Abstract
Objectives.
To review the role of Wnt pathways in the neurodevelopment of schizophrenia.
Methods:
Systematic PubMed search, using as keywords all the terms related to the Wnt pathways and crossing them with each of the following areas: normal neurodevelopment and physiology, neurodevelopmental theory of schizophrenia, schizophrenia, and antipsychotic drug action.
Results:
Neurodevelopmental, behavioural, genetic, and psychopharmacological data point to the possible involvement of Wnt systems, especially the canonical pathway, in the pathophysiology of schizophrenia and in the mechanism of antipsychotic drug action. The molecules most consistently found to be associated with abnormalities or in antipsychotic drug action are Akt1, glycogen synthase kinase3beta, and beta-catenin. However, the extent to which they contribute to the pathophysiology of schizophrenia or to antipsychotic action remains to be established.
Conclusions:
The study of the involvement of Wnt pathway abnormalities in schizophrenia may help in understanding this multifaceted clinical entity; the development of Wnt-related pharmacological targets must await the collection of more data.
Keywords: Antipsychotic Drugs, Neurodevelopment, Schizophrenia, Wingless (Wnt) signalling.
1. INTRODUCTION
1.1. Neurodevelopment in Schizophrenia
A neurodevelopmental hypothesis for schizophrenia finds its bases in observations and speculations dating back to the mid-sixties [1]. In the early eighties, Irwin Feinberg [2] proposed a defect in synaptic pruning, during a critical period spanning from childhood to adolescence, as the basis of many symptoms of schizophrenia. Further on, Daniel Weinberger [3] in the United States and Robin Murray in the United Kingdom [4] advanced neurodevelopmental hypotheses to explain the coexistence of both positive and negative symptoms in the same patients, symptoms which were previously attributed to antithetic neurochemical mechanisms [5]. These were refined through the years [6-28], while Feinberg’s original hypothesis was revisited and enriched with new ideas [29]. According to the neurodevelopmental hypothesis, schizophrenia represents the outcome of faulty brain development and maturation (and possibly, programming) occurring prior to clinical onset. The multitude of symptoms would arise from an interaction between genetic and environmental factors. The neurodevelopmental model of schizophrenia is supported by several genetic, preclinical, clinical, histological, and neuroimaging data. More precisely, perinatal and obstetric complications [30], prenatal infections [31, 32], cognitive-behavioural and motor signs and symptoms during childhood and adolescence [33], and the persistence of developmental markers beyond the time for which they are expected to be active [34], were reported in many patients with schizophrenia, although their relationship with the development of schizophrenia might be more complex than a simple association (for example, see references [35 and 36]). There is evidence that abnormal brain differentiation and development already occur during pregnancy. In fact, using prenatal ultrasound scans, as well as neonatal structural magnetic resonance imaging (MRI) and diffusion tensor imaging (DTI), differences in cerebrospinal fluid (CSF)-occupied spaces were found between male neonates with and without genetic risk for schizophrenia [37]. However, highly complex biological processes, like neurodevelopment, are unlikely to be driven by one single gene or factor.
1.2. The Wnt Signalling Pathway and Development
Although molecules of the Wnt signalling pathway are best known for their roles in embryogenesis and tumori-genesis, they are also involved in several aspects of develop-ment and adult physiology, including cell differentiation and cell polarity generation. The Wnt pathways are conventionally classified as canonical and non-canonical according to whether β-catenin is the principal target of the cascade or not, respectively, but the functional subdivision of Wnt signalling has no basis, as the signalling through the various participating molecules is integrated [38]. The Wnt/β-catenin pathway plays a critical role in the regulation of cell proliferation and in cell fate specification during development, including neurodevelopment [39]. There are many non-canonical, β-catenin-independent pathways, which are activated by Wnt4, Wnt5a, and Wnt11; the best-studied pathways are the Planar Cell Polarity (PCP) and Wnt/Ca2+ pathways [40, 41]. In the Wnt/Ca2+ pathway, Wnt5a signalling through frizzled (Fzd) and dishevelled (Dvl) regulates intracellular Ca2+ levels determining Ca2+ release from intracellular stores and causing activation of phospholipase C (PLC), leading to the generation of diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), the function of which is to increase intracellular Ca2+ concentration [42]. The non-canonical ligand Wnt5a showed the ability to inhibit the canonical pathway through its binding and internalisation of Ror2, an orphan tyrosine kinase, and subsequent c-Jun-N-terminal kinase (JNK) activation; the whole process ensues in the inhibition of downstream nuclear effects of β-catenin, namely TCF-dependent transcriptional activity and cyclin D1 expression [43]. In the PCP pathway, ligands bind also to Fzd and Dvl to promote scaffold protein-Daam1 complex formation, which activates the G-protein Rho. This protein activates ROCK (Rho-associated kinase), a major cytoskeletal regulator. The most important effects on the cytoskeleton are represented by ROCK binding to actin, which leads to its polymerisation [44-47]. PCP signalling and Wnt/β-catenin signallings are central in the development of the organism. They are strongly associated with body axis elongation and neural tube formation through the process of convergent extension [48, 49]. There are four major classes of secreted extracellular signalling molecules which are expressed in the developing brain during embryogenesis and participate in the patterning of the nervous system, i.e., Wnts, fibroblast growth factors (FGFs), Sonic Hedgehog (SHH), and Bone Morphogenetic Proteins (BMPs) [50]. Wnts, like BMPs, stem from the cortical hem, comprising the medial margin of each hemisphere. Wnt signalling has proven to be essential for neural development at various stages; the first evidence came from a study reporting hypotrophy of the cerebellum and midbrain in knockout-for-the-Wnt–1-gene mice [51]. Subsequently, a general principle of Wnt signalling during brain development has been discovered, namely, that regional specification needs an anteroposterior gradient of Wnt signalling, i.e., low for the anterior structures and high for the posterior ones [52, 53]. Wnt molecules are involved not only in patterning, but also in morphogenesis, neuronal migration and specification [54-56], neuronal precursor proliferation [55], synaptic differentiation [57, 58], and mature synapse modulation [59-61]. Their proliferation-promoting role is central in stem cell maintenance and progenitor pool expansion. Furthermore, Wnt signalling is involved in differentiation processes and lineage decision events during both central and peripheral nervous system development. In the adult brain, it may select neural circuitry formation through the regulation of long-term potentiation [62-64]. This matches its action in axon guidance, neurite outgrowth, and synaptic plasticity (recently reviewed in ref. [65]). The Wnt signalling pathway is also essential in maintaining proper communication between neuronal and non-neuronal cells; a distruption of this balance in advanced age may lead to neurodegeneration and related disorders, such as Alzheimer’s disease [66, 67].
The purpose of our review was to identify possible connections between schizophrenia and molecules related to the Wnt pathway in order to establish the hierarchy of this developmental pathway in the pathophysiology of this complex disorder.
2. METHODS
To identify studies of Wnt-related molecules, we searched the PubMed database using all Wnt-related terms and crossing them with each of the specific targets of our review, i.e., all antipsychotics and all disorders of the schizophrenic spectrum. Identified papers’ reference lists were searched subsequently to identify further relevant literature.
We proceeded similarly for other sections of this review. To review the role of the Wnt pathway in human physiology and normal development, we crossed the above Wnt-related set with other key words, like neurodevelopment, development, apoptosis, cellular death, proliferation, oncogenesis, physiology, nervous system, embryogenesis etc. To review the neurodevelopmental theory of schizophrenia we crossed the key word schizophrenia with terms like neurodevelop-ment, neurodevelopmental, development, developmental, CNS, and neurodegeneration. There were no timeframe limitations in our searches.
3. ROLE OF INDIVIDUAL WNT SIGNALLING MOLECULES IN DEVELOPMENT AND NEURO-DEVELOPMENT
3.1. Wnt1: Wnt1 is a Secreted Glycoprotein and a Member of the Wnt Protein Family
In the human, it is encoded by the evolutionally highly conserved WNT1 gene. Wnt1 acts through the Frizzled (Fdz) family of receptors. Originally, Wnt1 has been identified as a proto-oncogene whose overexpression contributed to mammary oncogenesis [68]. Its interaction with Wnt3a is suggested to be important for neural tube patterning [69]. In vivo data from transgenic mouse lines and explant cultures show that Wnt1 is required for establishment and differentiation of midbrain dopaminergic neurons during embryogenesis [70].
3.2. Wnt2: Wnt2 is a Member of the Wnt Family
The human gene encoding Wnt-2 has been identified through its sequence similarity with the murine protoonco-gene Wnt1 gene and its Drosophila ‘wingless’ homologue [71]. However, its role in schizophrenia is unclear, since its polymorphisms were not found to be associated with this disorder [72].
3.3. Frizzled Family
The Frizzled gene family, mostly through the mediation of Wnt signaling, is involved in the development of various structures. There are several kinds of Wnt receptors, principally belonging to the Frizzled (FZD) family, that regulate both the canonical and non-canonical pathways [73, 74]. The Frizzled family receptors comprise ten different receptors that mainly modulate the canonical pathway [75]. The FZD3 gene consists of eight exons and is approximately 70 kb long; it encodes for the FZD3 receptor, one of the major components of the Wnt signaling pathway, which is involved in the regulation of early neurodevelopmental processes. FZD3 is an almost ubiquitous transmembrane receptor for Wnt ligands implicated in the specification of several structures, like neural crest derivatives, and in the correct axonal targeting during the development of major forebrain fibre tracts [76-80]. The FZD3 gene spans approximately 60 kb of DNA and about 40 SNPs are registered in the SNP database of the US National Center for Biotechnology Information.
Frizzled proteins possess an outer N-terminus and seven transmembrane domains, concluding with a C-terminal intracytoplasmic tail involved in cellular signalling [81]. Only for a few of these multiple FZDs has their role in development been elucidated, primarily in invertebrates. In the nematode, Caenorhabditis elegans, FZDs regulate cell fate, asymmetric cell divisions, and spindle pole orientation [82]. Much less is known about their roles in vertebrate development. In the highly aquatic amphibian, Xenopus, Xfz-10 promotes sensory neuron development [83]; maternal Xfz7 is required for dorsal axis development and for response to ectopic Xwnt8b [84], while Xfz8, which is expressed in the Spemann organizer, appears to organise cell movements during gastrulation [85], in dorsal development [86]. It is also required for oligodendrocyte development in the vertebrate zebrafish spinal cord [87]. Wnt/Frizzled signalling is involved through Xfz3 in early entraining neural crest development and specification [77-79]. Xfz3-interacting protein Kermit [88] also shows that Xfz3 signaling is required for neural crest formation. Last, Xfz3, mediates eye development in the Xenopus [89]. In the Cyprinida zebrafish, FzA, which is closely related to Xfz8, Xfz3 may play a role in dorsoventral patterning [90]; Fz7a shows both maternal and zygotic strong expression within the anterior neuroectoderm and is more weakly expressed within the lateral mesoderm. During somitogenesis, it is detected in the developing central nervous system (CNS), including the somatic and posterior lateral mesoderm, and the migrating lateral line primordium [91, 92]. Finally, F8 is required for oligodendrocyte development in the zebrafish spinal cord [87].
However, it is unclear which endogenous ligands activate these or any other vertebrate FZDs. Furthermore, ligands related to canonical and non-canonical pathways may compete for FZDs and exert opposite effects to Wnt-mediated intracellular translation [93], thus forming a complex network of Wnt molecules and FZDs to control different actions [94] and novel ligands are continuously discovered to render the whole picture extremely complex [95].
In mammals, two of the most complex roles are the specification of neurons within the CNS and the correct targeting of their axons. Despite the large number of FZDs and their widespread expression within the CNS [96-98], only a few reports investigated their role in specification and axon targeting. Fz3 knock-out mice express massive defects in the development of fibre tracts within the forebrain, while leaving other aspects of CNS development mostly unaffected [80].
3.4. Low-density Lipoprotein Receptor 5/6 (LRP5/6)
The low-density lipoprotein (LDL) receptor is a member of a family composed of seven structurally closely related transmembrane proteins (LRP1, LRP1b, megalin/LRP2, LDL receptor, very low-density lipoprotein receptor, MEGF7/LRP4, and LRP8/apolipoprotein E receptor2). Wnt proteins bind Frizzled (FZD) and LDL-receptor related protein (LRP)5/6 cell surface receptors and prevent glycogen synthase kinase (GSK)-dependent phosphorylation of β-catenin, thus leading to its stabilisation [99]. Regarding neurodevelopment, LRPs may be involved in signal transduction during early neuronal differentiation [100]. In Xenopus embryos LRP6 activated Wnt-Fz signalling and induced Wnt responsive genes, dorsal axis duplication and neural crest formation. In addition, a LRP6 mutant lacking the carboxyl intracellular domain blocked signalling through Wnt or Wnt-Fz, but not through Dishevelled or β-catenin, thus inhibiting neural crest development [101]. Anti-LRP antibody inhibited neurite outgrowth of primary hippocampal neurons, cultured in either serum-containing medium or in cortical astrocyte monolayers in serum-free medium [102].
3.5. Dishevelled-1 (DVL1)
Signalling through Frizzled and Ryk requires the cytoplasmic scaffold protein Dishevelled [103]. However, the discovery of new Wnt receptors [104] casts doubts on whether Dvl is an absolute requirement for all Wnt-mediated functions. In the canonical pathway, Wnt molecules activate Dvl to inhibit the serine/threonine kinase GSK3β, with subsequent cytoplasmic elevation and translocation of β-catenin to the nucleus, to regulate transcription [105]. Dvl is localised in various cellular compartments, like the cytosol, the nucleus, and the presynapse [106-108] and can bind to microtubules to increase their stability in both dividing cells and differentiating neurons [109]. In addition, Dvl regulates dendritic morphology in hippocampal neurons [110]. Loss of function of Dvl1, one of the three mouse Dvl genes, results in social interaction deficits [111].
3.6. Glycogen Synthase Kinase-3 (GSK3)
Glycogen synthase kinase-3 (GSK3) proteins are serine/threonine kinases that were originally identified as key regulatory enzymes in glucose metabolism [112, 113]. In rodents and humans, an alternative splice variant of GSK3β, GSK3β2, is specifically expressed in the nervous system; its highest levels are found during development [114]. Recent studies suggest that GSK3β2 plays a specific part in neuronal morphogenesis in vitro [115, 116]. GSK3 proteins and their upstream and downstream regulators play key roles during neurodevelopment and neuroplasticity, including neuro-genesis, neuronal migration, neuronal polarisation and axon growth and guidance. Disruption of GSK3 signalling adversely affects brain development and is associated with several neurodevelopmental disorders [117]. GSK3 is at the crossroads of many a pathway of intracellular signalling, including Wnt–β-catenin, Sonic Hedgehog, Notch, PI3K, ρ-GTPases, mTOR, Akt, PKA-LKB1, thus affecting intracellular events in a very complex way. As a result, it plays multiple roles in both neurodevelopment and morphogenesis, in form maintenance and remoulding, in homoeostatic function during adult life [118, 119], and also in response to various treatments, including mood stabilising and antipsychotic drugs [120, 121].
3.7. Adenomatous Polyposis Coli (APC)
APC was originally identified as a tumour suppressor gene mutated in familial adenomatous polyposis, an autosomal dominant condition with predisposition to colorectal cancers [122] and brain tumours [123]. It is a key component of the APC/AXIN/CKI/GSK3β destruction complex and a negative regulator of the Wnt signalling. APC protein facilitates growth and proliferation of neurons in vitro. Mutations in APC have also been linked to a case of mental retardation [124]. Many genes whose dysfunction is associated with tumour formation have important functions in the normal development of the nervous system [125]. Apc (the murine homologue of APC) is known to be involved in regulating a variety of cellular processes, including mitosis, cytoskeletal dynamics, axonogenesis, cell polarity, and apoptosis [126, 127]. Given the multiplicity of Apc functions, it affects the development of cerebral cortical cells in a multitude of ways. Apc is central to the Wnt signalling pathway, where it mediates the breakdown of cytoplasmic β-catenin protein [105], thus countering Wnt signals which stabilise β-catenin and allowing it to translocate to the nucleus and activate the transcription of target genes in collaboration with TCF/LEF. With cerebral cortical Apc depletion, important changes in cell polarity and cytoskeletal organisation occur in combination with changes in β-catenin activity; these result in reduced cortical size, disorganised cell connectivity, shift in cell identity (neurons lose their prefrontal characteristics to assume those of posterior brain) [128]. Interestingly, Apc heterozygous knockout mice display working memory deficits (a condition shared by people with schizophrenia) when they are 11-12 weeks-old, an age roughly corresponding to 16-18 years of age in humans, but not at 6-7 weeks of age, which is about 10 years for humans [129]; the parallel with age of onset of schizophrenia is impressive.
3.8. β-Catenin
β-catenin is a protein that in humans is encoded by the CTNNB1 gene. The nomenclature of the homologous protein in Drosophila is armadillo. β-Catenin is a subunit of the cadherine protein complex and is an integral component in the Wnt signaling pathway [130]. Tyrosine-142 phosphorylation of β-catenin is the initial event promoting the release of this protein from the cell membrane, thus rendering it available as a transcription factor [131]. The enzyme which reverses this step is the tyrosine phosphatase, RPTP β/ζ [132]. Pleiotrophin is a natural inhibitory ligand for RPTP β/ζ [132]. Pleiotrophin thus acts as a growth factor by enabling β-catenin release from the plasma membrane. β-Catenin is involved in developmental processes such as embryonic pattern formation, determination of cell fate, and articular cartilage function [133-135].
3.9. Transcription Factor-4 (TCF4)
Transcription factor-4 (TCF-4) is a protein member of the basic helix-loop-helix (bHLH) transcription factor family, binding DNA at CANNTG motifs, also termed E-Boxes [136]. TCF-4 is broadly expressed during early human development throughout the CNS, the sclerotome, peribronchial and kidney mesenchyme, and the genital bud [137]. TCF-4 isoforms control the development of different parts of the brain, i.e., midbrain and isthmus in the developing Xenopus; in this context, canonical Wnt signalling autoregulation and alternative expression of different isoforms of XTcf-4 are essential for developing CNS specification [138].
3.10. Dickkopf-1 (DKK1)
The secreted protein Dickkopf1 (Dkk1), a member of a multigene family, which induces head formation in amphibian embryos [139], was first found to inhibit Wnt in the late nineties [140]. Later, its inhibition of Wnt/β-catenin signalling was found to occur through binding to the LRP6/arrow Frizzled coreceptor moiety [141]. DKK1, together with Wnt/β-catenin signalling, plays an important role in antero-posterior (a-p) patterning of the primary embryonic axis. The different expression of Wnt pathway molecules and their antagonists, such as DKK1, create signalling gradients, which induce correct axial structure formation, in particular, head development. In the mouse embryo, DKK1 is expressed from the gastrula to the neurula in the anterior visceral endoderm, the anterior mesendoderm, and foregut endoderm. The expression of DKK1 in the anterior mesendoderm of the mouse is required for early anterior development, but is not essential in the anterior visceral endoderm for proper anterior morphogenesis [139]. Canonical Wnt signals and DKK1 function as repulsive and attractive guidance cues, respectively, during visceral endoderm cell migration. This supports a model where a signalling gradient of Wnt/β-catenin and DKK1 mediates a-p axis polarization by guiding anterior cell migration in the pregastrula mouse embryo.
3.11. Dikkopf-3 (DKK3)
Despite genetic and structural similarities between Dikkopf-3 (DKK3) and other Dikkopfs, its relation to Wnt signalling is unclear [142]. In situ hybridisation showed significant DKK3 expression in mouse cortex, hippocampus, and brain stem [143]. The role of DKK3 in the nervous system is far from clear. Interestingly, elevated plasma and cerebrospinal fluid DKK3 levels have been observed in Alzheimer’s disease [144].
3.12. Dickkopf-4 (Dkk4)
Dickkopf-4 (DKK4), like other members of this family, is a negative regulator of Wnt signaling. It appears to act like Dkk1, moderating Wnt signaling in the developing forebrain [145]. Its role during embryogenesis, despite specifically-focused studies, remains obscure. DKK4 acts in a negative feedback loop to attenuate canonical Wnt signalling, and may facilitate switch to the non-canonical Wnt planar cell polarity (PCP) pathway, which is involved in cell movements during morphogenesis [146].
3.13. Secreted Frizzled Related Protein-1 (sFRP1)
The sFRPs belong to the largest family of Wnt inhibitors. sFRP-1, an antagonist of Wnt signalling which is under control of the Hedgehog pathway [147] and inhibits β-catenin accumulation through a GSK-3-dependent mechanism [148], binds directly to Wnt molecules to interfere with its binding to Frizzled receptors [149]. It regulates cell proliferation, differentiation and apoptosis, but its deletion was not shown to have any obvious effect on the gross anatomy of the nervous system in mice [150]. sFRP-1 mRNA expression in the developing mouse neocortex occurs during the entire developmental period and it is spatially restricted to the proliferative zones [151]. Furthermore, sFRP-1 and sFRP-3 are expressed in anterolateral to caudomedial gradients within the telencephalic ventricular zone during corticogenesis [152]. The human chromosomal region 8p, which is involved in the genetics of neuropsychiatric and other diseases [153, 154], contains at least three Wnt signalling-related genes (FZD3, DKK4, and SFRP1). Summarising, sFRP-1 influences many processes around the body, but is essential for none. This could be related to functional redundancy and to the vicarious functions of other similar proteins involving sFRPs and Dkks [155-156]. Furthermore, it appears to have functions related to both growth and breakdown, thus contributing to morphogenesis and remoulding, but its effects are temporally limited and developmental state-related [157], and in a later stage its actions may oppose its earlier ones [158].
Wnt signaling appears to be very important during neurodevelopment, but it also affects synaptic plasticity and neurogenesis during adult life and controls excitatory synaptic transmission in a key area, like the hippocampus [61, 159, 160], which is believed to be involved in the pathophysiology of schizophrenia [161, 162]. Presynaptic effects of canonical Wnt signaling and postsynaptic Wnt non-canonical signalling concur in excitatory neuro-transmitter release in adult hippocampus and in postsynaptic density 95 (PSD-95)-glutamate receptor incorporation [67] and both these mechanisms contribute to maintenance of long-term potentiation, the first through mTOR activation secondary to GSK3 suppression [163], the second with a Ca2+-dependent non-canonical Wnt activation [63]. It should be recalled that schizophrenia is accompanied by long-term potentiation deficit [164]. Hence, it is possible that deficits which first appeared early in development and which depended on faulty molecular communication on a possible genetic basis, continue to be involved in maladaptive behaviour in adult life. There is evidence from a ground-breaking paper that such defects regard the genoma of people with psychotic disorders, as they are present even in fibroblasts reprogrammed to become neurons derived from such patients [165]. Interestingly, in this paper, altered gene expression was found in both NMDA receptor genes and in several genes encoding Wnt-related molecules and other molecules which are closely related to pathways interacting with Wnt signaling. These exciting discoveries pave the way for the identification of genetic markers of abnormal behaviour and may provide a clue in the future as to the nature of what we currently call schizophrenia.
4. THE WNT SIGNALLING PATHWAY AND SCHIZOPHRENIA
An alteration in one or more of Wnt pathway genes may affect brain development and be related to several neurodevelopmental disorders [117]. In particular, the role of the Wnt pathway in the regulation of neuronal migration during development suggests its possible involvement in the cytoarchitectural defects observed in schizophrenia [166]. In fact, the Wnt pathway (and GSK3 in particular) is also involved in programmed cell death, an essential component of normal brain development. The abnormal brain neuronal distribution reported from post-mortem samples of patients with schizophrenia may result from aberrant programmed cell death [167]. Preventing apoptosis through GSK3 inhibition has been advocated as a possible therapeutic target to protect against psychiatric disorder-related neuro-pathology [168]. GSK3β is involved in serotonin deficiency-related aberrant behaviour, since its inhibition alleviates such behaviour [169], while it plays a part in dopamine-mediated hyperlocomotion, an effect which is blocked by anti-psychotic treatment [170]; the interplay between these neuro-transmitters could prove to be crucial in schizophrenia [171, 172], and many psychiatric treatments active in schizophrenia target both monoamine neurotransmission and GSK3 [173]. Lithium and valproate directly inhibit GSK3, whereas clozapine, haloperidol, risperidone, fluoxetine and imipramine increase its inhibitory phosphorylation. Atypical and typical antipsychotic drugs alter GSK3β activity, as do psychosis-inducing drugs; some psychoactive substances decrease GSK3 activity while chronic antipsychotic treatment increases GSK3 activity [174, 175]. The inhibition of GSK3 could represent a key factor in the behavioural and neurotrophic effects of psychotropic drugs and a possible target for treatment [176, 177].
4.1. Wnt1
Mutations in Wnt cause neurodevelopmental alterations in mice. Late-gestation foetuses homozygous for mutated Wnt-1 alleles show a virtual absence of midbrain and cerebellum and newborns die within 24 hours [39]. The only early immunohistochemical post-mortem study performed to date, showed increased Wnt-1 immunoreactivity in hippocampus, in particular in the CA3 and CA4 subregions, in patients with schizophrenia versus controls with no clinical or morphologic evidence of brain pathology [34].
4.2. Wnt2
The WNT2 gene is located at chromosome 7q31.2; the 7q31 region showed in the Genome Scan of European-American Schizophrenia Pedigrees a significant linkage to schizophrenia [178], but this is not a sufficiently strong point for the involvement of Wnt2 in schizophrenia. Six studies to date investigated whether the WNT2 gene is associated with autism, another neurodevelopmental disorder [179, 181-184], with three studies finding rare mutations in the WNT2 gene to increase significantly the susceptibility to autism [179, 182, 183], while another found a possible interaction between two SNPs, one for WNT2 and one for EN2, in influencing language development in autism [184]. Given the similarities between autistic disorders and schizophrenia [as a neurodevelopmental disorder], H.J. Kim et al. [72] investigated whether WNT2 gene variations are risk factors for schizophrenia in a Korean sample; however, their results suggested that Wnt2 may not be involved in the pathogenesis of schizophrenia. Of all the SNPs and haplotypes analysed, only one SNP, rs4730775, showed a weak association with the disorder [72]. Overall, these results give little support to the existence of a genetic association between Wnt2 and schizophrenia.
4.3. Frizzled Receptor
Studies in the last decade focused on the association between SNPs of the human Frizzled 3 gene (FZD3) and schizophrenia. The first studies analysed eight SNPs in candidate gene association studies, finding significant associations for SNP rs2323019 (SspI site), rs960914, and rs880481 in Chinese Han populations through a transmission disequilibrium and chi-square test [185, 186], while neither allele-wise nor genotype-wise analysis revealed any evidence of association in SNP rs181277, rs352222, rs352226, rs2874940, rs3757884, and rs3757888 [185-188]. A Japanese study found an association between schizophrenia and IVS3+258T>C polymorphism (rs960914) or IVS3+258T−435G haplotype [187]. However, a study in Caucasians, which investigated the SNP rs960914, did not confirm the FZD3 as a risk locus for schizophrenia [188]. Regarding SNPs rs352203 (NlaIII site) and rs2241802 (AluI site), they were first significantly associated with schizophrenia in a Chinese Han population [185], but this was not confirmed in Japanese, British, South Korean and other Chinese populations [186-192]. The frequencies of the rs2241802 SNP allele or genotype were found to differ between healthy controls and patients with schizophrenia in a Va Chinese population [193]. Finally, a recent study analysed the rs7836920 SNP in a Brazilian sample, but found no significant association with schizophrenia [194].
FZD3 gene SNPs have been also studied in the context of the methamphetamine psychosis model, which shows significant association between the FDZ3 gene and vulnerability to schizophrenia; two haplotypes comprising SNP rs2241802, rs2323019, rs352203, and rs880481 have been identified as potential risk factors [195], suggesting a role for FZD3 in both schizophrenia and drug-induced psychosis.
4.4. Dishevelled1
Dishevelled1 (DVL1), the human homologue of the Drosophila dishevelled gene (Dsh), encodes a cytoplasmic phosphoprotein, Dvl-1, which is integrated with the Wnt pathway, regulating cell proliferation, transducing developmental processes like segmentation and neuroblast specification [196], cell fate decisions, cell polarity, and microtubular stabilisation [109]. A gene-targeting study of Dvl1, showed this mouse gene to be a genetic factor influencing social behaviour and sensorimotor gating in mice; Dvl1-deficient mice are viable, fertile, and structurally normal but they exhibit reduced social interaction, including differences in whisker trimming, deficits in nest-building, less huddling contact during home cage sleeping, and subordinate responses in a social dominance test [111, 197]. Sensorimotor gating is abnormal, as measured by deficits in prepulse inhibition of acoustic and tactile startle. Thus, Dvl1 mutant mice may provide a model for aspects of several human psychiatric disorders [111]. In humans, hemizygous deletion of a 1.5-3 Mb region of chromosome 22q11.2 may lead to DiGeorge syndrome, which is characterised by learning disabilities, craniofacial abnormalities and congenital heart defects, and who may develop schizophrenia in about one-fourth of cases in adult life [198]. However, caution is needed, because none of the known DVL structural genes map to human chromosome 22 or to syntenic mouse regions, and not even to any of the loci presently known to map schizophrenia [111].
Sensorimotor-gating dysfunction (impaired startling response or abnormality in pre-pulse inhibition) is characteristic of schizophrenia, among other psychiatric disorders [199, 200]. Impaired pre-pulse inhibition patients with schizophrenia appears to be a stable trait, independent from treatment and course of the disease [201]. Social impairment is present in schizophrenia and may relate to autistic traits and structural brain abnormalities [202]. It is tempting to speculate that DVL1 gene abnormalities may underlie some of the complex behavioural abnormalities encountered in schizophrenia, such as impaired response to startling and social interaction deficit, but this is currently unlikely to apply to all patients with schizophrenia.
4.5. Akt
Emamian et al. [203] showed a reduction in AKT1 protein levels and phosphorylation of GSK3β in Ser9 in post-mortem frontal cortex and hippocampus of patients with schizophrenia; similar changes were observed also in peripheral lymphocytes of patients with schizophrenia, compared to controls. The same study also showed that treatment with haloperidol in mice is associated with increased total Akt phosphorylation. Further studies showed decreased Akt expression in the dorsolateral prefrontal cortex (Brodmann’s area 46) of patients with schizophrenia [204, 205]. Van Beveren et al. [206] not only found decreased expression of AKT1, 2, and 3 genes in periferal momonucleate blood cells (PMBCs) of recent-onset (<5 years), male patients with schizophrenia, but also differences between patients and healthy controls in other genes involved in several cellular processes. A total of 1224 genes were found to be dysregulated in patients, with most transcripts down-regulated (N=952), and involving many pathways, like JAK/Stat, ERK/MAPK, Wnt/β-Catenin, neuregulin, and VEGF signalling. Interestingly, Akt is involved in many of these processes.
Lower phosphorylated Akt levels were also associated to a decrease in hippocampal volume [207], one of the most frequent structural brain abnormalities in schizophrenia [208, 209]. Another study [210] showed a decrease in phosphorylated Akt in the dentate gyrus, the neurogenetic zone of the hippocampus, in tissue samples from patients with schizophrenia. Structural abnormalities in the prefrontal cortex (PFC) such as impaired dendritic architecture of pyramidal neurons, alterations in the expression of PFC genes involved in neuronal development, altered synaptic transmission, abnormal myelination, and deficit in working memory retention under neurochemical challenge of three distinct neurotransmitter systems were reported in Akt1-deficient mice [211].
Despite the apparent importance of Akt in brain physiology and its abnormally low activity in schizophrenia, haloperidol, which is a classical neuroleptic drug, enhances the nuclear translocation of phosphatidylinositol 3'-kinase, which in turn disrupts Akt phosphorylation [212]. It is possible that this action mediates some side effects of this drug, because haloperidol, but not risperidone, was able to increase apoptosis of primary cultured rat cortical neurons through reduction of Akt activity [213].
Genetic linkage analyses showed co-segregation of Akt1 haplotypes with schizophrenia, suggesting that the AKT1 gene may be a schizophrenia susceptibility gene [203]. Other genetic studies demonstrated association of AKT1 gene polymorphisms with schizophrenia in Chinese [214], Irish [205], British [215], Japanese [206], Iranian [217], and European [218] populations. Pinheiro et al. [219] examined the potential association of genetic variation in the AKT1 gene with neurocognition in schizophrenia. Using Emamian’s et al. [203] five schizophrenia-associated SNPs in the AKT1 gene (Table 1), they found no significant association with multiple neurocognitive domains. More recently, the relationship between these five AKT1 SNPs and cognition in healthy subjects has been studied; gene variants were found to be associated with differences in specific domains of cognitive function including IQ, executive functioning, and processing speed [220]. SNP2 rs1130233 was found to be associated with reduced expression of Akt1 in peripheral lymphocytes and brain grey matter in the same study, while another study which investigated obstetric complications in patients with schizophrenia, failed to find associations surviving correction for multiple testing [35]. This SNP was also associated with brain volume reductions in the caudate bilaterally and in the right prefrontal cortex [220]. It is possible that disruption of Akt regulation of GSK3 activity in the brain plays a role in dysregulation of brain function in schizophrenia, and that its restoration by antipsychotic medications may contribute to the clinical efficacy of these drugs. Consistent with these data, recent work has linked GSK3 to DISC1 [221, 222; see below].
Table 1.
Summary of Post-mortem Studies of Wnt-related Molecules in Schizophrenia
| Study | Investigated Association Between | Subjects | Results |
|---|---|---|---|
| J. Yang et al., 2003 [185] | rs2241802, rs2323019, rs352203 SNPs of FZD3 gene and schizophrenia | 246 Chinese Han family trios | Positive association between FZD3 locus and schizophrenia |
| Katsu et al., 2003 [187] | rs3757888, rs960914, rs2241802 SNPs of FZD3 gene and schizophrenia | Japanese population; 200 unrelated schizophrenia patients and 218 healthy controls | Positive association between rs960914 and schizophrenia |
| Emamian et al., 2004 [203] | SNPs SNP5: rs2494732, SNP4: rs1130233A, SNP3: rs3730358, SNP2: rs1130214, SNP1: rs3803300 of AKT1 gene and schizophrenia | 210 proband-parent triads and 58 extended families, containing a total of 335 individuals affected with schizophrenia or schizoaffective disorder | One three-SNP haplotype (SNP2/3/4, TCG) is associated with schizophrenia |
| Ide et al., 2004 [189] | rs2241802, rs2323019, rs352203, rs352210, rs960914 SNPs of the FZD3 gene and schizophrenia | Japanese population; 212 schizophrenia families[family samples]; 540 unrelated schizophrenia patients [case-control samples] | No association |
| Scassellati et al., 2004 [248] | Two common SNPs at position -1727 A/T and -50 C/T and a (CAA)(n) repeat polymorphism localized in intron 1 of the GSK3β gene | 147 schizophrenia patients and 212 healthy controls | No association with schizophrenia but the (CAA)(3)/(CAA)(5) heterozygotes are more often represented in the subtype of paranoid schizophrenic patients |
| Wei and Hemmings, 2004 [190] | rs2241802, rs2323019, rs352203 SNPs of FZD3 gene and schizophrenia | British population; 120 family trios (fathers, mothers and schizophrenic offspring) | No association |
| Zhang et al., 2004 [186] | 2241802, rs2323019, rs352203, rs880481 SNPs of FZD3 gene and schizophrenia | Chinese Han Population; 236 schizophrenia patients | Positive association between rs2323019 and rs880481 and schizophrenia |
| Cui et al., 2005 [263] | rs2229992, rs42427, rs465899 SNPs at the exon region of APC gene and schizophrenia; measurement of APC mRNA in leukocytes of patients with schizophrenia | Chinese population; six schizophrenic patients and six healthy controls. 59 patients with schizophrenia and schizophrenia-like disorders and 30 healthy controls. 163 parent- offspring trios | Presence of association; an increase mRNA is found in leukocytes. |
| Hashimoto et al., 2005 [191] | rs960914, rs2241802, rs2323019, rs352203 SNPs of FZD3 gene and major psychosis | Japanese population; 427 patients with schizophrenia, 91 with bipolar disorder, 396 with major depression and 473 healthy controls | No association |
| Ikeda et al., 2005 [249] | rs4388007, rs2037547 SNPs of the GSK3β gene and schizophrenia, age-at-onset and subtypes of schizophrenia | Japanese population; 381 schizophrenic patients and 352 controls | No association |
| Lee et al., 2006 [250] | SNPs at position -1727 A/T, -50 C/T of GSK3β polymorphisms and schizophrenia | Korean population; 138 schizophrenia patients, 120 bipolar patients and 350 controls | No association |
| Szczepankiewicz et al., 2006 [251] | -50 T/C polymorphism of GSK3β gene and schizophrenia and bipolar disorder | 402 schizophrenia patients, 416 bipolar patients and 408 controls | No association with schizophrenia |
| Pinheiro et al., 2007 [219] | rs2494732, rs2498799, rs3730358, rs1130241, rs3803300 AKT1 SNPs and neurocognitive domains in schizophrenia | 641 individuals with schizophrenia | No association |
| Xu et al., 2007 [214] | Five SNPs (rs3803300, rs1130214, rs3730358, rs2498799, rs2494732) of the AKT1 gene and schizophrenia | Chinese population; 384 patients with schizophrenia vs. 384 healthy controls | G allele of rs3803300 SNP is associated; haplotype A-G-C-G-A constructed by 5 SNPs is associated |
| Kishimoto et al., 2008 [195] | rs3757888, rs960914, rs2241802, rs2323019, rs352203, rs880481 SNPs of the FZD3 gene and methamphetamine psychosis | Japanese population; 188 patients with methamphetamine psychosis and 240 controls. | Haplotype frequency of G-A-T-G and A-G-C-A of rs2241802, rs2323019 and rs352203 SNP were significantly lower in patients with methamphetamine psychosis than in controls |
| J. Meng et al., 2008 [252] | rs334563, rs12630592, rs4688054, rs10934503, rs2319398 and rs4624596 SNPs of GSK3β gene and schizophrenia (case-control study) | Chinese population; 329 schizophrenia patients and 288 healthy controls | No association |
| Proitsi et al., 2008 [300] | 50 SNPs in 28 Wnt signaling genes and schizophrenia | 307 family-trios of Chinese origin; case-control samples of over 500 schizophrenia cases and 500 controls from Hong Kong | rs2073665 SNPs in DKK4 gene is associated with schizophrenia |
| Souza et al., 2008 [254] | rs7624540, rs4072520, and rs6779828 SNPs of GSK-3β and schizophrenia | Caucasian population; 150 schizophrenia patients, 85 small nuclear families and 185 healthy controls | Positive association |
| Tan et al., 2008 [220] | rs2494732, rs1130233A, rs3730358, rs1130214, rs3803300, SNPs of AKT1 gene and cognition in healthy subjects; effects of rs1130233 SNP of AKT1 gene on AKT1 protein in peripheral lymphoblasts of healthy subjects; association between rs1130233 SNP of AKT1 gene and prefrontal function and cortical DA function using fMRI in healthy subjects; association between rs1130233 SNP of AKT1 gene and brain structure using MRI Family-based and a non-independent case-control association study: association between rs2494732, rs1130233A, rs3730358, rs1130214, rs3803300, SNPs of AKT1 gene and schizophrenia | European population 319 individuals for cognitive tests 32 healthy controls for AKT1 protein expression in lymphoblasts 46 and 68 healthy subjects for fMRI 171 individuals for structural brain MRI 358 unrelated probands and 370 unrelated healthy controls for family-based and a non-independent case-control association study | Positive association is found between rs 1130233 SNP and brain volume reductions in the caudate bilaterally and right prefrontal cortex in patients with schizophrenia rs1130233 is associated with reduced performance on cognitive functions, with reduced AKT1 protein levels, with increased prefrontal activation (inefficient activation) rs1130233 is associated with relatively reduced gray-matter volumes in the bilateral caudate and right prefrontal cortex rs1130233 is associated with increased risk for schizophrenia in the case control dataset but not in the non-independent family-based association study |
| Thiselton et al., 2008 [205] | rs1130214, rs2494732, rs2498799, rs3730358 rs2494746, rs3803304, rs2498802, rs2498804 SNPs of the AKT1 gene and schizophrenia | Irish population; 265 high-density schizophrenia families with 1408 individuals available for genotyping. | No association |
| Gregorio et al., 2009 [194] | G/C (intron 2) of Wnt7A gene, SNP1 rs3730358, SNP2 rs2498784, of AKT1 gene and MRI morphological alterations observed in 25 patients with schizophrenia; associations between impaired left/right gyrification index and protocadherin 12 gene SNP rs164515, and between ventricular enlargement and reelin gene SNP rs362691 | Twenty-five schizophrenic patients | No association |
| Stefansson et al., 2009 [281] | rs9960767 SNP of TCF4 gene and sensorimotor gating in schizophrenia | European population; 2663 patients with schizophrenia and 13498 controls | Positive association between rs9960767 SNP and schizophrenia |
| Aleksic et al., 2010 [298] | rs713526 SNP in the promoter region of KREMEN1 and schizophrenia | Japanese population; 1624 patients with schizophrenia and 1621 healthy volunteers | Presence of association |
| Benedetti et al., 2010 [168] | rs334558 SNP of GSK3β gene and grey matter volumes (voxel-based morphometry) in schizophrenic patients | 57 schizophrenia patients | Carriers of C allele variant showed higher brain volumes in an area comprising posterior regions of right middle and superior temporal gyri |
| H.J. Kim et al., 2010 [72] | rs2024233, rs1051751, rs733154, rs3779548, rs17132543, rs6948009, rs4730775, rs39315 SNPs of the Wnt2 gene and schizophrenia | Korean population; 288 patients with schizophrenia and 305 healthy controls | Weak positive association between rs4730775 and schizophrenia only, none for other SNPs |
| C. Kang et al., 2011 [193] | FZD3 SNPs rs2241802, rs2323019, rs352203, rs880481, and rs3757888 and schizophrenia | 81 Va Chinese patients with schizophrenia vs. 210 healthy controls | Haplotype analyses showed significant differences between patients and controls at rs2241802 only, after Bonferroni correction, and differences at at rs2241802-rs2323019-rs352203 haplotype |
| Lennertz et al., 2011 [284, 285] | C allele of the rs9960767 polymorphism of the TCF4 gene and verbal declarative memory (VDM) functions in schizophrenia | 401 patients with schizophrenia | Carriers of the C allele were less impaired in recognition compared to those carrying the AA genotype |
| J. Li et al., 2011 [271] | BCL-9 SNPs rs9326555, rs10494251, rs1240083, rs672607, rs688325, and rs3766512 and schizophrenia, bipolar disorder or major depressive disorder | 4187 Chinese Han patients with schizophrenia vs. 5772 healthy controls vs. 1135 patients with bipolar disorder vs. 1135 patients with major depressive disorder | Association between SNPs rs9326555, rs10494251, rs1240083, rs672607 (strongest), rs688325, and rs3766512 and schizophrenia, rs672607 and bipolar disorder, and rs672607, rs1541187, rs688325, rs10494251, rs946903 and major depressive disorder |
C=Controls; ELISA=Enzyme-linked immuno sorbent assay.
In addition to rs1130233, another AKT1 SNP, rs2494732, was found to mediate long-lasting negative effects of psychosis on cognition (reduced performance on the continuous performance test) in people who had used cannabis, even if they had stopped using cannabis [223]. Both SNPs doubled the risk of being diagnosed with a psychotic disorder after cannabis use [224]. The authors speculated that cannabinoids may act down-stream the dopamine receptor to affect Akt-GSK3 interaction.
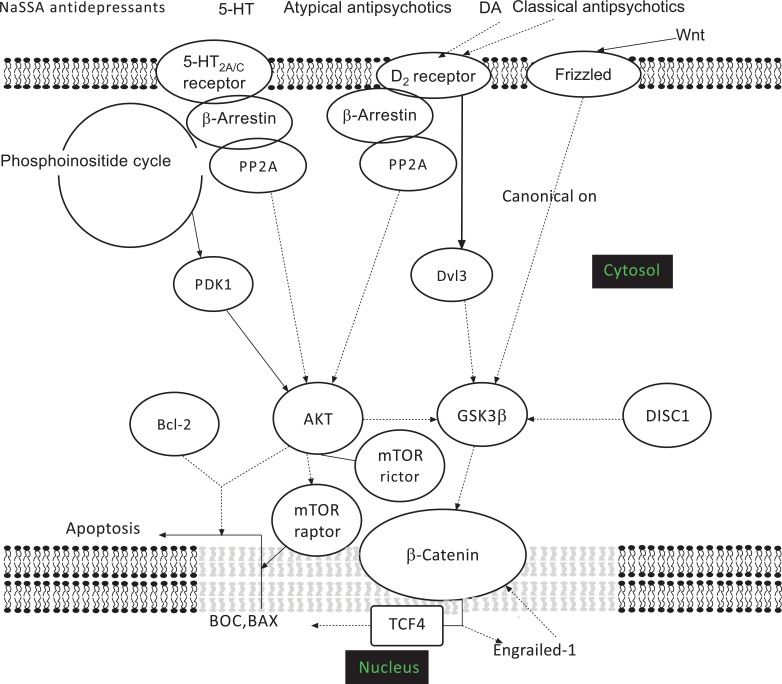
Gregorio et al. [194] investigated the possible association between structural brain alterations in patients with schizophrenia and 32 gene polymorphisms on 30 genes involved in neurogenesis and brain maturation (DNA extracted from peripheral blood mononuclear cells). Ventricular enlargement on 1.5 T magnetic resonance imaging (MRI) was associated with the reelin gene SNP rs362691 and impaired lateralisation of cortical gyrification was associated with the protocadherin 12 gene SNP rs164515; no significant associations were found for other structural abnormalities and gene polymorphisms, including the Wnt signalling-related Wnt7A SNP G/C (intron 2), AKT1 SNP1 rs3730358, and AKT1 SNP2 rs2498784. Taken together, these data indicate that schizophrenia may be associated with impaired neuronal migration and signalisation, and synaptic network specification. While the reelin pathway does not directly interact with Wnt signalling [225, 226], the cadherins display mutual interactions with the canonical Wnt pathway [227; Fig. 1].
Fig. (1).
Simplified drawing of putative interactions between canonical Wnt and other pathways to illustrate their complex interplay. Dashed lines indicate inhibition, solid lines activation.
4.6. GSK3
Animal schizophrenia models are consistent with altered GSK activity. Significant reduction in GSK3β protein levels was found in the frontal cortex of the pre-puberty lesioned with excitotoxic hippocampal injury rats compared to post-pubertal rats and controls [228]. Another study showed increased inhibitory phosphorylation of GSK3β and β-catenin, but no significant changes in mRNA or protein levels in PFC and VTA of rats treated with MK-801, an NMDA glutamate-receptor antagonist which induces schizophrenia-like behaviours [229]. In the same study, increased APC mRNA and protein levels were found and matched enhanced β-catenin phosphorylation, suggesting that the latter is mediated through an APC effect on GSK3β. Doble et al. [230] produced mouse embryonic stem cells in which both alleles of GSK3α and GSK3-β were disrupted. Only in cells lacking three or all four alleles did a measurable increase in Wnt signaling occur; this indicates that a considerable proportion of GSK3 inhibition is required to increase the activity of the Wnt pathway.
A human study, which did not focus on brain alterations of GSK content or activity, also pointed to an alteration of GSK3 in schizophrenia. In fact, it found a reduction in both cellular activities and protein levels of kinase FA/GSK3α in the lymphocytes of patients with schizophrenia, as compared to healthy controls [231].
Post-mortem studies showed some inconsistencies; nevertheless they suggested GSK3 alterations in schizophrenia. A significant reduction in GSK3β [232], but not in β-catenin or Dvl-2 levels [233], was found post-mortem in the prefrontal cortex of 10 patients with schizophrenia, compared to 10 controls; however, this was not replicated when the same group of investigators extended their sample to 15 and compared it with 15 patients with bipolar disorder, 15 with unipolar depression and 15 with non-psychiatric conditions, with GSK-3β, β-catenin and Dvl-2 not differing among the groups [234]. In contrast, a subsequent post-mortem study failed to find changes in GSK3β expression in the occipital cortex of patients with schizophrenia, compared to bipolar and unipolar patients, and controls, but found decreased GSK activity in the frontal cortex of patients with schizophrenia [235]. Kozlovsky et al. [236] showed borderline reduction in dorsolateral prefrontal cortical mRNA levels of GSK3β, but not GSK3α, in patients with schizophrenia compared to other comparison groups [bipolar disorder, unipolar depression, and normal controls]. Another study failed to show significant differences in GSK3α and GSK3β mRNA levels in the frontal cortex of patients with schizophrenia, compared to healthy controls, but showed significantly lower hippocampal GSK3α and GSK3β mRNA levels in the patients with schizophrenia [237]. Based on the above findings, we may not infer as to the level of a basic alteration that may account for the multitude of the Wnt alterations found in post-mortem brain of patients with schizophrenia.
Dopamine receptor activation may have multiple effects on GSK3 function, which depend upon the receptor involved and whether they act through D2 receptor-mediated β-arrestin2/protein phosphatase 2A (PP2A) recruitment (increased Akt phosphorylation and release of GSK3 from Akt regulation) or through D1 and/or D2-mediated tyrosine kinase transactivation (enhanced Akt action and consequent GSK3 phosphorylation/inactivation) [238]. Antipsychotic drugs, who are believed to act through inhibiting mainly the D2 dopamine receptor, should be expected to inhibit the β-arrestin2/PP2A recruitment, thus enhancing the action of Akt and the phosphorylation of GSK3. However, the differential effects of these drugs on D1-D2 dopamine groups of receptors, as well as on other neurotransmitter receptors, account for a multitude of effects that have been described for this class of drugs.
Antipsychotic drugs like haloperidol, clozapine, risperidone and the experimental substituted benzoamide raclopride, a selective D2 blocker, induced an amphetamine-reversible increase in GSK3β levels in rat medial prefrontal cortex, striatum, ventral midbrain, and hippocampus without affecting its phosphorylation status [239, 240], as well as an increase in Dvl-3 levels in rat ventral forebrain and hippocampus [240]. However, another study found haloperidol and clozapine to differ in their ability to phosphorylate rat frontal cortical GSK3β, in that the effect of haloperidol was transient on Akt and poor on Dvl, while clozapine stably increased the amount of GSK3α/β phophorylation and Dvl [241]. Similarly, clozapine was found to activate Akt- and Dvl-mediated Ser9 phosphorylation of GSK-3β in SH-SY5Y human neuroblastoma cells [242]. The atypical drug paliperidone (9-hydroxyrisperidone) was also found to act through the canonical Wnt pathway, inasmuch it inhibited the detrimental effect of the NMDA antagonist dizocilpine (MK-801) on Akt1 and GSK3β [243]. The differential effects of atypical vs. classical antipsychotics have been attributed to a preferential inhibition of the D2 receptor-mediated β-arrestin2/PP2A recruitment by the atypical antipsychotics, while classical neuroleptics like haloperidol would equipotently act through both the β-arrestin2/PP2A pathway and G protein signalling; this view was supported by the fact that GSK3β knock-out of D2 dopamine receptor-bearing mouse neurones mimicked the antipsychotic effect of aripiprazole, leaving unaffected the motor effect elicited by haloperidol (catalepsy) [244].
While the effect of antipsychotics on GSK-3β phosphorylation status is unclear, it is possible that some of the effects of antipsychotics are mediated through their actions on the Wnt pathway. Regretably, the post-mortem studies did not distinguish among the various antipsychotics received during their life by these patients, and this might have influenced the obtained results, as the effects of the various antipsychotics on the Wnt pathway vary [238, 245].
GSK-β function is under the control of DISC1 signalling, hence altered DISC1 could result in impaired Wnt signalling. DISC1 physically inhibits GSK3β activity, thus promoting subsequent phosphorylation and stabilization of β-catenin [220, 246, 247, Fig. 1]. DISC1-mediated inhibition of GSK3β regulates neuronal precursor cell proliferation in both embryonic and adult mouse brains, further stressing the role of GSK3 and the Wnt pathway signalling in neurodevelop-ment [220]. While wildtype DISC1 effectively interferes with GSK3β signalling, some variants in many species are associated with the loss of GSK3β-regulating activity; for example, human lymphoblast cell lines endogenously expressing the R264Q DISC1 variant showed impairment in Wnt signalling and the S704C DISC1 variant was associated with impaired neuronal neocortical migration [221], pointing to a conjoint action of the DISC1 and Wnt signalling in neurodevelopment.
Genetic studies are not uniform in their conclusions about the involvement of GSK mutations or polymorphisms in schizophrenia. Several studies failed to find an association between mutations or polymorphisms of the GSK3-β gene and schizophrenia [248-253]. However, a case-control study examined GSK3β polymorphisms of the GSK gene SNPs rs7624540, rs4072520, and rs6779828 in patients with schizophrenia compared to healthy controls and showed a genotypic association between these SNPs and schizophrenia, suggesting that that this gene might be involved in the risk for schizophrenia [254]. The results of these studies were unfairly criticised [255], since the other studies claiming no association did not investigate the same SNPs as Souza et al. [254, 256]. Another study, analysing two common SNPs at position -1727 A/T and -50 C/T and a (CAA)(n) repeat polymorphism localized in intron1 of the GSK3-β gene in a group of patients with schizophrenia compared to healthy controls, showed that the allele, genotype, and haplotype distributions for the three polymorphisms investigated do not differ between patients and controls. However, (CAA)(3)/(CAA)(5) heterozygotes with the paranoid subtype of schizophrenia were more represented. These results support the reports that GSK3β appears to be involved in a subtype of schizophrenic patients, but not in schizophrenia in general [248]. Studying the effect of a GSK3-β SNP rs334558 on grey matter volumes of a group of patients affected by chronic schizophrenia, a recent study showed significantly higher brain volumes in an area encompassing posterior regions of right middle and superior temporal gyrus, within the boundaries of Brodmann area 21 in carriers of the rs334558 C allele, which is associated with reduced activity [168]. The temporal lobe is consistently associated with morphometric abnormalities in schizophrenia [168, 257, 258]. Summarising, it appears that the function of GSK3 is decreased in schizophrenia and its levels or phosphorylation increased by antipsychotic drugs, but neither the regions of decreased GSK activity are consistent (cortex and hippocampus are mostly affected, but the findings differ among studies), nor the effects of drugs are consistent, with benzoamides increasing levels, and butyrophenones and dibenzoazepines increasing phosphorylation, which ensues in decreased GSK3 activity and enhanced catenin formation. Genetic studies also show similar inconsistencies, across populations and across the SNPs reported to be associated. It is possible that GSK3 activity alterations in schizophrenia appear to be so inconsistent because the important in the genetic expression of this disorder is gene interaction [259] or because GSK3 is under some sort of epistatic control by other genes [260].
4.7. Adenomatous Polyposis Coli (APC)
A first post-mortem study [261] showed no significant differences in APC immunoreactivity (used as an oligodendrocyte marker) in any brain area among people with schizophrenia, bipolar disorder or healthy controls. A link with the glutamatergic hypothesis of schizophrenia [262] is provided by an animal study using the NMDA glutamate-receptor antagonist, MK-801 (dizocilpine), to induce schizophrenia-related behaviours in mice [229]. This study showed a significant increase in both APC mRNA and protein levels in the prefrontal cortex and the ventral tegmental area of mice displaying dizocilpine-induced schizophrenia-related behaviour, compared to controls. This may suggest that increased expression of APC is associated to attenuated Wnt pathway signalling in this animal model of schizophrenia. Backing the increased APC-induced Wnt down-regulation hypothesis, another study found an increased APC mRNA expression in leukocytes from patients with schizophrenia, which was independent from antipsychotic treatment [263]. This study also reported a significant association between APC mutations and schizophrenia using the transmission disequilibrium test (TDT) in Chinese trio families with schizophrenia; three SNPs at the exon region in the APC gene were found to be associated with schizophrenia, i.e., rs2229992, rs42427, and rs465899 [261]. Combining the above, it is likely that increased APC could impair Wnt signalling in schizophrenia and contribute to its pathology [263].
4.8. β-Catenin
A micro-array study of biopsies from olfactory epithelium from patients with schizophrenia, bipolar disorder, and healthy volunteers found a more than 50% reduction in the expression of pleiotrophin, a growth factor which enables β-catenin release from the plasma membrane, in schizophrenia [265]. The functional consequence of a strong reduction in pleiotrophin would be a reduction in β-catenin/Wnt signaling. B-cell CLL/lymphoma 9 protein is a protein that in humans is encoded by the BCL9 gene [266]. BCL-9 contributes to efficient β-catenin-mediated transcription. In fact, it was shown that BCL9/Legless increased Wnt signal by promoting the transcriptional activity of β-catenin/Armadillo in normal and malignant cells [267].
Three studies examined copy number variations (CNVs) in schizophrenia cases and matched controls [268-270]. All these studies reported hemizygous de novo deletions at chromosome 1q21.1, an area comprising BCL-9. Although it is not yet known whether the haplo-insufficiency affects BCL-9 protein levels, it is conceivable that it leads or contributes to diminished Wnt signalling. However, a recent study showed various BCL-9 CNVs to be associated with schizophrenia in a Han Chinese population [271]. Mutations of an armadillo repeat gene (ARVCF), which is known to interact with β-catenin, was associated with the velo-cardio-facial syndrome [272], which is a disorder associated with 22q11.2 deletion and heart defects, facial dysmorphism and psychosis, which is a counterpart to the 22q11.2 deletion/DiGeorge syndrome, adding to the genetic complexity of 22q11.2 deletion.
Immunohistochemistry showed significantly reduced β- and γ-catenin post-mortem levels in the hippocampus of patients with schizophrenia compared to controls; such changes were not related to antipsychotic treatment [166]. However, another post-mortem study found no decrease in β-catenin levels in the prefrontal cortex of patients with schizophrenia, compared to healthy controls and to patients affected by other psychiatric disorders, such as bipolar disorder or unipolar depression [233].
Antipsychotic drugs like haloperidol, clozapine, raclopride, and risperidone induce increases in β-catenin levels in the rat medial prefrontal cortex, striatum [239], ventral midbrain, and hippocampus [240]; these effects were countered by amphetamine, a psychotogenic drug, indicating that D2 antagonism is important in promoting the effects of antipsychotic drugs on the Wnt pathway in these areas.
4.9. TCF4
The TCF4 gene, which maps in 18q21.2, encodes a basic helix-loop-helix (bHLH) transcription factor that interacts with other transcription factors to activate or repress gene expression. This protein belongs to the E-protein subfamily, which is known to be involved in neurodevelopment, but its function in the adult brain is still obscure [138, 273, 274]. TCF has Wnt/β-catenin-activated transcription factor properties [275]. Tcf4 transgenic mice with brain TCF4 overexpression display profound deficits in contextual and cued fear conditioning and sensorimotor gating, memory deficits and, importantly, deficits in prepulse inhibition; the latter is a neurophysiological correlate of schizophrenia and other psychiatric disorders [274]. On the other hand, TCF4-null/knock-out mice die in the first 24 h after birth [276-277]. In the human, TCF4 haploinsufficiency causes the Pitt–Hopkins syndrome, an autosomal dominant neuro-developmental disorder characterized by severe motor and mental retardation, microcephaly, epilepsy, and facial dysmorphisms [137, 278]. TCF4, like other MHC class I molecule alleles (VRK2 and ZNF804A), were found in genome-wide association studies (GWAS) to be associated with schizophrenia in an Irish population [279]. The TCF4 SNP rs2958182 was found to interact with diagnosis of schizophrenia on IQ and attention-related task performance in a Chinese Han population [280].
Investigating the impact of the rs9960767 SNP in intron 4 of the TCF4 gene on sensorimotor gating measured by prepulse inhibition (PPI) of the of the acoustic startle response in healthy humans and in patients with schizophrenia spectrum disorders, a statistically significant association between this SNP and schizophrenia was found [281]. Furthermore, in both samples, PPI was strongly decreased in carriers of the schizophrenia risk allele C of the TCF4 gene, whereas startle reactivity and habituation were unaffected by the TCF4 genotype [282]. TCF4 genotype sensorimotor gating modulation indicates a role for TCF4 gene variations in the development of early information-processing deficits in schizophrenia. If PPI is an appropriate endophenotype of schizophrenia, as proposed, a combination of these SNPs might also contribute to the risk of schizophrenia [281]. Patients with schizophrenia also displayed strong impairments of verbal declarative memory (VDM) function [283]. Carriers of the C allele of the rs9960767 polymorphism of the TCF4 gene are less impaired in recognition, compared to those carrying the AA genotype. A trend toward higher scores in patients with the risk allele was found for delayed recall. The TCF4 genotype did not affect intelligence or total verbal learning score, but decreased VDM performance in patients with schizophrenia who carried the C allele of the rs9960767 polymorphism of the TCF4 gene. However, the elevated risk for schizophrenia was not conferred by TCF4-mediated VDM impairment [284]. The rs9960767 polymorphism of the TCF4 gene was shown in a GWAS to increase the risk of schizophrenia only slightly [285].
Seeking the mechanism at the TCF gene which could account for the association between the SNP rs9960767 and schizophrenia, a recent study examined TCF4 for coding variants, and for cis-regulated variation in TCF4 gene expression correlated with the associated SNP; this study used a SNaPshot assay to detect differential allelic expression measuring the relative expression levels of mRNA containing each exonic allele [286]. This study failed to identify any non-synonymous coding variants at the locus. Allele-specific expression analysis using human post-mortem brain samples obtained no evidence for cis-regulated mRNA expression related to genotype at this schizophrenia-associated SNP. This points to the association between schizophrenia and TCF4 not being mediated by a relatively common non-synonymous variant, or by a variant that alters mRNA expression, as measured in adult human brain; however, it is still possible that the risk allele at this locus exerts effects on expression exclusively in a developmental context, in cell types or brain regions not adequately represented in Williams’ et al. analysis [286], or through post-transcriptional effects, for example, in the abundance of the protein or its sub-cellular distribution. However, the relationship between the rs9960767 SNP and schizophrenia is complex, since this SNP interacts with nicotine in modulating auditory sensory gating [287], and nicotine use is widespread among patients with schizophrenia [288]. A recent study identified a new schizophrenic risk factor at 18q21.2 rs4309482, which lies approximately equidistant from CCDC68 and TCF4, and could act through one of these genes or both [289]. Another mechanism, which may unify several genetic data obtained in schizophrenia, may be through the SNP rs1625579 of the MIR137 gene, at 1p21.3 [290], which controls the production of microarray RNA miR-137, which in turn affects the function of other molecules, like CSMD1, C10orf26, CACNA1C, TCF4 [291], and ZNF804A [292], whose genes were found to be significantly associated with schizophrenia.
TCF gene variants which appear to be associated with schizophrenia are common variants, as opposed to the rare variants associated with neurodevelopmental disorders, and sometimes with the Pitt-Hopkins syndrome [293]. TCF appears not to mediate psychosis in general, as it is not involved in bipolar disorder, but rather psychotic traits specifically associated to schizophrenia [294, 295].
4.10. Dickkopf1
Dkk1 is encoded by the DKK1 gene, which maps on chromosome 10 region q11.2, a region with linkage evidence for schizophrenia [178]. Dkk1 inhibits Wnt signaling by triggering LRP5/6 internalization through formation of a ternary complex with Kremen receptors [141, 296]. Kremen1, a high-affinity dickkopf homolog1, is KREMEN1 gene-encoded transmembrane receptor mapping to chromosome 22q12.1, another region with linkage evidence for schizophrenia [297]. The involvement in schizophrenia of the KREMEN1 locus appears to be mediated through the rs713526 single nucleotide polymorphism (SNP), which is located in the promoter region of KREMEN1 and showed association with schizophrenia [298].
4.11. Dickkopf3
Gene expression analysis of the Wnt signalling antagonist Dickkopf-3 (Dkk3) has shown that its mRNA is down regulated in the temporal cortex of elderly people with schizophrenia [299]. The functional consequence regarding Wnt signalling is, in this particular case, unclear, since DKK3 is the only Dickkopf protein which apparently does not act as a Wnt antagonist [139].
4.12. Dickkopf4
An association study of 28 Wnt signalling genes found the polymorphic variation rs2073665 in the Dkk4 gene (DKK4) to be associated with schizophrenia [300]. Interestingly, the DKK4 SNP, rs2073665 was found to be significantly associated with brain volume under both additive and dominant genetic models in 961 healthy Chinese individuals [301], thus strengthening the evidence for a neurodevelopmental hypothesis of schizophrenia.
4.13. Secreted Frizzled Related Protein1 (Sfrp1)
A possible connection of the SFRP1 gene and neuropsychiatric disorders, including schizophrenia, has been advanced because of the location of the gene chromosome 8p, but to date no direct evidence for an association has been obtained [154].
Summarising genetic and post-mortem neuropathological findings of molecules or genes of Wnt-related signalling molecules in schizophrenia, we have to admit that genetic studies are much variable, despite the identification of many risk-conferring SNPs of Wnt-related genes, that replication studies all often do not confirm initial studies, that identified SNPs are not universal (i.e., they differ from population to population) and the added risk is generally low, while neuropathological studies are a little more consistent, with some studies identifying and other not confirming alterations of the same molecules, but at least showing involvement of the same molecules in the same direction, although the brain regions involved may differ. Neuropathological studies show increased Wnt1 in the hippocampus, decreased catenins and Akt1 in the hippocampus and decreased GSK function in cortex and hippocampus, although some of them did not find differences in GSK or GSK3 mRNA in cortex. Whether these differences may reflect the effect of long-term antipsychotic drug intake, is difficult to say based on reported data, as antipsychotics were usually reported as lifetime chlorpromazine equivalents with no differentiation among the various antipsychotic drugs. It should be emphasised that antipsychotics differ in the kinetics of Akt/GSK-3 phosphorylation, in the amount of post-drug exposure proteins, and in the proportion of pathway activation [245].
5. CONCLUSIONS
Schizophrenia is a multifactorial disease with onset in late adolescence-early adulthood. Among the genes putatively involved in its pathogenesis, many are those regulating the Wnt pathway and other pathways crossing their routes with the Wnt pathway. The strict connections of the Wnt signalling system with other intracellular pathways produces a sort of buffering that may dampen possible defects in few specific molecules, thereby putting the whole pathway lower in the pathophysiological hierarchy of schizophrenia. Although the entire picture is unclear, many Wnt-related molecules show abnormalities in schizophrenia, and may become a specific target for pharmacological intervention. The entire sytem, however, is so well buffered by other intracellular systems, that is not easy to anticipate the final effect of such a drug. It is much more reasonable to believe that a substance will facilitate the effect of currently available antipschotics, thus improving our ability to control schizophrenia.
However, it is possible that studies of the involvement of Wnt pathways in schizophrenia and in the action of antipsychotic drugs will be of heuristic value in our attempt to “better characterize and subtype” this psychiatric entity. Current evidence supports that the concomitant expression of specific SNPs of various genes concur in determining specific behavioural traits, some of which are related to the expression of “schizophrenic” phenotypes. Some of these involve ErbB4, NRG1, and KCNH2 isoforms; it should be recalled that the former two interact with the Wnt pathway in neural tissues. Future studies should focus on gene interaction in the expression of behaviours related to the heterogeneous entity we now call schizophrenia and on how antipsychotic treatment affects these interactions.
ACKNOWLEDGEMENTS
We gratefully acknowledge the contribution of the Librarians of the Sant’Andrea Hospital – Sapienza University, School of Medicine and Psychology Library, the late Tiziana Mattei, and Ms. Mimma Ariano, Ms Ales Casciaro, Ms Teresa Prioreschi, and Ms Susanna Rospo, Librarians of the Sant’Andrea Hospital, School of Medicine and Psychology, Sapienza University, Rome, for rendering precious bibliographical material accessible, as well as their Secretary Lucilla Martinelli for her assistance during the writing of the manuscript.
CONFLICT OF INTERESTS
Paolo Girardi has received in the past two years research support from Lilly and Janssen, has participated in Advisory Boards for Lilly, Otsuka, Pfizer, and Schering, and has received honoraria from Lilly and Organon; Giorgio D. Kotzalidis is recipient of an Italian Ministry of Education Ph.D. Grant for Early Intervention in the Psychoses.
All other authors of this paper have no relevant affiliations or financial involvement with any organisation or entity with a financial interest in, or financial conflict with the subject matter or materials discussed in the manuscript. This includes employment, consultancies, honoraria, stock ownership or options, expert testimony, grants or patents received or pending, or royalties. No writing assistance was utilised in the production of this manuscript.
ABBREVIATIONS
- ACP1
= acid phosphatase 1
- Akt
= Protein kinase B/Non-specific serine/threonine protein kinase family
- APC
= Adenomatous poliposys coli
- BP
= Bipolar disorder
- BMPs
= Bone Morphogenetic Proteins
- Ca2+
= Calcium ion
- Cdc42
= Cell division control protein 42
- CNV
= Copy number variation
- CUS
= Chronic unpredictable stress
- DAG
= Diacylglycerol
- Dvl
= Dishevelled
- EPM
= Elevated plus maze
- FGF
= Fibroblast growth factor
- Fzd
= FZD, Frizzled
- GM
= Grey matter
- GSK3(α/β)
= Glycogen synthase kinase-3(alpha, beta))
- HDAC
= Histone deacetylase
- InsP3
= Inositol-1,4,5-trisphosphate
- LRP5/6
= Low Density Lipoprotein receptor-related protein 5 and -6
- LTP
= Long-term potentiation
- MAPK
= Mitogen-activated protein kinase
- Mg2+
= magnesium ions
- MDD
= Major Depressive Disorder
- NLK
= Nemo-like kinase
- PPARD
= Peroxisome proliferators-activated receptor type-δ
- PCP
= Planar Cell Polarity
- PI3K
= Phosphatidylinositol-3-kinase
- PKC
= Protein kinase C
- ROCK
= Rho-associated kinase
- SHH
= Sonic Hedgehog
- SNP
= Single Nucleotide Polymorphism
- TAK1
= Transforming growth factor-β-activated protein kinase
- TCF/LEF
= T-cell factor/lymphoid enhancer factor family of transcription factors
- Wnt
= Wingless
REFERENCES
- 1.Huber G. Reine Defektsyndrome und Basisstadien endogener Psychosen. Fortschr. Neurol. Psychiatr. 1966;34:409–426. [Google Scholar]
- 2.Feinberg I. Schizophrenia caused by a fault in programmed synaptic elimination during adolescence. J. Psychiatr. Res. 1982. 1983;17:319–334. doi: 10.1016/0022-3956(82)90038-3. [DOI] [PubMed] [Google Scholar]
- 3.Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Arch. Gen. Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- 4.Murray RM, Lewis SW. Is schizophrenia a neurodevelopmental disorder. BMJ. 1987;295:681–682. doi: 10.1136/bmj.295.6600.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crow TJ. Positive and negative schizophrenic symptoms and the role of dopamine. Br. J. Psychiatry. 1980;137:383–386. [PubMed] [Google Scholar]
- 6.Murray RM, O'Callaghan E, Castle DJ, Lewis SW. A neurodevelopmental approach to the classification of schizophrenia. Schizophr. Bull. 1992;18:319–332. doi: 10.1093/schbul/18.2.319. [DOI] [PubMed] [Google Scholar]
- 7.Dean K, Bramon E, Murray RM. The causes of schizophrenia neurodevelopment and other risk factors. J. Psychiatr. Pract. 2003;9:442–454. doi: 10.1097/00131746-200311000-00007. [DOI] [PubMed] [Google Scholar]
- 8.Arnold SE, Talbot K, Hahn CG. Neurodevelopment neuroplasticity and new genes for schizophrenia. Prog. Brain Res. 2005;147:319–345. doi: 10.1016/S0079-6123(04)47023-X. [DOI] [PubMed] [Google Scholar]
- 9.Rehn AE, Rees SM. Investigating the neurodevelopmental hypothesis of schizophrenia. Clin. Exp. Pharmacol. Physiol. 2005;32:687–696. doi: 10.1111/j.1440-1681.2005.04257.x. [DOI] [PubMed] [Google Scholar]
- 10.Malaspina D. Schizophrenia a neurodevelopmental or a neurodegenerative disorder. J. Clin. Psychiatry. 2006;67:e07–0. doi: 10.4088/jcp.0806e07. [DOI] [PubMed] [Google Scholar]
- 11.Harrison PJ. Schizophrenia susceptibility genes and their neurodevelopmental implications focus on neuregulin 1. Novartis Found. Symp. 2007;288:246–255. [PubMed] [Google Scholar]
- 12.Harrison PJ. Schizophrenia susceptibility genes and neurodevelopment. Biol. Psychiatry. 2007;61:1119–1120. doi: 10.1016/j.biopsych.2007.02.021. [DOI] [PubMed] [Google Scholar]
- 13.Kato T. Bridging pharmacology and neurodevelopment in schizophrenia. Int. J. Neuropsychopharmacol. 2007;10:713–716. doi: 10.1017/S1461145707007936. [DOI] [PubMed] [Google Scholar]
- 14.Venkatasubramanian G, Jayakumar PN, Reddy VV, Reddy US, Gangadhar BN, Keshavan MS. Corpus callosum deficits in antipsychotic-naïve schizophrenia evidence for neuro- developmental pathogenesis. Psychiatry Res. 2010;182:141–145. doi: 10.1016/j.pscychresns.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 15.Fatjó-Vilas M, Gourion D, Campanera S, Mouaffak F, Levy-Rueff M, Navarro ME, Chayet M, Miret S, Krebs MO, Fañanás L. New evidences of gene and environment interactions affecting prenatal neurodevelopment in schizophrenia-spectrum disorders a family dermatoglyphic study. Schizophr. Res. 2008;103:209–217. doi: 10.1016/j.schres.2008.04.015. [DOI] [PubMed] [Google Scholar]
- 16.Gross G, Huber G. Schizophrenia neurodevelopmental disorder or degenerative brain process. Fortschr. Neurol. Psychiatr. 2008;76[Suppl 1]:S57–62. doi: 10.1055/s-2008-1038153. [DOI] [PubMed] [Google Scholar]
- 17.Fatemi SH, Folsom TD. The neurodevelopmental hypothesis of schizophrenia, revisited. Schizophr. Bull. 2009;35:528–548. doi: 10.1093/schbul/sbn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sivkov ST, Akabaliev VH, Kaleva NN. Comparative dermatoglyphic study of schizophrenic patients evidence of the neurodevelopmental model of schizophrenia. Folia Med [Plovdiv] 2009;51:25–30. [PubMed] [Google Scholar]
- 19.Andreasen NC. The lifetime trajectory of schizophrenia and the concept of neurodevelopment. Dialogues Clin. Neurosci. 2010;12:409–415. doi: 10.31887/DCNS.2010.12.3/nandreasen. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Deutsch SI, Burket JA, Katz E. Does subtle disturbance of neuronal migration contribute to schizophrenia and other neurodevelopmental disorders.Potential genetic mechanisms with possible treatment implications. Eur. Neuropsychopharmacol. 2010;20:281–287. doi: 10.1016/j.euroneuro.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 21.Gupta S, Kulhara P. What is schizophrenia A neuro- developmental or neurodegenerative disorder or a combination of both.A critical analysis. Indian J. Psychiatry. 2010;52:21–27. doi: 10.4103/0019-5545.58891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayashi-Takagi A, Sawa A. Disturbed synaptic connectivity in schizophrenia convergence of genetic risk factors during neurodevelopment. Brain. Res. Bull. 2010;83:140–146. doi: 10.1016/j.brainresbull.2010.04.007. [DOI] [PubMed] [Google Scholar]
- 23.Owen MJ, O'Donovan MC, Thapar A, Craddock N. Neuro- developmental hypothesis of schizophrenia. Br. J. Psychiatry. 2011;198:173–175. doi: 10.1192/bjp.bp.110.084384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uhlhaas PJ, Singer W. The development of neural synchrony and large-scale cortical networks during adolescence relevance for the pathophysiology of schizophrenia and neurodevelopmental hypothesis. Schizophr. Bull. 2011;37:514–523. doi: 10.1093/schbul/sbr034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rapoport JL, Addington A, Frangou S. The neuro- developmental model of schizophrenia what can very early onset cases tell us. Curr. Psychiatry. Rep. 2005;7:81–82. doi: 10.1007/s11920-005-0001-z. [DOI] [PubMed] [Google Scholar]
- 26.Rapoport JL, Addington AM, Frangou S, Psych MRC. The neurodevelopmental model of schizophrenia update 2005. Mol. Psychiatry. 2005;10:434–449. doi: 10.1038/sj.mp.4001642. [DOI] [PubMed] [Google Scholar]
- 27.Rapoport JL, Gogtay N. Childhood onset schizophrenia support for a progressive neurodevelopmental disorder. Int. J. Dev. Neurosci. 2011;29:251–258. doi: 10.1016/j.ijdevneu.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Demjaha A, Maccabe JH, Murray RM. How Genes and Environmental Factors Determine the Different Neuro- developmental Trajectories of Schizophrenia and Bipolar Disorder. Schizophr. Bull. 2012;38:209–214. doi: 10.1093/schbul/sbr100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keshavan MS, Anderson S, Pettegrew JW. Is schizophrenia due to excessive synaptic pruning in the prefrontal cortex.The Feinberg hypothesis revisited. J. Psychiatr. Res. 1994;28:239–265. doi: 10.1016/0022-3956(94)90009-4. [DOI] [PubMed] [Google Scholar]
- 30.Matheson SL, Shepherd AM, Laurens KR, Carr VJ. A systematic meta-review grading the evidence for non-genetic risk factors and putative antecedents of schizophrenia. Schizophr. Res. 2011;133:133–142. doi: 10.1016/j.schres.2011.09.020. [DOI] [PubMed] [Google Scholar]
- 31.Yolken RH, Torrey EF. Are some cases of psychosis caused by microbial agents.A review of the evidence. Mol. Psychiatry. 2008;13:470–479. doi: 10.1038/mp.2008.5. [DOI] [PubMed] [Google Scholar]
- 32.Brown AS, Derkits EJ. Prenatal infection and schizophrenia a review of epidemiologic and translational studies. Am. J. Psychiatry. 2010;167:261–280. doi: 10.1176/appi.ajp.2009.09030361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Crow TJ, Done DJ, Sacker A. Childhood precursors of psychosis as clues to its evolutionary origins. Eur. Arch. Psychiatry Clin. Neurosci. 1995;245:61–69. doi: 10.1007/BF02190732. [DOI] [PubMed] [Google Scholar]
- 34.Miyaoka T, Seno H, Ishino H. Increased expression of Wnt-1 in schizophrenic brains. Schizophr. Res. 1999;38:1–6. doi: 10.1016/s0920-9964(98)00179-0. [DOI] [PubMed] [Google Scholar]
- 35.Joo EJ, Lee KY, Jeong SH, Roh MS, Kim SH, Ahn YM, Kim YS. AKT1 gene polymorphisms and obstetric complications in the patients with schizophrenia. Psychiatry Investig. 2009;6:102–107. doi: 10.4306/pi.2009.6.2.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clarke MC, Tanskanen A, Huttunen M, Leon DA, Murray RM, Jones PB, Cannon M. Increased risk of schizophrenia from additive interaction between infant motor developmental delay and obstetric complications evidence from a population-based longitudinal study. Am. J. Psychiatry. 2011;168:1295–1302. doi: 10.1176/appi.ajp.2011.11010011. [DOI] [PubMed] [Google Scholar]
- 37.Gilmore JH, Kang C, Evans DD, Wolfe HM, Smith JK, Lieberman JA, Lin W, Hamer RM, Styner M, Gerig G. Prenatal and neonatal brain structure and white matter maturation in children at high risk for schizophrenia. Am. J. Psychiatry. 2010;167:1083–1091. doi: 10.1176/appi.ajp.2010.09101492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao TP, Kühl M. An updated overview on Wnt signaling pathways a prelude for more. Circ. Res. 2010;106:1798–1806. doi: 10.1161/CIRCRESAHA.110.219840. [DOI] [PubMed] [Google Scholar]
- 39.McMahon AP, Bradley A. The Wnt-1 [int-1] proto-oncogene is required for development of a large region of the mouse brain. Cell. 1990;62:1073–1085. doi: 10.1016/0092-8674(90)90385-r. [DOI] [PubMed] [Google Scholar]
- 40.Komiya Y, Habas R. Wnt signal transduction pathways. Organogenesis. 2008;4:68–75. doi: 10.4161/org.4.2.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sugimura R, Li L. Noncanonical Wnt signaling in vertebrate development stem cells and diseases. Birth Defects Res. C. Embryo Today. 2010;90:243–256. doi: 10.1002/bdrc.20195. [DOI] [PubMed] [Google Scholar]
- 42.vanAmerongen R, Nusse R. Towards an integrated view of Wnt signaling in development. Development. 2009;136:3205–3214. doi: 10.1242/dev.033910. [DOI] [PubMed] [Google Scholar]
- 43.Yuan Y, Niu CC, Deng G, Li ZQ, Pan J, Zhao C, Yang ZL, Si WK. The Wnt5a/Ror2 noncanonical signaling pathway inhibits canonical Wnt signaling in K562 cells. Int. J. Mol. Med. 2011;27:63–69. doi: 10.3892/ijmm.2010.560. [DOI] [PubMed] [Google Scholar]
- 44.Habas R, Dawid IB, He X. Coactivation of Rac and Rho by Wnt/Frizzled signaling is required for vertebrate gastrulation. Genes. Dev. 2003;17:295–309. doi: 10.1101/gad.1022203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohn AD, Moon RT. Wnt and calcium signaling beta-catenin-independent pathways. Cell Calcium. 2005;38:439–446. doi: 10.1016/j.ceca.2005.06.022. [DOI] [PubMed] [Google Scholar]
- 46.Habas R, He X. Activation of Rho and Rac by Wnt/frizzled signaling. Methods Enzymol. 2006;406:500–511. doi: 10.1016/S0076-6879(06)06038-1. [DOI] [PubMed] [Google Scholar]
- 47.Gao C, Chen YG. Dishevelled The hub of Wnt signaling. Cell. Signal. 2010;22:717–727. doi: 10.1016/j.cellsig.2009.11.021. [DOI] [PubMed] [Google Scholar]
- 48.Mlodzik M. Planar cell polarization do the same mechanisms regulate Drosophila tissue polarity and vertebrate gastrulation. Trends Genet. 2002;18:564–571. doi: 10.1016/s0168-9525(02)02770-1. [DOI] [PubMed] [Google Scholar]
- 49.Simons M, Mlodzik M. Planar cell polarity signaling from fly development to human disease. Annu. Rev. Genet. 2008;42:517–540. doi: 10.1146/annurev.genet.42.110807.091432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stevens HE, Smith KM, Rash BG, Vaccarino FM. Neural stem cell regulation fibroblast growth factors and the developmental origins of neuropsychiatric disorders. Front. Neurosci. 2010;4:59–0. doi: 10.3389/fnins.2010.00059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas KR, Capecchi MR. Targeted disruption of the murine int-1 proto-oncogene resulting in severe abnormalities in midbrain and cerebellar development. Nature. 1990;346:847–850. doi: 10.1038/346847a0. [DOI] [PubMed] [Google Scholar]
- 52.Patapoutian A, Reichardt LF. Roles of Wnt proteins in neural development and maintenance. Curr. Opin. Neurobiol. 2000;10:392–399. doi: 10.1016/s0959-4388(00)00100-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Machon O, Backman M, Machonova O, Kozmik Z, Vacik T, Andersen L, Krauss S. A dynamic gradient of Wnt signaling controls initiation of neurogenesis in the mammalian cortex and cellular specification in the hippocampus. Dev. Biol. 2007;311:223–237. doi: 10.1016/j.ydbio.2007.08.038. [DOI] [PubMed] [Google Scholar]
- 54.Lange C, Mix E, Rateitschak K, Rolfs A. Wnt signal pathways and neural stem cell differentiation. Neurodegener. Dis. 2006;3:76–86. doi: 10.1159/000092097. [DOI] [PubMed] [Google Scholar]
- 55.Zhou CJ, Borello U, Rubenstein JL, Pleasure SJ. Neuronal production and precursor proliferation defects in the neocortex of mice with loss of function in the canonical Wnt signaling pathway. Neuroscence. 2006;, 142:1119–1131. doi: 10.1016/j.neuroscience.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 56.Silhankova M, Korswagen HC. Migration of neuronal cells along the anterior-posterior body axis of C.elegans Wnts are in control. Curr. Opin. Genet. Dev. 2007;17:320–325. doi: 10.1016/j.gde.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 57.Hall AC, Lucas FR, Salinas PC. Axonal remodeling and synaptic differentiation in the cerebellum is regulated by WNT-7a signaling. Cell. 2000;100:525–535. doi: 10.1016/s0092-8674(00)80689-3. [DOI] [PubMed] [Google Scholar]
- 58.Varela-Nallar L, Alfaro IE, Serrano FG, Parodi J, Inestrosa NC. Wingless-type family member 5A (Wnt-5a) stimulates synaptic differentiation and function of glutamatergic synapses. Proc. Natl. Acad. Sci. U.S.A. 2010;107(49 ):21164–21169. doi: 10.1073/pnas.1010011107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Jensen M, Brockie PJ, Maricq AV. Wnt signaling regulates experience-dependent synaptic plasticity in the adult nervous system. Cell Cycle. 2012;11(14 ):2585–2586. doi: 10.4161/cc.21138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu MV, Hen R. The young and the restless regulation of adult neurogenesis by Wnt signaling. Cell Stem Cell. 2013;12(2 ):139–140. doi: 10.1016/j.stem.2013.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wu Q, Li Y, Xiao B. DISC1-related signaling pathways in adult neurogenesis of the hippocampus. Gene. 2013;518(2 ):223–230. doi: 10.1016/j.gene.2013.01.015. [DOI] [PubMed] [Google Scholar]
- 62.Chen J, Park CS, Tang SJ. Activity-dependent synaptic Wnt release regulates hippocampal long term potentiation. J. Biol. Chem. 2006;281:11910–11916. doi: 10.1074/jbc.M511920200. [DOI] [PubMed] [Google Scholar]
- 63.Cerpa W, Gambrill A, Inestrosa NC, Barria A. Regulation of NMDA-receptor synaptic transmission by Wnt signaling. J. Neurosci. 2011;31:9466–9471. doi: 10.1523/JNEUROSCI.6311-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ma T, Tzavaras N, Tsokas P, Landau EM, Blitzer RD. Synaptic stimulation of mTOR is mediated by Wnt signaling and regulation of Glycogen Synthetase Kinase-3. J. Neurosci. 2011;31:17537–17546. doi: 10.1523/JNEUROSCI.4761-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Okerlund ND, Cheyette BN. Synaptic Wnt signaling.A contributor to major psychiatric disorders. J. Neurodevelop. Disord. 2011;3:162–174. doi: 10.1007/s11689-011-9083-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ille F, Sommer L. Wnt signaling multiple functions in neural development. Cell. Mol. Life Sci. 2005;62:1100–1108. doi: 10.1007/s00018-005-4552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cerpa W, Toledo EM, Varela-Nallar L, Inestrosa NC. The role of Wnt signaling in neuroprotection. Drug News Perspect. 2009;22:579–591. doi: 10.1358/dnp.2009.10.1436817. [DOI] [PubMed] [Google Scholar]
- 68.Nusse R, vanOoyen A, Cox D, Fung YK, Varmus H. Mode of proviral activation of a putative mammary oncogene [int-1] on mouse chromosome 15. Nature. 1984;307:131–136. doi: 10.1038/307131a0. [DOI] [PubMed] [Google Scholar]
- 69.Augustine KA, Liu ET, Sadler TW. Interactions of Wnt-1 and Wnt-3a are essential for neural tube patterning. Teratology. 1995;51:107–119. doi: 10.1002/tera.1420510209. [DOI] [PubMed] [Google Scholar]
- 70.Prakash N, Brodski C, Naserke T, Puelles E, Gogoi R, Hall A, Panhuysen M, Echevarria D, Sussel L, Weisenhorn DM, Martinez S, Arenas E, Simeone A, Wurst W. A Wnt1-regulated genetic network controls the identity and fate of midbrain-dopaminergic progenitors in vivo. Development. 2006;133:89–98. doi: 10.1242/dev.02181. [DOI] [PubMed] [Google Scholar]
- 71.Wainwright BJ, Scambler PJ, Stanier P, Watson EK, Bell G, Wicking C, Estivill X, Courtney M, Bour A, Pedersen PS, Williamson R, Farrall M. Isolation of a human gene withprotein sequence similarity to human and murine int-1 and the Drosophila segment polarity mutant wingless. EMBO J. 1988;7:1743–1748. doi: 10.1002/j.1460-2075.1988.tb03003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kim HJ, Park JK, Kim SK, Kang SW, Kim JW, Park HK, Cho AR, Song JY, Chung JH. No association between polymorphisms of Wnt2 and schizophrenia in a Korean population. BMC Med. Genet. 2010;11:78–0. doi: 10.1186/1471-2350-11-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Adler PN, Lee H. Frizzled signaling and cell-cell interactions in planar polarity. Curr. Opin. Cell. Biol. 2001;13:635–640. doi: 10.1016/s0955-0674(00)00263-5. [DOI] [PubMed] [Google Scholar]
- 74.Wu J, Klein TJ, Mlodzik M. Subcellular localization of frizzled receptors mediated by their cytoplasmic tails regulates signaling pathway specificity. PLoS Biol. 2004;2:E158–0. doi: 10.1371/journal.pbio.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huang HC, Klein PS. The Frizzled family receptors for multiple signal transduction pathways. Genome Biol. 2004;5:234–0. doi: 10.1186/gb-2004-5-7-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sala CF, Formenti E, Terstappen GC, Caricasole A. Identification gene structure and expression of human frizzled-3 [FZD3] Biochem. Biophys. Res. Commun. 2000;273:27–34. doi: 10.1006/bbrc.2000.2882. [DOI] [PubMed] [Google Scholar]
- 77.Deardorff MA, Tan C, Saint-Jeannet JP, Klein PS. A role for frizzled 3 in neural crest development. Development. 2001;128:3655–3663. doi: 10.1242/dev.128.19.3655. [DOI] [PubMed] [Google Scholar]
- 78.Kiecker C, Niehrs C. A morphogen gradient of Wnt/beta-catenin signalling regulates anteroposterior neural patterning in Xenopus. Development. 2001;128:4189–4201. doi: 10.1242/dev.128.21.4189. [DOI] [PubMed] [Google Scholar]
- 79.Wilson SI, Rydstrom A, Trimborn T, Willert K, Nusse R, Jessell TM, Edlund T. The status of Wnt signaling regulates neural and epidermal fates in the chick embryo. Nature. 2001;411:325–330. doi: 10.1038/35077115. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y, Thekdi N, Smallwood PM, Macke JP, Nathans J. Frizzled-3 is required for the development of major fiber tracts in the rostral CNS J. Neurosci. 2002;22:8563–8573. doi: 10.1523/JNEUROSCI.22-19-08563.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vinson CR, Conover S, Adler PN. A Drosophila tissue polarity locus encodes a protein containing seven potential transmembrane domains. Nature. 1989;338:263–264. doi: 10.1038/338263a0. [DOI] [PubMed] [Google Scholar]
- 82.Thorpe CJ, Schlesinger A, Bowerman B. Wnt signalling in Caenorhabditis elegans regulating repressors and polarizing the cytoskeleton. Trends Cell. Biol. 2000;10:10–17. doi: 10.1016/s0962-8924(99)01672-4. [DOI] [PubMed] [Google Scholar]
- 83.Garcia-Morales C, Liu CH, Abu-Elmagd M, Hajihosseini MK, Wheeler GN. Frizzled-10 promotes sensory neuron development in Xenopus embryos. Dev. Biol. 2009;335:143–155. doi: 10.1016/j.ydbio.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 84.Sumanas S, Strege P, Heasman J, Ekker SC. The putative wnt receptor Xenopus frizzled-7 functions upstream of ß-catenin in vertebrate dorsoventral mesoderm patterning. Development. 2000;127: 1981–1990. doi: 10.1242/dev.127.9.1981. [DOI] [PubMed] [Google Scholar]
- 85.Deardorff MA, Tan C, Conrad LJ, Klein PS. Frizzled 8 is expressed in the Spemann organizer and plays a role in early morphogenesis. Development. 1998;125:2687–2700. doi: 10.1242/dev.125.14.2687. [DOI] [PubMed] [Google Scholar]
- 86.Itoh K, Jacob JY, Sokol S. A role for Xenopus Frizzled 8 in dorsal development. Mech. Dev. 1998;74:145–157. doi: 10.1016/s0925-4773(98)00076-8. [DOI] [PubMed] [Google Scholar]
- 87.Kim S, Kim SH, Kim H, Chung AY, Cha YI, Kim CH, Huh TL, Park HC. Frizzled 8a function is required for oligodendrocyte development in the zebrafish spinal cord. Dev. Dyn. 2008;237:3324–3331. doi: 10.1002/dvdy.21739. [DOI] [PubMed] [Google Scholar]
- 88.Tan C, Deardorff MA, Saint-Jeannet JP, Yang J, Arzoumanian A, Klein PS. Kermit a frizzled interacting protein regulates frizzled-3 signaling in neural crest development. Development. 2001;128:3665–3674. doi: 10.1242/dev.128.19.3665. [DOI] [PubMed] [Google Scholar]
- 89.Rasmussen JT, Deardorff MA, Tan C, Rao MS, Klein PS, Vetter ML. Regulation of eye development by frizzled signaling in Xenopus. Proc. Natl. Acad. Sci. U.S.A. 2001;98:3861–3866. doi: 10.1073/pnas.071586298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nasevicius A, Hyatt T, Kim H, Guttman J, Walsh E, Sumanas S, Wang Y, Ekker SC. Evidence for a frizzled-mediated wnt pathway required for zebrafish dorsal mesoderm formation. Development. 1998;125:4283–4292. doi: 10.1242/dev.125.21.4283. [DOI] [PubMed] [Google Scholar]
- 91.El-Messaoudi S, Renucci A. Expression pattern of the frizzled 7 gene during zebrafish embryonic development. Mech. Dev. 2001;102:231–234. doi: 10.1016/s0925-4773(01)00291-x. [DOI] [PubMed] [Google Scholar]
- 92.Sumanas S, Kim HJ, Hermanson SB, Ekker SC. Lateral line nervous system and maternal expression of Frizzled 7a during zebrafish embryogenesis. Mech. Dev. 2002;115:107–111. doi: 10.1016/s0925-4773(02)00084-9. [DOI] [PubMed] [Google Scholar]
- 93.Grumolato L, Liu G, Mong P, Mudbhary R, Biswas R, Arroyave R, Vijayakumar S, Economides AN, Aaronson SA. Canonical and noncanonical Wnts use a common mechanism to activate completely unrelated coreceptors. Genes Dev. 2010;24(22 ):2517–2530. doi: 10.1101/gad.1957710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Sonderegger S, Husslein H, Leisser C, Knöfler M. Complex expression pattern of Wnt ligands and frizzled receptors in human placenta and its trophoblast subtypes. Placenta. 2007;28 Suppl A:S97–102. doi: 10.1016/j.placenta.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. J. Biol. Chem. 2006;281(19 ):13247–13257. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- 96.Wang Y, Macke JP, Abella BS, Andreasson K, Worley P, Gilbert DJ, Copeland NG, Jenkins NA, Nathans J. A large family of putative transmembrane receptors homologous to the product of the Drosophila tissue polarity gene frizzled. J. Biol. Chem. 1996;271:4468–4476. doi: 10.1074/jbc.271.8.4468. [DOI] [PubMed] [Google Scholar]
- 97.Grove EA, Tole S, Limon J, Yip Y, Ragsdale CW. The hem of the embryonic cerebral cortex is defined by the expression of multiple Wnt genes and is compromised in Gli3-deficient mice. Development. 1998;125:2315–2325. doi: 10.1242/dev.125.12.2315. [DOI] [PubMed] [Google Scholar]
- 98.Borello U, Buffa V, Sonnino C, Melchionna R, Vivarelli E, Cossu G. Differential expression of the Wnt putative receptors Frizzled during mouse somitogenesis. Mech. Dev. 1999;89:173–177. doi: 10.1016/s0925-4773(99)00205-1. [DOI] [PubMed] [Google Scholar]
- 99.Moon RT, Kimelman D. From cortical rotation to organizer gene expression toward a molecular explanation of axis specification in Xenopus. Bioessays. 1998;20:536–545. doi: 10.1002/(SICI)1521-1878(199807)20:7<536::AID-BIES4>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 100.Sahin M, Dowling JJ, Hockfield S. Seven protein tyrosine phosphatases are differentially expressed in the developing rat brain. J. Comp. Neurol. 1995;351:617–631. doi: 10.1002/cne.903510410. [DOI] [PubMed] [Google Scholar]
- 101.Tamai K, Semenov M, Kato Y, Spokony R, Liu C, Katsuyama Y, Hess F, Saint-Jeannet JP, He X. LDL-receptor-related proteins in Wnt signal transduction. Nature. 2000;407:530–535. doi: 10.1038/35035117. [DOI] [PubMed] [Google Scholar]
- 102.Narita M, Bu G, Holtzman DM, Schwartz AL. The low-density lipoprotein receptor-related protein a multifunctional apolipoprotein E receptor modulates hippocampal neurite development. J. Neurochem. 1997;68:587–595. doi: 10.1046/j.1471-4159.1997.68020587.x. [DOI] [PubMed] [Google Scholar]
- 103.Cadigan KM, Liu YI. Wnt signaling complexity at the surface. J. Cell. Sci. 2006;119 (Pt 3 ):395–402. doi: 10.1242/jcs.02826. [DOI] [PubMed] [Google Scholar]
- 104.Mikels AJ, Nusse R. Purified Wnt5a protein activates or inhibits beta-catenin-TCF signaling depending on receptor context. PLoS Biol. 2006;4:e115–0. doi: 10.1371/journal.pbio.0040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ciani L, Salinas PC. WNTs in the vertebrate nervous system from patterning to neuronal connectivity. Nat. Rev. Neurosci. 2005;6:351–362. doi: 10.1038/nrn1665. [DOI] [PubMed] [Google Scholar]
- 106.Ciani L, Krylova O, Smalley MJ, Dale TC, Salinas PC. A divergent canonical WNT-signaling pathway regulates microtubule dynamics dishevelled signals locally to stabilize microtubules. J. Cell. Biol. 2004;164:243–253. doi: 10.1083/jcb.200309096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Itoh K, Brott BK, Bae GU, Ratcliffe MJ, Sokol SY. Nuclear localization is required for Dishevelled function in Wnt/ß-catenin signaling. J. Biol. 2005;4:3–0. doi: 10.1186/jbiol20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ahmad-Annuar A, Ciani L, Simeonidis I, Herreros J, Fredj NB, Rosso SB, Hall A, Brickley S, Salinas PC. Signaling across the synapse a role for Wnt and Dishevelled in presynaptic assembly and neurotransmitter release. J. Cell. Biol. 2006;174:127–139. doi: 10.1083/jcb.200511054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Krylova O, Messenger MJ, Salinas PC. Dishevelled-1 regulates microtubule stability a new function mediated by glycogen synthase kinase-3ß. J. Cell. Biol. 2000;151:83–94. doi: 10.1083/jcb.151.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Rosso SB, Sussman D, Wynshaw-Boris A, Salinas PC. Wnt signaling through Dishevelled, Rac and JNK regulates dendritic development. Nat. Neurosci. 2005;8:34–42. doi: 10.1038/nn1374. [DOI] [PubMed] [Google Scholar]
- 111.Lijam N, Paylor R, McDonald MP, Crawley JN, Deng CX, Herrup K, Stevens KE, Maccaferri G, McBain CJ, Sussman DJ, Wynshaw-Boris A. Social interaction and sensorimotor gating abnormalities in mice lacking Dvl1. Cell. 1997;90:895–905. doi: 10.1016/s0092-8674(00)80354-2. [DOI] [PubMed] [Google Scholar]
- 112.Woodgett JR, Cohen P. Multisite phosphorylation of glycogen synthase.Molecular basis for the substrate specificity of glycogen synthase kinase-3 and casein kinase-II [glycogen synthase kinase-5] Biochim. Biophys. Acta. 1984;788:339–347. doi: 10.1016/0167-4838(84)90047-5. [DOI] [PubMed] [Google Scholar]
- 113.Wang Y, Roach PJ. Inactivation of rabbit muscle glycogen synthase by glycogen synthase kinase-3.Dominant role of the phosphorylation of Ser-640 [site-3a] J. Biol. Chem. 1993;268:23876–23880. [PubMed] [Google Scholar]
- 114.Mukai F, Ishiguro K, Sano Y, Fujita SC. Alternative splicing isoform of tau protein kinase I/glycogen synthase kinase 3ß. J. Neurochem. 2002;81:1073–1083. doi: 10.1046/j.1471-4159.2002.00918.x. [DOI] [PubMed] [Google Scholar]
- 115.Castano Z, Gordon-Weeks PR, Kypta RM. The neuron-specific isoform of glycogen synthase kinase-3ß is required for axon growth. J. Neurochem. 2010;113:117–130. doi: 10.1111/j.1471-4159.2010.06581.x. [DOI] [PubMed] [Google Scholar]
- 116.Wood-Kaczmar A, Kraus M, Ishiguro K, Philpott KL, Gordon-Weeks PR. An alternatively spliced form of glycogen synthase kinase-3ß is targeted to growing neurites and growth cones. Mol. Cell. Neurosci. 2009;42:184–194. doi: 10.1016/j.mcn.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 117.Hur EM, Zhou FQ. GSK3 signalling in neural development. Nat. Rev. Neurosci. 2010;11:539–51. doi: 10.1038/nrn2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim WY, Snider WD. Functions of GSK-3 signaling in development of the nervous system. Front. Mol. Neurosci. 2011;4:44–0. doi: 10.3389/fnmol.2011.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim YT, Hur EM, Snider WD, Zhou FQ. Role of GSK3 signaling in neuronal morphogenesis. Front. Mol. Neurosci. 2011;4:48–0. doi: 10.3389/fnmol.2011.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Li X, Liu M, Cai Z, Wang G, Li X. Regulation of glycogen synthase kinase-3 during bipolar mania treatment. Bipolar Disord. 2010;12:741–752. doi: 10.1111/j.1399-5618.2010.00866.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Kalinichev M, Dawson LA. Evidence for antimanic efficacy of glycogen synthase kinase-3 (GSK3) inhibitors in a strain-specific model of acute mania. Int. J. Neuropsychopharmacol. 2011;14:1051–1067. doi: 10.1017/S1461145710001495. [DOI] [PubMed] [Google Scholar]
- 122.Groden J, Thliveris A, Samowitz W, Carlson M, Gelbert L, Albertsen H, Joslyn G, Stevens J, Spirio L, Robertson M, Sargeant L, Krapcho K, Wolff E, Burt R, Hughes JP, Warrington J, McPherson J, Wasmuth J, LePaslier D, Abderrahim H, Cohen D, Leppert M, White R. Identification and characterization of the familial adenomatous polyposis coli gene. Cell. 1991;66:589–600. doi: 10.1016/0092-8674(81)90021-0. [DOI] [PubMed] [Google Scholar]
- 123.Hamilton SR, Liu B, Parsons RE, Papadopoulos N, Jen J, Powell SM, Krush AJ, Berk T, Cohen Z, Tetu B, Burger PC, Wood PA, Taqi F, Booker SV, Petersen GM, Offerhaus GJA, Tersmette AC, Giardiello FM, Vogelstein B, Kinzler KW. The molecular basis of Turcot's syndrome. N. Engl. J. Med. 1995;332:839–847. doi: 10.1056/NEJM199503303321302. [DOI] [PubMed] [Google Scholar]
- 124.Heald B, Moran R, Milas M, Burke C, Eng C. Familial adenomatous polyposis in a patient with unexplained mental retardation. Nat. Clin. Pract. Neurol. 2007;3:694–700. doi: 10.1038/ncpneuro0658. [DOI] [PubMed] [Google Scholar]
- 125.Baker SJ, McKinnon PJ. Tumour-suppressor function in the nervous system. Nat. Rev. Cancer. 2004;4:184–196. doi: 10.1038/nrc1297. [DOI] [PubMed] [Google Scholar]
- 126.Fodde R, Kuipers J, Rosenberg C, Smits R, Kielman M, Gaspar C, vanEs JH, Breukel C, Wiegant J, Giles RH, Clevers H. Mutations in the APC tumour suppressor gene cause chromosomal instability. Nat. Cell Biol. 2001;3:433–438. doi: 10.1038/35070129. [DOI] [PubMed] [Google Scholar]
- 127.Dikovskaya D, Schiffmann D, Newton IP, Oakley A, Kroboth K, Sansom O, Jamieson TJ, Meniel V, Clarke A, Nathke IS. Loss of APC induces polyploidy as a result of a combination of defects in mitosis and apoptosis. J. Cell Biol. 2007;176:183–195. doi: 10.1083/jcb.200610099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ivaniutsin U, Chen Y, Mason JO, Price DJ, Pratt T. Adenomatous polyposis coli is required for early events in the normal growth and differentiation of the developing cerebral cortex. Neural. Dev. 2009;4:3–0. doi: 10.1186/1749-8104-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Koshimizu H, Fukui Y, Takao K, Ohira K, Tanda K, Nakanishi K, Toyama K, Oshima M, Taketo MM, Miyakawa T. Adenomatous polyposis coli heterozygous knockout mice display hypoactivity and age-dependent working memory deficits. Front. Behav. Neurosci. 2011;5:85–0. doi: 10.3389/fnbeh.2011.00085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Kraus C, Liehr T, Hülsken J, Behrens J, Birchmeier W, Grzeschik KH, Ballhausen WG. Localization of the human ß-catenin gene [CTNNB1] to 3p21 a region implicated in tumor development. Genomics. 1994;23:272–274. doi: 10.1006/geno.1994.1493. [DOI] [PubMed] [Google Scholar]
- 131.Brembeck FH, Rosário M, Birchmeier W. Balancing cell adhesion and Wnt signaling the key role of ß-catenin. Curr. Opin. Genet. Dev. 2006;16:51–59. doi: 10.1016/j.gde.2005.12.007. [DOI] [PubMed] [Google Scholar]
- 132.Meng K, Rodriguez-Peña A, Dimitrov T, Chen W, Yamin M, Noda M, Deuel TF. Pleiotrophin signals increased tyrosine phosphorylation of ß-catenin through inactivation of the intrinsic catalytic activity of the receptor-type protein tyrosine phosphatase ß/? Proc. Natl. Acad. Sci. USA. 2000;97:2603–2608. doi: 10.1073/pnas.020487997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kolpakova E, and Olsen BR. Wnt/beta-catenin-a canonical tale of cell-fate choice in the vertebrate skeleton. Dev. Cell. 2005;8:626–627. doi: 10.1016/j.devcel.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 134.Chen M, Zhu. M, Awad H, Li TF, Sheu TJ, Boyce BF, Chen D, O'Keefe RJ. Inhibition of beta-catenin signaling causes defects in postnatal cartilage development. J. Cell Sci. 2008;121:1455–1465. doi: 10.1242/jcs.020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Selvadurai HJ, Mason JO. Wnt/ß-catenin signalling is active in a highly dynamic pattern during development of the mouse cerebellum. PLoS One. 2011;6:e23012–0. doi: 10.1371/journal.pone.0023012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ephrussi A, Church GM, Tonegawa S, Gilbert WB. lineage-specific interactions of an immunoglobulin enhancer with cellular factors in vivo. Science. 1985;227:134–140. doi: 10.1126/science.3917574. [DOI] [PubMed] [Google Scholar]
- 137.dePontual L, Mathieu Y, Golzio C, Rio M, Malan V, Boddaert N, Soufflet C, Picard C, Durandy A, Dobbie A, Heron D, Isidor B, Motte J, Newburry-Ecob R, Pasquier L, Tardieu M, Viot G, Jaubert F, Munnich A, Colleaux L, Vekemans M, Etchevers H, Lyonnet S, Amiel J. Mutational functional and expression studies of the TCF4 gene in Pitt-Hopkins syndrome. Hum. Mutat. 2009;30:669–676. doi: 10.1002/humu.20935. [DOI] [PubMed] [Google Scholar]
- 138.Kunz M, Herrmann M, Wedlich D, Gradl D. Autoregulation of canonical Wnt signaling controls midbrain development. Dev. Biol. 2004;273:390–401. doi: 10.1016/j.ydbio.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 139.Niehrs C. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 2006;25:7469–7481. doi: 10.1038/sj.onc.1210054. [DOI] [PubMed] [Google Scholar]
- 140.Glinka A, Wu W, Delius H, Monaghan AP, Blumenstock C, Niehrs C. Dickkopf-1 is a member of a new family of secreted proteins and functions in head induction. Nature. 1998;391:357–362. doi: 10.1038/34848. [DOI] [PubMed] [Google Scholar]
- 141.Bafico A, Liu G, Yaniv A, Gazit A, Aaronson SA. Novel mechanism of Wnt signalling inhibition mediated by Dickkopf-1 interaction with LRP6/Arrow. Nat. Cell. Biol. 2001;3:683–686. doi: 10.1038/35083081. [DOI] [PubMed] [Google Scholar]
- 142.Nakamura RE, Hunter DD, Yi H, Brunken WJ, Hackam AS. Identification of two novel activities of the Wnt signaling regulator Dickkopf 3 and characterization of its expression in the mouse retina. BMC Cell. Biol. 2007;8:52–0. doi: 10.1186/1471-2121-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Zhang J, Moseley A, Jegga AG, Gupta A, Witte DP, Sartor M, Medvedovic M, Williams SS, Ley-Ebert C, Coolen LM, Egnaczyk G, Genter MB, Lehman M, Lingrel J, Maggio J, Parysek L, Walsh R, Xu M, Aronow BJ. Neural system-enriched gene expression relationship to biological pathways and neurological diseases. Physiol. Genomics. 2004;18:167–183. doi: 10.1152/physiolgenomics.00220.2003. [DOI] [PubMed] [Google Scholar]
- 144.Zenzmaier C, Marksteiner J, Kiefer A, Berger P, Humpel C. Dkk-3 is elevated in CSF and plasma of Alzheimer's disease patients. J. Neurochem. 2009;110:653–661. doi: 10.1111/j.1471-4159.2009.06158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Diep DB, Hoen N, Backman M, Machon O, Krauss S. Characterisation of the Wnt antagonists and their response to conditionally activated Wnt signaling in the developing mouse forebrain. Dev. Brain Res. 2004;153:261–270. doi: 10.1016/j.devbrainres.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 146.Bazzi H, Fantauzzo KA, Richardson GD, Jahoda CAM, Christiano AM. The Wnt inhibitor Dickkopf 4 is induced by canonical Wnt signaling during ectodermal appendage morphogenesis. Dev. Biol. 2007;305:498–507. doi: 10.1016/j.ydbio.2007.02.035. [DOI] [PubMed] [Google Scholar]
- 147.He J, Sheng T, Stelter AA, Li C, Zhang X, Sinha M, Luxon BA, Xie J. Suppressing Wnt signaling by the hedgehog pathway through sFRP-1. J. Biol. Chem. 2006;281(47 ):35598–35602. doi: 10.1074/jbc.C600200200. [DOI] [PubMed] [Google Scholar]
- 148.Galli LM, Barnes T, Cheng T, Acosta L, Anglade A, Willert K, Nusse R, Burrus LW. Differential inhibition of Wnt-3a by Sfrp-1 Sfrp-2 and Sfrp-3. Dev. Dyn. 2006;235(3 ):681–90. doi: 10.1002/dvdy.20681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kawano Y, Kypta R. Secreted antagonists of the Wnt signalling pathway. J. Cell. Sci. 2003;116(Pt 13 ):2627–2634. doi: 10.1242/jcs.00623. [DOI] [PubMed] [Google Scholar]
- 150.Trevant B, Gaur T, Hussain S, Symons J, Komm BS, Bodine PV, Stein GS, Lian JB. Expression of secreted frizzled related protein 1 a Wnt antagonist in brain kidney and skeleton is dispensable for normal embryonic development. J. Cell. Physiol. 2008;217:113–126. doi: 10.1002/jcp.21482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Augustine C, Gunnersen J, Spirkoska V, Tan SS. Place- and time-dependent expression of mouse sFRP-1 during development of the cerebral neocortex. Mech. Dev. 2001;109:395–397. doi: 10.1016/s0925-4773(01)00533-0. [DOI] [PubMed] [Google Scholar]
- 152.Kim AS, Lowenstein DH, Pleasure SJ. Wnt receptors and Wnt inhibitors are expressed in gradients in the developing telencephalon. Mech. Dev. 2001;103:167–172. doi: 10.1016/s0925-4773(01)00342-2. [DOI] [PubMed] [Google Scholar]
- 153.Harrison PJ, Weinberger DR. Schizophrenia genes gene expression and neuropathology on the matter of their convergence. Mol. Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 154.Tabarés-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders implications for schizophrenia autism and cancer. Mol. Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- 155.Leaf I, Tennessen J, Mukhopadhyay M, Westphal H, Shawlot W. Sfrp5 is not essential for axis formation in the mouse. Genesis. 2006;44:573–578. doi: 10.1002/dvg.20248. [DOI] [PubMed] [Google Scholar]
- 156.Satoh W, Gotoh T, Tsunematsu Y, Aizawa S, Shimono A. Sfrp1 and Sfrp2 regulate anteroposterior axis elongation and somite segmentation during mouse embryogenesis. Development. 2006;133:989–999. doi: 10.1242/dev.02274. [DOI] [PubMed] [Google Scholar]
- 157.Tebar M, Destrée O, deVree WJ, Ten Have-Opbroek AA. Expression of Tcf/Lef and sFrp and localization of beta-catenin in the developing mouse lung. Mech. Dev. 2001;109:437–440. doi: 10.1016/s0925-4773(01)00556-1. [DOI] [PubMed] [Google Scholar]
- 158.Foronjy R, Imai K, Shiomi T, Mercer B, Sklepkiewicz P, Thankachen J, Bodine P, D'Armiento J. The divergent roles of secreted frizzled related protein-1 (SFRP1) in lung morphogenesis and emphysema. Am. J. Pathol. 2010;177:598–607. doi: 10.2353/ajpath.2010.090803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Beaumont V, Thompson SA, Choudhry F, Nuthall H, Glantschnig H, Lipfert L, David GR, Swain CJ, McAllister G, Munoz-Sanjuan I. Evidence for an enhancement of excitatory transmission in adult CNS by Wnt signaling pathway modulation. Mol. Cell. Neurosci. 2007;35(4 ):513–524. doi: 10.1016/j.mcn.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 160.Jang MH, Bonaguidi MA, Kitabatake Y, Sun J, Song J, Kang E, Jun H, Zhong C, Su Y, Guo JU, Wang MX, Sailor KA, Kim JY, Gao Y, Christian KM, Ming GL, Song H. Secreted frizzled-related protein 3 regulates activity-dependent adult hippocampal neurogenesis. Cell Stem Cell. 2013;12(2 ):215–223. doi: 10.1016/j.stem.2012.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Talbot K, Eidem WL, Tinsley CL, Benson MA, Thompson EW, Smith RJ, Hahn CG, Siegel SJ, Trojanowski JQ, Gur RE, Blake DJ, Arnold SE. Dysbindin-1 is reduced in intrinsic, glutamatergic terminals of the hippocampal formation in schizophrenia. J. Clin. Invest. 2004;113(9 ):1353–1363. doi: 10.1172/JCI20425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.vanElst LT, Valerius G, Büchert M, Thiel T, Rüsch N, Bubl E, Hennig J, Ebert D, Olbrich HM. Increased prefrontal and hippocampal glutamate concentration in schizophrenia evidence from a magnetic resonance spectroscopy study. Biol. Psychiatry. 2005;58(9 ):724–730. doi: 10.1016/j.biopsych.2005.04.041. [DOI] [PubMed] [Google Scholar]
- 163.Ma T, Tzavaras N, Tsokas P, Landau EM, Blitzer RD. Synaptic stimulation of mTOR is mediated by Wnt signaling and regulation of glycogen synthetase kinase-3. J. Neurosci. 2011;31(48 ):17537–17546. doi: 10.1523/JNEUROSCI.4761-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Frantseva MV, Fitzgerald PB, Chen R, Möller B, Daigle M, Daskalakis ZJ. Evidence for impaired long-term potentiation in schizophrenia and its relationship to motor skill learning. Cereb. Cortex. 2008;18(5 ):990–996. doi: 10.1093/cercor/bhm151. [DOI] [PubMed] [Google Scholar]
- 165.Brennand KJ, Simone A, Jou J, Gelboin-Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473(7346 ):221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cotter D, Kerwin R, al-Sarraji S, Brion JP, Chadwich A, Lovestone S, Anderton B, Everall I. Abnormalities of Wnt signalling in schizophrenia-evidence for neurodevelopmental abnormality. Neuroreport. 1998;9:1379–1383. doi: 10.1097/00001756-199805110-00024. [DOI] [PubMed] [Google Scholar]
- 167.Kozlovsky N, Belmaker RH, Agam G. GSK-3 and the neurodevelopmental hypothesis of schizophrenia. Eur. Neuro-psychopharmacol. 2002;12:13–25. doi: 10.1016/s0924-977x(01)00131-6. [DOI] [PubMed] [Google Scholar]
- 168.Benedetti F, Poletti S, Radaelli D, Bernasconi A, Cavallaro R, Falini A, Lorenzi C, Pirovano A, Dallaspezia S, Locatelli C, Scotti G, Smeraldi E. Temporal lobe grey matter volume in schizophrenia is associated with a genetic polymorphism influencing glycogen synthase kinase-3ß activity. Genes. Brain Behav. 2010;9:365–371. doi: 10.1111/j.1601-183X.2010.00566.x. [DOI] [PubMed] [Google Scholar]
- 169.Beaulieu JM, Zhang X, Rodriguiz RM, Sotnikova TD, Cools MJ, Wetsel WC, Gainetdinov RR, Caron MG. Role of GSK-3ß in behavioral abnormalities induced by serotonin deficiency. Proc. Natl. Acad. Sci. USA. 2008;105:1333–1338. doi: 10.1073/pnas.0711496105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Beaulieu JM, Sotnikova TD, Yao WD, Kockeritz L, Woodgett JR, Gainetdinov RR, Caron MG. Lithium antagonizes dopamine-dependent behaviors mediated by an AKT/glycogen synthase kinase-3 signaling cascade. Proc. Natl. Acad. Sci. USA. 2004;101:5099–5104. doi: 10.1073/pnas.0307921101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Meltzer HY. Clinical studies on the mechanism of action of clozapine the dopamine-serotonin hypothesis of schizophrenia. Psychopharmacology [Berl.] 1989;99[Suppl]:S18–27. doi: 10.1007/BF00442554. [DOI] [PubMed] [Google Scholar]
- 172.Ichikawa J, Meltzer HY. Relationship between dopaminergic and serotonergic neuronal activity in the frontal cortex and the action of typical and atypical antipsychotic drugs. Eur. Arch. Psychiatry Clin. Neurosci. 1999;249(Suppl 4):90–98. doi: 10.1007/pl00014190. [DOI] [PubMed] [Google Scholar]
- 173.Sani G, Napoletano F, Forte AM, Kotzalidis GD. The Wnt pathway in mood disorders. Curr. Neuropharmacol. 2012; 10:1570–159. doi: 10.2174/157015912803217279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Li X, Rosborough KM, Friedman AB, Zhu W, Roth KA. Regulation of mouse brain glycogen synthase kinase-3 by atypical antipsychotics. Int. J. Neuropsychopharmacol. 2007;10:7–19. doi: 10.1017/S1461145706006547. [DOI] [PubMed] [Google Scholar]
- 175.Lovestone S, Killick R, DiForti M, Murray R. Schizophrenia as a GSK-3 dysregulation disorder. Trends Neurosci. 2007;30:142–149. doi: 10.1016/j.tins.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 176.Beaulieu JM, Gainetdinov RR, Caron MG. Akt/GSK-3 signaling in the action of psychotropic drugs. Annu. Rev. Pharmacol. Toxicol. 2009;49:327–347. doi: 10.1146/annurev.pharmtox.011008.145634. [DOI] [PubMed] [Google Scholar]
- 177.Gould TD, Picchini AM, Einat H, Manji HK. Targeting glycogen synthase kinase-3 in the CNS implications for the development of new treatments for mood disorders. Curr. Drug Targets. 2006;7:1399–1409. doi: 10.2174/1389450110607011399. [DOI] [PubMed] [Google Scholar]
- 178.Faraone SV, Matise T, Svrakic D, Pepple J, Malaspina D, Suarez B, Hampe C, Zambuto CT, Schmitt K, Meyer J, Markel P, Lee H, Harkavy Friedman J, Kaufmann C, Cloninger CR, Tsuang MT. Genome scan of European-American schizophrenia pedigrees results of the NIMH Genetics Initiative and Millennium Consortium. Am. J. Med. Genet. 1998;81:290–295. [PubMed] [Google Scholar]
- 179.Wassink TH, Piven J, Vieland VJ, Huang J, Swiderski RE, Pietila J, Braun T, Beck G, Folstein SE, Haines JL, Sheffield VC. Evidence supporting Wnt2 as an autism susceptibility gene. Am. J. Med. Genet. 2001;105:406–413. doi: 10.1002/ajmg.1401. [DOI] [PubMed] [Google Scholar]
- 180.McCoy PA, Shao Y, Wolpert CM, Donnelly SL, Ashley-Koch A, Abel HL, Ravan SA, Abramson RK, Wright HH, DeLong GR, Cuccaro ML, Gilbert JR, Pericak-Vance MA. No association between the Wnt2 gene and autistic disorder. Am. J. Med. Genet. 2002;114:106–109. doi: 10.1002/ajmg.10182. [DOI] [PubMed] [Google Scholar]
- 181.Li J, Nguyen L, Gleason C, Lotspeich L, Spiker D, Risch N, Myers RM. Lack of evidence for an association between Wnt2 and RELN polymorphisms and autism. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;126B:51–57. doi: 10.1002/ajmg.b.20122. [DOI] [PubMed] [Google Scholar]
- 182.Marui T, Funatogawa I, Koishi S, Yamamoto K, Matsumoto H, Hashimoto O, Jinde S, Nishida H, Sugiyama T, Kasai K, Watanabe K, Kano Y, Kato N. Association between autism and variants in the wingless-type MMTV integration site family member 2 (WNT2) gene. Int. J. Neuropsychopharmacol. 2010;13(4 ):443–449. doi: 10.1017/S1461145709990903. [DOI] [PubMed] [Google Scholar]
- 183.Chien YL, Wu YY, Chiu YN, Liu SK, Tsai WC, Lin PI, Chen CH, Gau SS, Chien WH. Association study of the CNS patterning genes and autism in Han Chinese in Taiwan. Prog. Neuropsychopharmacol. Biol. Psychiatry. 2011;35(6 ):1512–1517. doi: 10.1016/j.pnpbp.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 184.Lin PI, Chien YL, Wu YY, Chen CH, Gau SS, Huang YS, Liu SK, Tsai WC, Chiu YN. The WNT2 gene polymorphism associated with speech delay inherent to autism. Res. Dev. Disabil. 2012;33(5 ):1533–1540. doi: 10.1016/j.ridd.2012.03.004. [DOI] [PubMed] [Google Scholar]
- 185.Yang J, Si T, Ling Y, Ruan Y, Han Y, Wang X, Zhang H, Kong Q, Li X, Liu C, Zhang D, Zhou M, Yu Y, Liu S, Shu L, Ma D, Wei J, Zhang D. Association study of the human FZD3 locus with schizophrenia. Biol Psychiatry. 2003;54:1298–1301. doi: 10.1016/s0006-3223(03)00291-9. [DOI] [PubMed] [Google Scholar]
- 186.Zhang Y, Yu X, Yuan Y, Ling Y, Ruan Y, Si T, Lu T, Wu S, Gong X, Zhu Z, Yang J, Wang F, Zhang D. Positive association of the human frizzled 3 [FZD3] gene haplotype with schizophrenia in Chinese Han population. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2004;129B:16–19. doi: 10.1002/ajmg.b.30076. [DOI] [PubMed] [Google Scholar]
- 187.Katsu T, Ujike H, Nakano T, Tanaka Y, Nomura A, Nakata K, Takaki M, Sakai A, Uchida N, Imamura T, Kuroda S. The human frizzled-3 [FZD3] gene on chromosome 8p21 a receptor gene for Wnt ligands is associatedwith the susceptibility to schizophrenia. Neurosci. Lett. 2003;353:53–56. doi: 10.1016/j.neulet.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 188.Reif A, Melchers M, Strobel A, Jacob CP, Herterich S, Lesch KP, Zimmer M. FZD3 is not a risk gene for schizophrenia a case-control study in a Caucasian sample. J. Neural. Transm. Suppl. 2007;72:297–301. doi: 10.1007/978-3-211-73574-9_36. [DOI] [PubMed] [Google Scholar]
- 189.Ide M, Muratake T, Yamada K, Iwayama-Shigeno Y, Iwamoto K, Takao H, Toyota T, Kaneko N, Minabe Y, Nakamura K, Kato T, Mori N, Asada T, Someya T, Yoshikawa T. Genetic and expression analyses of FZD3 in schizophrenia. Biol. Psychiatry. 2004;56:462–465. doi: 10.1016/j.biopsych.2004.06.039. [DOI] [PubMed] [Google Scholar]
- 190.Wei J, Hemmings GP. Lack of a genetic association between the frizzled-3 gene and schizophrenia in a British population. Neurosci. Lett. 2004;366:336–338. doi: 10.1016/j.neulet.2004.05.063. [DOI] [PubMed] [Google Scholar]
- 191.Hashimoto R, Suzuki T, Iwata N, Yamanouchi Y, Kitajima T, Kosuga A, Tatsumi M, Ozaki N, Kamijima K, Kunugi H. Association study of the frizzled-3 [FZD3] gene with schizophrenia and mood disorders. J. Neural. Transm. 2005;112:303–307. doi: 10.1007/s00702-004-0264-2. [DOI] [PubMed] [Google Scholar]
- 192.Jeong SH, Joo EJ, Ahn YM, Lee KY, Kim YS. Investigation of genetic association between human Frizzled homolog 3 gene [FZD3] and schizophrenia results in a Korean population and evidence from meta-analysis. Psychiatry Res. 2006;143:1–11. doi: 10.1016/j.psychres.2005.07.026. [DOI] [PubMed] [Google Scholar]
- 193.Kang C, Zhou L, Liu H, Yang J. Association study of the frizzled 3 gene with Chinese Va schizophrenia. Neurosci. Lett. 2011;505:196–199. doi: 10.1016/j.neulet.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 194.Gregório SP, Sallet PC, Do KA, Lin E, Gattaz WF, Dias-Neto E. Polymorphisms in genes involved in neurodevelopment may be associated with altered brain morphology in schizophrenia preliminary evidence. Psychiatry Res. 2009;165:1–9. doi: 10.1016/j.psychres.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 195.Kishimoto M, Ujike H, Okahisa Y, Kotaka T, Takaki M, Kodama M, Inada T, Yamada M, Uchimura N, Iwata N, Sora I, Iyo M, Ozaki N, Kuroda S. The Frizzled 3 gene is associated with methamphetamine psychosis in the Japanese population. Behav. Brain Funct. 2008;4:37–0. doi: 10.1186/1744-9081-4-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Pizzuti A, Amati F, Calabrese G, Mari A, Colosimo A, Silani V, Giardino L, Ratti A, Penso D, Calzà L, Palka G, Scarlato G, Novelli G, Dallapiccola B. cDNA characterization and chromosomal mapping of two human homologues of the Drosophila dishevelled polarity gene. Hum. Mol. Genet. 1996;5:953–958. doi: 10.1093/hmg/5.7.953. [DOI] [PubMed] [Google Scholar]
- 197.Long JM, LaPorte P, Paylor R, Wynshaw-Boris A. Expanded characterization of the social interaction abnormalities in mice lacking Dvl1. Genes Brain. Behav. 2004;3:51–62. doi: 10.1046/j.1601-183x.2003.00045.x. [DOI] [PubMed] [Google Scholar]
- 198.Pizzuti A, Novelli G, Mari A, Ratti A, Colosimo A, Amati F, Penso D, Sangiuolo F, Calabrese G, Palka G, Silani V, Gennarelli M, Mingarelli R, Scarlato G, Scambler P, Dallapiccola B. Human homologue sequences to the Drosophila dishevelled segment-polarity gene are deleted in the DiGeorge syndrome. Am. J. Hum. Genet. 1996;58:722–729. [PMC free article] [PubMed] [Google Scholar]
- 199.Geyer MA, Braff DL. Startle habituation and sensorimotor gating in schizophrenia and related animal models. Schizophr. Bull. 1987;13:643–668. doi: 10.1093/schbul/13.4.643. [DOI] [PubMed] [Google Scholar]
- 200.Braff DL. Prepulse inhibition of the startle reflex a window on the brain in schizophrenia. Curr. Top. Behav. Neurosci. 2010;4:349–371. doi: 10.1007/7854_2010_61. [DOI] [PubMed] [Google Scholar]
- 201.Hammer TB, Oranje B, Fagerlund B, Bro H, Glenthøj BY. Stability of prepulse inhibition and habituation of the startle reflex in schizophrenia a 6-year follow-up study of initially antipsychotic-naive, first-episode schizophrenia patients. Int. J. Neuropsychopharmacol. 2011;14:913–925. doi: 10.1017/S1461145711000034. [DOI] [PubMed] [Google Scholar]
- 202.Sasamoto A, Miyata J, Hirao K, Fujiwara H, Kawada R, Fujimoto S, Tanaka Y, Kubota M, Sawamoto N, Fukuyama H, Takahashi H, Murai T. Social impairment in schizophrenia revealed by Autism-Spectrum Quotient correlated with gray matter reduction. Soc. Neurosci. 2011;6:548–558. doi: 10.1080/17470919.2011.575693. [DOI] [PubMed] [Google Scholar]
- 203.Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3ß signaling in schizophrenia. Nat. Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- 204.Zhao Z, Ksiezak-Reding H, Riggio S, Haroutunian V, Pasinetti GM. Insulin receptor deficits in schizophrenia and in cellular and animal models of insulin receptor dysfunction. Schizophr. Res. 2006;84:1–14. doi: 10.1016/j.schres.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 205.Thiselton DL, Vladimirov VI, Kuo PH, McClay J, Wormley B, Fanous A, O’Neill FA, Walsh D, VandenOord JCG, Kendler KS, Riley BP. AKT1 is associated with schizophrenia across multiple symptom dimensions in the Irish study of high density schizophrenia families. Biol. Psychiatry. 2008;63:449–457. doi: 10.1016/j.biopsych.2007.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.vanBeveren NJ, Buitendijk GH, Swagemakers S, Krab LC, Röder C, deHaan L, vanderSpek P, Elgersma Y. Marked reduction of AKT1 expression and deregulation of AKT1-associated pathways in peripheral blood mononuclear cells of schizophrenia patients. PLoS One. 2012;7:e32618–0. doi: 10.1371/journal.pone.0032618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Szamosi A, Kelemen O, Kéri S. Hippocampal volume and the AKT signaling system in first-episode schizophrenia. J. Psychiatr. Res. 2012;46:279–284. doi: 10.1016/j.jpsychires.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 208.Nelson MD, Saykin AJ, Flashman LA, Riordan HJ. Hippocampal volume reduction in schizophrenia as assessed by magnetic resonance imaging a meta-analytic study. Arch. Gen. Psychiatry. 1998;55:433–440. doi: 10.1001/archpsyc.55.5.433. [DOI] [PubMed] [Google Scholar]
- 209.Wright IC, Rabe-Hesketh S, Woodruff PW, David AS, Murray RM, Bullmore ET. Metaanalysis of regional brain volumes in schizophrenia. Am. J. Psychiatry. 2000;157:16–25. doi: 10.1176/ajp.157.1.16. [DOI] [PubMed] [Google Scholar]
- 210.Balu DT, Carlson GC, Talbot K, Kazi H, Hill-Smith TE, Easton RM, Birnbaum MJ, Lucki I. Akt1 deficiency in schizophrenia and impairment of hippocampal plasticity and function. Hippocampus. 2012;22:230–240. doi: 10.1002/hipo.20887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Lai WS, Xu B, Westphal KGC, Paterlini M, Olivier B, Pavlidis P, Karayiorgou M, Gogos JA. Akt1 deficiency affects neuronal morphology and predisposes to abnormalities in prefrontal cortex functioning. Proc. Natl. Acad. Sci. USA. 2006;103:16906–16911. doi: 10.1073/pnas.0604994103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Dai Y, Wei Z, Sephton CF, Zhang D, Anderson DH, Mousseau DD. Haloperidol induces the nuclear translocation of phosphatidylinositol 3'-kinase to disrupt Akt phosphorylation in PC12 cells. J. Psychiatry Neurosci. 2007;32:323–330. [PMC free article] [PubMed] [Google Scholar]
- 213.Ukai W, Ozawa H, Tateno M, Hashimoto E, Saito T. Neurotoxic potential of haloperidol in comparison with risperidone implication of Akt-mediated signal changes by haloperidol. J. Neural. Transm. 2004;111:667–681. doi: 10.1007/s00702-004-0109-z. [DOI] [PubMed] [Google Scholar]
- 214.Xu MQ, Xing QH, Zheng YL, Li S, Gao JJ, He G, Guo TW, Feng GY, Xu F, He L. Association of AKT1 gene polymorphisms with risk of schizophrenia and with response to antipsychotics in the Chinese population. J. Clin. Psychiatry. 2007;68:1358–1367. doi: 10.4088/jcp.v68n0906. [DOI] [PubMed] [Google Scholar]
- 215.Mathur A, Law MH, Megson IL, Shaw DJ, Wei J. Genetic association of the AKT1 gene with schizophrenia in a British population. Psychiatr. Genet. 2010;20:118–122. doi: 10.1097/YPG.0b013e32833a2234. [DOI] [PubMed] [Google Scholar]
- 216.Ikeda M, Iwata N, Suzuki T, Kitajima T, Yamanouchi Y, Kinoshita Y, Inada T, Ozaki N. Association of AKT1 with schizophrenia confirmed in a Japanese population. Biol. Psychiatry. 2004;56:698–700. doi: 10.1016/j.biopsych.2004.07.023. [DOI] [PubMed] [Google Scholar]
- 217.Bajestan SN, Sabouri AH, Nakamura M, Takashima H, Keikhaee MR, Behdani F, Fayyazi MR, Sargolzaee MR, Bajestan MN, Sabouri Z, Khayami E, Haghighi S, Hashemi SB, Eiraku N, Tufani H, Najmabadi H, Arimura K, Sano A, Osame M. Association of AKT1 haplotype with the risk of schizophrenia in Iranian population. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2006;141B:383–386. doi: 10.1002/ajmg.b.30291. [DOI] [PubMed] [Google Scholar]
- 218.Schwab SG, Hoefgen B, Hanses C, Hassenbach MB, Albus M, Lerer B, Trixler M, Maier W, Wildenauer DB. Further evidence for association of variants in the AKT1 gene with schizophrenia in a sample of European sib-pair families. Biol. Psychiatry. 2005;58:446–450. doi: 10.1016/j.biopsych.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 219.Pinheiro AP, Keefe RSE, Skelly T, Olarte M, Leviel K, Lange LA, Lange EM, Stroup TS, Lieberman J, Sullivan PF. AKT1 and neurocognition in schizophrenia. Aust. N. Z. J. Psychiatry. 2007;41:169–177. doi: 10.1080/00048670601109956. [DOI] [PubMed] [Google Scholar]
- 220.Tan HY, Nicodemus KK, Chen Q, Li Z, Brooke JK, Honea R, Kolachana BS, Straub RE, Meyer-Lindenberg A, Sei Y, Mattay VS, Callicott JH, Weinberger DR. Genetic variation in AKT1 is linked to dopamine-associated prefrontal cortical structure and function in humans. J. Clin. Invest. 2008;118:2200–2208. doi: 10.1172/JCI34725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Mao Y, Ge X, Frank CL, Madison JM, Koehler AN, Doud MK, Tassa C, Berry EM, Soda T, Singh KK, Biechele T, Petryshen TL, Moon RT, Haggarty SJ, Tsai LH. Disrupted in schizophrenia 1 regulates neuronal progenitor proliferation via modulation of GSK-3ß/ß-catenin signaling. Cell. 2009;136:1017–1031. doi: 10.1016/j.cell.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Singh KK, DeRienzo G, Drane L, Mao Y, Flood Z, Madison J, Ferreira M, Bergen S, King C, Sklar P, Sive H, Tsai LH. Common DISC1 polymorphisms disrupt Wnt/GSK3ß signaling and brain development. Neuron. 2011;72:545–558. doi: 10.1016/j.neuron.2011.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.vanWinkel R, vanBeveren NJ, Simons C. Genetic Risk and Outcome of Psychosis (GROUP) Investigators.AKT1 moderation of cannabis-induced cognitive alterations in psychotic disorder. Neuropsychopharmacology. 2011;36:2529–2537. doi: 10.1038/npp.2011.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.vanWinkel R. Genetic Risk and Outcome of Psychosis (GROUP) Investigators.Family-based analysis of genetic variation underlying psychosis-inducing effects of cannabis sibling analysis and proband follow-up. Arch. Gen. Psychiatry. 2011;68:148–157. doi: 10.1001/archgenpsychiatry.2010.152. [DOI] [PubMed] [Google Scholar]
- 225.Reiner O, Sapir T. Similarities and differences between the Wnt and reelin pathways in the forming brain. Mol. Neurobiol. 2005;31:117–134. doi: 10.1385/MN:31:1-3:117. [DOI] [PubMed] [Google Scholar]
- 226.Toro CT, Deakin JF. Adult neurogenesis and schizophrenia a window on abnormal early brain development. Schizophr. Res. 2007;90:1–14. doi: 10.1016/j.schres.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 227.Heuberger J, Birchmeier W. Interplay of cadherin-mediated cell adhesion and canonical Wnt signaling. Cold Spring Harb. Perspect. Biol. 2010;2:a002915–0. doi: 10.1101/cshperspect.a002915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Nadri C, Lipska BK, Kozlovsky N, Weinberger DR, Belmaker RH, Agam G. Glycogen synthase kinase (GSK)-3beta levels and activity in a neurodevelopmental rat model of schizophrenia. Dev. Brain Res. 2003;141:33–37. doi: 10.1016/s0165-3806(02)00639-9. [DOI] [PubMed] [Google Scholar]
- 229.Yu J, Qi D, Xing M, Li R, Jiang K, Peng Y, Cui D. MK-801 induces schizophrenic behaviors through down regulating Wnt signaling pathways in male mice. Brain Res. 2011;1385:281–292. doi: 10.1016/j.brainres.2011.02.039. [DOI] [PubMed] [Google Scholar]
- 230.Doble BW, Patel S, Wood GA, Kockeritz LK, Woodgett JR. Functional redundancy of GSK-3a and GSK-3ß in Wnt/ß -catenin signaling shown by using an allelic series of embryonic stem cell lines. Dev. Cell. 2007;12:957–971. doi: 10.1016/j.devcel.2007.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Yang SD, Yu JS, Lee TT, Yang CC, Ni MH, Yang YY. Dysfunction of protein kinase FA/GSK-3a in lymphocytes of patients with schizophrenic disorder. J. Cell. Biochem. 1995;59:108–116. doi: 10.1002/jcb.240590112. [DOI] [PubMed] [Google Scholar]
- 232.Kozlovsky N, Belmaker RH, Agam G. Low GSK-3ß immunoreactivity in postmortem frontal cortex of schizophrenic patients. Am. J. Psychiatry. 2000;157:831–833. doi: 10.1176/appi.ajp.157.5.831. [DOI] [PubMed] [Google Scholar]
- 233.Beasley C, Cotter D, Khan N, Pollard C, Sheppard P, Varndell I, Lovestone S, Anderton B, Everall I. Glycogen synthase kinase-3ß immunoreactivity is reduced in the prefrontal cortex in schizophrenia. Neurosci. Lett. 2001;302:117–120. doi: 10.1016/s0304-3940(01)01688-3. [DOI] [PubMed] [Google Scholar]
- 234.Beasley C, Cotter D, Everall I. An investigation of the Wnt-signalling pathway in the prefrontal cortex in schizophrenia bipolar disorder and major depressive disorder. Schizophr. Res. 2002;58:63–67. doi: 10.1016/s0920-9964(01)00376-0. [DOI] [PubMed] [Google Scholar]
- 235.Kozlovsky N, Belmaker RH, Agam G. Low GSK-3 activity in frontal cortex of schizophrenic patients. Schizophr. Res. 2001;52:101–105. doi: 10.1016/s0920-9964(00)00174-2. [DOI] [PubMed] [Google Scholar]
- 236.Kozlovsky N, Shanon-Weickert C, Tomaskovic-Crook E, Kleinman JE, Belmaker RH, Agam G. Reduced GSK-3ß mRNA levels in postmortem dorsolateral prefrontal cortex of schizophrenic patients. J. Neural. Transm. 2004;111:1583–1592. doi: 10.1007/s00702-004-0166-3. [DOI] [PubMed] [Google Scholar]
- 237.Nadri C, Dean B, Scarr E, Agam G. GSK-3 parameters in postmortem frontal cortex and hippocampus of schizophrenic patients. Schizophr. Res. 2004;71:377–382. doi: 10.1016/j.schres.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 238.Beaulieu JM, Del’Guidice T, Sotnikova TD, Lemasson M, Gainetdinov RR. Beyond cAMP The regulation of Akt and GSK3 by dopamine receptors. Front. Mol. Neurosci. 2011;4:38–0. doi: 10.3389/fnmol.2011.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Alimohamad H, Rajakumar N, Seah YH, Rushlow W. Antipsychotics alter the protein expression levels of ß-catenin and GSK-3 in the rat medial prefrontal cortex and striatum. Biol. Psychiatry. 2005;57:533–542. doi: 10.1016/j.biopsych.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 240.Alimohamad H, Sutton L, Mouyal J, Rajakumar N, Rushlow WJ. The effects of antipsychotics on beta-catenin, glycogen synthase kinase-3 and dishevelled in the ventral midbrain of rats. J. Neurochem. 2005;95:513–525. doi: 10.1111/j.1471-4159.2005.03388.x. [DOI] [PubMed] [Google Scholar]
- 241.Roh MS, Seo MS, Kim Y, Kim SH, Jeon WJ, Ahn YM, Kang UG, Juhnn YS, Kim YS. Haloperidol and clozapine differentially regulate signals upstream of glycogen synthase kinase 3 in the rat frontal cortex. Exp. Mol. Med. 2007;39:353–360. doi: 10.1038/emm.2007.39. [DOI] [PubMed] [Google Scholar]
- 242.Kang UG, Seo MS, Roh MS, Kim Y, Yoon SC, Kim YS. The effects of clozapine on the GSK-3-mediated signaling pathway. FEBS Lett. 2004;560:115–119. doi: 10.1016/S0014-5793(04)00082-1. [DOI] [PubMed] [Google Scholar]
- 243.Peng L, Zhu D, Feng X, Dong H, Yue Q, Zhang J, Gao Q, Hao J, Zhang X, Liu Z, Sun J. Paliperidone protects prefrontal cortical neurons from damages caused by MK-801 via Akt1/GSK3ß signaling pathway. Schizophr. Res. 2013. doi S0920-9964(13)00152-7. doi: 10.1016/j.schres.2013.03.006. [DOI] [PubMed] [Google Scholar]
- 244.Urs NM, Snyder JC, Jacobsen JP, Peterson SM, Caron MG. Deletion of GSK3ß in D2R-expressing neurons reveals distinct roles for ß-arrestin signaling in antipsychotic and lithium action. Proc. Natl. Acad. Sci. U.S.A. 2012;109:20732–20737. doi: 10.1073/pnas.1215489109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Freyberg Z, Ferrando SJ, Javitch JA. Roles of the Akt/GSK-3 and Wnt signaling pathways in schizophrenia and antipsychotic drug action. Am. J. Psychiatry. 2010;167:388–396. doi: 10.1176/appi.ajp.2009.08121873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Ishizuka K, Kamiya A, Oh EC, Kanki H, Seshadri S, Robinson JF, Murdoch H, Dunlop AJ, Kubo K, Furukori K, Huang B, Zeledon M, Hayashi-Takagi A, Okano H, Nakajima K, Houslay MD, Katsanis N, Sawa A. DISC1-dependent switch from progenitor proliferation to migration in the developing cortex. Nature. 2011;473:92–96. doi: 10.1038/nature09859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.DeRienzo G, Bishop JA, Mao Y, Pan L, Ma TP, Moens CB, Tsai LH, Sive H. Disc1 regulates both ß-catenin-mediated and noncanonical Wnt signaling during vertebrate embryogenesis. FASEB J. 2011;25:4184–4197. doi: 10.1096/fj.11-186239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Scassellati C, Bonvicini C, Perez J, Bocchio-Chiavetto L, Tura GB, Rossi G, Racagni G, Gennarelli M. Association study of -1727 A/T-50 C/T and [CAA]n repeat GSK-3ß gene polymorphisms with schizophrenia. Neuropsychobiology. 2004;50:16–20. doi: 10.1159/000077936. [DOI] [PubMed] [Google Scholar]
- 249.Ikeda M, Iwata N, Suzuki T, Kitajima T, Yamanouchi Y, Kinoshita Y, Ozaki N. No association of GSK-3ß gene (GSK-3ß) with Japanese schizophrenia. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;134B:90–92. doi: 10.1002/ajmg.b.30155. [DOI] [PubMed] [Google Scholar]
- 250.Lee KY, Ahn YM, Joo EJ, Jeong SH, Chang JS, Kim SC, Kim YS. No association of two common SNPs at position -1727 A/T-50 C/T of GSK-3ß polymorphisms with schizophrenia and bipolar disorder of Korean population. Neurosci. Lett. 2006;395:175–178. doi: 10.1016/j.neulet.2005.10.059. [DOI] [PubMed] [Google Scholar]
- 251.Szczepankiewicz A, Skibinska M, Hauser J, Slopien A, Leszczynska-Rodziewicz A, Kapelski P, Dmitrzak-Weglarz M, Czerski PM, Rybakowski JK. Association analysis of the GSK-3ß T-50C gene polymorphism with schizophrenia and bipolar disorder. Neuropsychobiology. 2006;53:51–56. doi: 10.1159/000090704. [DOI] [PubMed] [Google Scholar]
- 252.Meng J, Shi Y, Zhao X, Zhou J, Zheng Y, Tang R, Ma G, Zhu X, He Z, Wang Z, Xu Y, Feng G, He L. No significant association between the genetic polymorphisms in the GSK-3ß gene and schizophrenia in the Chinese population. J. Psychiatr. Res. 2008;42:365–370. doi: 10.1016/j.jpsychires.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 253.Olsen L, Hansen T, Jakobsen KD, Djurovic S, Melle I, Agartz I, Hall H, Ullum H, Timm S, Wang AG, Jönsson EG, Andreassen OA, Werge T. The estrogen hypothesis of schizophrenia implicates glucose metabolism association study in three independent samples. BMC Med. Genet. 2008;9:39–0. doi: 10.1186/1471-2350-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Souza RP, Romano-Silva MA, Lieberman JA, Meltzer HY, Wong AH, Kennedy JL. Association study of GSK-3 gene polymorphisms with schizophrenia and clozapine response. Psychopharmacology [Berl] 2008;200:177–186. doi: 10.1007/s00213-008-1193-9. [DOI] [PubMed] [Google Scholar]
- 255.Sand PG, Domani M, Smesny S. GSK3B and schizophrenia a case not closed. Psychopharmacology [Berl] 2010;208:333–334. doi: 10.1007/s00213-009-1728-8. [DOI] [PubMed] [Google Scholar]
- 256.Souza RP, Levy DL, Kennedy JL. “GSK3B and schizophrenia a case not closed” reply. Psychopharmacology [Berl] 2010;208:335–336. doi: 10.1007/s00213-009-1728-8. [DOI] [PubMed] [Google Scholar]
- 257.McCarley RW, Wible CG, Frumin M, Hirayasu Y, Levitt JJ, Fischer IA, Shenton ME. MRI anatomy of schizophrenia. Biol. Psychiatry. 1999;45:1099–1119. doi: 10.1016/s0006-3223(99)00018-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Honea R, Crow TJ, Passingham D, Mackay CE. Regional deficits in brain volume in schizophrenia a metanalysis of voxel-based morphometry studies. Am. J. Psychiatry. 2005;162:2233–2245. doi: 10.1176/appi.ajp.162.12.2233. [DOI] [PubMed] [Google Scholar]
- 259.Wider C, Vilariño-Güell C, Heckman MG, Jasinska-Myga B, Ortolaza-Soto AI, Diehl NN, Crook JE, Cobb SA, Bacon JA, Aasly JO, Gibson JM, Lynch T, Uitti RJ, Wszolek ZK, Farrer MJ, Ross OA. SNCA MAPT and GSK3B in Parkinson disease a gene-gene interaction study. Eur. J. Neurol. 2011;18:876–881. doi: 10.1111/j.1468-1331.2010.03297.x. [DOI] [PubMed] [Google Scholar]
- 260.Bikkavilli RK, Feigin ME, Malbon CC. p38 mitogen-activated protein kinase regulates canonical Wnt-ß-catenin signaling by inactivation of GSK3ß. J. Cell. Sci. 2008;121:3598–3607. doi: 10.1242/jcs.032854. [DOI] [PubMed] [Google Scholar]
- 261.Chambers JS, Perrone-Bizzozero NI. Altered myelination of the hippocampal formation in subjects with schizophrenia and bipolar disorder. Neurochem. Res. 2004;29:2293–2302. doi: 10.1007/s11064-004-7039-x. [DOI] [PubMed] [Google Scholar]
- 262.Carlsson A. Neurocircuitries and neurotransmitter interactions in schizophrenia. Int. Clin. Psychopharmacol. 1995;10 Suppl 3:21–8. [PubMed] [Google Scholar]
- 263.Cui DH, Jiang KD, Jiang SD, Xu YF, Yao H. The tumor suppressor adenomatous polyposis coli gene is associated with susceptibility to schizophrenia. Mol. Psychiatry. 2005;10:669–677. doi: 10.1038/sj.mp.4001653. [DOI] [PubMed] [Google Scholar]
- 264.Kalkman HO. Altered growth factor signaling pathways as the basis of aberrant stem cell maturation in schizophrenia. Pharmacol. Ther. 2009;121:115–122. doi: 10.1016/j.pharmthera.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 265.McCurdy RD, Féron F, Perry C, Chant DC, McLean D, Matigian N, Hayward NK, McGrath JJ, Mackay-Sim A. Cell cycle alterations in biopsied olfactory neuroepithelium in schizophrenia and bipolar I disorder using cell culture and gene expression analyses. Schizophr. Res. 2006;82:163–173. doi: 10.1016/j.schres.2005.10.012. [DOI] [PubMed] [Google Scholar]
- 266.Willis TG, Zalcberg IR, Coignet LJ, Wlodarska I, Stul M, Jadayel DM, Bastard C, Treleaven JG, Catovsky D, Silva ML, Dyer MJ. Molecular cloning of translocation t[1.4][q21.q32] defines a novel gene (BCL9) at chromosome 1q21. Blood. 1998;91:1873–1881. [PubMed] [Google Scholar]
- 267.Fiedler M, Sánchez-Barrena MJ, Nekrasov M, Mieszczanek J, Rybin V, Müller J, Evans P, Bienz M. Decoding of methylated histone H3 tail by the Pygo-BCL9 Wnt signaling complex. Mol. Cell. 2008;30:507–518. doi: 10.1016/j.molcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Stefansson H, Rujescu D, Cichon S, Pietiläinen OP, Ingason A, Steinberg S, Fossdal R, Sigurdsson E, Sigmundsson T, Buizer-Voskamp JE, Hansen T, Jakobsen KD, Muglia P, Francks C, Matthews PM, Gylfason A, Halldorsson BV, Gudbjartsson D, Thorgeirsson TE, Sigurdsson A, Jonasdottir A, Jonasdottir A, Bjornsson A, Mattiasdottir S, Blondal T, Haraldsson M, Magnusdottir BB, Giegling I, Möller HJ, Hartmann A, Shianna KV, Ge D, Need AC, Crombie C, Fraser G, Walker N, Lonnqvist J, Suvisaari J, Tuulio-Henriksson A, Paunio T, Toulopoulou T, Bramon E, DiForti M, Murray R, Ruggeri M, Vassos E, Tosato S, Walshe M, Li T, Vasilescu C, Mühleisen TW, Wang AG, Ullum H, Djurovic S, Melle I, Olesen J, Kiemeney LA, Franke B, Sabatti C, Freimer NB, Gulcher JR, Thorsteinsdottir U, Kong A, Andreassen OA, Ophoff RA, Georgi A, Rietschel M, Werge T, Petursson H, Goldstein DB, Nöthen MM, Peltonen L, Collier DA, StClair D, Stefansson K. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–236. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Sohail Z. International Schizophrenia Consortium.Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. International Schizophrenia Consortium Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Li J, Zhou G, Ji W, Feng G, Zhao Q, Liu J, Li T, Li Y, Chen P, Zeng Z, Wang T, Hu Z, Zheng L, Wang Y, Shen Y, He L, Shi Y. Common variants in the BCL9 gene conferring risk of schizophrenia. Arch. Gen. Psychiatry. 2011;68:232–240. doi: 10.1001/archgenpsychiatry.2011.1. [DOI] [PubMed] [Google Scholar]
- 272.Sirotkin H, O'Donnell H, DasGupta R, Halford S, StJore B, Puech A, Parimoo S, Morrow B, Skoultchi A, Weissman SM, Scambler P, Kucherlapati R. Identification of a new human catenin gene family member [ARVCF] from the region deleted in velo-cardio-facial syndrome. Genomics. 1997;41:75–83. doi: 10.1006/geno.1997.4627. [DOI] [PubMed] [Google Scholar]
- 273.Cho EA, Dressler GR. TCF-4 binds ß-catenin and is expressed in distinct regions of the embryonic brain and limbs. Mech. Dev. 1998;77:9–18. doi: 10.1016/s0925-4773(98)00131-2. [DOI] [PubMed] [Google Scholar]
- 274.Brzózka MM, Radyushkin K, Wichert SP, Ehrenreich H, Rossner MJ. Cognitive and sensorimotor gating impairments in transgenic mice overexpressing the schizophrenia susceptibility gene Tcf4 in the brain. Biol. Psychiatry. 2010;68:33–40. doi: 10.1016/j.biopsych.2010.03.015. [DOI] [PubMed] [Google Scholar]
- 275.Shitashige M, Hirohashi S, Yamada T. Wnt signaling inside the nucleus. Cancer Sci. 2008;99:631–637. doi: 10.1111/j.1349-7006.2007.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Zhuang Y, Cheng P, Weintraub H. B-lymphocyte development is regulated by the combined dosage of three basic helix-loop-helix genes E2A E2-2 and HEB. Mol. Cell. Biol. 1996;16:2898–2905. doi: 10.1128/mcb.16.6.2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Flora A, Garcia JJ, Thaller C, Zoghbi HY. The E-protein Tcf4 interacts with Math1 to regulate differentiation of a specific subset of neuronal progenitors. Proc. Natl. Acad. Sci. USA. 2007;104:15382–15387. doi: 10.1073/pnas.0707456104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Brockschmidt A, Todt U, Ryu S, Hoischen A, Landwehr C, Birnbaum S, Frenck W, Radlwimmer B, Lichter P, Engels H, Driever W, Kubisch C, Weber RG. Severe mental retardation with breathing abnormalities [Pitt-Hopkins syndrome] is caused by haploinsufficiency of the neuronal bHLH transcription factor TCF4. Hum. Mol. Genet. 2007;16:1488–1494. doi: 10.1093/hmg/ddm099. [DOI] [PubMed] [Google Scholar]
- 279.Sohail Z. Irish Schizophrenia Genomics Consortium and the Wellcome Trust Case Control Consortium 2.Genome-wide association study implicates HLA-C*0102 as a risk factor at the major histo- compatibility complex locus in schizophrenia. Biol. Psychiatry. 2012;72:620–628. doi: 10.1016/j.biopsych.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Zhu X, Gu H, Liu Z, Xu Z, Chen X, Sun X, Zhai J, Zhang Q, Chen M, Wang K, Deng X, Ji F, Liu C, Li J, Dong Q, Chen C. Associations between TCF4 gene poly- morphism and cognitive functions in schizophrenia patients and healthy controls. Neuropsychopharmacology. 2013;38:683–689. doi: 10.1038/npp.2012.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Werge T, Pietiläinen OP, Mors O, Mortensen PB, Sigurdsson E, Gustafsson O, Nyegaard M, Tuulio-Henriksson A, Ingason A, Hansen T, Suvisaari J, Lonnqvist J, Paunio T, Børglum AD, Hartmann A, Fink-Jensen A, Nordentoft M, Hougaard D, Norgaard-Pedersen B, Böttcher Y, Olesen J, Breuer R, Möller HJ, Giegling I, Rasmussen HB, Timm S, Mattheisen M, Bitter I, Réthelyi JM, Magnusdottir BB, Sigmundsson T, Olason P, Masson G, Gulcher JR, Haraldsson M, Fossdal R, Thorgeirsson TE, Thorsteinsdottir U, Ruggeri M, Tosato S, Franke B, Strengman E, Kiemeney LA, Melle I, Djurovic S, Abramova L, Kaleda V, Sanjuan J, deFrutos R, Bramon E, Vassos E, Fraser G, Ettinger U, Picchioni M, Walker N, Toulopoulou T, Need AC, Ge D, Yoon JL, Shianna KV, Freimer NB, Cantor RM, Murray R, Kong A, Golimbet V, Carracedo A, Arango C, Costas J, Jönsson EG, Terenius L, Agartz I, Petursson H, Nöthen MM, Rietschel M, Matthews PM, Muglia P, Peltonen L, StClair D, Goldstein DB, Stefansson K, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Quednow BB, Ettinger U, Mössner R, Rujescu D, Giegling I, Collier DA, Schmechtig A, Kühn KU, Möller HJ, Maier W, Wagner M, Kumari V. The schizophrenia risk allele C of the TCF4 rs9960767 polymorphism disrupts sensorimotor gating in schizophrenia spectrum and healthy volunteers. J. Neurosci. 2011;31:6684–6691. doi: 10.1523/JNEUROSCI.0526-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Egan GJ, Hasenkamp W, Wilcox L, Green A, Hsu N, Boshoven W, Lewison B, Keyes MD, Duncan E. Declarative memory and WCST-64 performance in subjects with schizophrenia and healthy controls. Psychiatry Res. 2011;188:191–196. doi: 10.1016/j.psychres.2011.02.026. [DOI] [PubMed] [Google Scholar]
- 284.Lennertz L, Rujescu D, Wagner M, Frommann I, Schulze-Rauschenbach S, Schuhmacher A, Landsberg MW, Franke P, Möller HJ, Wölwer W, Gaebel W, Häfner H, Maier W, Mössner R. Novel schizophrenia risk gene TCF4 influences verbal learning and memory functioning in schizophrenia patients. Neuropsychobiology. 2011;63:131–136. doi: 10.1159/000317844. [DOI] [PubMed] [Google Scholar]
- 285.Lennertz L, Quednow BB, Benninghoff J, Wagner M, Maier W, Mössner R. Impact of TCF4 on the genetics of schizophrenia. Eur. Arch. Psychiatry Clin. Neurosci. 2011;261(Suppl 2):161–165. doi: 10.1007/s00406-011-0256-9. [DOI] [PubMed] [Google Scholar]
- 286.Williams HJ, Moskvina V, Smith RL, Dwyer S, Russo G, Owen MJ, O'Donovan MC. Association between TCF4 and schizophrenia does not exert its effect by common nonsynonymous variation or by influencing cis-acting regulation of mRNA expression in adult human brain. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 2011;156B:781–784. doi: 10.1002/ajmg.b.31219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Quednow BB, Brinkmeyer J, Mobascher A, Nothnagel M, Musso F, Gründer G, Savary N, Petrovsky N, Frommann I, Lennertz L, Spreckelmeyer KN, Wienker TF, Dahmen N, Thuerauf N, Clepce M, Kiefer F, Majic T, Mössner R, Maier W, Gallinat J, Diaz-Lacava A, Toliat MR, Thiele H, Nürnberg P, Wagner M, Winterer G. Schizophrenia risk polymorphisms in the TCF4 gene interact with smoking in the modulation of auditory sensory gating. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6271–6276. doi: 10.1073/pnas.1118051109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Zhang XY, Liang J, Chenda C, Xiu MH, He J, Cheng W, Wu Z, Yang FD, Haile CN, Sun H, Lu L, Kosten TA, Kosten TR. Cigarette smoking in male patients with chronic schizophrenia in a Chinese population prevalence and relationship to clinical phenotypes. PLoS One. 2012;7:e30937–0. doi: 10.1371/journal.pone.0030937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Steinberg S, deJong S, Andreassen OA, Werge T, Børglum AD, Mors O, Mortensen PB, Gustafsson O, Costas J, Pietiläinen OP, Demontis D, Papiol S, Huttenlocher J, Mattheisen M, Breuer R, Vassos E, Giegling I, Fraser G, Walker N, Tuulio-Henriksson A, Suvisaari J, Lönnqvist J, Paunio T, Agartz I, Melle I, Djurovic S, Strengman E, Jürgens G, Glenthøj B, Terenius L, Hougaard DM, Ørntoft T, Wiuf C, Didriksen M, Hollegaard MV, Nordentoft M, vanWinkel R, Kenis G, Abramova L, Kaleda V, Arrojo M, Sanjuán J, Arango C, Sperling S, Rossner M, Ribolsi M, Magni V, Siracusano A, Christiansen C, Kiemeney LA, Veldink J, vandenBerg L, Ingason A, Muglia P, Murray R, Nöthen MM, Sigurdsson E, Petursson H, Thorsteinsdottir U, Kong A, Rubino IA, DeHert M, Réthelyi JM, Bitter I, Jönsson EG, Golimbet V, Carracedo A, Ehrenreich H, Craddock N, Owen MJ, O'Donovan MC, Ruggeri M, Tosato S, Peltonen L, Ophoff RA, Collier DA, StClair D, Rietschel M, Cichon S, Stefansson H, Rujescu D, Stefansson K. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum. Mol. Genet. 2011;20:4076–4081. doi: 10.1093/hmg/ddr325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Lin DY, Duan J, Ophoff RA, Andreassen OA, Scolnick E, Cichon S, StClair D, Corvin A, Gurling H, Werge T, Rujescu D, Blackwood DHR, Pato CN, Malhotra AK, Purcell S, Dudbridge F, Neale BM, Rossin L, Visscher PM, Posthuma D, Ruderfer DM, Fanous A, Stefansson H, Steinberg S, Mowry BJ, Golimbet V, DeHert M, Jönsson EG, Bitter I, Pietiläinen OPH, Collier DA, Tosato S, Agartz I, Albus M, Alexander M, Amdur RL, Amin F, Bass N, Bergen SE, Black DW, Børglum AD, Brown MA, Bruggeman R, Buccola NG, Byerley WF, Cahn W, Cantor RM, Carr VJ, Catts SV, Choudhury K, Cloninger CR, Cormican P, Craddock N, Danoy PA, Datta S, deHaan L, Demontis D, Dikeos D, Djurovic S, Donnelly P, Donohoe G, Duong L, Dwyer S, Fink-Jensen A, Freedman R, Freimer NB, Friedl M, Georgieva L, Giegling I, Gill M, Glenthøj B, Godard S, Hamshere M, Hansen M, Hansen T, Hartmann AM, Henskens FA, Hougaard DM, Hultman CM, Ingason A, Jablensky AV, Jakobsen KD, Jay M, Jürgens G, Kahn RS, Keller MC, Kenis G, Kenny E, Kim Y, Kirov GK, Konnerth H, Konte B, Krabbendam L, Krasucki R, Lasseter VK, Laurent C, Lawrence J, Lencz T, Lerer FB, Liang KY, Lichtenstein P, Lieberman JA, Linszen DH, Lönnqvist J, Loughland CM, Maclean AW, Maher BS, Maier W, Mallet J, Malloy P, Mattheisen M, Mattingsdal M, McGhee KA, McGrath JJ, McIntosh A, McLean DE, McQuillin A, Melle I, Michie PT, Milanova V, Morris DW, Mors O, Mortensen PB, Moskvina V, Muglia P, Myin-Germeys I, Nertney DA, Nestadt G, Nielsen J, Nikolov I, Nordentoft M, Norton N, Nöthen MM, O’Dushlaine CT, Olincy A, Olsen L, O’Neill FA, Ørntoft TF, Owen MJ, Pantelis C, Papadimitriou G, Pato MT, Peltonen L, Petursson H, Pickard B, Pimm J, Pulver AE, Puri V, Quested D, Quinn EM, Rasmussen HB, Réthelyi JM, Ribble R, Rietschel M, Riley BP, Ruggeri M, Schall U, Schulze TG, Schwab SG, Scott RJ, Shi J, Sigurdsson E, Silverman JM, Spencer CCA, Stefansson K, Strange A, Strengman E, Stroup TS, Suvisaari J, Terenius L, Thirumalai S, Thygesen JH, Timm S, Toncheva D, vandenOord E, vanOs J, vanWinkel R, Veldink J, Walsh D, Wang AG, Wiersma D, Wildenauer DB, Williams HJ, Williams NM, Wormley B, StanZammit S, Sullivan PF, O’Donovan MC, Daly MJ, Gejman PV. Genome-wide association study identifies five new schizophrenia loci. Nat. Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol. Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- 292.Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr. Res. 2012;141:60–64. doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Talkowski ME, Rosenfeld JA, Blumenthal I, Pillalamarri V, Chiang C, Heilbut A, Ernst C, Hanscom C, Rossin E, Lindgren AM, Pereira S, Ruderfer D, Kirby A, Ripke S, Harris DJ, Lee JH, Ha K, Kim HG, Solomon BD, Gropman AL, Lucente D, Sims K, Ohsumi TK, Borowsky ML, Loranger S, Quade B, Lage K, Miles J, Wu BL, Shen Y, Neale B, Shaffer LG, Daly MJ, Morton CC, Gusella JF. Sequencing chromosomal abnormalities reveals neuro- developmental loci that confer risk across diagnostic boundaries. Cell. 2012;149:525–537. doi: 10.1016/j.cell.2012.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Lee KW, Woon PS, Teo YY, Sim K. Genome wide association studies (GWAS) and copy number variation (CNV) studies of the major psychoses what have we learnt. Neurosci. Biobehav. Rev. 2012;36:556–571. doi: 10.1016/j.neubiorev.2011.09.001. [DOI] [PubMed] [Google Scholar]
- 295.Wirgenes KV, Snderby IE, Haukvik UK, Mattingsdal M, Tesli M, Athanasiu L, Sundet K, Rssberg JI, Dale AM, Brown AA, Agartz I, Melle I, Djurovic S, Andreassen OA. TCF4 sequence variants and mRNA levels are associated with neurodevelopmental characteristics in psychotic disorders. Transl. Psychiatry. 2012;2:e112–0. doi: 10.1038/tp.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.M B, Wu W, Davidson G, Marhold J, Li M, Mechler BM, Delius H, Hoppe D, Stannek P, Walter C, Glinka A, Niehrs C. Kremen proteins are Dickkopf receptors that regulate Wnt/β-catenin signalling. Nature. 2002;417:664–667. doi: 10.1038/nature756. [DOI] [PubMed] [Google Scholar]
- 297.Pulver AE, Karayiorgou M, Wolyniec PS, Lasseter VK, Kasch L, Nestadt G, Antonarakis S, Housman D, Kazazian HH, Meyers D, Ott J, Lamacz M, Liang KY, Hanfelt J, Ullrich G, DeMarchi N, Ramu E, McHugh PR, Adler L, Thomas M, Carpenter WT, Manschreck T, Gordon CT, Kimberland M, Babb R, Puck J, Childs B. Sequential strategy to identify a susceptibility gene for schizophrenia report of potential linkage on chromosome 22q12-q13. Part 1. Am. J. Med. Genet. 1994;54:36–43. doi: 10.1002/ajmg.1320540108. [DOI] [PubMed] [Google Scholar]
- 298.Aleksic B, Kushima I, Ito Y, Nakamura Y, Ujike H, Suzuki M, Inada T, Hashimoto R, Takeda M, Iwata N, Ozaki N. Genetic association study of KREMEN1 and DKK1 and schizophrenia in a Japanese population. Schizophr. Res. 2010;118:113–117. doi: 10.1016/j.schres.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 299.Ftouh S, Akbar MT, Hirsch SR, deBelleroche JS. Down-regulation of Dickkopf 3 a regulator of the Wnt signalling pathway in elderly schizophrenic subjects. J. Neurochem. 2005;94:520–530. doi: 10.1111/j.1471-4159.2005.03239.x. [DOI] [PubMed] [Google Scholar]
- 300.Proitsi P, Li T, Hamilton G, DiForti M, Collier D, Killick R, Chen R, Sham P, Murray R, Powell J, Lovestone S. Positional pathway screen of Wnt signaling genes in schizophrenia association with DKK4. Biol. Psychiatry. 2008;63:13–16. doi: 10.1016/j.biopsych.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 301.Zhang LH, Li M, Shi XD, Ren H, Su B, Xu Z. [Association of the schizophrenia susceptible gene DKK4 with brain volume in Chinese populations. [Article in Chinese]. Dongwuxue Yanjiu (Zoolog Res) 2011;32:62–65. doi: 10.3724/SP.J.1141.2011.01062. [DOI] [PubMed] [Google Scholar]