Abstract
Chromosome segregation during mitosis requires assembly of the kinetochore complex at the centromere. Key to kinetochore assembly is the specific recognition of the histone variant CENP-A in the centromeric nucleosome by centromere protein C (CENP-C). We have defined the determinants of this recognition mechanism and discovered that CENP-C binds a hydrophobic region in the CENP-A tail and docks onto the acidic patch of histone H2A/H2B. We further find that the more broadly conserved CENP-C motif uses the same mechanism for CENP-A nucleosome recognition. Our findings reveal a conserved mechanism for protein recruitment to centromeres and a histone recognition mode whereby a disordered peptide binds the histone tail through nucleosome-docking-facilitated hydrophobic interactions.
The accurate segregation of chromosomes during mitosis is essential for reproduction and development of all organisms. Chromosome segregation is facilitated by the attachment of mitotic spindle microtubules to the kinetochore, which is assembled on the chromosomal centromere (1). Human CENP-A chromatin is constitutively associated with a complex of sixteen centromere proteins (2, 3). Among these sixteen proteins, only CENP-C has been identified in all model organisms (4). CENP-C recognizes the carboxyl tail of CENP-A in the centromeric nucleosome (5, 6). Human CENP-C consists of four functional regions (fig. S1A). The N-terminal region interacts with the Mis12 complex (7). The central region and the CENP-C motif are required for targeting CENP-C to the centromere (5, 8-11). The central region directly binds to the CENP-A nucleosome while the targeting mechanism for the CENP-C motif is unknown. The C-terminal region is responsible for CENP-C dimerization (12).
To explore how CENP-C recognizes the CENP-A nucleosome, we first investigated the central region (CENP-C426-537) alone by nuclear magnetic resonance spectroscopy (NMR) and found that it is disordered (fig. S1B, C). We then examined CENP-C426-537 in complex with reconstituted Drosophila nucleosomes containing the H31-132-LEEGLG chimera. The residues in the CENP-A nucleosome that are exposed for binding of CENP-C are essentially conserved in the Drosophila H3 nucleosome (5, 13) (fig. S2A, B), and CENP-C426-537 binds to the CENP-A and Drosophila H31-132-LEEGLG nucleosomes with the same stoichiometry and similar affinity (figs. S2C-E, S3). We found that the residues 426-481 of CENP-C426-537 remain disordered while residues 482-537 fold into the chimeric nucleosome core (fig. S1B).
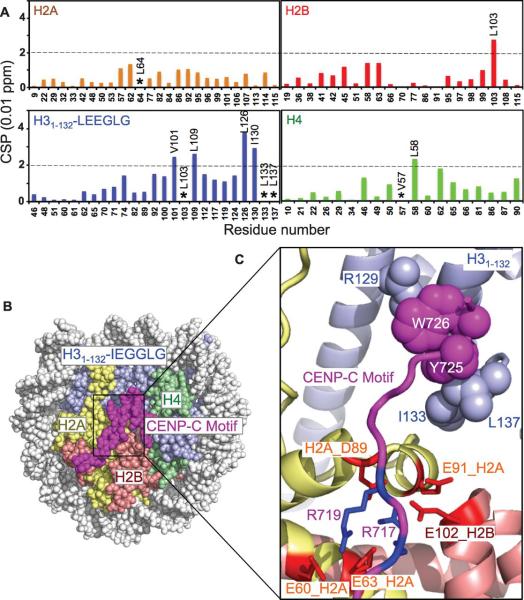
Using methyl-based chemical shift perturbation and paramagnetic relaxation enhancement (14, 15), we mapped the binding sites between CENP-C426-537 and the chimeric nucleosome. Upon binding of CENP-C426-537, the cross-peaks of methyl groups of residues Leu62 in H2A, Leu103 in H2B, Leu100, Leu103, Ile124, Ile126, and Ile130 in H3, and Ile50 and Leu58 in H4 displayed large chemical shift changes, while those of Leu64 in H2A and Val57, Val60, Ile62, and Val65 in H4 disappeared (Fig. 1A, C and fig. S4), suggesting close contact of these residues with CENP-C426-537. We next individually mutated each of the residues Phe500, Val509, Val517, Ile523, and Ser535 in CENP-C484-537 to Cys and linked them to a paramagnetic spin label. Binding of paramagnetic spin-labeled CENP-C484-537 reduced the peak intensities of the methyl groups in the nucleosome in accordance with their distance from the paramagnetic center. While spin labels of residues Val509 and Val517 mainly affected the methyl groups near the C-terminal region of the histone fold of H2B, the spin label of residue Ile523 affected methyl groups at an acidic residue-rich region (acidic patch) (Fig. 1B, C and fig. S5). In contrast, the spin label of residue Ser535 affected the methyl groups of H31-132-LEEGLG near the C-terminal tail (Fig. 1B, C). These results indicate that CENP-C residues 509-535 bind to the histone surface, using hydrophobic residues 530WWVV533 to recognize the LEEGLG tail and docking the positively charged region (Arg521, Arg522, Arg525 and Arg526) on the acidic patch region (fig. S5E, S6, and SOM text). The CENP-C residues 482-508, which include many positively charged residues, likely bind to DNA (fig. S6). The CENP-C binding region in the core histones of the Drosophila H3 nucleosome has the same structure as the corresponding region of the human CENP-A nucleosome (16) (fig. S7), further validating the use of the chimeric nucleosome.
Figure 1.
Binding sites between the CENP-C central region on the chimeric nucleosome mapped by methyl-based NMR. (A) Histone methyl chemical shift perturbation (CSP) upon CENP-C426-537 binding. (B) Effects on methyl groups by paramagnetic spin labels in CENP-C484-537. (C) Nucleosome structure showing representative side chains (black balls) whose methyl groups display large changes in (A) or (B), and approximate locations of CENP-C residues (arrows). E133 in H3 is used to represent L133 in H31-132-LEEGLG.
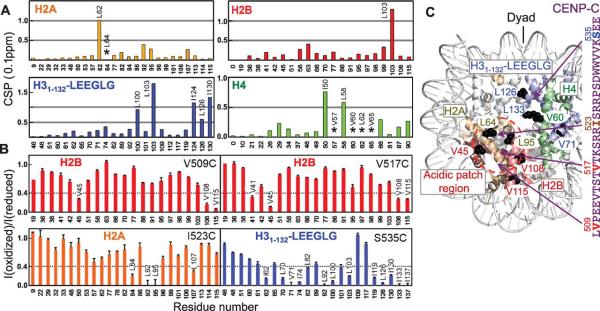
Deletion and isothermal titration calorimetry studies showed that CENP-C 520-537 region contains residues critical for binding to the chimeric nucleosome (table S1). Ala-scanning revealed that substitution of any one of the Arg522, Trp530, or Trp531 residues with Ala in CENP-C444-537 abolished binding (Fig. 2A, fig. S8A and table S2). Conversely, the single charge-reversal mutations Glu60Lys, Glu63Lys, and the neutralizing mutation Glu90Thr in H2A each essentially abolished the binding (fig. S8A and table S2). Individual substitution of the two Leu residues in the C-terminus of H31-132-LEEGLG with Ala reduced the dissociation equilibrium constant (Kd) by a factor of ~3 and ~4, respectively (fig. S8A and table S2). In contrast, Ala substitution of the two Glu residues increased the Kd by a factor of ~3 and ~10 (table S2), respectively, indicating that the CENP-A C-terminal tails have not evolved for high affinity binding with CENP-C. Mutation of CENP-C central region residues Arg522, Trp530 or Trp531 to Ala also substantially decreased the localization of GFP-fused CENP-C1-537 to centromeres in human cells, whereas the S520A or S524A mutation had smaller effects (Fig. 2B and fig. S8B). Mutation of the two acidic patch residues Glu61 and Glu64 residues in H2A to either Ala or positively charged Lys inhibited the recruitment of CENP-C from the X. laevis egg extract to the reconstituted CENP-A nucleosome array (Fig. 2C and fig. S8C) (6). Neutralization of the acidic patch by the mutations did not disrupt the formation of condensed CENP-A nucleosome arrays (fig. S9).
Figure 2.
Residues important for binding of the central region of CENP-C and the chimeric nucleosome. (A) Effects of mutations in CENP-C426-537 on its binding to the H31-132-LEEGLG nucleosome. No detectable binding (*). (B) Effect of CENP-C1-537 mutations on centromere targeting in human cells (fig. S8B). Error represents SEM n=4, Students t probability p≤0.02 for all mutants. (C) Effect of H2A acidic patch mutations on localization of CENP-C to the CENP-A nucleosome arrays. Error represents SEM, n=4, Students t probability p≤0.05 for both mutants.
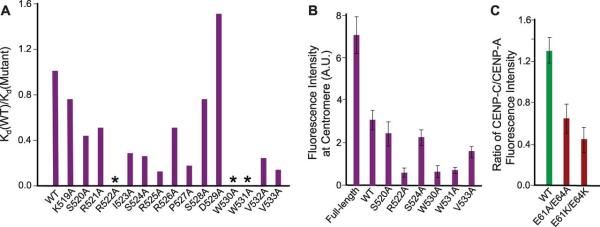
The central region of CENP-C is conserved in most mammals, whereas the CENP-C motif is conserved from budding yeast to human (fig. S10A). Residues Arg522, Trp530, and Trp531, which are most important for the binding of the CENP-C central region to the chimeric nucleosome, are essentially conserved in the CENP-C motif and correspond to residues Arg742, Tyr750 and Trp751, suggesting that the human CENP-C motif may also bind to CENP-A and H31-132-LEEGLG nucleosomes. Indeed, chemical shift perturbation studies indicate that the CENP-C motif binds to the similar sites on the core histones in the H31-132-LEEGLG nucleosome as the CENP-C central region (Fig. 3A and fig. S11). Furthermore, binding of the CENP-C motifs from human, rat, fission yeast, and budding yeast to the H3 chimeric nucleosomes containing the corresponding C-terminal residues of the CENP-A homolog displayed reasonably strong affinities with Kd of 1.4 μM, 0.1 μM, 1.4 μM, and 0.3 μM, respectively (fig. S12 and table S3). Charge reversal mutation of a single acidic residue in the acidic patch (Glu60Lys or Glu63Lys) abolished the binding of the CENP-C motif (table S3). Furthermore, mutation of the conserved sites within the rat CENP-C motif (Arg717Ala, Tyr725Ala or Trp726Ala) substantially decreased the binding affinity (fig. S12 and table S4). Mutation of the residues corresponding to Arg717 in frog, fly and fission yeast prevented targeting of CENP-C to the centromere (9-11).
Figure 3.
Broadly conserved CENP-C motifs bind to the H3 nucleosome chimera containing CenH3 C-terminal tails. (A) Nucleosome methyl CSP upon binding of the CENP-C motif. (B) The structure of the rat CENP-C motif in complex with the H31-132-IEGGLG nucleosome. (C) Enlarged region showing hydrophobic and electrostatic interactions.
We determined the crystal structure of the rat CENP-C motif in complex with the corresponding H31-132-IEGGLG nucleosome at 3.5 Å resolution (Fig. 3B, fig. S13 and table S5). In the structure, the conserved CENP-C motif residue Tyr725 recognizes the residues Ile133 and Leu137 in the IEGGLG tail through hydrophobic interactions, inducing the IEGGLG tail to fold to a turn structure and promoting intramolecular hydrophobic interactions between Ile133 and Leu137. CENP-C residue Trp726 forms intra-molecular hydrophobic interactions with CENP-C residue Tyr725, in addition to interacting with the hydrophobic region of the side chain of residue Arg129 in H31-132. CENP-C residues Arg717 and Arg719 form electrostatic interactions with the acidic patch residues Glu60, Glu63, Asp89, and Glu91 of H2A and Glu102 of H2B. Binding to the acidic patch was similarly found in structures of LANA (17), RCC1 (18), and the Sir3 BAH domain (19) in complex with the nucleosome.
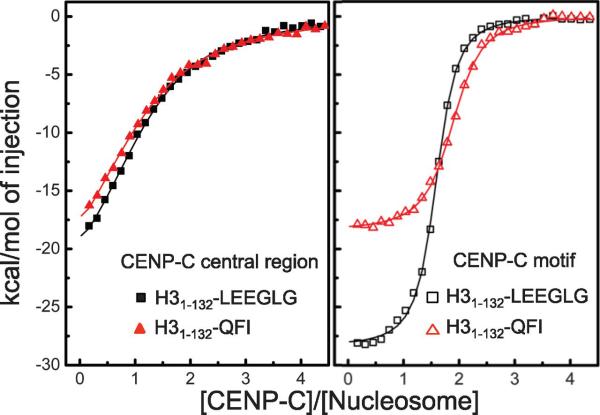
Whereas CENP-C motifs are well conserved among all species, the C-terminal tails of CENP-As are not (fig. S10B). We observed a common feature in the C-terminal tails of CENP-As: they are more hydrophobic than those of corresponding H3. In particular, the C-terminal tails of CENP-As have at least one bulky hydrophobic residue such as Ile, Leu, Val or aromatic residues (with the exception of fission yeast), which are absent in the C-terminal tails of H3. In addition, nucleosomes containing chimeric H31-132-QFI (C-terminal tail of CENP-A in budding yeast) bound to the central region and the CENP-C motif of human CENP-C with similar affinity as nucleosomes containing H31-132-LEEGLG (Fig. 4 and table S3). Thus, the higher hydrophobicity of the C-terminal tail of CENP-A rather than a specific amino acid sequence is the key determinant for specific recognition by CENP-C.
Figure 4.
Hydrophobicity of the CenH3 tail is the primary determinant for recognition of CenH3 by CENP-C. Isothermal titration calorimetric curves for binding of the human CENP-C central region (CENP-C444-537) and the CENP-C motif (CENP-C727-767) to nucleosomes containing H31-132-LEEGLG (human) and H31-132-QFI (budding yeast), respectively.
Our findings can explain diverse experimental results in the literature. In human cells, the CENP-C584-943 fragment, which contains the CENP-C motif and the dimerization domain, is capable of being targeted to the centromere when over-expressed (20). In Xenopus, chromatin in which H2A/H2B is replaced by protamines does not support CENP-C localization to the centromere of sperm (9, 21). However, CENP-C co-localizes with CENP-A upon addition of egg extract, as the histone chaperone nucleoplasmin catalyzes the eviction of protamines and the incorporation of H2A/H2B dimers into the sperm chromatin (21). The function of human CENP-A can be complemented by the budding yeast Saccharomyces cerevisiae homolog Cse4 (22) (Fig. 4). Our study also suggests that CENP-C can recognize CENP-A chromatin through multivalent interactions, allowing it to associate more strongly and selectively (figs. S14, S15) (20, 23). In addition, our results have implications for various structural models of the centromeric nucleosome, a widely discussed and controversial topic (24). The finding that the broadly conserved CENP-C motif in a dimeric CENP-C could bind to two H2A/H2B and two CENP-A/H4 molecules constraints these models since CENP-C is constitutively associated with the centromeric nucleosome. Our study provides the structural basis for the recognition of the CENP-A octameric nucleosome by CENP-C. Further understanding of how it engages chromatin should provide important insight into favored compositions of CENP-A nucleosomes.
In a broader perspective, our finding that the widely conserved CENP-C motif binds to the centromeric nucleosome, along with the recent identification of the budding yeast homologues of the vertebrate centromere proteins CENP-T and CENP-W (25, 26), supports a conserved mechanism of centromere targeting by the kinetochore. Furthermore, our study has revealed a histone recognition mode whereby an intrinsically disordered peptide binds to the histone tail through nucleosome-docking-facilitated hydrophobic interactions, which broadens the repertoire of the cell to “read” histone variations (27, 28), not only at the centromere but possibly also in other contexts.
Supplementary Material
Acknowledgments
We thank the staffs at the GM/CA-CAT, Advanced Photo Source for technical support, Drs. Christopher Carroll for initial participation of the project, Jemima Barrowman for manuscript editing, Alex Kelly, Carl Wu and Claude Klee for comments, Hitoshi Kurumizaka and Kosuke Morikawa for providing human histone plasmids and advice for CENP-A nucleosome reconstitution, and Song Tan for the ‘601’ DNA plasmid. This work was supported by the intramural programs of the National Cancer Institute (H.K., H.F., B-R.Z. and Y.B.), the National Institute of Allergy and Infectious Diseases (J.J. and T.S.X.), the National Institute of Diabetes and Digestive and Kidney Diseases (R.G.), NIH grant R01 GM074728 (M.R. and A.F.S.), and NCI (Y1-CO-1020), NIGMS (Y1-GM-1104) and DOE (DE-AC02-06CH11357) (APS). Structure PDB ID: 4INM.
References and notes
- 1.Henikoff S, Ahmad K, Malik HS. Science. 2001;293:1098. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- 2.Verdaasdonk JS, Bloom K. Nat. Rev. Mol. Cell Biol. 2011;12:320. doi: 10.1038/nrm3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hori T, et al. Cell. 2008;135:1039. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 4.Przewloka MR, et al. Curr. Biol. 2011;21:399. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 5.Carroll CW, Milks KJ, Straight AF, Cell Biol J. 2010;189:1143. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guse A, Carroll CW, Moree B, Fuller CJ, Straight AF. Nature. 2011;477:354. doi: 10.1038/nature10379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Screpanti E, et al. Curr. Biol. 2011;21:391. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Song K, Gronemeyer B, Lu W, Eugster E, Tomkiel JE. Exp. Cell Res. 2002;275:81. doi: 10.1006/excr.2002.5495. [DOI] [PubMed] [Google Scholar]
- 9.Milks KJ, Moree B, Straight AF. Mol. Biol. Cell. 2009;20:4246. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tanaka K, Chang HL, Kagami A, Watanabe Y. Dev. Cell. 2009;17:334. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 11.Heeger S, et al. Genes Dev. 2005;19:2041. doi: 10.1101/gad.347805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cohen RL, et al. Mol. Biol. Cell. 2008;19:4480. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Nat. Cell Biol. 2009;11:896. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tugarinov V, Kanelis V, Kay LE. Nat. Protoc. 2006;1:749. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- 15.Kato H, et al. Proc. Natl. Acad. Sci. U.S.A. 2011;108:12283. [Google Scholar]
- 16.Tachiwana H, et al. Nature. 2011;476:232. doi: 10.1038/nature10258. [DOI] [PubMed] [Google Scholar]
- 17.Barbera AJ, et al. Science. 2006;311:856. doi: 10.1126/science.1120541. [DOI] [PubMed] [Google Scholar]
- 18.Makde RD, England JR, Yennawar HP, Tan S. Nature. 2010;467:562. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Science. 2011;334:977. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sugimoto K, Kuriyama K, Shibata A, Himeno M. Chromosome Res. 1997;5:132. doi: 10.1023/a:1018422325569. [DOI] [PubMed] [Google Scholar]
- 21.Philpott A, Leno GH. Cell. 1992;69:759. doi: 10.1016/0092-8674(92)90288-n. [DOI] [PubMed] [Google Scholar]
- 22.Wieland G, Orthaus S, Ohndorf S, Diekmann S, Hemmerich P. Mol. Cell. Biol. 2004;24:6620. doi: 10.1128/MCB.24.15.6620-6630.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ando S, Yang H, Nozaki N, Okazaki T, Yoda K. Mol. Cell. Biol. 2002;22:2229. doi: 10.1128/MCB.22.7.2229-2241.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Black BE, Cleveland DW. Cell. 2011;144:471. doi: 10.1016/j.cell.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock LJ, et al. Nat. Cell Biol. 2012;14:614. doi: 10.1038/ncb2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schleiffer A, et al. Nat. Cell Biol. 2012;14:604. doi: 10.1038/ncb2493. [DOI] [PubMed] [Google Scholar]
- 27.Khorasanizadeh S. Curr. Opin. Struct. Biol. 2011;21:744. doi: 10.1016/j.sbi.2011.10.001. [DOI] [PubMed] [Google Scholar]
- 28.Musselman CA, Lalonde ME, Cote J, Kutateladze TG. Nat. Struct. Mol. Biol. 2012;19:1218. doi: 10.1038/nsmb.2436. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.