Abstract
A frequently used experimental model of chronic pancreatitis (CP) recapitulating human disease is repeated injection of cerulein into mice. C57BL/6 is the most commonly used inbred mouse strain for biomedical research, but widespread demand has led to generation of several substrains with subtly different phenotypes. In this study, two common substrains, C57BL/6J and C57BL/6NHsd, exhibited different degrees of CP, with C57BL/6J being more susceptible to repetitive cerulein-induced CP as assessed by pancreatic atrophy, pancreatic morphological changes, and fibrosis. We hypothesized that the deficiency of nicotinamide nucleotide transhydrogenase (NNT) protein in C57BL/6J is responsible for the more severe C57BL/6J phenotype but the parameters of CP in NNT-expressing transgenic mice generated on a C57BL6/J background do not differ with those of wild-type C57BL/6J. The highly similar genetic backgrounds but different CP phenotypes of these two substrains presents a unique opportunity to discover genes important in pathogenesis of CP. We therefore performed whole mouse genome Affymetrix microarray analysis of pancreatic gene expression of C57BL/6J and C57BL/6NHsd before and after induction of CP. Genes with differentially regulated expression between the two substrains that might be candidates in CP progression included Mmp7, Pcolce2, Itih4, Wdfy1, and Vtn. We also identified several genes associated with development of CP in both substrains, including RIKEN cDNA 1810009J06 gene (trypsinogen 5), Ccl8, and Ccl6.
Chronic pancreatitis (CP) is an irreversible process characterized by chronic inflammation, acinar cell loss, and progressive fibrosis of the pancreas.1 Identified risk factors for the development of CP include alcoholism, smoking, autoimmune injury, and hereditary factors.2 Activated pancreatic stellate cells (PSCs) play a major role in the development of pancreatic fibrosis.3 The mechanism of CP is an area of active investigation, and a variety of experimental animal models of CP have been described.4 Repeated episodes of acute pancreatitis induced by injecting the cholecystokinin analog cerulein in mice induces CP similar to that of human CP,5,6 and this model is widely used to study the causes and treatment of CP.
The C57BL/6 strain is the most commonly used mouse strain in biomedical research, including research into the mechanism of CP. In experiments using genetically modified C57BL/6 mice, wild-type C57BL/6 mice are typically used as experimental controls, on the assumption that the genetically engineered mice and control mice differ only in the artificially altered genes. The validity of this assumption may depend also on the substrain of the C57BL/6 mice used as comparators. Multiple substrains of C57BL/6 mice have been produced through decades of propagation in different mouse breeding facilities. Data have begun to accumulate that these substrains have important genetic and phenotypic differences and that they cannot be used interchangeably.7–10 Here, we report major differences in the parameters of CP induced by repetitive injection of cerulein between C57BL/6J mice (B6J) from the Jackson Laboratory (Bar Harbor, ME) and C57BL/6NHsd mice (B6N) from Harlan Laboratories (Indianapolis, IN). The C57BL/6 strain was used to establish the B6J colony at the Jackson Laboratory in 1948, and the B6J mice from the Jackson Laboratory were subsequently used to establish a C57BL/6 colony at the NIH in 1951. The B6N colony was established by Harlan Laboratories from NIH stock in 1974. Thus, the B6J and B6N colonies have been inbred separately for more than six decades, allowing genetic divergence over time.11 The most striking example of this divergence is a spontaneous deletion in the B6J substrain causing the loss of several exons and thereby loss of function of the nicotinamide nucleotide transhydrogenase (Nnt) gene in B6J mice.12,13
In the present study, observed differences in the CP phenotype of the closely related B6J and B6N substrains provided a unique opportunity to identify candidate genes that contribute to the development of CP. Because one difference between these two substrains is the absence of a functional NNT protein in B6J, we started by evaluating of the role of NNT in the CP model. NNT is a nuclear encoded mitochondrial inner membrane protein that plays a role in the detoxification of mitochondrial reactive oxygen species and might be an essential factor in different pathological conditions associated with mitochondrial dysfunction.13–15 Mitochondrial dysfunction has been shown to play a critical role in the pathogenesis of pancreatic acinar cell injury, resulting in pancreatitis.16,17 The NNT protein was therefore a rational candidate for a contributor to pancreatitis susceptibility. To analyze whether NNT expression is an important factor in CP, we compared the course of cerulein-induced chronic and acute pancreatitis of NNT-expressing transgenic mice on the B6J Jackson background versus wild-type B6 mice (Nnt−/−). We also sought to identify the spectrum of candidate genes that might contribute to the difference in the severity of CP between the two C57BL/6 substrains, and that may thus represent CP susceptibility genes, by performing transcriptome analysis of pancreatic tissues from B6J and B6N mice before and after CP induction.
Materials and Methods
Animals
C57BL/6J (B6J) mice were obtained from the Jackson Laboratory. C57BL/6NHsd (B6N) mice were obtained from Harlan Laboratories. NNT-expressing transgenic mice on the C57BL/6J background (Nnt+/+) and C57BL/6J (Nnt−/−) control mice were provided by MRC Harwell (Harwell, UK). All mice were housed in standard facilities under conditions of controlled temperature, humidity, and a 12-hour light/dark cycle; mice were maintained on standard rodent chow with free access to water. Animal care and all procedures were approved by the Institutional Animal Care and Use Committee of Saint Louis University. Mice obtained from commercial vendors were kept in the Saint Louis University animal facility for 2 weeks before the start of experiments. Nnt−/− and Nnt+/+ mice were propagated at the Saint Louis University animal facility to obtain sufficient numbers of mice for conducting the experiments. Genotyping was performed as described below.
Experimental Model of CP and Tissue Processing
CP was induced in 8-week-old B6J and B6N female mice (18 to 20 g body weight) by repeated intraperitoneal injections of cerulein (Sigma-Aldrich, St. Louis, MO), 50 μg/kg hourly, as described previously.18 Six hourly injections given in one day constituted one treatment. Treatments were given every other day for a total of three treatments. Sex- and age-matched control mice received comparable injections of saline. To allow resolution of acute changes, mice were euthanized by CO2 asphyxiation at 3 days after their final cerulein treatment. Each pancreas was removed, weighed, and divided into three sections. One section was fixed in 10% neutral buffered formalin solution (Sigma-Aldrich) for histological analysis, another was immediately frozen in liquid nitrogen and stored at −80°C for subsequent protein extraction and Western blot analysis, and the third portion was placed in an RNA-stabilizing solution (Ambion RNAlater; Life Technologies, Carlsbad, CA) and stored overnight at 4°C for RNA isolation and subsequent real-time quantitative PCR (qPCR) analysis.
Experimental Model of Acute Pancreatitis and Tissue Processing
To evaluate acute injury, 8-week-old female B6J and B6N mice were subjected to a single cerulein treatment (ie, six hourly injections of 50 μg/kg each), and at 9 hours after the first cerulein injection the mice were euthanized by CO2 asphyxiation. Sex- and age-matched control mice received comparable injections of saline solution. Each pancreas was removed, weighed, and placed in 10% neutral buffered formalin solution (Sigma-Aldrich) for histological analysis. Blood was collected at this time for measuring plasma amylase activity.
Histology Analysis
Formalin-fixed pancreatic tissues were embedded in paraffin, sectioned, and stained with H&E using standard protocols for microscopic evaluation. Slides were graded by an experienced pathologist (K.O.) masked to treatment group and substrain using a semiquantitative histopathology scoring system similar to one that we have described previously.18,19 In the grading system used for the analysis of slides after induction of CP, within the pancreatic sections any areas of abnormal architecture were estimated as a percentage of the normal area. Within these abnormal areas, necrosis was graded as 0 = absent, 1 = minimal (<10%), 2 = moderate (10% to 50%), and 3 = severe (>50%). In addition, the presence of acute inflammatory cells (mainly neutrophils) and chronic inflammatory cells (mononuclear cells) was graded as 0 = absent, 1 = minimal, 2 = mild, 3 = moderate, and 4 = severe. H&E-stained pancreatic sections obtained at 9 hours after induction of acute pancreatitis were graded on four criteria, vacuolization, necrosis, inflammation, edema, on the following scales. Vacuolization was graded as 0 = absent, 1 = 5% to 14%, 2 = 15% to 35%, 3 = 35% to 50%, and 4 = >50%. Necrosis was graded as 0 = absent, 1 = periductal necrosis <5%, 2 = focal necrosis 5% to 20%, and 3 = diffuse periductal necrosis 20% to 50%. Inflammation was graded as 0 = absence of inflammatory infiltrates, 1 = inflammatory infiltration in ducts, 2 = inflammatory infiltration in the parenchyma <50%, and 3 = inflammatory infiltration in the parenchyma >50%. Edema was graded as 0 = absent, 1 = focally increased between lobules, 2 = diffusely increased between lobules, and 3 = acini disrupted and separated.
To evaluate pancreatic collagen content, paraffin-embedded pancreatic sections were stained with Sirius Red (F3B) solution, as we have described previously.18 The extent of collagen accumulation was evaluated by morphometric analysis20 as we have detailed previously,18,19 relative to the amount of collagen in the saline-treated controls.
Western Blotting
To obtain pancreatic protein, frozen pancreatic tissues were homogenized in ice-cold radioimmunoprecipitation assay buffer containing 150 mmol/L NaCl, 50 mmol/L Tris-HCl, pH 7.4, 1 mmol/L EDTA, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholic acid, and a freshly added mixture of proteinase inhibitor cocktail (Sigma-Aldrich). Total protein extract (20 μg) was resolved by SDS-PAGE and blotted to Immobilon-P polyvinylidene difluoride transfer membranes (EMD Millipore, Billerica, MA). Blots were blocked in 5% nonfat dried milk in TBST buffer (10 mmol/L Tris-HCl, pH 7.4, 0.9% NaCl, and 0.05% Tween 20) and were probed with primary antibody. α-SMA protein was detected with monoclonal anti–α-SMA antibody (Sigma-Aldrich). For the loading control, blots were probed with an antibody to extracellular signal-regulated kinase 1/2 (ERK1/2) [ERK 1 antibody (C-16); Santa Cruz Biotechnology, Santa Cruz, CA]. ERK1/2 is an established loading control for quantification of pancreatic proteins after cerulein-induced chronic and acute pancreatitis.21,22 NNT protein was detected using a custom-made (Eurogentec, Liege, Belgium) polyclonal NNT antibody,23 and a polyclonal β-actin antibody (Santa Cruz Biotechnology) was used as a loading control. Signals were developed using horseradish peroxidase–conjugated anti-mouse IgG (Sigma-Aldrich) for α-SMA and anti-rabbit IgG (Santa Cruz Biotechnology) for NNT, ERK1/2, and β-actin, with Amersham ECL Plus Western blotting detection reagent (GE Healthcare, Little Chalfont, UK). Protein band intensities were quantified using a Personal Densitometer SI, Model 375 (Molecular Dynamics, Sunnyvale, CA) and ImageJ software version 1.37 (NIH, Bethesda, MD).
Genomic DNA Extraction and the NNT Mutation Detection
Genomic DNA was isolated from the mouse tail snips by overnight digestion in 500 μL of lysis buffer (100 mmol/L Tris-HCl, pH 8.5, 5 mmol/L EDTA, 0.2% SDS, 200 mmol/L NaCl) containing 0.6 mg/mL of proteinase K (Sigma-Aldrich) at 50°C. After the incubation, samples were centrifuged at 14,000 × g, and DNA was precipitated from the supernatant by isopropanol, washed with 70% ethanol, and resuspended in 200 μL of TE buffer (10 mmol/L Tris-HCl, pH 8.0, 1 mmol/L EDTA). Three microliters of the solution was used in each PCR reaction. Primer sequences for the detection of the Nnt mutation were based on the sequences in the Primer Bank (http://pga.mgh.harvard.edu/primerbank, last accessed March 2, 2010)24: forward primer, 5′-GGGTCAGTTGTTGTGGATTTAGC-3′; reverse primer, 5′-GCCTTCAGGAGCTTAGTGATGTT-3′. The corresponding amplicon is in exons 7 and 8, which are deleted in B6J mice. The expected size of the PCR product in B6N mice is 732 bp.
Mitochondria Isolation
Mitochondrial fractions were isolated from the frozen pancreatic tissues using a Pierce mitochondrial isolation kit for tissue (Thermo Fisher Scientific, Rockford, IL) according to the manufacturer’s instructions. Five micrograms of total mitochondrial protein was used for the Western blot analysis.
qPCR Analysis
Total RNA was prepared from pancreatic tissues stabilized in Ambion RNAlater solution (Life Technologies) by extraction with TRIzol reagent (Life Technologies) according to the manufacturer’s protocol. The concentration and purity of isolated RNA was estimated by measuring absorbances at 260 and 280 nm using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). For qPCR assay, 2 μg of total RNA was treated with Turbo DNase (Life Technologies) and reverse-transcribed to cDNA using a high-capacity RNA–to-cDNA kit (Life Technologies). qPCR was performed with a 7500 real-time PCR System (Life Technologies) using SYBR Green real-time PCR ReadyMix Low Rox (Sigma-Aldrich) according to the manufacturer’s instructions. PCR primers were synthesized by Life Technologies based on the sequences from Primer Bank.24 The primer sequences of transcripts evaluated by qPCR are listed in Table 1. Threshold cycle numbers were determined using Applied Biosystems 7500 software version 2.05 (Life Technologies). Amplification products were verified by melting curves. The efficiency of amplification for each set of primers was tested by a serial dilution of the templates and corresponded to a two-fold increase per amplification cycle. Control reactions in the absence of template were used as negative controls. Results were calculated with normalization to ribosomal protein, large, P0 (Rplp0) mRNA. Rplp0 [alias acidic ribosomal phosphoprotein P0 (Arbp)] was chosen as the housekeeping gene because it was previously demonstrated that its mRNA does not change significantly with single or multiple episodes of cerulein-induced pancreatitis.18,25–27 The comparative threshold cycle method28 was used to calculate changes in mRNA abundance.
Table 1.
GenBank Accession Numbers and Primer Sequences of Mouse Genes Evaluated by qPCR
| Gene | Accession no. | Forward sequence | Reverse sequence |
|---|---|---|---|
| 1810009J06Rik | NM_023707 | 5′-GTTCGCCTTGGTGAACACAAT-3′ | 5′-GGATGGCAGGTGACTTCAATTT-3′ |
| Rplp0 | NM_007475 | 5′-AGATTCGGGATATGCTGTTGGC-3′ | 5′-TCGGGTCCTAGACCAGTGTTC-3′ |
| C4b | NM_009780 | 5′-AATAACCTGGGTCGGACTTTGG-3′ | 5′-CTCCGTCAGGGACGATGTTG-3′ |
| Ccl8 | NM_021443 | 5′-CTGGGCCAGATAAGGCTCC-3′ | 5′-CATGGGGCACTGGATATTGTT-3′ |
| Itih4 | NM_018746 | 5′-CCTCCGTTGCAGCACAATATC-3′ | 5′-GCGGCAAGGTAATAGTAGGAAC-5′ |
| Pcolce2 | NM_029620 | 5′-TGTGGCGGCATTCTTACCG-3′ | 5′-CCCTCAGGAACTGTGATTTTCCA-3′ |
| Acta2 | NM_007392 | 5′-GTCCCAGACATCAGGGAGTAA-3′ | 5′-TCGGATACTTCAGCGTCAGGA-3′ |
| Vim | NM_011701 | 5′-TCCACACGCACCTACAGTCT-3′ | 5′-CCGAGGACCGGGTCACATA-3′ |
| Vtn | NM_011707 | 5′-CCCCTGAGGCCCTTTTTCATA-3′ | 5′-CAAAGCTCGTCACACTGACA-3′ |
| Wdfy1 | NM_027057 | 5′-ACCATCCGAGTATGGCTGAAA-3′ | 5′-CCTGCTGTCGTGGTGGTATG-3′ |
Acta2, alpha2, smooth muscle, aorta; C4b, complement component 4B (Chido blood group); Ccl8, chemokine (C-C motif) ligand 8; Itih4, inter alpha-trypsin inhibitor, heavy chain 4; Pcolce2, procollagen C-endopeptidase enhancer 2; 1810009J06Rik, RIKEN cDNA 1810009J06 gene (trypsinogen 5); Rplp0, ribosomal protein, large, P0 [alias acidic ribosomal protein P0 (Arbp)]; Vim, vimentin; Vtn, vitronectin; Wdfy1, WD repeat and FYVE domain containing 1.
Microarrays
Total RNA was isolated from pancreatic tissues of B6J and B6N mice at day 10 after induction of CP by repetitive cerulein treatment and from saline-treated B6J and B6N controls. To prepare total RNA, pancreatic tissue in RNA stabilization solution (Ambion RNAlater; Life Technologies) was first extracted with TRIzol reagent (Life Technologies) according to the manufacturer’s instructions and then further purified using an RNeasy mini kit (Qiagen, Valencia, CA). RNA integrity was confirmed with an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA), and concentrations were measured using a NanoDrop ND-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Samples of 200 μg of total RNA were reversed-transcribed and then amplified by in vitro transcription according to the Affymetrix (Santa Clara, CA) standard protocol. Mouse Affymetrix GeneChip Mouse Genome 430.2.0 arrays were used for all hybridizations. These arrays contain probes representing more than 39,000 mouse transcripts. Four groups (cerulein-treated B6J, cerulein-treated B6N, saline-treated B6J, and saline-treated B6N) were each analyzed in triplicate (12 microarrays total). Microarray data were normalized and statistically analyzed using the Affymetrix Expression Console version 1.2 (Santa Clara, CA), R/Bioconductor version 2.7 (http://www.bioconductor.org, last accessed October 19, 2011), and Partek Genomics Suite version 6.5 (Partek, St. Louis, MO) software packages. Microarray hybridization and analysis were performed at the microarray core facility of Saint Louis University. Pathway analysis was performed using DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatic Resources version 6.7 (http://david.abcc.ncifcrf.gov/home.jsp, last accessed January 12, 2012).29 The data discussed in this publication have been deposited in the NCBI Gene Expression Omnibus30 and are accessible through GEO series (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE41418; accession number GSE41418).
Pancreatic Stellate Cell Isolation and Culture
Pancreata of 7- to 9-week-old mice (n = 9 for each B6N and B6J substrain) were combined for PSC isolation. PSCs were isolated using collagenase digestion and gradient centrifugation as we have described previously.19 Isolated cells were plated on 100-mm plastic culture plates and were maintained in 10% fetal bovine serum in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) with antibiotics (Penicillin, 100 units, Streptomycin, 0.1 mg/mL) in a humidifying incubator with a 5% CO2–enriched atmosphere at 37°C. PSCs were cultured for 9 days, plated on 12-well plates (Corning, Corning, NY), and cultured for another 48 hours. Cells were serum starved for 48 hours; RNA was then extracted using TRIzol reagent (Life Technologies) and used for the qPCR analysis.
Statistical Analysis
Nonparametric U-test was used for statistical analysis of histological measures of chronic and acute pancreatitis. Statistical analysis of all other experiments was performed using one-way analysis of variance followed by two-tailed t-test. P values of <0.05 were considered to be statistically significant (SigmaStat software version 3.1, San Jose, CA). For qPCR results, ΔCT values were used for statistical analysis, as recommended.31 Data are expressed as means ± SEM or as means with confidence intervals.
Results
Cerulein-Induced CP Is More Severe in B6J than in B6N Mice
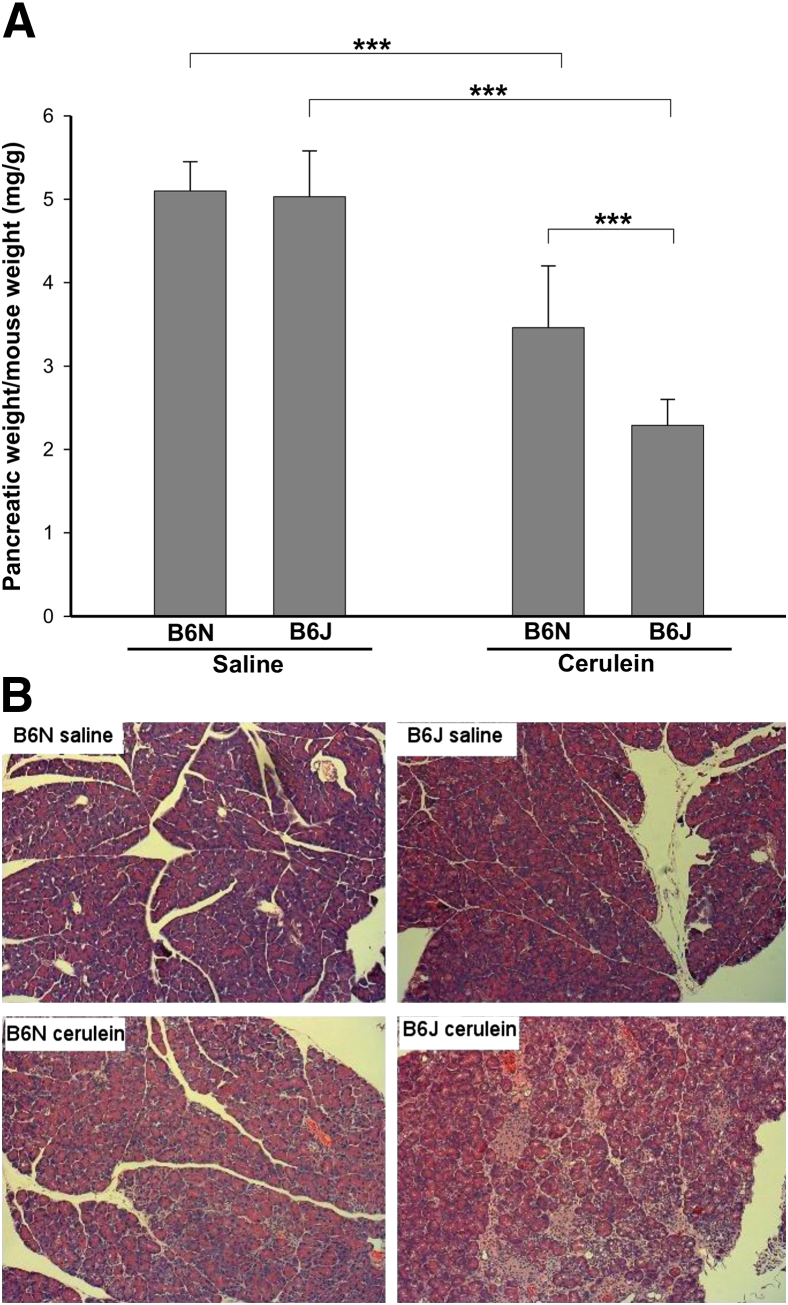
CP was induced in groups of B6J and B6N mice by repetitive cerulein treatment. Saline-treated mice were used as controls. The ratios of pancreatic weight to body weight, as well as histological changes, were evaluated to assess pancreatic atrophy, inflammation, and fibrosis. Pancreatic weights were significantly lower in cerulein-treated groups, compared with control mice, indicating significant organ atrophy with repetitive injury. B6J mice were found to have more pancreatic atrophy after cerulein treatment, compared with B6N mice (Figure 1A). Morphological changes in the pancreas were assessed by H&E staining (Figure 1B and Table 2). In the untreated mice, the acinar units were tightly packed, and there were no apparent differences between B6N and B6J mice (Figure 1B). Repetitive cerulein treatment caused significant morphological alterations, including disruption of acinar cell architecture and inflammatory cell infiltration. These alterations were more severe in cerulein-treated B6J mice than in B6N mice, as assessed by masked evaluation (Table 2 and Figure 1B).
Figure 1.
Severity of cerulein-induced CP in B6N and B6J substrains. Mice were subjected to three episodes of cerulein treatment over 5 days to induce CP and were sacrificed at 3 days after the last treatment. A: Loss of pancreatic weight relative to total body weight indicates significant pancreatic atrophy. B6J mice have significantly more pancreatic atrophy, compared with B6N mice. B: Histological changes in H&E-stained pancreas of B6N and B6J mice after induction of CP. Pancreas from B6N and B6J mice after control saline treatment shows no abnormalities. After cerulein treatment, pancreas from both B6J and B6N mice shows disrupted acinar architecture, dedifferentiation to tubular complexes, and interstitial inflammation; all of these parameters were more severe in B6J than B6N mice (Table 2). Data are expressed as means ± SEM. n = 6 mice per group. ∗∗∗P < 0.001. Original magnification, ×200.
Table 2.
Severity of Cerulein-Induced CP in B6N and B6J Mice
| Histopathology∗ | No cerulein |
Cerulein |
P | ||
|---|---|---|---|---|---|
| B6N | B6J | B6N | B6J | ||
| % Abnormal architecture | 0 ± 0 | 0 ± 0 | 14 ± 5.5 | 28 ± 2.7 | 0.008 |
| Necrosis | 0 ± 0 | 0 ± 0 | 1.8 ± 0.45 | 2.0 ± 0 | 1.00 |
| Acute inflammation∗ | 0 ± 0 | 0 ± 0 | 1 ± 0 | 2 ± 0 | 0.008 |
| Chronic inflammation† | 0 ± 0 | 0 ± 0 | 2 ± 0 | 2.6 ± 0.55 | 0.151 |
Histopathology was scored as described under Materials and Methods. Higher values indicate greater severity. Data are expressed as means ± SEM. n = 5 mice per group. P values were calculated between cerulein-treated B6N and B6J mice.
For acute inflammation, the content of acute inflammatory cells (neutrophils) was graded.
For chronic inflammation, the content of mononuclear cells was graded.
Fibrosis Associated with CP Is Greater in B6J than in B6N Mice
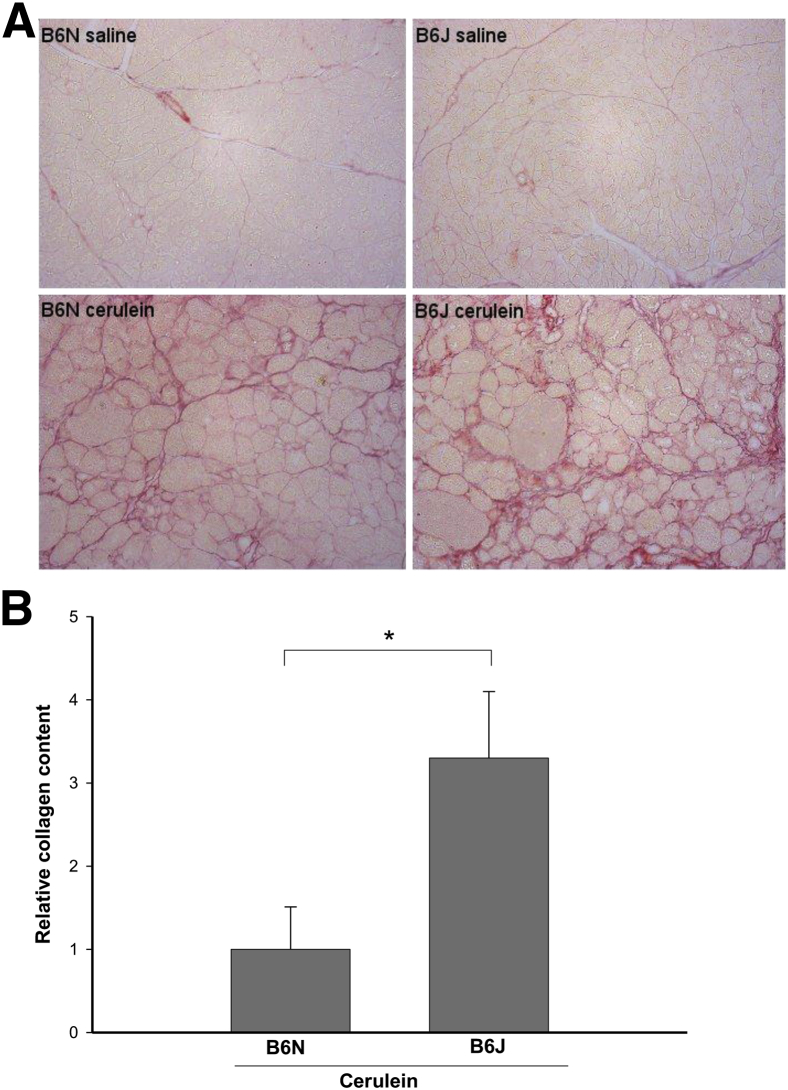
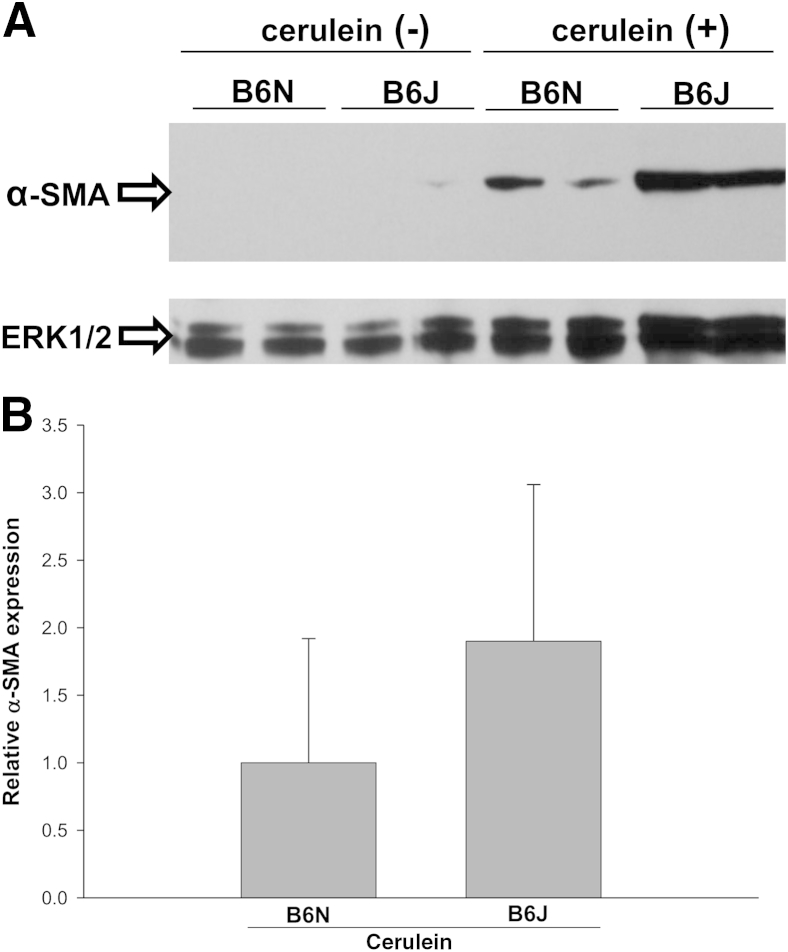
Pancreatic damage in CP is associated with pancreatic fibrogenesis, a process that may interfere with normal regeneration and organ function just as it does in the liver. We used Sirius Red staining of pancreatic sections as a measure of collagen deposition and found increased collagen content of cerulein-treated B6J mice, compared with B6N mice (Figure 2A). To quantify these fibrotic changes, the extent of Sirius Red staining was evaluated with morphometric image analysis (Figure 2B). This analysis demonstrated that the amount of pancreatic Sirius Red-positive staining was higher in B6J mice than in B6N mice. PSCs play a key role in extracellular matrix deposition and pancreatic fibrosis. In response to injury, PSCs develop an activated myofibroblast-like phenotype with increased α-SMA accumulation. The level of α-SMA protein varied among similarly treated mice and showed a trend toward being greater in B6J mice than in B6N mice (Figure 3).
Figure 2.
Collagen content in the pancreas assessed by Sirius Red staining after induction of CP by repetitive cerulein injections. A: Representative pancreatic sections of pancreas from saline-treated control B6N and B6J mice and from cerulein-treated B6N and B6J mice. Control mice exhibit some perilobular collagen, with no difference between substrains. Cerulein-treated mice demonstrate extensive interlobular and periacinar collagen staining, indicating robust fibrogenesis in this model of CP. Collagen staining was more pronounced in B6J than in B6N mice. B: Quantification by morphometric analysis showed that the relative amount of pancreatic collagen was greater in B6J than in B6N mice. Data are expressed as means ± SEM. n = 6. ∗P = 0.0005. Original magnification, ×200.
Figure 3.
Activation of PSCs after induction of CP. A: Western blotting of pancreatic extracts demonstrated increased expression of α-SMA, a marker of PCS activation, in both B6N and B6J mice. ERK1/2 was used as a loading control. B: Densitometric analysis of α-SMA protein expression after induction of CP. An increase in α-SMA expression was observed for B6J mice, compared with B6N mice, but the difference did not reach statistical significance (P = 0.9). Data are expressed as means ± SEM. n = 5 mice per group.
Degree of Acute Pancreatic Injury Induced by Cerulein Is Similar in B6J and B6N Mice
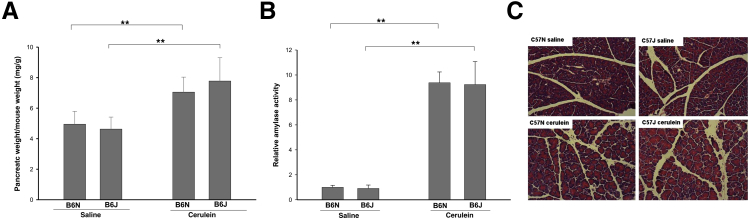
To assess the possibility that the difference in the degree of CP between B6J and B6N mice arises from differences in the degree of acute cerulein-induced injury, we compared the parameters of acute pancreatic injury after a single episode of cerulein treatment. As a measure of acute pancreatic injury, the following parameters were evaluated: pancreatic edema (ratio of pancreatic weight to mouse weight), plasma level of amylase, and histological scoring of H&E-stained pancreatic sections at 9 hours after the first cerulein injection (Figure 4). Pancreatic edema and plasma amylase levels increased significantly after acute pancreatic injury with no differences between B6J and B6N substrains (Figure 4, A and B). Histological manifestations of acute pancreatic injury as assessed by masked scoring were also not significantly different between B6N and B6J mice (Figure 4C and Table 3).
Figure 4.
Parameters of acute pancreatic injury in B6J and B6N substrains. Mice were subjected to a single episode of acute pancreatitis (six hourly injections in one day) and were sacrificed at 9 hours after the first cerulein injection. Saline-injected mice were used as controls. A: Pancreatic edema, measured as the ratio of pancreatic weight to body weight, showed similar increases in B6N and B6J mice. B: Plasma amylase levels were also elevated similarly in B6N and B6J mice. C: Histological changes in the pancreas were evaluated by H&E staining. Pancreas from saline-treated mice appeared normal, whereas the pancreas from cerulein-treated mice showed acute injury including vacuolization, inflammatory cell infiltration, and necrosis. Data are expressed as means ± SEM. n = 6 mice per group. ∗∗P < 0.01. Original magnification, ×200.
Table 3.
Similar Severity of Cerulein-Induced Acute Pancreatic Injury in B6N and B6J Mice
| Histopathology | No cerulein |
Cerulein |
||
|---|---|---|---|---|
| B6N | B6J | B6N | B6J | |
| Vacuolization | 0 ± 0 | 0 ± 0 | 3.0 ± 0 | 3.0 ± 0 |
| Necrosis | 0 ± 0 | 0 ± 0 | 2.0 ± 0.82 | 2.25 ± 0.5 |
| Inflammatory infiltration | 0 ± 0 | 0 ± 0 | 1.5 ± 0.58 | 2 ± 0 |
| Edema | 0 ± 0 | 0 ± 0 | 1.25 ± 0.5 | 1.0 ± 0 |
Histopathology was scored as described under Materials and Methods. Higher values indicate greater severity. Data are expressed as means ± SEM. n = 5 mice per group. No significant differences were detected.
Absence of NNT Protein in B6J Mice Is Not Responsible for the More Severe CP
We confirmed the deletion of the Nnt gene in B6J mice obtained from the Jackson Laboratory by PCR amplification of genomic DNA using primers designed to span the deleted exons (Figure 5A). The expression of NNT protein in the pancreas of B6N mice and the absence of such expression in the pancreas of B6J mice was confirmed by Western blot analysis (Figure 5B).
Figure 5.
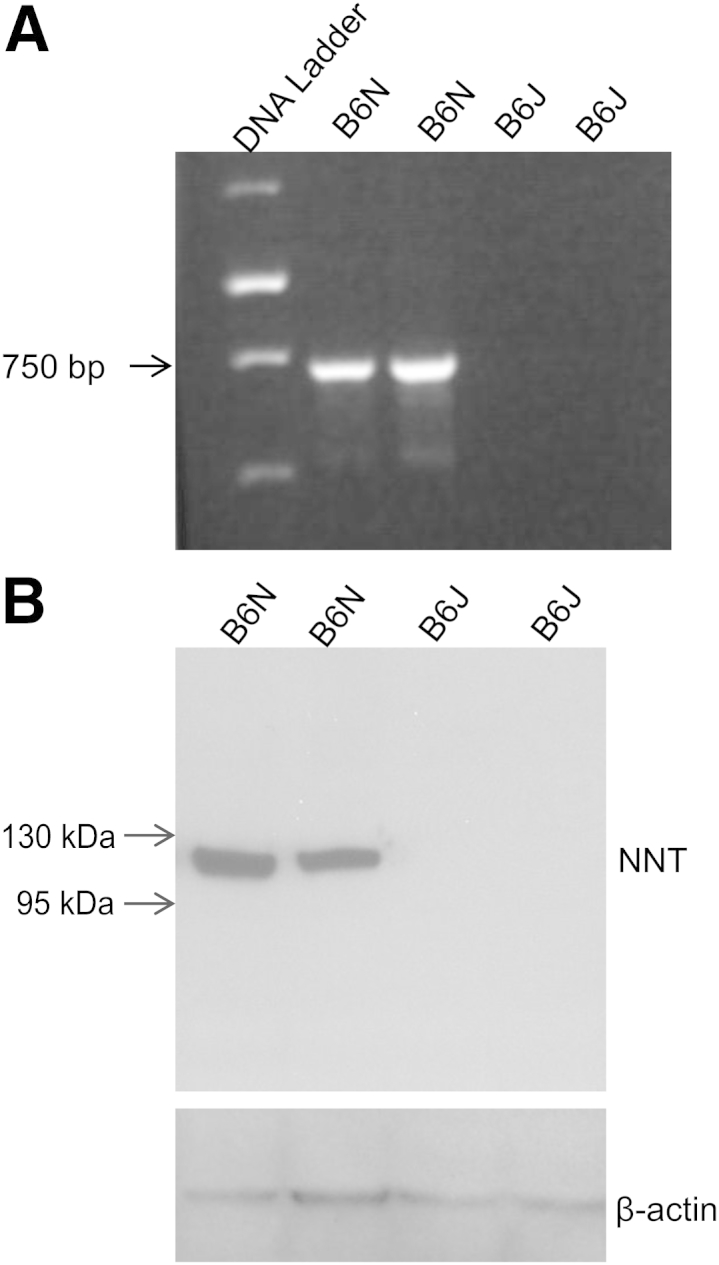
B6J mice are nicotinamide nucleotide transhydrogenase (NNT) deficient. A: PCR analysis of genomic DNA from B6J and B6N mice confirmed that B6J mice lack the Nnt gene. B: Western blotting of pancreatic extracts demonstrated the presence of NNT protein in B6N and its absence in B6J mice. β-Actin was used as in internal control.
To determine whether the absence of NNT expression in B6J mice contributes to the more severe CP phenotype in this substrain, we compared the degree of cerulein-induced CP in B6J mice and transgenic mice expressing NNT protein on the B6J background. The NNT transgenic mice were generated previously at MRC Harwell, to investigate the role of NNT protein in glucose intolerance in B6J mice.23,32 Those studies demonstrated that transgenic expression of NNT in B6J mice rescued the B6J glucose-intolerance phenotype, indicating that transgenic mice express functional NNT protein. To confirm that transgenic B6J mice express NNT protein in mitochondria at a level similar to that of B6N mice, we performed Western blot analysis of NNT protein from isolated pancreatic mitochondria. The results confirmed that similar levels of NNT protein are expressed in NNT transgenic mice and B6N mice (Figure 6).
Figure 6.
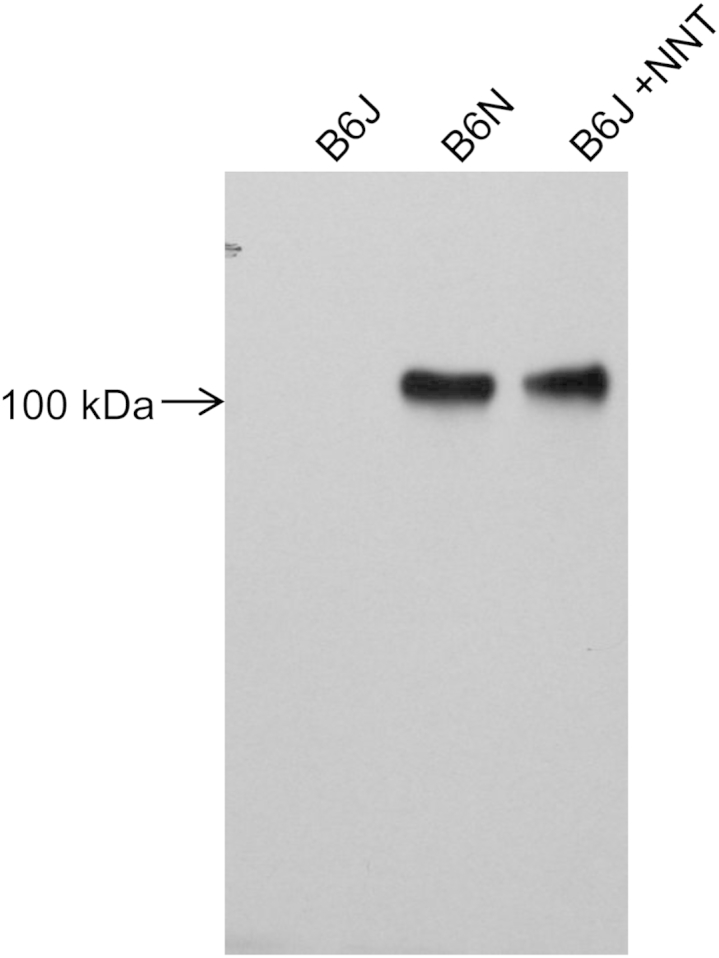
Western blot of pancreatic mitochondrial nicotinamide nucleotide transhydrogenase (NNT) protein, confirming absence of expression in B6J mice and similar expression in B6J Nnt+/+ transgenic mice and B6J mice.
To investigate the role of NNT in CP, CP was induced in wild-type B6J mice (Nnt−/−) and NNT transgenic mice (Nnt+/+) on the same B6J background using repetitive cerulein treatment (6 or 7 mice per group). Sex- and age-matched groups of Nnt−/− and Nnt+/+ mice were used as saline-treated controls. Although repetitive cerulein treatment caused significant pancreatic damage and fibrosis, we did not find any differences in any of the parameters (measured as described in the previous section for B6J and B6N mice) between cerulein-treated Nnt−/− and Nnt+/+ mice (data not shown), indicating that the presence of NNT in the transgenic mice was not protective against CP and that the substrain difference in CP severity is not due to the absence of NNT in B6J mice.
Substrain Differences in mRNA Expression of Other Genes in the Normal Pancreas
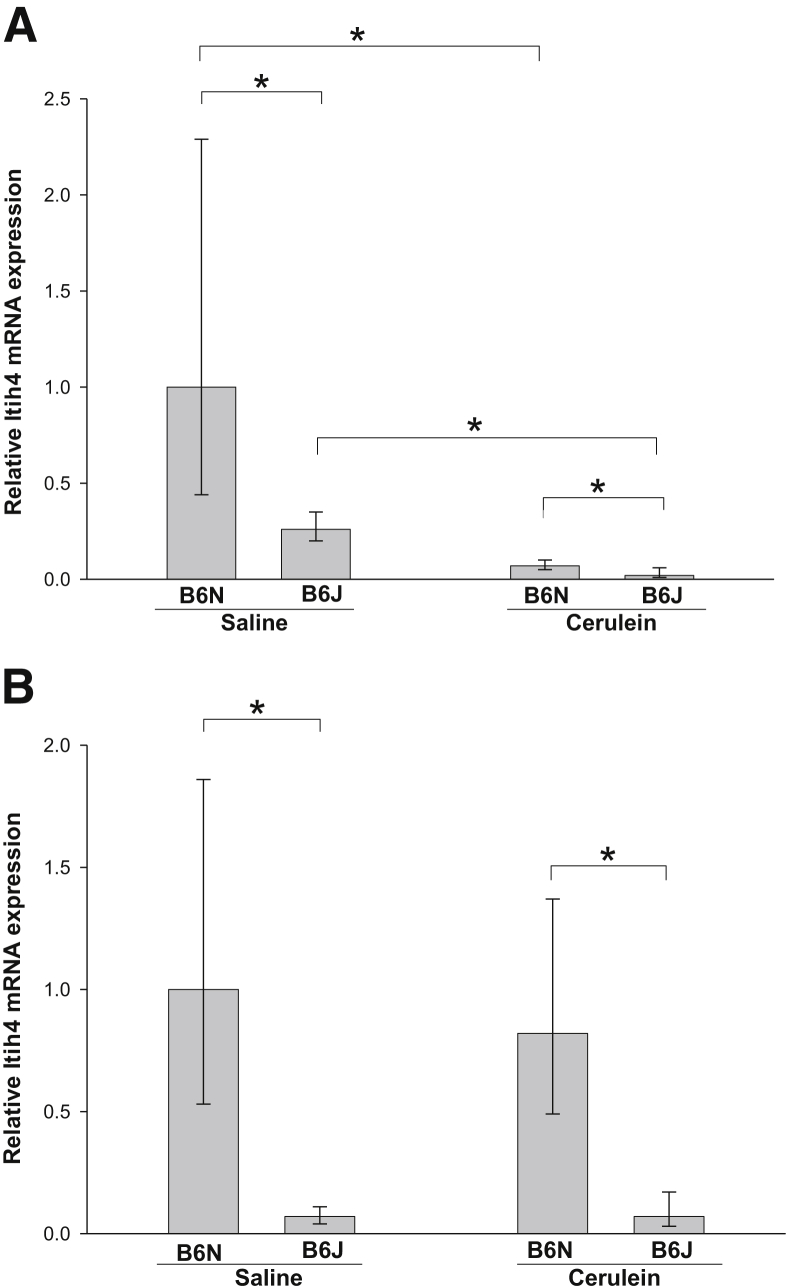
To explore the differences in expression of other genes in the two substrains after injury, we first determined whether there were differences in gene expression in these mice under basal conditions without any treatment. Affymetrix microarray analysis of pancreatic mRNA from normal pancreas of B6N and B6J mice revealed that most of the mRNAs from these two substrains were expressed at similar levels. Nevertheless, seven mRNAs (including NNT) exhibited significant differences between substrains, with more than twofold change (P < 0.01) in their expression as assessed by microarray (Table 4). The presence of detectable NNT mRNA on the microarray in B6J mice has been attributed to the presence of a short truncated unstable mRNA transcript that does not lead to production of functional NNT protein in this substrain.13 The difference in gene expression under basal conditions identified on the microarray was confirmed by qPCR for Itih4 (Figure 7A).
Table 4.
Differential Expression of Pancreatic Genes in B6J and B6N Mouse Substrains in the Normal Pancreas
| Gene name | Gene symbol | EntrezGene ID | Chr | Fold difference B6J vs B6N |
|---|---|---|---|---|
| UDP glucuronosyltransferase 2 family, polypeptide B34 | Ugt2b34 | 100727 | 5 | 3.0 |
| Ectonucleoside triphosphate diphosphohydrolase 4 | Entpd4 | 67464 | 14 | 2.3 |
| Immunoglobulin lambda variable 1 | Iglv1 | 16142 | 16 | −8.0 |
| Complement component 4B (Chido blood group)∗ | C4b | 12268 | 17 | −5.7 |
| Nicotinamide nucleotide transhydrogenase | Nnt | 18115 | 13 | −2.8 |
| Inter alpha-trypsin inhibitor, heavy chain 4∗ | Itih4 | 16427 | 14 | −2.8 |
| WD repeat and FYVE domain containing 1∗ | Wdfy1 | 69368 | 1 | −2.0 |
A transcript was considered to be highly significantly divergently expressed between B6J and B6N substrains if the P value between strains was <0.01 and the average fold change was >2. A minus sign in the fold difference column indicates lower mRNA abundance in B6J versus B6N. Chr, chromosome.
Significantly altered after cerulein treatment in the CP mouse model.
Figure 7.
Pancreatic Itih4 expression in B6J and B6N substrains. A: Itih4 mRNA expression in the pancreas after induction of CP was much lower in B6J mice than in B6N mice under control conditions of saline injections. After induction of CP, pancreatic Itih4 expression was markedly down-regulated in both substrains, but its expression was higher in B6N than in B6J. B: Itih4 mRNA expression in the pancreas 9 hours after the acute injury induced by one cerulein treatment. Acute injury did not decrease Itih4 mRNA expression. Itih4 mRNA expression was normalized to Rplp0 and expressed as fold increase over the level in B6N saline-treated control mice. Data are expressed as means with 95% confidence intervals. Confidence intervals were calculated using the ΔCT values before exponential transformation to fold increase in mRNA and are therefore asymmetric about the means. n = 6. ∗P < 0.05.
Pancreatic Gene Expression Profile Associated with Cerulein-Induced CP in B6N and B6J Substrains
Induction of CP caused significant changes in the pattern of pancreatic gene expression in both B6N and B6J substrains, as determined by comparing pancreatic mRNA expression before and after the repetitive cerulein treatment, using microarray analysis. On average, close to 7% of genes were up-regulated and approximately 0.7% of genes were down-regulated more than twofold (P < 0.01) after repetitive cerulein treatment in both substrains. The genes identified by this analysis were involved mainly in eight cellular pathways: cell cycle, DNA replication and repair, focal adhesion, fibrosis, ubiquitin-mediated proteolysis, cancer, apoptosis, and immune response. Genes that were differentially regulated by more than 10-fold (P < 0.01) in one or both of the substrains after induction of CP are listed in Table 5. Genes associated with fibrosis that were differentially regulated to the greatest degree included collagen, type V, alpha 2 (Col1a2); collagen, type III, alpha 1 (Col3a1); collagen, type V, alpha 2 (Col5a2); CD44 antigen (Cd44); inter alpha-trypsin inhibitor heavy chain 4 (Itih4), and matrix metalloproteinases 2 and 7 (Mmp2 and Mmp7). To determine whether gene expression profiles associated with cerulein-induced CP in mouse correspond to human CP profiles, we compared the results of our microarray analysis with a published list of 107 pancreatic human genes up-regulated in stromal compartments of patients with CP and pancreatic cancer.33 Of the human genes on that list, 40% were also up-regulated in our cerulein model of mouse CP. Among these mouse genes were apolipoprotein E (Apoe); cathepsin K (Ctsk); cathepsin S (Ctss); Fc receptor, IgE, high affinity I, gamma polypeptide (Fcer1g); lumican (Lum); lysyl oxidase (Lox); Mmp2; periostin, osteoblast specific factor (Postn); and Col1a2, Col3a1, Col5a2, and Col1a1.
Table 5.
Highly Differentially Regulated Pancreatic Genes in the Cerulein-Induced Model of CP
| Gene | Gene symbol | EntrezGene ID | Chr | Fold change, CP vs control B6J; B6N |
|---|---|---|---|---|
| Adenylate cyclase 7 | Adcy7 | 11513 | 8 | 11.3; 3.5 |
| Alpha 1 microglobulin; bikunin | Ambp | 11699 | 4 | 12.9; 10.0 |
| Annexin A10∗ | Anxa10 | 26359 | 8 | 28.0; 10.8 |
| Anterior gradient 2 | Agr2 | 23795 | 12 | 38.8; 21.9 |
| Antigen identified by monoclonal antibody Ki 67 | Mki67 | 17345 | 7 | 10.0; 4.5 |
| Apolipoprotein E | Apoe | 11816 | 7 | 13.0; 14.4 |
| Asporin∗ | Aspn | 66695 | 13 | 11.4; 2.5 |
| Baculoviral IAP repeat-containing 5 | Birc5 | 11799 | 11 | 30.3; 20.3 |
| Biglycan | Bgn | 12111 | X | 10.9; 3.2 |
| Carbonic anhydrase 9 | Car9 | 230099 | 4 | 11.0; 6.8 |
| Cartilage intermediate layer protein, nucleotide pyrophosphohydrolase | Cilp | 214425 | 9 | 23.7; 27.0 |
| Cathepsin K | Ctsk | 13038 | 3 | 11.3; 12.4 |
| Cathepsin S | Ctss | 13040 | 3 | 13.9; 11.9 |
| CD14 antigen | Cd14 | 12475 | 18 | 10.3; 8.2 |
| CD44 antigen | Cd44 | 12505 | 2 | 13.6; 12 |
| CD52 antigen | Cd52 | 23833 | 4 | 10.9; 5.8 |
| CDC28 protein kinase 1b | Cks1b | 54124 | 3 | 14.4; 11.8 |
| CDC28 protein kinase regulatory subunit 2 | Cks2 | 66197 | 4 | 10.3; 11.3 |
| Cell division cycle 20 | Cdc20 | 107995 | 4 | 10.8; 7.5 |
| Centromere protein A | Cenpa | 12615 | 5 | 10.0; 8.0 |
| Ceruloplasmin | Cp | 12870 | 3 | 4.8; 11.5 |
| Chemokine (C-C motif) ligand 6 | Ccl6 | 20305 | 11 | 11.9; 3.5 |
| Chemokine (C-C motif) ligand 8∗ | Ccl8 | 20307 | 11 | 61.5; 12.9 |
| Claudin 2 | Cldn | 12738 | X | 20.1; 23.4 |
| Complement component 1, q subcomponent, beta polypeptide | C1qb | 12260 | 4 | 13.7; 7.2 |
| Complement component 1, q subcomponent, C chain | C1qc | 12262 | 4 | 14.6; 13.4 |
| Complement component 3a receptor 1 | C3ar1 | 12267 | 6 | 4.1; 10.1 |
| Complement component factor i | Cfi | 12630 | 3 | 8.4; 13.7 |
| Cyclin-dependent kinase 1 | Cdk1 | 12534 | 10 | 11.3; 7.4 |
| DNA segment, Chr 17, human D6S56E5 | D17H6S56E-5 | 110956 | 17 | 20.0; 35.3 |
| EGF-like module containing, mucin-like, hormone receptor-like sequence 1 | Emr1 | 13733 | 17 | 11.0; 8.5 |
| Expressed sequence AU020206 | AU020206 | 101757 | 7 | 7.1; 12.5 |
| Fc receptor, IgE, high affinity I, gamma polypeptide | Fcer1g | 14127 | 1 | 12.3; 6.1 |
| Fc receptor, IgG, low affinity Iib | Fcgr2b | 14130 | 1 | 11.6; 7.6 |
| Flap structure specific endonuclease 1 | Fen1 | 14156 | 19 | 5.6; 10.3 |
| Gastric intrinsic factor | Gif | 14603 | 19 | 15.2; 16.3 |
| Group specific component | Gc | 14473 | 5 | 17.1; 9.4 |
| Growth arrest specific 5 | Gas5 | 14455 | 1 | 8.2; 17.7 |
| Helicase, lymphoid specific | Hells | 15201 | 19 | 10.7; 4.5 |
| Hexosaminidase B | Hexb | 15212 | 13 | 10.4; 9.9 |
| Integral membrane protein 2A | Itm2a | 16431 | X | 15.9; 17.3 |
| Inter-alpha (globulin) inhibitor H5 | Itih5 | 209378 | 2 | 4.6; 11.5 |
| Interferon activated gene 203 | Ifi203 | 15950 | 1 | 10.3; 8.3 |
| Interferon, alpha-inducible protein 27 like 2A∗ | Ifi27l2a | 76933 | 12 | 10.0; 2.6 |
| Interferon activated gene 205 | Ifi205 | 226695 | 1 | 10.5; 6.7 |
| Lectin, galactose binding, soluble 3 | Lgals3 | 16854 | 14 | 16.8; 12.3 |
| Legumain | Lgmn | 19141 | 12 | 10.0; 8.4 |
| Lipocalin 2 | Lcn2 | 16819 | 2 | 25.0; 19.1 |
| Lumican | Lum | 17022 | 10 | 12.6; 14.1 |
| Lymphocyte antigen 86 | Ly86 | 17084 | 13 | 14.3; 8.0 |
| Lysozyme 1 | Lyz1 | 17110 | 10 | 11.6; 9.6 |
| Lysozyme 2 | Lyz2 | 17105 | 10 | 15.3; 11.3 |
| Lysyl oxidase | Lox | 16948 | 18 | 10.1; 8.6 |
| Macrophage expressed gene 1 | Mpeg1 | 17476 | 19 | 11.1; 15.4 |
| Major urinary protein 2 | Mup2 | 17841 | 4 | 31.2; 53.4 |
| Matrix Gla protein | Mgp | 17313 | 15 | 9.6; 11.1 |
| Matrix metallopeptidase 2 | Mmp2 | 17390 | 8 | 10.0; 16.2 |
| Matrix metallopeptidase 7∗ | Mmp7 | 17393 | 9 | 16.0; 3.8 |
| Membrane-spanning 4-domains, subfamily A, member 6B | Ms4a6b | 69774 | 19 | 14.3; 8.3 |
| Membrane-spanning 4-domains, subfamily A, member 7 | Ms4a7 | 109225 | 19 | 29.5; 25.9 |
| Mesoderm specific transcript | Mest | 17294 | 6 | 10.8; 12.7 |
| Minichromosome maintenance deficient 4 homolog (S. cerevisiae) | Mcm4 | 17217 | 16 | 8.8; 19.1 |
| Minichromosome maintenance deficient 5, cell division cycle 46 (S. cerevisiae) | Mcm5 | 17218 | 8 | 23.0; 17.8 |
| Minichromosome maintenance deficient 6 (MIS5 homolog, S. pombe) (S.cerevisiae) | Mcm6 | 17219 | 1 | 22.8; 19.5 |
| Minichromosome maintenance deficient 7 (S. cerevisiae) | Mcm7 | 17220 | 16 | 10.4; 6.6 |
| Non-SMC condensin II complex, subunit G2 | Ncapg2 | 76044 | 12 | 10.5; 7.2 |
| Osteoglycin | Ogn | 18295 | 13 | 12.7; 6.7 |
| PDZ binding kinase | Pbk | 52033 | 14 | 12.3; 9.7 |
| Periostin, osteoblast specific factor | Postn | 50706 | 3 | 37.8; 40.7 |
| Placenta-specific 8 | Plac8 | 231507 | 5 | 11.8; 5.8 |
| Pleckstrin homology-like domain, family B, member 2 | Phldb2 | 208177 | 16 | 10.2; 13.2 |
| Pleiotrophin | Ptn | 19242 | 6 | 10.0; 12.3 |
| Procollagen, type I, alpha 2 | Col1a2 | 12843 | 6 | 6.3; 11.2 |
| Procollagen, type III, alpha 1 | Col3a1 | 12825 | 1 | 7.7; 10.4 |
| Procollagen, type V, alpha 2 | Col5a2 | 12832 | 1 | 6.2; 10.0 |
| Receptor transporter protein 4 | Rtp4 | 67775 | 16 | 11.4; 6.1 |
| Regenerating islet-derived 3 gamma∗ | Reg3g | 19695 | 6 | 7.9; 21.2 |
| Retinol binding protein 1, cellular | Rbp1 | 19659 | 9 | 13.5; 7.6 |
| RIKEN cDNA 1810009J06e | 1810009J06Rik | 73626 | 6 | 160.9; 97.1 |
| Serine (or cysteine) peptidase inhibitor, clade A, member 6 | Serpina6 | 12401 | 12 | 18.6; 6.1 |
| Similar to histone 2a | MGC73635 | 665433 | 13 | 12.3; 21.3 |
| Stathmin 1 | Stmn1 | 16764 | 4 | 11.1; 10.2 |
| Sulfatase 2 | Sulf2 | 72043 | 2 | 9.1; 17.3 |
| Tetraspanin 1 | Tspan1 | 66805 | 4 | 10.8; 6.0 |
| Thymosin, beta 10 | Tmsb10 | 19240 | 7 | 10.1; 8.1 |
| Topoisomerase (DNA) II alpha | Top2a | 21973 | 11 | 11.5; 13.4 |
| TYRO protein tyrosine kinase binding protein | Tyrobp | 22177 | 7 | 22.6; 11.6 |
| Ubiquitin-conjugating enzyme E2C | Ube2c | 68612 | 2 | 12.9; 13.4 |
| Widely-interspaced zinc finger motif | Wiz | 22404 | 17 | 15.8; 10.0 |
| Galanin | Gal | 14419 | 19 | −21.1; −11.7 |
| Hepcidin antimicrobial peptide 2 | Hamp2 | 66438 | 7 | −42.3; −21.0 |
| Inter alpha-trypsin inhibitor, heavy chain 4∗ | Itih4 | 16427 | 14 | −16.8; −12.7 |
| RIKEN cDNA 1810007D17 gene | 1810007D17Rik | 69055 | 19 | −17.7; −10.5 |
| Two pore segment channel 2 | Tpcn2 | 233979 | 7 | −14.2; −6.3 |
Only transcripts that were regulated more than 10-fold in at least one of the substrains (B6J or B6N) are shown. P < 0.01, cerulein-treated versus control probes. A minus sign in the fold change column indicates downregulation in B6J versus B6N.
Expressed at significantly different level (fold change >2) between B6J and B6N after cerulein treatment in the CP mouse model. P < 0.05.
Trypsinogen 5 mRNA (Homolog of Human Mesotrypsinogen) Is Strongly Up-Regulated in the Pancreas of Mice with Cerulein-Induced Chronic and Acute Pancreatitis
Microarray analysis revealed that the 1810009J06Rik transcript was the most highly up-regulated gene (up to 160-fold) in the pancreas of mice with CP, in both the B6J and the B6N substrain. Using NCBI BLAST homology search of human transcripts, we found that 1810009J06Rik is most closely related to the human mesotrypsinogen gene (protease, serine, 3; PSSR3), with 75% identity. Mesotrypsinogen is a minor trypsinogen isoform that is resistant to naturally occurring trypsin inhibitors. The functional ortholog of human mesotrypsin in the rat is a protein identified as P23, and P23 expression in rats is also up-regulated by cerulein. To determine whether mouse 1810009J06Rik is similar to rat P23, we compared the sequences of these transcripts and found that they have 95% sequence identity. All active sites of rat P23 are preserved in mouse 1810009J06Rik, including the sequence differences that distinguish it from other trypsinogen isoforms and make P23 resistant to endogenous trypsin inhibitors. These findings indicate that 1810009J06Rik is the mouse homolog of rat P23.
There are 20 trypsinogen genes (T1 to T20) in mouse, 12 of which express trypsinogen proteins.34,35 According to this classification, 1810009J06Rik is identified as T5. We did not find any significant induction in the mRNA expression of other trypsinogens with repetitive cerulein treatment. The mRNA of T7, an isoform demonstrated to be involved in pathological intra-acinar trypsinogen activation in mice,36 was slightly (1.4-fold) but significantly (P = 0.006) down-regulated. In the pancreas of mice with CP, T5 and T7 mRNA were expressed at similar levels. The result of microarray analysis of T5 mRNA expression was validated by RT-qPCR analysis. The expression of T5 mRNA was induced more than 1000-fold in the repetitive cerulein model of CP (Figure 8A). Because the severity of cerulein-induced acute pancreatitis episodes may determine the severity of the resulting CP phenotype, we evaluated whether cerulein induces T5 expression in the acute pancreatitis model and found that T5 mRNA is also strongly up-regulated, in the acute pancreatitis model, similar to the CP model (Figure 8B).
Figure 8.
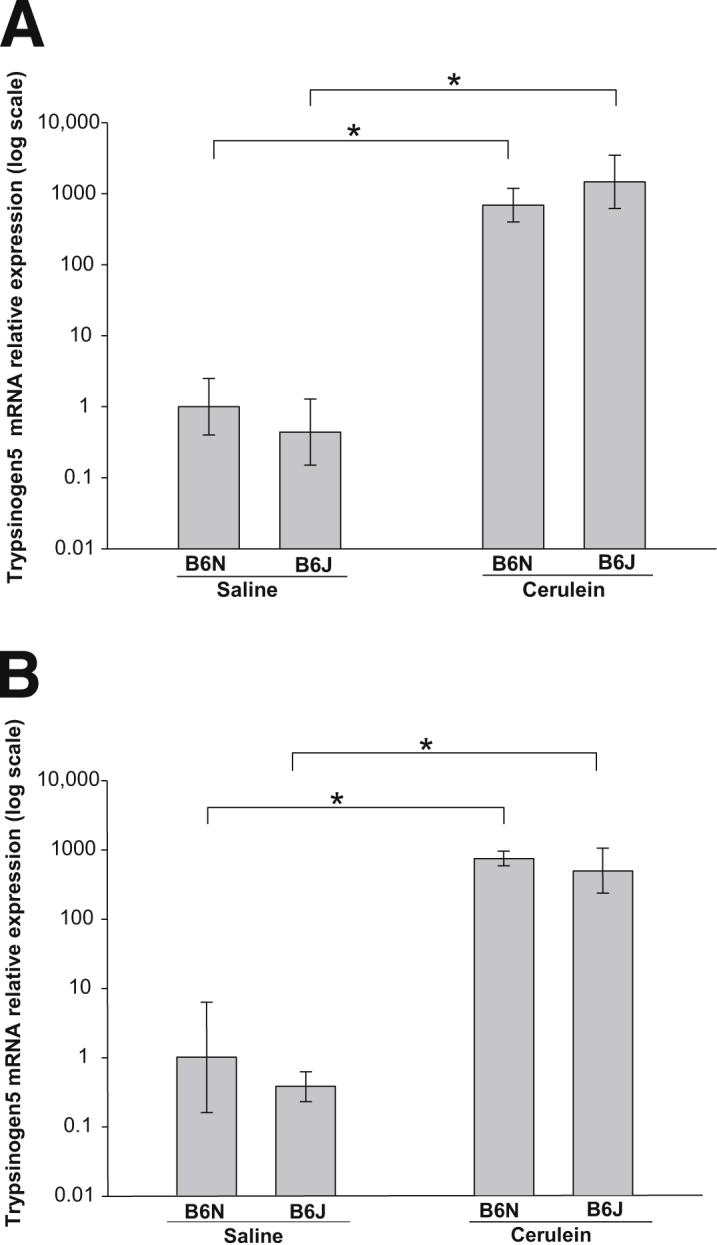
Pancreatic T5 mRNA expression in B6J and B6N substrains. A: Pancreatic T5 expression in saline-injected control mice and in repetitive cerulein-injected mice after induction of CP. T5 expression was significantly induced after induction of CP in both B6N and B6J substrains. B: Pancreatic T5 expression in mice after acute cerulein-induced injury. Similar to the increase in CP, T5 mRNA expression was significantly induced in both substrains after acute injury. T5 mRNA expression was normalized to Rplp0 and expressed as fold increase over the level in B6N saline-treated control mice. Data are expressed as means with 95% confidence intervals. Confidence intervals were calculated using the ΔCT values before exponential transformation to fold increase in mRNA and are therefore asymmetric about the means. n = 6. ∗P < 0.05.
Two Members of the Chemokine (C-C Motif) Family of Ligands, Ccl8 and Ccl6, Are Strongly Up-Regulated in the Pancreas of Mice with Cerulein-Induced CP
Chemokines are involved in the pathogenesis of inflammatory and fibrotic diseases, including CP. A number of cytokines and chemokines were differentially regulated after repetitive cerulein treatment, as determined by microarray analysis. Among these, two chemokines were up-regulated the most: chemokine (C-C motif) ligand 8 [Ccl8; alias monocyte chemoattractant protein 2 (MCP-2)]) and chemokine (C-C motif) ligand 6 (Ccl6). In fact, Ccl8 was the second most highly regulated gene in the microarray (after T5) (Table 5). Microarray data for Ccl8 were evaluated by qPCR analysis (Figure 9A). The B6J substrain demonstrated higher Ccl8 mRNA expression than the B6N substrain in microarray analysis (P < 0.05), but with only a trend toward higher expression in qPCR analysis (P = 0.2). The expression of Ccl8 mRNA after induction of acute pancreatitis was much lower, with only an approximately 2.5-fold increase, only in the B6J substrain (Figure 9B).
Figure 9.
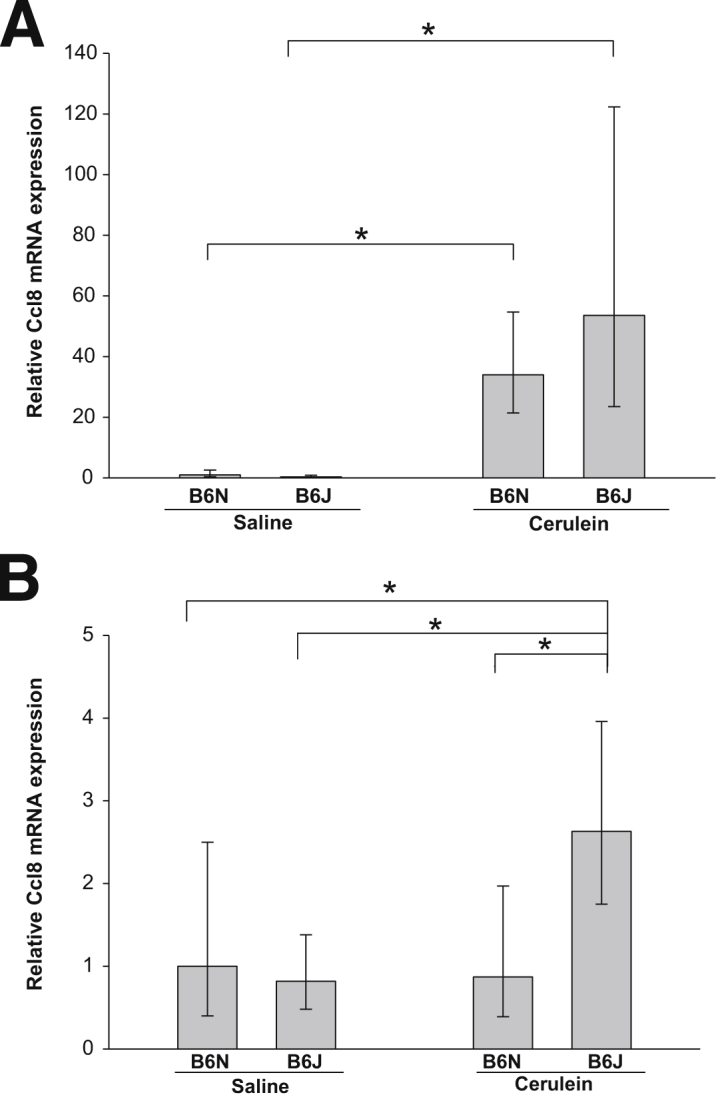
Pancreatic Ccl8 expression in B6J and B6N substrains. A: Ccl8 expression in the pancreas after induction of CP. Ccl8 was strongly up-regulated in the pancreas of mice with CP, compared with control mice. The up-regulation in B6J mice was not significantly greater than in B6N mice (P = 0.02). B: Ccl8 expression in the pancreas after acute cerulein-induced injury. Acute injury induced Ccl8 mRNA increase approximately 2.5-fold in B6J mice, but not in B6N mice. Ccl8 mRNA expression was normalized to Rplp0 and expressed as fold increase over the level in B6N saline-treated control mice. Data are expressed as means with 95% confidence intervals. Confidence intervals were calculated using the ΔCT values before exponential transformation to fold increase in mRNA and are therefore asymmetric about the means. n = 6. ∗P < 0.05.
Genes with Differentially Regulated Expression between B6J and B6N Mice with CP Are Identified as Potential Candidate Genes in the Development of CP
Based on the differences in the course of CP between B6J and B6N substrains, we sought to identify genes within the most differentially regulated microarray data (defined by P < 0.01 and average fold change of >2) that were associated with the differences in the disease severity. Using this approach, we identified nine genes differentially expressed between normal pancreas and CP and between B6J and B6N substrains after induction of CP (Table 6). Of these genes, five were up-regulated in the B6J substrain that develops more severe CP, compared with the B6N substrain, and the other four were down-regulated.
Table 6.
Differential Expression of Pancreatic Genes Between B6J and B6N Mouse Substrains after the Induction of CP
| Gene | Gene symbol | EntrezGene ID | Chr | Fold difference B6J vs B6N |
|---|---|---|---|---|
| Interferon-induced protein with tetratricopeptide repeats 1 | Ifit1 | 15957 | 19 | 3.8 |
| Matrix metallopeptidase 7 | Mmp7 | 17393 | 9 | 3.5 |
| Placenta specific 9 | Plac9 | 211623 | 14 | 3.4 |
| Transmembrane protein 27 | Tmem27 | 57394 | X | 2.3 |
| Procollagen C-endopeptidase enhancer 2 | Pcolce2 | 76477 | 9 | 2.0 |
| Inter alpha-trypsin inhibitor, heavy chain 4 | Itih4 | 16427 | 14 | −4.8 |
| WD repeat and FYVE domain containing 1 | Wdfy1 | 69368 | 1 | −4.6 |
| Neurofascin | Nfasc | 269116 | 1 | −2.7 |
| Vitronectin | Vtn | 22370 | 11 | −2.6 |
A transcript was considered to be highly significantly divergently expressed between B6J and B6N substrains if the P value between strains was <0.01 and the average fold change was >2. A minus sign in the fold difference column indicates lower mRNA abundance in B6J versus B6N.
One of the genes that was up-regulated in CP and further up-regulated in the B6J substrain was procollagen C-endopeptidase enhancer 2 (Pcolce2). Collagen is secreted as a soluble procollagen molecule with C- and N-terminal propeptides attached. Enzymatic cleavage of C-terminal propeptide by bone morphogenic protein 1 (BMP-1) is required for collagen incorporation into insoluble collagen fibrils.37 The Pcolce2 protein enhances catalytic activity of BMP-1 for fibrillar collagen formation.38 Thus, Pcolce2 could be a factor responsible for the increased collagen accumulation in B6J mice. Although there are no data in the literature on the role of Pcolce2 in pancreatic fibrosis, Pcolce2-deficient mice exhibited decreased collagen deposition in myocardium of pressure overloaded mice.39
Another highly induced and differentially regulated gene identified on the microarrays, Mmp7 (which encodes matrilysin) has been reported to play an essential role in the development of CP in an obstructive pancreatitis mouse model.40 Mmp7 protein levels are also highly up-regulated in human and mouse pancreatic ductal adenocarcinomas and noninvasive ductal precursors, and Mmp7 expression is necessary for pancreatic ductal adenocarcinoma progression.41
We identified inter α-trypsin inhibitor, heavy chain 4 (Itih4) as a gene differentially expressed between substrains in both normal pancreas (Table 4) and in the pancreas of mice with CP (Table 6) (B6N > B6J). Itih4 mRNA expression was markedly down-regulated after repetitive cerulein treatment (Table 5). The precise function of the Itih4 protein is not known. Studies have shown that Itih4 can bind to hyaluronan and thereby stabilize extracellular matrix,42 and Itih4 has also been proposed as a new serum biomarker for liver fibrosis in hepatitis C patients, because it was strongly down-regulated in the blood in patients with cirrhosis.43 To validate our microarray results, we performed qPCR analysis of Itih4 mRNA expression in mouse pancreas before and after the repetitive cerulein injury (Figure 7A). The results confirmed differential expression of Itih4 mRNA between the substrains and significant down-regulation of Itih4 expression in the pancreas of mice with CP. To evaluate whether Itih4 mRNA expression changes in acute pancreatitis, we performed qPCR analysis of mouse pancreas mRNA before and after acute pancreatic injury (Figure 7B). Acute pancreatic injury did not change pancreatic expression of Itih4 mRNA.
Another potential candidate gene identified in our screen that could be essential for the development of pancreatic fibrosis was vitronectin (Vtn). We demonstrated strong down-regulation of Vtn in the pancreas of mice with CP, with more down-regulation in B6J than B6N mice. Vtn is an abundant component of extracellular matrix and promotes cell adhesion and spreading. Vtn binds to plasminogen activator inhibitor 1 (PAI-1), an important modulator of fibrotic diseases, and stabilizes its activity.44 Its role in pancreatic fibrogenesis, however, is not known. In support of our finding of striking decreases of Vtn mRNA expression in CP mouse pancreas, recent proteomic studies of human formalin-fixed, paraffin-embedded tissues from normal pancreas and pancreatic tissues from patients with CP identified Vtn as a protein expressed exclusively in normal tissues.45
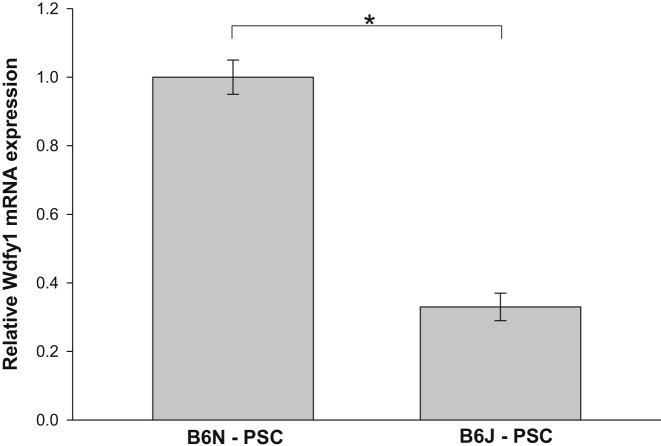
One more gene of potential importance identified in our screening is the WD repeat and FYVE domain containing 1 gene (Wdfy1) (Tables 4 and 6). Little information is available in the literature about the functions of Wdfy1, but recently Wdfy1 was identified in microarray studies as a candidate gene influencing alcohol consumption in mice.9
Expression of CP Candidate Genes in PSCs Isolated from B6N and B6J Substrains
Because the differences in the severity of CP between B6N and B6J substrains were not related to the degree of acute injury, but rather from subsequent processes, and that PSCs are the main cell type responsible for fibrogenesis in the pancreas, substrain-specific differential gene expression in PSCs could be responsible for the observed alterations. We therefore examined whether genes identified in the whole pancreas as potential candidates important in CP development are differentially expressed in PSCs isolated from B6N and B6J substrains. To obtain enough cells for the RT-qPCR mRNA analysis, cells were propagated and activated by culturing on plastic culture plates. mRNA expression of Mmp7, Pcolce2, Itih4, Vtn, Wdfy1, C4b, and Ccl8 were measured in PSCs isolated from B6N and B6J mice. Expression of Itih4 and Mmp7 was essentially undetectable in PSCs by qPCR. The other genes were all expressed at similar levels in the two substrains (data not shown), except Wdfy1. PSCs isolated from B6N expressed threefold more Wdfy1 mRNA, compared with PSCs isolated from B6J (Figure 10), which is similar to the differential expression found in the whole-pancreas microarray data. We also assessed mRNA expression of vimentin and α-SMA as markers of PSC identity and activation, respectively, and found no difference in the expression of these genes between B6N and B6J (data not shown).
Figure 10.
Wdfy1 mRNA expression in PSCs isolated from B6N and B6J substrains. Wdfy1 mRNA expression was normalized to Rplp0 and expressed as fold change over the level in PSCs isolated from B6N mice. Data are expressed as means (n = 3) with 95% confidence intervals. Confidence intervals were calculated using the ΔCT values before exponential transformation to fold increase in mRNA and are therefore asymmetric about the means. ∗P < 0.05.
Discussion
CP is a devastating disease, and the mechanisms involved in CP development are actively being studied. A well-developed model of CP recapitulating human disease is the use of repetitive cerulein injection in mice. The C57BL/6 mouse is the most common inbred mouse strain used in biomedical research, and it is a preferred strain for the propagation of knockout and transgenic mice. The wide demand for this strain has led to the creation and maintenance of multiple separate colonies of C57BL/6 in different facilities for many decades, resulting in substrains of C57BL/6. These substrains are often used interchangeably in experiments, on the assumption that they are genetically identical. Recently, however, data have begun to accumulate that important differences exist in the phenotypes and genotypes of these very similar substrains. Some of the reported phenotypic differences between C57BL/6 substrains include variations in behavior,8 alcohol consumption and alcohol preference,9 glucose homeostasis,12 and response to liver injury.7 Genetic differences among substrains include genomic copy number variations,10 single-nucleotide polymorphisms,11,46 and spontaneous deletions and mutations.12,13,47
Here, we have reported striking differences in the course of cerulein-induced CP between the B6J substrain from the Jackson Laboratory and the B6N substrain from Harlan Laboratories. C57BL/6 mouse colonies have been bred in these separate facilities for more than six decades. Features of repetitive cerulein-induced CP such as pancreatic atrophy, inflammatory cell infiltration, and fibrosis were significantly more severe in B6J than in B6N mice (Figures 1, 2, and 3), whereas the response to acute injury was similar between the substrains (Figure 4). This observation led us to test specific hypotheses and obtain a large body of data to explore additional genotype–phenotype associations. These important findings are also relevant to pancreatitis research and to investigations in other fields, because most investigators treat different C57BL/6 substrains as identical, often reducing the identifier simply to C57, B6, or C57 black. If transgenic or knockout mice used in studies are generated on a C57BL/6 background and control mice on a C57BL/6 substrain, then the experimental results could be misleading. It is also critically important to identify the exact genetic background of genetically engineered mice; some mice may have mixed backgrounds, because C57BL/6 mice obtained from different commercial sources sometimes coexist in animal core facilities.
Although the presence of genotypic and phenotypic differences between substrains may, if ignored, lead to misinterpretation of experimental results, the divergence among genetically closely related mouse strains or substrains also presents an opportunity to identify genes that may contribute to disease development and progression.9,48 In the present study, we examined the potential role of NNT in chronic and acute pancreatitis, because B6J mice lack functional NNT12,13 and because NNT may play an important role in preventing mitochondrial oxidative stress.13 Because mitochondrial oxidative stress plays a pathological role in pancreatitis,16,17 we hypothesized that B6J mice have a more severe CP phenotype because of the absence of NNT. However, we did not find any differences in the parameters of CP between transgenic Nnt+/+ B6J mice and the control mice lacking NNT. Because the level of expression of NNT in the mitochondria of transgenic mice was similar to that in B6N mice (Figure 5), and because transgenic NNT had been previously demonstrated to be a functional protein,23 we concluded that the loss of NNT is not the sole factor underlying the observed phenotypic differences between B6J and B6N mice. However, our experiment cannot exclude the possibility of more complex gene–gene interactions, in which loss of NNT and other differences in gene expression between the two substrains combine to cause the more severe phenotype in the B6J substrain. Interestingly, two substrains of C57BL/6 mice, one with an Nnt mutation and the other without, were recently compared in two models of liver injury; the authors reported that, to their surprise, the substrain with functional NNT expression had more liver injury.7 Further work is clearly needed to better understand the functional consequences of the loss of NNT in B6J mice.
To identify genes associated with cerulein-induced CP and to understand the genetic factors responsible for observed differences in the degree of CP between B6J and B6N substrains, we performed a microarray gene expression analysis of pancreatic mRNAs from B6J and B6N substrains with and without CP using a whole mouse genome Affymetrix chip. Comparison of pancreatic mRNAs of B6J and B6N substrains treated only with saline revealed that, although most genes were expressed at similar levels between the two substrains, several genes in addition to Nnt were differentially expressed (Table 4). Corroborating our findings, two genes with differential expression between the substrains in normal pancreas in our microarray analysis (Entpd4 and Wdfy1) were also found in another study to be differentially regulated in the brain in B6J and C57BL/6NCrl (Charles River Laboratories, Wilmington, MA) substrains.9
Induction of CP with repeated cerulein treatment caused significant differential regulation (∼7.7%) of a large number of genes in both substrains. Our pancreatic gene expression profile associated with CP in the mouse can serve as a valuable data set for understanding some of the contributing factors to CP development and progression. The most highly differentially regulated genes are listed in Table 5. An interesting result was the finding that mouse T5 (1810009J06Rik) is strongly up-regulated in CP. This mouse trypsinogen isoform is expressed at very low levels in normal pancreas, but it was increased almost 1000-fold after induction of chronic as well as acute pancreatitis (Figure 8). We identified T5 as the mouse ortholog of human mesotrypsinogen and of the P23 protein in rats. Importantly, mesotrypsin and P23 are trypsins that are resistant to naturally occurring trypsin inhibitors.49 The identification of a new trypsin-resistant form of trypsinogen in the mouse during pancreatic injury could be important, because an imbalance of pancreatic proteases, and in particular the trypsin isoforms and their specific inhibitors, is thought to play a central role in the pathogenesis of both chronic and acute pancreatitis.1 Patients with acute pancreatitis were recently found to have significantly increased serum levels of mesotrypsinogen.50 The mRNA for T5 was also reported by Hayashi et al35 to be up-regulated approximately 1000-fold in mouse pancreas lacking interferon regulatory factor 2 (Irf2). Interestingly, Irf2−/− mice were reported to have a defect in pancreatic endocytosis,51 and treatment of these mice with synthetic double-stranded DNA caused severe acute pancreatitis and death within 1 week of treatment. Further studies are needed to elucidate the role of T5 in chronic and acute pancreatitis.
Another important group of genes involved in the mechanism of CP encode chemokines. Chemokines play an important role in the inflammatory response to tissue injury, and changes in serum chemokine levels could also be useful as diagnostic markers in early detection of CP.52 A number of chemokines were up-regulated in the microarray; among these, two members of chemokine (C-C motif) family of ligands, Ccl8 and Ccl6, were highly up-regulated (up to 60-fold in B6J mice) (Table 4). Moreover, Ccl8 was up-regulated to a greater extent in the pancreas of B6J mice than in that of B6N mice, indicating the possibility of its involvement in the mechanism of the more severe CP phenotype of B6J substrain mice (Figure 9A). Acute injury did not cause up-regulation of Ccl8 mRNA in B6N; in B6J, it caused slight, approximately twofold, up-regulation. Ccl8 and Ccl6 are not well characterized functionally in the mouse and have not previously been associated with CP.
Because the severity of CP differed between B6J and B6N but there were no differences in the parameters of acute injury between the substrains (Figures 3 and 4), we hypothesize that the genes that are differentially regulated in CP between the two substrains could be important in the pathogenesis of pancreatic fibrogenesis, either as factors contributing to causation or the consequence of the severity of injury. In our microarray analysis, we identified nine genes significantly differentially regulated in CP and with different expression levels between the two substrains (Table 6). In support of our findings, one gene from this list, Mmp7, was reported to be involved in the pathology of CP in a CP model caused by surgical ligation of the main pancreatic duct.40 Several other genes have been reported to participate in the fibrotic mechanisms in other organs. For example, the Pcolce2 gene product was found to be required for the efficient collagen processing and deposition of myocardial fibrillar collagen in a chronic pressure overload model.39 Pcolce2 is an enhancer of collagen synthesis, and its role in pancreatic fibrogenesis is not known. The mRNA of Pcolce2 was induced approximately fivefold after induction of CP in B6J.
Another of our nine genes, Vtn, is reported to be associated with fibrotic diseases. An abundant protein found in serum and in extracellular matrix, Vtn promotes cell adhesion and spreading by multiple mechanisms. Vtn has a high binding affinity for the potent fibrosis-promoting molecule PAI-1, and its expression was found to be closely associated with fibrosis in fibrotic liver and lung diseases.53,54 In the kidney, Vtn can both increase and attenuate fibrogenesis, depending on the model, suggesting that its net effect is regulated by local environmental factors.55 In contrast to liver fibrosis, in our repetitive cerulein model of CP the expression of Vtn mRNA was strongly down-regulated (∼6.6-fold in B6J). In support of our findings, human pancreatic tissues from CP patients were also reported to have decreased Vtn expression.45
Another interesting candidate gene identified in our microarray analysis is Itih4. Its mRNA expression is higher in normal pancreas of B6N, compared with B6J (Table 4), raising the possibility that Itih4 could play a protective role. In both substrains, pancreatic expression of Itih4 decreased dramatically after CP induction (Table 5), with B6N expression remaining higher than B6J (Table 6). Itih4 belongs to the subfamily of Kunitz-type protease inhibitors. Five members of this family (Itih1 through Itih5) have been isolated in mice, and all have human homologs. Itih1, Itih2, and Itih3 are plasma glycoproteins assembled in the liver from heavy chains encoded by separate genes and from a light chain, designated bikunin. Proteinase activity of Itih proteins resides solely in the bikunin part of the molecule.56 Itih4 does not have a site for binding with bikunin and therefore exists as a free heavy chain polypeptide. Although Itih5 has all of the necessary features for bikunin binding, the existence of the complex with bikunin has not yet been demonstrated. In our microarray analysis, Itih5 mRNA level significantly increased after induction of CP, with no alteration for the other Itih genes, indicating possibly different roles of these proteins in pancreatitis. The functions of these proteins are not clear, but they may be involved in the stabilization of extracellular matrix by binding hyaluronan.42 Itih4 was reported to be highly inducible by IL-6 in regenerating mouse livers, and it is considered to be an acute-phase protein in several animal models for liver.57 At the same time, Itih4 has been shown to be decreased in the serum of patients with hepatitis C cirrhosis43 and in multiple human solid tumors.58 The role of Itih4 in the pancreas has not been studied, although Itih4 mRNA is expressed in human pancreas at a level second only to the liver in abundance.58
Because we established that the differences in the course of CP arose during the process of fibrogenesis and not from the degree of acute injury, and given that PSCs are the primary source of pancreatic fibrosis, differences in the expression of genes in PSCs might be responsible for the observed substrain differences in CP. In the normal pancreas, PSCs comprise only approximately 4% of the total pancreatic cells; after induction of CP, however, PSCs are activated, proliferate, and acquire a myofibroblast phenotype.3 Analysis of the microarray results after induction of CP did not reveal any difference between B6N and B6J substrains in the mRNA expression of genes commonly attributed to PSC activation, such as α-SMA or the procollagens. To determine whether differentially expressed genes in the whole pancreas would be differentially expressed in PSCs isolated from B6N and B6J mice, we isolated and cultured activated PSCs from these substrains. Wdfy1 mRNA was expressed at significantly lower levels in PSCs from B6J than from B6N (Figure 10). Further research will be needed to evaluate whether changes in Wdfy1 mRNA expression lead to the changes in protein expression, morphology, proliferation, or fibrotic responses of PSCs.
In conclusion, with the present study we have identified substantial phenotypic differences in the course of cerulein-induced CP in B6J and B6N substrains of C57BL/6 mice. Although NNT is absent in B6J mice, we could not implicate its absence in causing a more severe CP phenotype. Using whole mouse genome Affymetrix microarray analysis, we identified pancreatic transcripts that are associated with the development of CP in mice. Using the B6J and B6N substrains, we have provided a data set for future studies of pancreatic fibrogenesis. Novel transcripts associated with CP identified in the present study include T5, Ccl8, and Ccl6. Finally, by comparing B6J and B6N microarrays, we identified a small subset of potential gene candidates implicated in a more severe phenotype of CP, including Mmp7, Pcolce2, Itih4, Wdfy1, and Vtn. The phenotypic substrain differences and differential gene expression demonstrated in this study also highlight the need to accurately recognize and report the substrain background of C57BL/6 mice used in research and also to avoid the substrain mispairing (likely a common error in the past) that could lead to incorrect interpretation of results.
Footnotes
Supported by the President’s Research Fund (3-07821 to B.U. and B.A.N.) and by a seed grant from the Frank R. Burton Memorial Fund (B.U.), Saint Louis University.
References
- 1.Witt H., Apte M.V., Keim V., Wilson J.S. Chronic pancreatitis: challenges and advances in pathogenesis, genetics, diagnosis, and therapy. Gastroenterology. 2007;132:1557–1573. doi: 10.1053/j.gastro.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 2.Nair R.J., Lawler L., Miller M.R. Chronic pancreatitis. Am Fam Physician. 2007;76:1679–1688. [PubMed] [Google Scholar]
- 3.Omary M.B., Lugea A., Lowe A.W., Pandol S.J. The pancreatic stellate cell: a star on the rise in pancreatic diseases. J Clin Invest. 2007;117:50–59. doi: 10.1172/JCI30082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aghdassi A.A., Mayerle J., Christochowitz S., Weiss F.U., Sendler M., Lerch M.M. Animal models for investigating chronic pancreatitis. Fibrogenesis Tissue Repair. 2011;4:26. doi: 10.1186/1755-1536-4-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuschwander-Tetri B.A., Burton F.R., Presti M.E., Britton R.S., Janney C.G., Garvin P.R., Brunt E.M., Galvin N.J., Poulos J.E. Repetitive self-limited acute pancreatitis induces pancreatic fibrogenesis in the mouse. Dig Dis Sci. 2000;45:665–674. doi: 10.1023/a:1005423122127. [DOI] [PubMed] [Google Scholar]
- 6.Neuschwander-Tetri B.A., Bridle K.R., Wells L.D., Marcu M., Ramm G.A. Repetitive acute pancreatic injury in the mouse induces procollagen alpha1(I) expression colocalized to pancreatic stellate cells. Lab Invest. 2000;80:143–150. doi: 10.1038/labinvest.3780018. [DOI] [PubMed] [Google Scholar]
- 7.Bourdi M., Davies J.S., Pohl L.R. Mispairing C57BL/6 substrains of genetically engineered mice and wild-type controls can lead to confounding results as it did in studies of JNK2 in acetaminophen and concanavalin A liver injury. Chem Res Toxicol. 2011;24:794–796. doi: 10.1021/tx200143x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bryant C.D., Zhang N.N., Sokoloff G., Fanselow M.S., Ennes H.S., Palmer A.A., McRoberts J.A. Behavioral differences among C57BL/6 substrains: implications for transgenic and knockout studies. J Neurogenet. 2008;22:315–331. doi: 10.1080/01677060802357388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mulligan M.K., Ponomarev I., Boehm S.L., 2nd, Owen J.A., Levin P.S., Berman A.E., Blednov Y.A., Crabbe J.C., Williams R.W., Miles M.F., Bergeson S.E. Alcohol trait and transcriptional genomic analysis of C57BL/6 substrains. Genes Brain Behav. 2008;7:677–689. doi: 10.1111/j.1601-183X.2008.00405.x. [DOI] [PubMed] [Google Scholar]
- 10.Watkins-Chow D.E., Pavan W.J. Genomic copy number and expression variation within the C57BL/6J inbred mouse strain. Genome Res. 2008;18:60–66. doi: 10.1101/gr.6927808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zurita E., Chagoyen M., Cantero M., Alonso R., Gonzalez-Neira A., Lopez-Jimenez A., Lopez-Moreno J.A., Landel C.P., Benitez J., Pazos F., Montoliu L. Genetic polymorphisms among C57BL/6 mouse inbred strains. Transgenic Res. 2011;20:481–489. doi: 10.1007/s11248-010-9403-8. [DOI] [PubMed] [Google Scholar]
- 12.Toye A.A., Lippiat J.D., Proks P., Shimomura K., Bentley L., Hugill A., Mijat V., Goldsworthy M., Moir L., Haynes A., Quarterman J., Freeman H.C., Ashcroft F.M., Cox R.D. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia. 2005;48:675–686. doi: 10.1007/s00125-005-1680-z. [DOI] [PubMed] [Google Scholar]
- 13.Huang T.T., Naeemuddin M., Elchuri S., Yamaguchi M., Kozy H.M., Carlson E.J., Epstein C.J. Genetic modifiers of the phenotype of mice deficient in mitochondrial superoxide dismutase. Hum Mol Genet. 2006;15:1187–1194. doi: 10.1093/hmg/ddl034. [DOI] [PubMed] [Google Scholar]
- 14.Arkblad E.L., Tuck S., Pestov N.B., Dmitriev R.I., Kostina M.B., Stenvall J., Tranberg M., Rydström J. A Caenorhabditis elegans mutant lacking functional nicotinamide nucleotide transhydrogenase displays increased sensitivity to oxidative stress. Free Radic Biol Med. 2005;38:1518–1525. doi: 10.1016/j.freeradbiomed.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 15.Freeman H., Shimomura K., Horner E., Cox R.D., Ashcroft F.M. Nicotinamide nucleotide transhydrogenase: a key role in insulin secretion. Cell Metab. 2006;3:35–45. doi: 10.1016/j.cmet.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 16.Mukherjee R., Criddle D.N., Gukovskaya A., Pandol S., Petersen O.H., Sutton R. Mitochondrial injury in pancreatitis. Cell Calcium. 2008;44:14–23. doi: 10.1016/j.ceca.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Odinokova I.V., Sung K.F., Mareninova O.A., Hermann K., Gukovsky I., Gukovskaya A.S. Mitochondrial mechanisms of death responses in pancreatitis. J Gastroenterol Hepatol. 2008;23(Suppl 1):S25–S30. doi: 10.1111/j.1440-1746.2007.05271.x. [DOI] [PubMed] [Google Scholar]
- 18.Ulmasov B., Xu Z., Talkad V., Oshima K., Neuschwander-Tetri B.A. Angiotensin II signaling through the AT1a and AT1b receptors does not have a role in the development of cerulein-induced chronic pancreatitis in the mouse. Am J Physiol Gastrointest Liver Physiol. 2010;299:G70–G80. doi: 10.1152/ajpgi.00006.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulmasov B., Xu Z., Tetri L.H., Inagami T., Neuschwander-Tetri B.A. Protective role of angiotensin II type 2 receptor signaling in a mouse model of pancreatic fibrosis. Am J Physiol Gastrointest Liver Physiol. 2009;296:G284–G294. doi: 10.1152/ajpgi.90409.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.French S.W., Miyamoto K., Wong K., Jui L., Briere L. Role of the Ito cell in liver parenchymal fibrosis in rats fed alcohol and a high fat-low protein diet. Am J Pathol. 1988;132:73–85. [PMC free article] [PubMed] [Google Scholar]
- 21.Mareninova O.A., Sung K.F., Hong P., Lugea A., Pandol S.J., Gukovsky I., Gukovskaya A.S. Cell death in pancreatitis: caspases protect from necrotizing pancreatitis. J Biol Chem. 2006;281:3370–3381. doi: 10.1074/jbc.M511276200. [DOI] [PubMed] [Google Scholar]
- 22.Bai H., Chen X., Zhang L., Dou X. The effect of sulindac, a non-steroidal anti-inflammatory drug, attenuates inflammation and fibrosis in a mouse model of chronic pancreatitis. BMC Gastroenterol. 2012;12:115. doi: 10.1186/1471-230X-12-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Freeman H.C., Hugill A., Dear N.T., Ashcroft F.M., Cox R.D. Deletion of nicotinamide nucleotide transhydrogenase: a new quantitive [sic] trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes. 2006;55:2153–2156. doi: 10.2337/db06-0358. [DOI] [PubMed] [Google Scholar]
- 24.Wang X., Seed B. A PCR primer bank for quantitative gene expression analysis. Nucleic Acids Res. 2003;31:e154. doi: 10.1093/nar/gng154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lugea A., Gukovsky I., Gukovskaya A.S., Pandol S.J. Nonoxidative ethanol metabolites alter extracellular matrix protein content in rat pancreas. Gastroenterology. 2003;125:1845–1859. doi: 10.1053/j.gastro.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 26.Gukovsky I., Gukovskaya A.S., Blinman T.A., Zaninovic V., Pandol S.J. Early NF-kappaB activation is associated with hormone-induced pancreatitis. Am J Physiol. 1998;275:G1402–G1414. doi: 10.1152/ajpgi.1998.275.6.G1402. [DOI] [PubMed] [Google Scholar]
- 27.Perides G., Tao X., West N., Sharma A., Steer M.L. A mouse model of ethanol dependent pancreatic fibrosis. Gut. 2005;54:1461–1467. doi: 10.1136/gut.2004.062919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 30.Edgar R., Domrachev M., Lash A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan J.S., Reed A., Chen F., Stewart C.N., Jr. Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimomura K., Galvanovskis J., Goldsworthy M., Hugill A., Kaizak S., Lee A., Meadows N., Quwailid M.M., Rydstrom J., Teboul L., Ashcroft F., Cox R.D. Insulin secretion from beta-cells is affected by deletion of nicotinamide nucleotide transhydrogenase. Methods Enzymol. 2009;457:451–480. doi: 10.1016/S0076-6879(09)05025-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Binkley C.E., Zhang L., Greenson J.K., Giordano T.J., Kuick R., Misek D., Hanash S., Logsdon C.D., Simeone D.M. The molecular basis of pancreatic fibrosis: common stromal gene expression in chronic pancreatitis and pancreatic adenocarcinoma. Pancreas. 2004;29:254–263. doi: 10.1097/00006676-200411000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Spicuglia S., Pekowska A., Zacarias-Cabeza J., Ferrier P. Epigenetic control of Tcrb gene rearrangement. Semin Immunol. 2010;22:330–336. doi: 10.1016/j.smim.2010.07.002. [DOI] [PubMed] [Google Scholar]
- 35.Hayashi H., Kohno T., Yasui K., Murota H., Kimura T., Duncan G.S., Nakashima T., Yamamoto K., Katayama I., Ma Y., Chua K.J., Suematsu T., Shimokawa I., Akira S., Kubo Y., Mak T.W., Matsuyama T. Characterization of dsRNA-induced pancreatitis model reveals the regulatory role of IFN regulatory factor 2 (Irf2) in trypsinogen5 gene transcription. Proc Natl Acad Sci USA. 2011;108:18766–18771. doi: 10.1073/pnas.1116273108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dawra R., Sah R.P., Dudeja V., Rishi L., Talukdar R., Garg P., Saluja A.K. Intra-acinar trypsinogen activation mediates early stages of pancreatic injury but not inflammation in mice with acute pancreatitis. Gastroenterology. 2011;141:2210–2217.e2. doi: 10.1053/j.gastro.2011.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kessler E., Takahara K., Biniaminov L., Brusel M., Greenspan D.S. Bone morphogenetic protein-1: the type I procollagen C-proteinase. Science. 1996;271:360–362. doi: 10.1126/science.271.5247.360. [DOI] [PubMed] [Google Scholar]
- 38.Steiglitz B.M., Keene D.R., Greenspan D.S. PCOLCE2 encodes a functional procollagen C-proteinase enhancer (PCPE2) that is a collagen-binding protein differing in distribution of expression and post-translational modification from the previously described PCPE1. J Biol Chem. 2002;277:49820–49830. doi: 10.1074/jbc.M209891200. [DOI] [PubMed] [Google Scholar]
- 39.Baicu C.F., Zhang Y., Van Laer A.O., Renaud L., Zile M.R., Bradshaw A.D. Effects of the absence of procollagen C-endopeptidase enhancer-2 on myocardial collagen accumulation in chronic pressure overload. Am J Physiol Heart Circ Physiol. 2012;303:H234–H240. doi: 10.1152/ajpheart.00227.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Crawford H.C., Scoggins C.R., Washington M.K., Matrisian L.M., Leach S.D. Matrix metalloproteinase-7 is expressed by pancreatic cancer precursors and regulates acinar-to-ductal metaplasia in exocrine pancreas. J Clin Invest. 2002;109:1437–1444. doi: 10.1172/JCI15051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fukuda A., Wang S.C., Morris J.P., 4th, Folias A.E., Liou A., Kim G.E., Akira S., Boucher K.M., Firpo M.A., Mulvihill S.J., Hebrok M. Stat3 and MMP7 contribute to pancreatic ductal adenocarcinoma initiation and progression. Cancer Cell. 2011;19:441–455. doi: 10.1016/j.ccr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhuo L., Hascall V.C., Kimata K. Inter-alpha-trypsin inhibitor, a covalent protein-glycosaminoglycan-protein complex. J Biol Chem. 2004;279:38079–38082. doi: 10.1074/jbc.R300039200. [DOI] [PubMed] [Google Scholar]
- 43.Gangadharan B., Antrobus R., Dwek R.A., Zitzmann N. Novel serum biomarker candidates for liver fibrosis in hepatitis C patients. Clin Chem. 2007;53:1792–1799. doi: 10.1373/clinchem.2007.089144. [DOI] [PubMed] [Google Scholar]
- 44.Hertig A., Rondeau E. Plasminogen activator inhibitor type 1: the two faces of the same coin. Curr Opin Nephrol Hypertens. 2004;13:39–44. doi: 10.1097/00041552-200401000-00006. [DOI] [PubMed] [Google Scholar]
- 45.Paulo J.A., Lee L.S., Banks P.A., Steen H., Conwell D.L. Proteomic analysis of formalin-fixed paraffin-embedded pancreatic tissue using liquid chromatography tandem mass spectrometry. Pancreas. 2012;41:175–185. doi: 10.1097/MPA.0b013e318227a6b7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mekada K., Abe K., Murakami A., Nakamura S., Nakata H., Moriwaki K., Obata Y., Yoshiki A. Genetic differences among C57BL/6 substrains. Exp Anim. 2009;58:141–149. doi: 10.1538/expanim.58.141. [DOI] [PubMed] [Google Scholar]
- 47.Specht C.G., Schoepfer R. Deletion of the alpha-synuclein locus in a subpopulation of C57BL/6J inbred mice. BMC Neurosci. 2001;2:11. doi: 10.1186/1471-2202-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grottick A.J., Bagnol D., Phillips S., McDonald J., Behan D.P., Chalmers D.T., Hakak Y. Neurotransmission- and cellular stress-related gene expression associated with prepulse inhibition in mice. Brain Res Mol Brain Res. 2005;139:153–162. doi: 10.1016/j.molbrainres.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 49.Szmola R., Kukor Z., Sahin-Tóth M. Human mesotrypsin is a unique digestive protease specialized for the degradation of trypsin inhibitors. J Biol Chem. 2003;278:48580–48589. doi: 10.1074/jbc.M310301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Oiva J., Itkonen O., Koistinen R., Hotakainen K., Zhang W.M., Kemppainen E., Puolakkainen P., Kylänpää L., Stenman U.H., Koistinen H. Specific immunoassay reveals increased serum trypsinogen 3 in acute pancreatitis. Clin Chem. 2011;57:1506–1513. doi: 10.1373/clinchem.2011.167965. [DOI] [PubMed] [Google Scholar]
- 51.Mashima H., Sato T., Horie Y., Nakagawa Y., Kojima I., Ohteki T., Ohnishi H. Interferon regulatory factor-2 regulates exocytosis mechanisms mediated by SNAREs in pancreatic acinar cells. Gastroenterology. 2011;141:1102–1113.e1–e8. doi: 10.1053/j.gastro.2011.05.051. [DOI] [PubMed] [Google Scholar]
- 52.Ito T. Can measurement of chemokines become useful biological and functional markers of early-stage chronic pancreatitis? J Gastroenterol. 2007;42(Suppl 17):72–77. doi: 10.1007/s00535-006-1929-4. [DOI] [PubMed] [Google Scholar]
- 53.Eklund A.G., Sigurdardottir O., Ohrn M. Vitronectin and its relationship to other extracellular matrix components in bronchoalveolar lavage fluid in sarcoidosis. Am Rev Respir Dis. 1992;145:646–650. doi: 10.1164/ajrccm/145.3.646. [DOI] [PubMed] [Google Scholar]
- 54.Koukoulis G.K., Shen J., Virtanen I., Gould V.E. Vitronectin in the cirrhotic liver: an immunomarker of mature fibrosis. Hum Pathol. 2001;32:1356–1362. doi: 10.1053/hupa.2001.29675. [DOI] [PubMed] [Google Scholar]
- 55.López-Guisa J.M., Rassa A.C., Cai X., Collins S.J., Eddy A.A. Vitronectin accumulates in the interstitium but minimally impacts fibrogenesis in experimental chronic kidney disease. Am J Physiol Renal Physiol. 2011;300:F1244–F1254. doi: 10.1152/ajprenal.00701.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bost F., Diarra-Mehrpour M., Martin J.P. Inter-alpha-trypsin inhibitor proteoglycan family–a group of proteins binding and stabilizing the extracellular matrix. Eur J Biochem. 1998;252:339–346. doi: 10.1046/j.1432-1327.1998.2520339.x. [DOI] [PubMed] [Google Scholar]
- 57.Bhanumathy C.D., Tang Y., Monga S.P., Katuri V., Cox J.A., Mishra B., Mishra L. Itih-4, a serine protease inhibitor regulated in interleukin-6-dependent liver formation: role in liver development and regeneration. Dev Dyn. 2002;223:59–69. doi: 10.1002/dvdy.1235. [DOI] [PubMed] [Google Scholar]
- 58.Hamm A., Veeck J., Bektas N., Wild P.J., Hartmann A., Heindrichs U., Kristiansen G., Werbowetski-Ogilvie T., Del Maestro R., Knuechel R., Dahl E. Frequent expression loss of Inter-alpha-trypsin inhibitor heavy chain (ITIH) genes in multiple human solid tumors: a systematic expression analysis. BMC Cancer. 2008;8:25. doi: 10.1186/1471-2407-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]