Abstract
Exactly controlled conditional gene expressing systems are crucial for genomic functional research, animal transgenesis and gene therapy. Bacterial artificial chromosomes (BACs) are optimal for harboring long fragments of genomic DNA or large cDNA up to 300 kb in size. Therefore, BACs are available to produce transgenic cells and animals for the functional studies of genes. However, BAC can insert DNA randomly into the host genome, possibly causing unpredicted expression. We previously developed a human artificial chromosome (HAC) vector from human chromosome 21 using chromosome engineering. The HAC vector has several important characteristics desired for an ideal gene delivery vector, including stable episomal maintenance, and the ability to carry large genomic DNA containing its own regulatory element, thus allowing physiological regulation of the transgene in a manner similar to that of the native chromosome. In this study, we develop a system fusing BAC library and HAC technology together to allow tight control of gene expression. This system enables BAC to be cloned into the defined locus on the HAC vector by the Cre/loxP system. In addition, the genome in the BAC is possible to be engineered freely by the BAC recombineering technology. This system is a highly efficient tool for the rapid generation of stringently controlled gene expression system on the HAC vector.
Keywords: bacterial artificial chromosome, bacterial artificial chromosome recombineering, human artificial chromosome
The development of genomic functional studies, transgenic animal production and gene therapy requires stringently controlled gene expression systems. Bacterial artificial chromosomes (BACs) have become important tools in functional genomics (Brune et al., 2000; Heintz,2001; Sparwasser and Eberl, 2007; Tunster et al., 2011). BACs have made genomic fragments from humans, mice and many other species available through public genomic databases (O’Connor et al., 1989; Shizuya et al., 1992; Osoegawa et al., 2000). pBACe3.6 is used as a backbone vector in the majority of currently available BAC libraries (Frengen et al., 1999). Specific BACs are readily obtainable from several bioresource centers (e. g., Children’s Hospital Oakland Research Institute). As a result, the tedious screening for genomic DNA clones in complex libraries is reduced. Therefore, BACs have greater convenience in handing over other large cloning vectors like P1 phage-derived artificial chromosomes (PACs) and yeast artificial chromosomes. BACs cover average 100- to 300-kb genomic DNA and so most mammalian genes can be encompassed by a single BAC. Given their large size, BACs encode the complete open reading frame and most regulatory sequences, which recapitulate the regulation of endogenous genes much better than shorter transgenes (Yang et al., 1997). In addition, a homologus recombination system was developed that requires only short homology arms and enables the full range of BAC modifications to be quickly and easily engineered in Escherichia coli (E. coli) (Zhang et al., 1998; Muyrers et al., 1999). This BAC engineering methodology is now termed “BAC recombineering” (Yu et al., 2000; Copeland et al., 2001) and includes a broad range of applications including subcloning via gap repair (Zhang et al., 2000), point mutagenesis in BACs (Muyrers et al., 2000), high throughput DNA engineering (Sarov et al., 2006; Poser et al., 2008) and a variety of other, often complex, applications. Furthormore, BAC transgenic mice expressing a variety of reporters (Gong et al., 2003; Lobo et al., 2006) or epitope tagged-proteins (Bateup et al., 2008) driven by specific promoters become widely used in various in vivo functional applications. Because of these advantages, BACs have been used for transgenesis in higher model cells and organisms. To produce a target BAC transgene expression cells, random integration of foreign DNA is frequently used. However, the expression of BAC transgene can not only be affected by positional effects at the insertion site of the host genome but also by the number of copies inserted (Gong et al., 2003; Alexander et al., 2004; Heaney and Bronson, 2006; Chandler et al., 2007; Le et al., 2010). As a result, the expression level of genes varies greatly in the obtained clones.
Human artificial chromosomes (HACs) have several advantages over conventional gene delivery systems (Larin and Mejia, 2002; Basu and Willard, 2006). We recently developed HAC vectors from human chromosome 21 by deleting all of the endogenous genes (Kazuki et al., in press). Like natural chromosomes, the HAC vector has the capacity to replicate and segregate freely from the host genome. Therefore, the HAC vector can avoid the disruption of the host genome by transgene insertion (Pravtcheva and Wise, 1995). In addition, as reported previously, the HAC vector enables stable expression of the inserted gene, and is not affected by position in the genome (Katoh et al., 2004; Kakeda et al., 2005; Kazuki et al., in press). The HAC vector is a powerful gene delivery vector capable of carrying large genes such as the dystrophin gene (2.4 Mb) (Oshimura and Katoh, 2008; Hoshiya et al., 2009; Kazuki et al., 2010). Conceptually and experimentally, any circular DNA such as BACs can be inserted into the HAC vector. Therefore, we considered that a more precise evaluation of large DNA inserts for genetic complementation should be possible by incorporating the BAC system into the HAC vector. Site-specific insertion of BAC into the HAC vector may eliminate unwanted position effects caused by the random integration of exogenously introduced DNA. In this study, we describe the generation of modified BAC by the BAC recombineering for their introduction into the defined locus on the HAC vector by the Cre/loxP system. Also, the genome of interest in the BAC is possible to be engineered by the BAC recombineering.
Materials and Methods
Vector construction
The modification cassette vector I for engineering of the genome on the BAC was constructed as follows. RP11-67L14 (cloned in the pBACe3.6 vector, GenBank accession number: AC079753) was obtained from Invitrogen (Carlsbad, CA) and used as a model for BAC modification. As a gene of interest, the coding sequence of human interleukin (IL)-1β was contained in the BAC. Fragment of the genome specific homology arms to target the cassette into the start codon of the genome in the BAC was amplified by blunt PCR from RP11-67L14 using the primers IL-1β hm F and IL-1β hm R. This PCR product was cloned into the pSTblue1 blunt vector (Novagen, Madison, WI), creating pIL-1β hm vector. There was an internal NcoI restriction site within the IL-1β insert which is the ATG start-site of the first exon of the IL-1β genome. The reporter gene of interest (stable luciferase green; SLG) and an ampicillin-resistance (AmpR) gene were amplified by PCR from the pSLG test (Toyobo, Osaka, Japan) using the primers SLG-Amp F and SLG-Amp R. The PCR product was digested with NcoI and cloned into the equivalent site of pIL-1β hm vector, creating the modification cassette vector I. Sequence and orientation of the genome specific homology arms, SLG gene and an AmpR gene were confirmed by DNA sequencing and restriction enzyme pattern analysis.
The modification cassette vector II was constructed using the Multisite Gateway kit (Invitrogen) as previously described (Sasaki et al., 2004; Sone et al., 2008). The PCR fragments for loxP-3' HPRT, a zeocin-resistance (ZeoR) gene and G3PDH promoter flanked by the appropriate gateway attB sites were created. V901-3'HPRT-loxP (Kazuki et al., in press) was used as a template for loxP-3' HPRT amplification with primer pair B5- loxP-3' HPRT-B4 F and B5- loxP-3' HPRT-B4 R. The ZeoR gene was amplified from plasmid containing a ZeoR gene using the primers B4r-zeocin-B3r F and B4r-zeocin-B3r R. Human G3PDH promoter (GenBank accession number: AY340484 region 473–1974 bp) was amplified with primer pair B3-G3PDH-B2 F and B3-G3PDH-B2 R. The gateway attB-flanked PCR product was recombined with a donor vector containing the corresponding gateway attP signals in a BP reaction to generate an entry clone. Approximately 150 ng of PCR fragment and a donor vector (pDONR 221 P5-P4, pDONR 221 P4r-P3r or pDONR 221 P3-P2) were mixed with 2 μL of BP clonase II enzyme mix, and adjusted to 10 μL with TE buffer. The mixture was then incubated at 25°C for 1 h to create 3 entry clones (pENTR L5-loxP-3' HPRT-L4, pENTR R4-zeocin-R3, pENTR L3-G3PDH promoter-L2). After the BP reaction, the enzymes were inactivated by treatment with proteinase K for 10 min at 37°C. Competent Mach1 T1RE. coli (Invitrogen) was used according to the supplier's instruction. After transformation, the cell solution was diluted with super optimal broth with catabolite regression (SOC) medium and incubated at 37°C for 1 h. The transformation reactions were then spread onto lysogeny broth (LB) agar plates containing 50 μg/mL kanamycin and incubated for 14 h at 37°C. The sequence of these pENTRs was confirmed by DNA sequencing. The destination vector (pSLR-BAC hm DEST) contained the R1-ccdB-CmR-R2 cassette (Invitrogen), homology arms of BAC and a stable luciferase red (SLR) gene. pBACe3.6 (GenBank accession number: U80929) is used as a backbone vector for human RPCI-11 BAC library. Regions 4205–4793 bp and 4946–5833 bp of pBACe3.6 were used as BAC homology arms and cloned into the pGEM-T vector (Promega, Madison, WI), creating pBAC hm T vector. This vector contained KpnI and XbaI cleavage sites in exchange of original loxP site located in the pBACe3.6 region 4831–4864 bp. The R1-ccdB-CmR-R2 cassette was ligated into the pSLR test (Toyobo), creating pSLR DEST. To construct the destination vector containing a toxic ccdB gene, ccdB survival competent cells (Invitrogen) were used for propagation according to the supplier's instruction. After transformation, the cell solution was diluted with SOC medium and incubated at 37°C for 1 h. The transformation reactions were then spread onto LB agar plates containing 12.5 μg/mL chloramphenicol and incubated for 18 h at 37°C. The R1-ccdB-CmR-R2 cassette and the SLR gene were digested from pSLR DEST with KpnI and SacI (blunted). This fragment was cloned into the KpnI and XbaI (blunted) sites of pBAC hm T vector, creating a pSLR-BAC hm DEST. Sequence and orientation of this vector was confirmed by DNA sequencing and restriction enzyme pattern analysis. Three entry clones and pENTR L1-pLac-lacZ-R5 (Invitrogen) were recombined with a pSLR-BAC hm DEST to generate the modification cassette vector II. Before this reaction, the pSLR-BAC hm DEST was linearized at the BssHII site, between a ccdB gene and a chloramphenicol-resistance (CmR) gene. Around 20 ng of each entry clone and 80 ng of destination vector were mixed with 2 μL of LR clonase II Plus enzyme mix, and made up to 10 μL with TE buffer. The mixture was then incubated at 25°C for 16 h. After this reaction, the enzyme was inactivated by treatment with proteinase K for 10 min at 37°C. Competent Mach1 T1RE. coli was used for transformation. Following transformation, the cell solution was diluted with SOC medium and incubated at 37°C for 1 h. The transformation reactions were then spread onto LB agar plates containing 50 μg/mL ampicillin and 25 μg/mL zeocin. These plates were incubated for 14 h at 37°C. The modification cassette vector II carried the homology arms of pBACe3.6 and the non-functional loxP-3' HPRT for targeting the HAC vector. Cre recombinase-catalyzed integration required a functional HPRT gene.
BAC recombineering
A homologous recombination-proficient E. coli strain (DY380) was used for the BAC recombineering. For BAC modification, overnight cultures containing the BAC were grown from single colonies, diluted 10-fold in LB medium, and grown to an optical density at 600 nm of 0.4 to 0.6 at 32°C. Fifty-milliliter cultures were then induced for the expression of recombineering factors by shifting the cells to 42°C for 15 min followed by chilling on ice for 10 min. Cells were then centrifuged for 5 min at 5500 × g at 4°C and washed with 10 mL of ice-cold 1 mM HEPES 2 times. Cells were then resuspended in 100 μL of ice-cold 1 mM HEPES and electroporated. Cell transformation was performed by electroporation of 1 μg linear DNA into 100 μL of ice-cold competent cells in cuvettes (0.1 cm) using a Bio-Rad gene pulser set at 1.75 kV, 25 μF with a pulse controller set at 200 ohms. One milliliter of SOC medium was added after electroporation. Cells were incubated at 32°C for 1 h with shaking and spread on appropriate selective agar media.
PCR analyses
PCR analyses were carried out using standard techniques. The primer pairs used for confirmation of the IL-1β seque nce were IL-1β 1S/IL-1β 3000 AS, IL-1β 2611 S/IL-1β 5180 AS, IL-1β 4031 S/IL-1β 7151 AS and IL-1β 8051 S/IL-1β 11332 AS. The primer pairs used for confirmation of BAC recombination of the modification cassette I were amp2500 S and IL-1β 12100 AS, along with IL-1β 10930 S and slg200 AS.
The primer pairs utilized for detection of BAC recombination of the modification cassette II were SLR 1650 S and BACe3.6 5839 AS, along with BACe3.6 4168 S and LacZ 770 AS.
Results
Construction of the modification cassette vectors
BACs contain all of the endogenous control elements in their natural expression of the gene of interest and therefore almost always recapitulate all of the endogenous control mechanisms, including alternative splicing. BAC libraries based on pBACe3.6 have now been generated from various species (Table 1) and used for reporter assay of gene expression. The modification cassette vector I was created for engineering of the genome of interest on the BAC. This vector consists of 3 regions; i) the region encoding the reporter gene such as a fluorescent protein gene and a luciferase gene (SLG gene in our study), ii) AmpR gene and iii) homology arms (A' and B') to target the cassette into the start codon of chosen genome on the BAC (Fig. 1a). The homology arms were obtained by PCR from a BAC containing the target genome. In our current study, we used a BAC clone RP11-67L14 as a recipient for BAC modification. As a gene of interest, the coding sequence of IL-1β was contained in the BAC. The primers were designed to target precisely the cassette downstream of the IL-1β start codon (Table 2).
Table 1. A list of BAC libraries constructed in the pBACe3.6 .
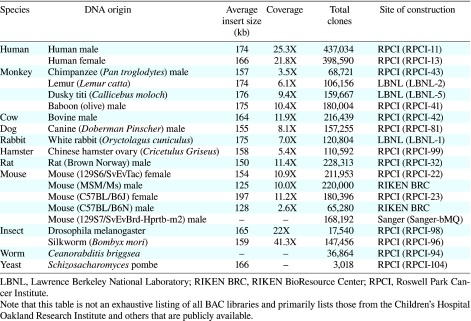
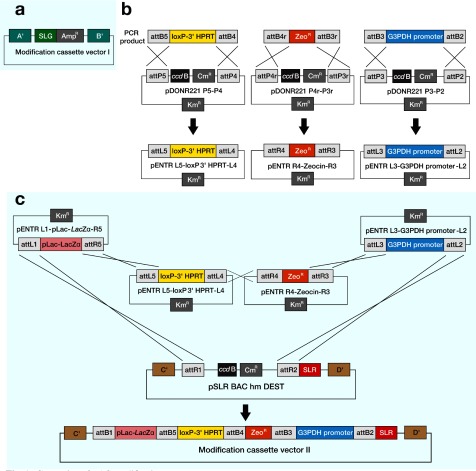
Fig. 1.
Generation of BAC modification cassette vectors.
a:The structure of the modification cassette vector I for engineering of the genome on the BAC. Features of the modification cassette vector I include genome homology arms (A' and B') that are identical to sequences A and B in the target genome for homologous recombination, and a modification cassette (SLG-AmpR) that carries the reporter gene to be inserted into the genome.
b:Construction of entry clones (pENTRs) from gateway attB-flanked PCR products and donor vectors (pDONRs).
c:Construction of the modification cassette vector II from 4 entry clones and a destination vector (pSLR-BAC hm DEST). Features of the modification cassette vector II include: pBACe3.6 homology arms (C' and D') that are identical to sequences C and D in the pBACe3.6, and the non-functional loxP-3' HPRT for targeting the HAC vector.
AmpR, ampicillin-resistance
BAC, bacterial artificial chromosome
CmR, chloramphenicol-resistance
HAC, human artificial chromosome
KmR, kanamycin-resistance
IL, interleukin
SLG, stable luciferase green
SLR, stable luciferase red
ZeoR, zeocin-resistance
Table 2. The PCR primers used in this study.

In attempts to clone BAC into the HAC vector, the modification cassette vector II was constructed. This vector carried a loxP sequence and the 3' hypoxanthine phosphoribosyl transferase (HPRT) sequence. The HAC vector used was 21HAC1 containing the 5' HPRT-loxP site (Kazuki et al., in press). The HPRT gene expressed in the HAC vector conferred HAT-resistance after site-specific recombination with the Cre/loxP system. The modification cassette vector II consists of 5 regions; i) loxP-3' HPRT region for introducing BAC into the HAC vector, ii) ZeoR gene, iii) lacZα gene which contributed for selection in E. coli, iv) G3PDH promoter-SLR region which is used as a marker in the mammalian cells because of its convenience for luciferase expression and v) homology arms (C' and D') to target the cassette into the pBACe3.6 region. The homology arms are obtained by PCR from pBACe3.6 (Table 2). The modification cassette vector II was constructed in E. coli using the Multiple Gateway system, which is useful in high-throughput construction of plasmids carrying multiple DNA sequences (Sasaki et al., 2004; Sone et al., 2008). The PCR fragments for loxP-3'HPRT, ZeoR gene and G3PDH promoter contained the appropriate gateway attB sites (Table 2). The 3 fragments were recombined into 3 different donor vectors (pDONR221 P5-P4, pDONR221 P4r-P3r, pDONR221 P3-P2) to create 3 entry clones (pENTR L5- loxP-3'HPRT-L4, pENTR R4-zeocin-R3, pENTR L3-G3PDH promoter-L2) (Fig 1b). DNA sequencing showed that 95.0% of kanamycin-resistant E. coli transformant colonies were correctly targeted. These 3 pENTR vectors, a pENTR L1-pLac-lacZα-R5 and a pSLR BAC hm DEST were mixed in the presence of LR clonase enzyme to generate the modification cassette vector II (Fig. 1c). A recombination event would incorporate the lacZα gene and the ZeoR gene, resulting in a blue colony which resistances to ampicillin and zeocin. Unrecombined plasmid would result in a white colony. As a result, almost all colonies were blue and had the AmpR and ZeoR. Twenty-four of the resulting blue colonies were analyzed by restriction enzyme pattern analysis, and 10 of these colonies had the expected recombinant bands (41.6%). Plasmid DNA extracted from 2 of these colonies was sequenced in the area of the recombination junction. All had the precise expected crossover event.
BAC modifications
We tested the activation and efficiency of BAC modification system for the HAC vector. The modification cassette I was targeted to the start codon locus of IL-1β carried on a BAC clone RP11-67L14 (Fig. 2a). The purpose was to create a BAC transgene that expresses reporter genes in the control of target genome for use in genomic functional studies. RP11-67L14 was electroporated into DY380 cells, and 13 chloramphenicol-resistant colonies were selected. To determine the transfer of the BAC, we employed PCR analysis. Using the appropriate primers (IL-1β 11041 S/IL-1β 11851 AS) (Table 2), the expected amplicon was detected in 11 of 13 colonies (84.6%). The modification cassette I was released from the modification cassette vector I by EcoRI digestion and used for BAC recombineering. DY380 cells carrying the BAC were then shifted to 42 ˚C to induce recombination. The cells were then electroporated with the modification cassette I, and the ampicillin and chloramphenicol-resistant colonies were selected. Approximately, 500 double resistant colonies were obtained. No colonies were obtained from uninduced cells. PCR analysis of 7 selected colonies using the primers (IL-1β 10930 S/slg200 AS and amp2500 S/IL-1β 12100 AS) (Table 2) that flanked the targeted locus indicated that all colonies were correctly targeted (100%). This recombineering efficiency was very high. This means that SLG-AmpR cassette was recombined into the 1st ATG locus of IL-1β carried on a RP11-67L14. This BAC was termed modified BAC1 (mBAC1).
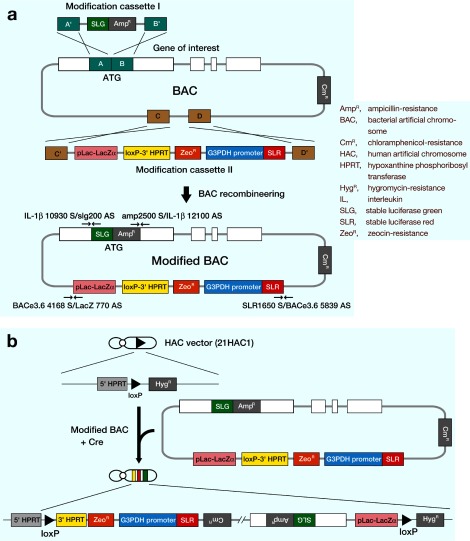
Fig. 2.
Strategy of BAC modification and introduction into the HAC vector.
a:The 1st step of the process is the homologus recombination of the BAC using the modification cassette I. It occurs by crossing over between the homology arms and the genome in the BAC. In this case, recombination between A B and A' B' results in incorporation of the modification cassette sequences into the genome to yield the mBAC1. The second step of the process is the homologus recombination of the mBAC1 using the modification cassette II. It occurs by a second homologous recombination event that occurs between C D and C' D' of the mBAC1. Recombination yields the precisely modified BAC2 (mBAC2), with the modification cassette II inserted at the correct position in the BAC. This modified BAC carries loxP-3'HPRT region for introduction to the HAC vector.
b:Construction of the HAC vector containing the modified BAC using Cre/loxP system.
Next, to insert the loxP-3'HPRT region into the mBAC1, the modification cassette II was targeted to the chosen site on the pBACe3.6 (Fig. 2a). This modification cassette II was released from the modification cassette vector II by SacII-NdeI digestion and used for recombineering. DY380 cells carrying the mBAC1 were then shifted to 42 °C. The cells were then electroporated with the modification cassette II, and the ampicillin-, chloramphenicol- and zeocin-resistant colonies were selected. As a result, 34 colonies were obtained. These colonies were blue and had the AmpR, CmR and ZeoR. PCR analysis of 11 selected colonies using the primers (SLR1650 S/BACe3.6 5839 AS and BACe3.6 4168 S/LacZ 770 AS) (Table 2) that flanked the targeted locus indicated that 6 colonies were correctly targeted (54.5%). This indicates that the modification cassette containing the loxP-3'HPRT region was inserted into the mBAC1. This BAC was termed modified BAC2 (mBAC2). Finally, mBAC2 had the correct insert genome as determined by PCR analysis using primers (IL-1β 1 S/IL-1β 3000 AS, IL-1β 2611 S/IL-1β 5180 AS, IL-1β 4031 S/IL-1β 7151 AS and IL-1β 8051 S/IL-1β 11332 AS) (Table 2). This modified BAC carries a loxP and 3'HPRT sequence, and has the capacity of site-specific insertion into the HAC vector via Cre/loxP system (Fig. 2b).
Discussion
In this study, we constructed a BAC modification system for the HAC vector which can be used to integrate the reporter gene into a chosen region of the genome and the loxP-3'HPRT cassette into a specific site in the BAC. Our system will enable insertion of large DNA fragments of interest at predetermined locations in the HAC vector. This system may thereby allow avoidance of the position effects and actualize intrinsic expression of transgenes. Furthermore, BAC libraries have now been generated from several species. Table 1 lists BAC libraries that were constructed in the pBACe3.6. These ready-made genomic BAC libraries can be loaded onto the HAC vector using our system.
Our system is based on BAC recombineering. The recombination activity was assessed in using the modification cassettes I and II. The results of these recombination assays indicated that BAC recombineering conferred high recombination activity (cassette I; 100%, cassette II; 54.5%). This technology may be suited for cassette exchange reactions in the BAC. In addition, BACs can be modified rapidly in vitro, with a construction time of less than 2 weeks in this study. As the HAC vector contains the 5' HPRT-loxP, the modified BAC could be introduced into the HAC vector. By introducing all candidate BAC clones into the same genomic locus, site-specific integration of large genomic clones can be used to streamline the positional cloning and transfer of genes into various mammalian cells. This ability of BAC-HAC system will reduce the number of subclones and transgenic cells required to determine which clones contain the target gene. This system will allow generation of an isogenic series of assays that would be expected to display comparable expression profiles due to their placement in identical genomic and cellular backgrounds. In addition, the HAC vector can be used for the generation of transgenic animals (trans-chromosomic mice) for the functional analysis of a desired gene in mice (Hoshiya et al., 2009; Kazuki et al., in press). Therefore, our BAC-HAC system could provide significant advantages for comparing the gene function of species at a specific locus, and allow for a better understanding of the correlation between cell (in vitro) and animal (in vivo). These types of assays are difficult to achieve with current techniques that rely on random integration of the target genes.
In summary, our system enables BAC modifications for the HAC vector to be quickly and easily engineered. The use of BAC clones in conjunction with the HAC vector opens the way to many additional applications, especially for investigating gene function, gene therapy and animal transgenesis.
Acknowledgments
The authors thank all colleagues in our laboratory for valuable discussion and encouragement through this study. We also thank Mr. Daigo Mori for technical assistance. We are grateful to Professor Mitsuo Oshimura, Professor Masako Tada, Associate Professor Toshiaki Inoue, Assistant Professors Mitsuhiko Osaki and Masaharu Hiratsuka for their helpful suggestions.
This study was supported in part by the New Energy and Industrial Technology Development Organization from the Ministry of Economy, Trade and Industry of Japan. The first author was supported by Research Fellowships from the Japan Society for the Promotion of Science for Young Scientists.
References
- 1.Alexander GM, Erwin KL, Byers N, Deitch JS, Augelli BJ, Blankenhorn EP.Effect of transgene copy number on survival in the G93A SOD1 transgenic mouse model of ALS. Brain Res Mol Brain Res 2004; 130: 7–15 [DOI] [PubMed] [Google Scholar]
- 2.Basu J, Willard HF. Human artificial chromosomes: potential applications and clinical considerations. Pediatr Clin North Am 2006; 53: 843–853 [DOI] [PubMed] [Google Scholar]
- 3.Bateup HS, Svenningsson P, Kuroiwa M, Gong S, Nishi A, Heintz N.Cell type-specific regulation of DARPP-32 phosphorylation by psychostimulant and antipsychotic drugs. Nat Neurosci 2008; 11: 932–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brune W, Messerle M, Koszinowski UH. Forward with BACs: new tools for herpesvirus genomics. Trends Genet 2000; 16: 254–259 [DOI] [PubMed] [Google Scholar]
- 5.Chandler KJ, Chandler RL, Broeckelmann EM, Hou Y, Southard-Smith EM, Mortlock DP. Relevance of BAC transgene copy number in mice: transgene copy number variation across multiple transgenic lines and correlations with transgene integrity and expression. Mamm Genome 2007; 18: 693–708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Copeland NG, Jenkins NA, Court DL. Recombineering: a powerful new tool for mouse functional genomics. Nat Rev Genet 2001; 2: 769–779 [DOI] [PubMed] [Google Scholar]
- 7.Frengen E, Weichenhan D, Zhao B, Osoegawa K, van Geel M, de Jong PJ. A modular, positive selection bacterial artificial chromosome vector with multiple cloning sites. Genomics 1999; 58: 250–253 [DOI] [PubMed] [Google Scholar]
- 8.Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB.A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 2003; 425: 917–925 [DOI] [PubMed] [Google Scholar]
- 9.Heaney JD, Bronson SK. Artificial chromosome-based transgenes in the study of genome function. Mamm Genome 2006; 17: 791–807 [DOI] [PubMed] [Google Scholar]
- 10.Heintz N. BAC to the future: the use of bac transgenic mice for neuroscience research. Nat Rev Neurosci 2001; 2: 861–870 [DOI] [PubMed] [Google Scholar]
- 11.Hoshiya H, Kazuki Y, Abe S, Takiguchi M, Kajitani N, Watanabe Y.A highly stable and nonintegrated human artificial chromosome (HAC) containing the 2.4 Mb entire human dystrophin gene. Mol Ther 2009; 17: 309–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kakeda M, Hiratsuka M, Nagata K, Kuroiwa Y, Kakitani M, Katoh M.Human artificial chromosome (HAC) vector provides long-term therapeutic transgene expression in normal human primary fibroblasts. Gene Ther 2005; 12: 852–856 [DOI] [PubMed] [Google Scholar]
- 13.Katoh M, Ayabe F, Norikane S, Okada T, Masumoto H, Horike S.Construction of a novel human artificial chromosome vector for gene delivery. Biochem Biophys Res Commun 2004; 321: 280–290 [DOI] [PubMed] [Google Scholar]
- 14.Kazuki Y, Hiratsuka M, Takiguchi M, Osaki M, Kajitani N, Hoshiya H.Complete genetic correction of ips cells from Duchenne muscular dystrophy. Mol Ther 2010; 18: 386–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kazuki Y, Hoshiya H, Takiguchi M, Abe S, Iida Y, Osaki M.Refined human artificial chromosome vectors for gene therapy and animal transgenesis. Gene Ther., . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larin Z, Mejia JE. Advances in human artificial chromosome technology. Trends Genet 2002; 18: 313–319 [DOI] [PubMed] [Google Scholar]
- 17.Le Saux A, Houdebine LM, Jolivet G. Chromosome integration of BAC (bacterial artificial chromosome): evidence of multiple rearrangements. Transgenic Res 2010; 19: 923–931 [DOI] [PubMed] [Google Scholar]
- 18.Lobo MK, Karsten SL, Gray M, Geschwind DH, Yang XW. FACS-array profiling of striatal projection neuron subtypes in juvenile and adult mouse brains. Nat Neurosci 2006; 9: 443–452 [DOI] [PubMed] [Google Scholar]
- 19.Muyrers JP, Zhang Y, Benes V, Testa G, Ansorge W, Stewart AF. Point mutation of bacterial artificial chromosomes by ET recombination. EMBO Rep 2000; 1: 239–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muyrers JP, Zhang Y, Testa G, Stewart AF. Rapid modification of bacterial artificial chromosomes by ET-recombination. Nucleic Acids Res 1999; 27: 1555–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O’Connor M, Peifer M, Bender W. Construction of large DNA segments in Escherichia coli. Science 1989; 244: 1307–1312 [DOI] [PubMed] [Google Scholar]
- 22.Oshimura M, Katoh M. Transfer of human artificial chromosome vectors into stem cells. Reprod Biomed Online 2008; 16: 57–69 [DOI] [PubMed] [Google Scholar]
- 23.Osoegawa K, Tateno M, Woon PY, Frengen E, Mammoser AG, Catanese JJ.Bacterial artificial chromosome libraries for mouse sequencing and functional analysis. Genome Res 2000; 10: 116–128 [PMC free article] [PubMed] [Google Scholar]
- 24.Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A.BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods 2008; 5: 409–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pravtcheva DD, Wise TL. A postimplantation lethal mutation induced by transgene insertion on mouse chromosome 8. Genomics 1995; 30: 529–544 [DOI] [PubMed] [Google Scholar]
- 26.Sarov M, Schneider S, Pozniakovski A, Roguev A, Ernst S, Zhang Y.A recombineering pipeline for functional genomics applied to Caenorhabditis elegans. Nat Methods 2006; 3: 839–844 [DOI] [PubMed] [Google Scholar]
- 27.Sasaki Y, Sone T, Yoshida S, Yahata K, Hotta J, Chesnut JD.Evidence for high specificity and efficiency of multiple recombination signals in mixed DNA cloning by the Multisite Gateway system. J Biotechnol 2004; 107: 233–243 [DOI] [PubMed] [Google Scholar]
- 28.Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y.Cloning and stable maintenance of 300-kilobase-pair fragments of human DNA in Escherichia coli using an F-factor-based vector. Proc Natl Acad Sci U S A 1992; 89: 8794–8797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sparwasser T, Eberl G. BAC to immunology – bacterial artificial chromosome-mediated transgenesis for targeting of immune cells. Immunology 2007; 121: 308–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sone T, Yahata K, Sasaki Y, Hotta J, Kishine H, Chesnut JD.Multi-gene gateway clone design for expression of multiple heterologous genes in living cells: modular construction of multiple cDNA expression elements using recombinant cloning. J Biotechnol 2008; 136: 113–121 [DOI] [PubMed] [Google Scholar]
- 31.Tunster SJ, Van De Pette M, John RM. BACs as tools for the study of genomic imprinting. J Biomed Biotechnol 2011;2011:283013. [DOI] [PMC free article] [PubMed]
- 32.Yang XW, Model P, Heintz N. Homologous recombination based modification in Escherichia coli and germline transmission in transgenic mice of a bacterial artificial chromosome. Nat Biotechnol 1997; 15: 859–865 [DOI] [PubMed] [Google Scholar]
- 33.Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG, Court DL. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 2000; 97: 5978–5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Buchholz F, Muyrers JP, Stewart AF. A new logic for DNA engineering using recombination in Escherichia coli. Nat Genet 1998; 20: 123–128 [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y, Muyrers JP, Testa G, Stewart AF. DNA cloning by homologous recombination in Escherichia coli. Nat Biotechnol 2000; 18: 1314–1317 [DOI] [PubMed] [Google Scholar]