4q25 | Viviani Anselmi 2008 [23] |
| Boston | 790/1,330 | 63.4 (14.6) | 66.4 (12.8) | 69.2 | 53.5 | KCNH2 | Sinner 2011 [24] |
| Cross-sectional studies | Age (mean, SD) | Sex (% male) | |||||
|
|
|||||||
| AGES | 241/2,718 | 76.5 (5.5) | 39.0 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| CHS | 66/3,201 | 72.3 (5.4) | 39.1 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| FHS | 280/4,184 | 65.6 (12.7) | 44.9 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| MDCS | 287/26,659 | 58.1 (7.6) | 39.4 | 4q25, 16q22, KCNH2 | Smith, in press | ||
| RS | 309/5,665 | 69.4 (9.1) | 40.6 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| Prospective cohort studies | |||||||
| AGES | 138/2,580 | 76.3 (5.5) | 37.2 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| ARIC | 731/7,355 | 57.0 (5.7) | 47.2 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| CHS | 763/2,438 | 72.2 (5.3) | 38.8 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| FHS | 343/3,841 | 64.7 (12.6) | 43.7 | 4q25, 16q22 | Benjamin 2009 [7] | ||
| MDCS | 2,050/24,609 | 58.1 (7.6) | 39.4 | 4q25, 16q22, KCNH2 | Smith, in press | ||
| RS | 542/5,123 | 69.1 (9.0) | 40.3 | 4q25, 16q22 | Benjamin 2009 [7] | ||
Characteristics at diagnosis for cases are presented separately characteristics at recruitment for controls, for case-control studies. Case-control samples are referred to based on geographic region of origin. Baseline characteristics are presented for the entire cohort for cross-sectional and prospective studies. AGES, Age, Gene/Environment Susceptibility study. ARIC, Atherosclerosis Risk in Communities study. CHS, Cardiovascular Health Study. FHS, Framingham Heart Study. MDCS, Malmö Diet and Cancer Study. NA, not available. RS, Rotterdam Study.
Sample characteristics retrieved from Sinner et al 2011 [23].
The association of 4q25 and 16q22 with AF was robust when analyzed in random-effects meta-analyses both overall (4q25, p=2×10−21; 16q22, p=1×10−8) and by study design as shown in Figure 1 and 2. No association was observed in a random-effects meta-analysis of KCNH2 with AF (p=0.15) as shown in Figure 3.
Significant heterogeneity of relative risk estimates across samples was observed for all three SNPs (I2=57–82%). When assessed by study design, heterogeneity for 4q25 was observed in case-control (I2=71%) and cross-sectional samples (I2=78%) but not prospective samples (I2=39%, p=0.15). Heterogeneity for 16q22 was observed for case-control samples (I2=65%). When meta-regressed on risk estimates for cross-sectional samples, no association was observed with mean sample age for the SNP on chromosome 4q25 (β [−0.01] per year, 95% CI=[−0.135] −0.115, p=0.81) or 16q22 (β 0.02 per year, 95% CI=[−0.04] −0.07, p=0.43).
No sample inclusion bias was detected for 4q25 or 16q22 (all p>0.05) overall or for any study design. Funnel plots including all samples are shown in Supplementary Figure 1 and 2.
Meta-analyses in samples of Asian and African ancestry
Minor allele frequencies were different in populations of African or Asian ancestry compared to European ancestry, as shown in Supplementary Table 2. Importantly, the minor allele for the SNP on chromosome 4q25 was was more highly prevalent in cohorts of Asian or African ancestry and was consistently the opposite allele (C) in populations of Asian ancestry compared to European or African ancestry (T). However, the SNP on chromosome 4q25 was significantly associated with AF in both individuals of Asian (p=2×10−4) and African (p=0.002) ancestry, as shown in Table 2. No significant heterogeneity was detected, but the number of studies for these ancestries were small. Only one study reported association of the SNP on chromosome 16q22 with AF in individuals of Asian ancestry and only one small study reported lack of association in individuals of African ancestry.
Table 2.
Results in samples of Asian and African ancestry
| Asian ancestry | 4q25 | 16q22 | Sample size |
|---|---|---|---|
| Hong Kong [6] | 1.42 (1.16–1.73, p=6×10−4) | - | 333/2836 |
| Han chinese [25] | 1.81 (1.21–3.20, p=4×10−11) | - | 383/851 |
| Taiwan [26] | 1.30 (0.95–1.78, p=0.10) | - | 200/158 |
| Han chinese [27] | - | 1.32 (1.15–1.51, p=7×10−5) | 650/1447 |
| Combined effect | 1.41 (1.15–1.67, p=2×10−4) | 916/3845 | |
| Heterogeneity | 0%, p=0.60 | ||
|
African ancestry
| |||
| USA [14] | 1.36 (1.12–1.66, p=0.002) | - | 263/3399 |
| Nashville [28] | 3.28 (1.50–7.21, p=0.003) | 1.05 (0.56–1.96, p=0.88) | 73/71 |
| Combined effect | 1.40 (1.10–1.70, p=0.002) | 336/3470 | |
| Heterogeneity | 50%, p=0.37 | ||
All studies were case-control studies, with the exception of the two population-based, African american cohorts in [14]. Results for the SNPs rs2200733 on chromosome 4q25 and rs2106261 on 16q22 are presented as odds ratio per risk allele with corresponding 95% confidence interval and p-value. Combined results from fixed-effects meta-analyses are presented where multiple samples were available, including odds ratio with corresponding 95% confidence interval, p-value and heterogeneity statistics including the I2 statistic and p-value for Cochran’s Q test. Sample size is presented as cases over controls.
Genetic inheritance models
In the prospective Malmö Diet and Cancer study, the lowest AIC and BIC statistics were consistently observed for additive genetic models for all three polymorphisms indicating best model fit, with the exception that the dominant model showed similar statistics to the additive model for 4q25 as shown in Table 3.
Table 3.
Genetic inheritance models
| Recessive | Dominant | Additive | ||||
|---|---|---|---|---|---|---|
| AIC | BIC | AIC | BIC | AIC | BIC | |
| 4q25 | 38898 | 38904 | 38852 | 38858 | 38852 | 38858 |
| 16q22 | 40502 | 40508 | 40506 | 40511 | 40502 | 40508 |
| KCNH2 | 38802 | 38808 | 38802 | 38807 | 38801 | 38806 |
Global model fit for each genetic model in the prospective Malmö Diet and Cancer study is presented as the Akaike information criterion (AIC) and the Schwarz bayesian information criterion (BIC).
Discussion
In this set of meta-analyses including up to 150,000 individuals, robust association with AF was observed for two genetic polymorphisms, both across all studies and within each individual study design. Both polymorphisms are common with population minor allele frequencies of 0.11 and 0.18. Relative risk estimates varied widely in magnitude across published studies and across case-control studies and cross-sectional studies, with 50–78% of variation due to heterogeneity rather than chance, but not across prospective cohort studies. Overall odds ratios were modest, 1.67 per risk allele for the polymorphism on chromosome 4q25 and 1.21 for 16q22. The very high odds ratios reported in some studies were confined to individual case-control or cross-sectional samples.
Few studies have addressed between-study heterogeneity of genetic risk estimates. Our findings stand in contrast to previous studies which observed minimal heterogeneity for polymorphisms associated with coronary artery disease [2,3] and highlight the fact that GWA studies constitute a discovery tool and that risk estimates need to be considered in the context of the population in which they were identified. The origin of the observed heterogeneity remains uncertain, but is likely to relate to the etiologic heterogeneity of the AF diagnosis. Although a stronger association with a family history of AF has been reported for AF presenting at a young age [11,12], we did not detect any effect interaction with age in meta-regression analyses. A particularly strong association with family history has been reported in individuals presenting with lone AF [13] and a previous study supports a larger effect of at least the polymorphism on chromosome 4q25 with lone AF [9]. In the light of these observations, it was not entirely unexpected that case-control studies varied widely in effect estimates, as they vary greatly in exclusion and inclusion criteria and ascertainment method. For example, cases with concomitant heart failure, valvular disease or hyperthyreoidism have been excluded from some case-control studies. The observation of heterogeneity across cross-sectional studies was more unexpected. Cohorts differ in age distribution but while age-dependent genotypic effects have been described for genetic polymorphisms associated with other traits [29], meta-regression analyses did not support any association between sample age and effect size. Effect heterogeneity across populations could also be due to varying linkage disequilibrium between genotyped polymorphisms with an ungenotyped, causal variant. However, a recent fine-mapping study of the 4q25 region did not find any stronger association than with the polymorphism genotyped here [30]. More likely, the observed heterogeneity across cross-sectional studies is explained by differences in population characteristics based on differences in recruitment, exclusion criteria and endpoint ascertainment. For example, participants with prevalent heart failure, valvular disease or coronary artery disease were excluded from genotyping in the Cardiovascular Health Study. These observations have implications for the interpretation of risk estimates from genetic studies of AF and other complex diseases.
Calls have been raised to add information about family history and genetic polymorphisms to current prediction models for AF in the general population [31]. Although guidelines from international arrhythmologist societies do not recommend genotyping for these polymorphisms [32], direct-to-consumer genotyping is available from commercial companies. Our findings suggest the feasibility of genetic risk prediction based on risk estimates from prospective cohort studies. Heterogeneity across prospective cohort studies was limited despite different age distributions at baseline, ascertainment methods and follow-up times. However, risk estimates are relatively modest and unlikely to improve predictive accuracy. Commercial companies offering genotyping need to carefully consider the context of estimates provided to customers. Furthermore, the lack of effective preventive interventions argue against clinical utility for these polymorphisms.
Our meta-analysis did not support the association of AF with the K897T missense variant in KCNH2. Although replication was claimed in the initial report, the association recently failed to replicate in another large case-control sample with similar age distribution and clinical characteristics to the sample in which the association was first identified [24], and in a large, prospective population-based study from our group [Smith JG et al, Arch Intern Med 2012: in press]. We estimated the power to detect (p<0.05) an additive OR of 1.25 as in the original report [10] or of 1.10 as in the meta-analysis to approaching 100% for both effect sizes, based on the current sample size and observed minor allele frequency for the K897T variant, using the genetic power calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/). Thus, the present meta-analysis does not support the large effect described in the initial report [10], but cannot rule out a small effect.
When this meta-analysis was performed, only the initial discovery studies with lone AF had examined the association of SNPs on 1q21 and GJA5 with AF, and only the initial discovery study had studied the association of SNPs near IL6R with AF. A number of SNPs associated with electrocardiographic PR interval have also recently been associated with AF, although this finding has not been replicated [33]. Even more recently, large meta-analysis increased the number of loci associated with AF from six to twelve [34]. Future studies will be needed to evaluate the robustness of effect estimates for these SNPs and the association with AF in the general population. Furthermore, only limited data was available in populations of primarily non-European ancestry. Additional studies in such populations are warranted.
In conclusion, two genetic polymorphisms are robustly associated with AF across study designs with consistent effect estimates across prospective cohort studies, but vary substantially across case-control and cross-sectional studies. These findings have general implications for the interpretation of risk estimates from genome-wide association studies and establish the feasibility of individual risk prediction of AF based on genetic polymorphisms in a prospective setting, although effect estimates are modest.
Supplementary Material
Supplementary Figure 1: Funnel plot of individual study samples for the SNP on chromosome 4q25 and AF.
Supplementary Figure 2: Funnel plot of individual study samples for the SNP on chromosome 16q22 and AF.
Supplementary Table 1: Minor allele frequencies in samples of European ancestry included in meta-analyses
Supplementary Table 2: Minor allele frequencies in samples of Asian and African ancestry
Acknowledgments
Drs Smith, Hedblad, Engström, Melander and Platonov gratefully acknowledge financial support from the Swedish Heart-Lung Foundation. Dr Smith was also supported by Skåne University Hospital, the Medical Faculty of Lund University and the Thorsten Westerström Foundation. Dr Newton-Cheh was supported by NIH grant K23-HL-080025, a Doris Duke Charitable Foundation Clinical Scientist Development Award, and a Burroughs Wellcome Fund Career Award for Medical Scientists. Drs Hedblad and Melander were supported by the Swedish Medical Research Council. Dr Melander was supported by grants from the European Research Council (StG-282255), the Medical Faculty of Lund University, Skåne University Hospital in Malmö, the Albert Påhlsson Research Foundation, the Crafoord Foundation, the Swedish National Health Service, the Hulda and Conrad Mossfelt Foundation, the Ernhold Lundströms Research Foundation, the King Gustaf V and Queen Victoria Fund, the Lennart Hanssons Memorial Fund, the Marianne and Marcus Wallenberg Foundation, and the Knut and Alice Wallenberg Foundation. Dr Platonov was supported by the Swedish National Health Service, Skåne University Hospital and the Craaford Foundation.
Footnotes
Financial disclosure: None.
Conflict of interest:
None.
References
- 1.Ng PC, Murray SS, Levy S, Venter JC. An agenda for personalized medicine. Nature. 2009;461:724–6. doi: 10.1038/461724a. [DOI] [PubMed] [Google Scholar]
- 2.Palomaki GE, Melillo S, Bradley LA. Association between 9p21 genomic markers and heart disease: a meta-analysis. JAMA. 2010;303:648–56. doi: 10.1001/jama.2010.118. [DOI] [PubMed] [Google Scholar]
- 3.Ripatti S, Tikkanen E, Orho-Melander M, et al. A multilocus genetic risk score for coronary heart disease: case-control and prospective cohort analyses. Lancet. 2010;376:1393–400. doi: 10.1016/S0140-6736(10)61267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009;150:65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pepe MS, Janes H, Longton G, Leisenring W, Newcomb P. Limitations of the odds ratio in gauging the performance of a diagnostic, prognostic, or screening marker. Am J Epidemiol. 2004;159:882–90. doi: 10.1093/aje/kwh101. [DOI] [PubMed] [Google Scholar]
- 6.Gudbjartsson DF, Arnar DO, Helgadottir A, et al. Variants conferring risk of atrial fibrillation on chromosome 4q25. Nature. 2007;448:353–7. doi: 10.1038/nature06007. [DOI] [PubMed] [Google Scholar]
- 7.Benjamin EJ, Rice KM, Arking DE, et al. Variants in ZFHX3 are associated with atrial fibrillation in individuals of European ancestry. Nat Genet. 2009;41:879–81. doi: 10.1038/ng.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gudbjartsson DF, Holm H, Gretarsdottir S, et al. A sequence variant in ZFHX3 on 16q22 associates with atrial fibrillation and ischemic stroke. Nat Genet. 2009;41:876–8. doi: 10.1038/ng.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellinor PT, Lunetta KL, Glazer NL, et al. Common variants in KCNN3 are associated with lone atrial fibrillation. Nat Genet. 2010;42:240–4. doi: 10.1038/ng.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sinner MF, Pfeufer A, Akyol M, et al. The non-synonymous coding IKr-channel variant KCNH2-K897T is associated with atrial fibrillation: results from a systematic candidate gene-based analysis of KCNH2 (HERG) Eur Heart J. 2008;29:907–14. doi: 10.1093/eurheartj/ehm619. [DOI] [PubMed] [Google Scholar]
- 11.Fox CS, Parise H, D’Agostino RB, Sr, et al. Parental atrial fibrillation as a risk factor for atrial fibrillation in offspring. JAMA. 2004;291:2851–5. doi: 10.1001/jama.291.23.2851. [DOI] [PubMed] [Google Scholar]
- 12.Arnar DO, Thorvaldsson S, Manolio TA, et al. Familial aggregation of atrial fibrillation in Iceland. Eur Heart J. 2006;27:708–12. doi: 10.1093/eurheartj/ehi727. [DOI] [PubMed] [Google Scholar]
- 13.Ellinor PT, Yoerger DM, Ruskin JN, MacRae CA. Familial aggregation in lone atrial fibrillation. Hum Genet. 2005;118:179–84. doi: 10.1007/s00439-005-0034-8. [DOI] [PubMed] [Google Scholar]
- 14.Schnabel RB, Kerr KF, Lubitz SA, et al. Large-scale candidate gene analysis in whites and African Americans identifies IL6R polymorphism in relation to atrial fibrillation: the National Heart, Lung, and Blood Institute’s Candidate Gene Association Resource (CARe) project. Circ Cardiovasc Genet. 2011;4:557–64. doi: 10.1161/CIRCGENETICS.110.959197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wirka RC, Gore S, Van Wagoner DR, et al. A common connexin-40 gene promoter variant affects connexin-40 expression in human atria and is associated with atrial fibrillation. Circ Arrhythm Electrophysiol. 2011;4:87–93. doi: 10.1161/CIRCEP.110.959726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cochran WG. The combination of estimates from different experiments. Biometrics. 1954;10:101–29. [Google Scholar]
- 17.Higgins JP, Thompson SG. Quantifying heterogeneity in a meta-analysis. Stat Med. 2002;21:1539–58. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- 18.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–88. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 19.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–23. [Google Scholar]
- 21.Schwarz GE. Estimating the dimension of a model. Ann Stat. 1978;6:461–4. [Google Scholar]
- 22.Kääb S, Darbar D, van Noord C, et al. Large scale replication and meta-analysis of variants on chromosome 4q25 associated with atrial fibrillation. Eur Heart J. 2009;30:813–9. doi: 10.1093/eurheartj/ehn578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Viviani Anselmi C, Novelli V, Roncarati R, et al. Association of rs2200733 at 4q25 with atrial flutter/fibrillation diseases in an Italian population. Heart. 2008;94:1394–6. doi: 10.1136/hrt.2008.148544. [DOI] [PubMed] [Google Scholar]
- 24.Sinner MF, Lubitz SA, Pfeufer A, et al. Lack of Replication in Polymorphisms Reported to be Associated with Atrial Fibrillation. Heart Rhythm. 2011;8:403–9. doi: 10.1016/j.hrthm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi L, Li C, Wang C, et al. Assessment of association of rs2200733 on chromosome 4q25 with atrial fibrillation and ischemic stroke in a Chinese Han population. Hum Genet. 2009;126:843–9. doi: 10.1007/s00439-009-0737-3. [DOI] [PubMed] [Google Scholar]
- 26.Lee KT, Yeh HY, Tung CP, et al. Association of rs2200733 but not rs10033464 on 4q25 with atrial fibrillation based on the recessive model in a Taiwanese population. Cardiology. 2010;116:151–6. doi: 10.1159/000318172. [DOI] [PubMed] [Google Scholar]
- 27.Li C, Wang F, Yang Y, et al. Significant association of SNP rs2106261 in the ZFHX3 gene with atrial fibrillation in a Chinese Han GeneID population. Hum Genet. 2011;129:239–46. doi: 10.1007/s00439-010-0912-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Delaney JT, Jeff JM, Brown NJ, et al. Characterization of genome-wide association-identified variants for atrial fibrillation in African Americans. PLoS One. 2012;7:e32338. doi: 10.1371/journal.pone.0032338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lasky-Su J, Lyon HN, Emilsson V, et al. On the replication of genetic associations: timing can be everything! Am J Hum Genet. 2008;82:849–58. doi: 10.1016/j.ajhg.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lubitz SA, Sinner MF, Lunetta KL, et al. Independent susceptibility markers for atrial fibrillation on chromosome 4q25. Circulation. 2010;122:976–84. doi: 10.1161/CIRCULATIONAHA.109.886440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnar DO, Holm H, Gudbjartsson DF. Predicting atrial fibrillation. Lancet. 2009;373:1523. doi: 10.1016/S0140-6736(09)60857-6. [DOI] [PubMed] [Google Scholar]
- 32.Ackerman MJ, Priori SG, Willems S, et al. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 33.Pfeufer A, van Noord C, Marciante KD, et al. Genome-wide association study of PR interval. Nat Genet. 2010;42:153–9. doi: 10.1038/ng.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellinor PT, Lunetta KL, Albert CM, et al. Meta-analysis identifies six new loci for atrial fibrillation. Nat Genet. 2012 doi: 10.1038/ng.2261. epub Apr 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1: Funnel plot of individual study samples for the SNP on chromosome 4q25 and AF.
Supplementary Figure 2: Funnel plot of individual study samples for the SNP on chromosome 16q22 and AF.
Supplementary Table 1: Minor allele frequencies in samples of European ancestry included in meta-analyses
Supplementary Table 2: Minor allele frequencies in samples of Asian and African ancestry