Abstract
There is limited and sometimes contradictory information about the genotoxicity of polycyclic aromatic hydrocarbon benzo[ghi]perylene (B[ghi]P). Using recently developed metabolic toxicity screening arrays and a biocolloid reactor-LC-MS/MS approach, both featuring films of DNA and human metabolic enzymes, we demonstrated relatively low reactivity of metabolically activated B[ghi]P towards DNA. Electro-optical toxicity screening arrays showed that B[ghi]P metabolites damage DNA at a 3-fold lower rate than benzo[a]pyrene (B[a]P), whose metabolites have a strong and well-understood propensity for DNA damage. Metabolic studies using magnetic bead biocolloid reactors coated with microsomal enzymes in 96-well plates showed that cyt P450s 1A1 and 1B1 provide high activity for B[ghi]P and B[a]P conversion. Consistent with published results, the major metabolism of B[ghi]P involved oxidations at 3,4 and 11,12 positions, leading to formation of B[ghi]P 3,4-oxide and B[ghi]P 3,4,11,12-bisoxide. B[ghi]P 3,4-oxide was synthesized and reacted with deoxyadenosine at N6 and N7 positions and with deoxyguanosine at the N2 position. B[ghi]P 3,4-oxide is hydrolytically unstable and transforms into the 3,4-diol or converts to 3- or 4-hydroxy B[ghi]P. LC-MS/MS of reaction products from the magnetic biocolloid reactor particles coated with DNA and human enzymes revealed for the first time that a major DNA adduct results from reaction between B[ghi]P 3,4,11,12-bisoxide and deoxyguanosine. Results also demonstrated 5-fold lower formation rates of the major DNA adduct for B[ghi]P metabolites compared to B[a]P. Overall, results from both ECL array and biocolloid reactor-LC-MS/MS consistently suggest a lower human genotoxicity profile of B[ghi]P than B[a]P.
INTRODUCTION
Polycyclic aromatic hydrocarbons (PAHs) are ubiquitous environmental pollutants, and some are carcinogenic in animals and humans.1-3 PAHs are lipophilic and are normally transformed into more hydrophilic products by metabolic enzymes, such as cytochrome (cyt) P450s. Certain metabolites of PAH (e.g. benzo[a]pyrene 7,8-dihydrodiol 9,10-epoxide (BPDE), are highly reactive towards nucleophilic sites of biomolecules such as DNA, producing DNA adducts that may induce mutations that potentially initiate tumor formation or other diseases.4-6 Correlations of DNA adduct levels with tumor-inducing potencies exist for a number of PAHs, including benzo[a]pyrene and dibenzo[a,l]pyrene in rodents.7-9 These data relate to relative toxicity of the PAH itself, and DNA adducts can serve as biomarkers for individual exposure to PAHs.10,11
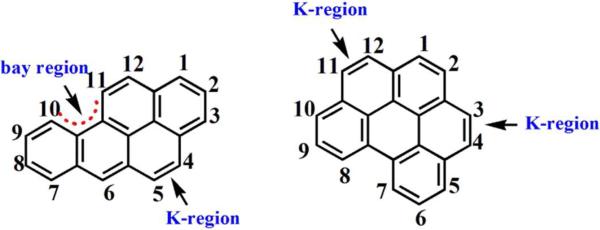
The formation of DNA-reactive PAH metabolites generally begins with oxidations catalyzed by cyt P450s.4,12,13 Oxidations can occur in the so-called K or bay regions (Scheme 1), producing phenols, dihydrodiols and/or epoxides.3 Structural variations may contribute to variation in reactivity and tumorigenic potencies.14
Scheme 1.
Structures of B[a]P, and B[ghi]P showing the bay and K regions.
B[a]P, perhaps the most well understood PAH in terms of genotoxicity, undergoes metabolic pathways producing diol epoxide and radicals that react with DNA in vitro and in vivo. 15-17 The active bay region (Scheme 1) diol epoxide BPDE is most significant as it forms stable DNA adducts that can lead to incorrect DNA replication and tumor initiation.18,19
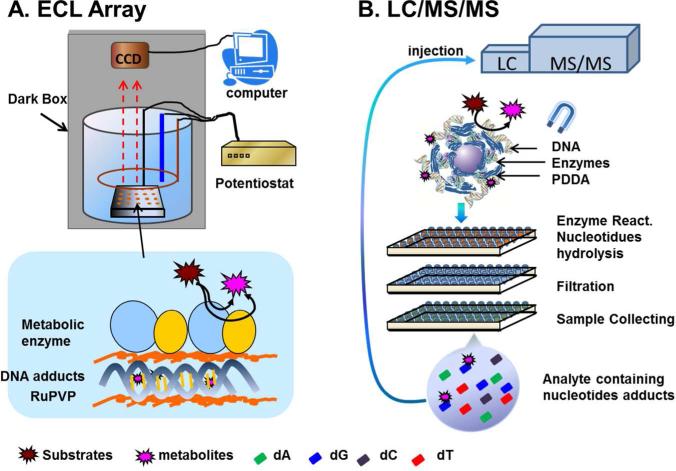
Benzo[ghi]perylene (B[ghi]P) is widely distributed in petroleum and coal tar.20,21 Its structure features two K regions, and represents PAHs that lack the very reactive bay region. Formation of a vicinal diol epoxide like BPDE from B[ghi]P is not possible.22 B[ghi]P displays mutagenicity in the Ames test with exogenous activation, but was negative in tumor initiating tests in mouse skin.23 Another investigation of B[ghi]P showed that active metabolites can bind to DNA in vivo and in vitro.24 Platt et. al. using mouse microsomal biotransformation of B[ghi]P and 32P-postlabeling suggested that 3,4-epoxy-3,4-dihydro-B[ghi]P (B[ghi]P 3,4-oxide) might be a carcinogenic metabolite with DNA reactivity.25 Comet assays yielded DNA damage for both B[a]P and B[ghi]P when activated by light without metabolic activation.26 However, there is no conclusive evidence to classify B[ghi]P as a carcinogen in humans, nor is the chemistry of DNA damage well understood. The goal of the present study is to establish the relative reactivity and chemistry of B[ghi]P metabolites towards DNA compared to that of B[a]P, employing in-vitro metabolite toxicity screening approaches developed in our laboratory using DNA-damage as detection endpoints.27,28 First, an electrochemiluminscent (ECL) array utilizing human enzyme/DNA films screening metabolite reactivity with DNA was employed (Figure 1A). 29,30 Then, biocolloid reactor particles coated with similar enzyme/DNA films were used to produce metabolites and DNA adducts for LC-MS/MS analysis (Figure 1B).31,32 Both of these approaches indicate the formation of metabolites that can possibly react with DNA, and that may be linked to genotoxicity.27,28
Figure 1.
Methods for DNA damage detection and identification: (A) ECL screening array showing enzyme/DNA film assembled on array spots and ECL detection utilizing a dark box equipped with a CCD camera; (B) magnetic particle biocolloid reactors and 96-well plate for high-throughput LC-MS/MS detection of DNA adducts. Incubation of assembled biocolloid reactors with damage agent and subsequent DNA hydrolysis is done in a 96-well filter plate. Solutions containing hydrolyzed products of DNA and hydrolysis enzymes are transferred into a second filter plate. A third plate is used to collect filtered samples containing the nucleoside adducts for LC-MS/MS analyses.
Both approaches use thin films of enzymes and DNA deposited on either a graphite chip for the ECL array or on 1 μm magnetic beads processed in 96-well plates for LC-MS/MS analyses. Specifically, ECL arrays feature multiple spots composed of ~40 nm thick composite films of double-stranded DNA, metabolic enzymes, and a ruthenium metallopolymer (RuPVP) that emits ECL light upon reaction with guanines in the DNA. These films are grown layer-by-layer via alternate electrostatic absorption.29 In the assay, parent compounds are converted into metabolites that react with nucleobases in the films leading to the partial disruption of the DNA double helix. Disorder in the DNA structure from the damage reactions provides guanines that are more accessible to the RuPVP centers compared with intact DNA. Therefore, the reaction between guanine and RuPVP is accelerated, producing an enhanced ECL signal in the detection step.27-29
The LC-MS/MS assays utilize magnetic biocolloid reactor particles coated with enzymes for metabolite profiling, or with enzymes and double stranded (ds)-DNA to determine of DNA adducts. These magnetic biocolloids bring the reaction components into close proximity, which greatly accelerates production of active metabolites and DNA adducts compared with conventional solution phase enzyme reactions.31,32 A magnet facilitates sample preparation in 96-well plates (Fig. 1B).32 Similar to ECL arrays, active metabolites generated in enzyme reactions are initially trapped by DNA in the thin films on the magnetic beads, and DNA adducts along with intact bases are subsequently released by enzyme hydrolysis followed by LC-MS/MS analyses (Fig. 1B). Using these approaches, this paper demonstrates significantly lower B[ghi]P metabolite reactivity towards DNA than B[a]P metabolites, and identifies a new adduct of B[ghi]P from reaction of B[ghi]P 3,4,11,12-bisoxide and deoxyguanosine.
EXPERIMENTAL
Chemicals and materials
Metallopolymer [Ru(bpy)2(PVP)10](ClO4)2 (RuPVP) was prepared and characterized according to previous work.33 Human liver supersomes 1A1 (Cyt P4501A1OR), human liver supersomes 1B1 (Cyt P4501B1OR), and human liver supersomes 1A2 (Cyt P4501A2OR), all containing reductase and epoxy hydrolase,34, were from BD Biosciences. Carboxylated magnetic particles were from Polysciences (Warrington, PA; 1 μm diameter; particle concentration 20 mg mL−1). B[a]P was from Toronto Research Chemicals. Water was purified with a Hydro Nanopure system to specific resistance >16 MΩ. B[ghi]P and all other chemicals were from Sigma-Aldrich. NADPH regenerating system: glucose 6-phosphate (G6P), glucose 6-phosphate dehydrogenase (G6PDH) and nicotinamide adenine dinucleotide phosphate (NADP) and other chemicals were from Sigma. Pyrolytic graphite was from Advanced Ceramics.
Film fabrication on ECL arrays
Arrays were fabricated layer-by-layer (LbL) with water washing between adsorption steps as previously described.29 Briefly, calf thymus ds-DNA (2 mg mL−1 in 50mM Tris buffer, pH 7.4), RuPVP (2 mg mL−1 in 12% ethanol and 88% water), and supersomes (20 mg mL−1 in sucrose) were deposited alternatively onto 2 × 2 inch PG blocks. Final film architectures represented as order of layer deposition on the spots are DNA/(RuPVP/DNA)2/(RuPVP/DNA/supersomes)2. Supersomes were cyt P4501A1OR, cyt P4501A2OR and cyt P4501B1OR. For simplicity, these films are referred to as DNA/RuPVP/1A1, DNA/RuPVP/1A2, DNA/RuPVP/1B1 in the text below.
Films on magnetic particles
LbL enzyme-DNA film fabrication on 1 μm particles was similar to that reported previously.31-32,35 Briefly, polycation poly(diallyldimethylammoniumchloride) (PDDA), supersomes and salmon testes dsDNA were assembled in alternate successive steps on the negatively charged magnetic particle surface.36 Steady-state adsorption times were 20 min for PDDA and DNA solutions and 30 min for supersomes while kept on ice. After each layer adsorption, the particles were first separated from the solution using aligned magnets, then washed and redispersed 10mM pH 7.4 Tris buffer. Final film architectures on the magnetic biocolloid reactors were PDDA/supersomes/PDDA/DNA. For generation of PAH metabolites, final films were PDDA/supersomes.
Synthesis of 3,4-epoxy-3,4-dihydro-B[ghi]P (B[ghi]P 3,4-oxide)
B[ghi]P 3,4-oxide was prepared by epoxidation of B[ghi]P with dimethyldioxirane (DMDO).37,38 To a solution of B[ghi]P (6 mg, 22 mmol) dissolved in acetone (1.5 ml) and dichloromethane (1.0 ml), DMDO (120 mmol) in 2 mL acetone was added. After overnight stirring, the reaction mixture was dried under vacuum and the resulting orange solid was dissolved in CDCl3 and analyzed by NMR (SI, Fig.S4)
Reaction of B[ghi]P 3,4-oxide with nucleosides
6 mg of unpurified product from the aforementioned synthesis containing ~ 2.4 mg B[ghi]P 3,4-oxide was dissolved in 1 mL DMSO and 100 μL of the resulting solution was added to 1 mL sodium phosphate buffer (pH 7.5, 10 mM) containing 1 mg of dG or dA. Reaction was stirred for 12 h both at 37 °C and 70 °C, and the resultant mixture had a dark brown color. Solvent was removed by vacuum, and the residue was dissolved in 200 μL of DMSO: MeOH (1:1), filtered, and characterized by LC-MS/MS.
Array protocol
Safety Note: B[ghi]P and B[a]P are suspected human carcinogens. Procedures were done wearing gloves in a closed hood. Stock solutions of B[ghi]P and B[a]P were prepared in DMSO and diluted in incubating solution to <0.5% (v/v). The incubating solution was 0.4 mL of 10 mM pH 7.0 MES buffer containing 1 mM DL-dithiothreitol (DTT), 1 mM ethylenediaminetetraacetic acid disodium salt (EDTA), 5 mM MgCl2 and an NADPH regenerating system (10 mM G6P, 5 unit G6PDH, 0.8 mM NADP). Incubations were done on arrays by spotting 50 μL of these solutions onto four of the RuPVP/DNA/enzyme spots at 37 °C for up to 60 seconds in the dark. The carbon chip was rinsed rapidly with water to stop the reaction.
After the enzyme reaction, the array was placed in a 150-mL open top electrochemical cell containing ~ 40 mL of pH 5.5 10 mM sodium acetate buffer with 0.15 M NaCl in a dark box. The counter electrode was a platinum wire ring placed directly above the array, and a Ag/AgCl electrode was used as a reference electrode (Fig. 1A).29 A constant potential of 1.25 V vs Ag/AgCl was applied to the array for 30s with a CH 1232 electrochemical analyzer. ECL signal was acquired by the CCD camera. Data analysis was done using GeneSnap and GeneTools software (SynGene).
Protocols for magnetic biocolloid reactors
Three 96-well plates were employed to produce either metabolites or DNA adducts samples for LC-MS/MS analysis. In general, 100 μL of biocolloid reactor particles dispersed in 10 mM phosphate buffer (pH 7.4) containing an NADPH-regenerating system was dispensed in each well in the reaction plate. Enzyme reactions were initiated in each well in the dark by adding either 1 μL of 2.5 mM B[a]P or 5 mM B[ghi]P in DMSO. The final concentration of PAHs is 25 μM for B[a]P and 50 μM for B[ghi]P.
For production of PAH metabolites, reactions were done for 10, 20, 30 and 60 mins in triplicate using biocolloid reactor particles coated with PDDA/supersome films. After the reaction, 100 μL DMSO was added to each of the reaction wells to increase the solubility of PAH metabolites in the solution phase from reactor particles. Biocolloid reactors were separated magnetically32, and solutions in the reaction plate were supplemented with 1 μM internal standard 6-hydroxychrysene (final concentration) and then transferred to the second filter plate where samples were filtered. The collecting plate underneath the filter plate was used to collect filtered sample, which was later injected to LC for analysis.
To generate PAH metabolite-DNA adducts, 1 μm magnetic particles coated with PDDA/supersomes/PDDA/DNA were used. Reactions of magnetic biocolloids and B[a]P or B[ghi]P were conducted in the reaction plate for 5, 10, 15, and 20 min in quintuplicate for each time point at 37 °C and stopped by adding 20 μL cold acetonitrile with 2 μL formic acid. The resulting solutions from 5 wells for the same time point were later combined. The biocolloid reactors were separated by a magnet, washed and reconstituted in 150 μL 10 mM Tris buffer pH 7.4 containing 1 mM CaCl2, 1 mM ZnCl2, and 10 mM MgCl2. Enzyme hydrolysis of DNA was then done in the reaction plate at 37 °C, following the previous protocol32 with slight modification. Briefly, biocolloid reactors in each well were incubated with deoxyribonuclease I (400 unit mg−1 of DNA) for 3 h, followed by incubation with phosphodiesterase I (0.2 unit mg−1 of DNA), phosphodiesterase II (0.1 unit mg−1) and alkaline phosphatase, (1.2 unit mg−1 of DNA) for 12 h at 37 °C, with a plate cover. After hydrolysis, samples were spiked with 0.1 μM 7-methylguanosine as an internal standard and transferred to the filtration plate. Upon filtration, samples were collected in the collecting plates and injected into LC-MS/MS for analysis.
LC-MS/MS analysis of PAH metabolites mixtures
4 μL of PAH metabolites sample was injected and analyzed using a capillary Luna C18-2 column (0.5 mm × 150 mm Phenomenex) coupled with a photodiode array (PDA) detector. Separation was achieved using a gradient of ammonium acetate buffer (10 mM, pH 5.5 with 0.1% formic acid) and methanol (0.1% formic acid), with methanol compositions 50% for 10 min, 50%-100% for 30 min, 100% for 10 min, 100%-50% for 2 min, and 50% for 3 mins at flow rate 15 μL min−1.
LC-MS/MS analysis DNA adducts from PAH-metabolites
A conventional LC (Waters, 2970) and a capillary LC (Waters, Capillary LC-XE) were used as previously described.32 A binary separation gradient composed of A: ammonium acetate buffer (10 mM, pH 5.5 with 0.1% formic acid) and B: acetonitrile (0.1% formic acid) was used. 20 μL reaction product of B[ghi]P 3,4-oxide and nucleosides sample was injected and analyzed using conventional LC with Luna C18-2 column (4.6 mm × 250 mm Phenomenex) using following gradient: 30% B for 10 min, 30%-50% B for 10 min, 50%B for 10 min, 50%-95% B for 15 min, 95% B for 10 min, 95%-30% B for 10 min and 30% B for 5 mins at flow rate 0.8 mL min−1. For metabolite-DNA adducts using magnetic biocolloid reactors, a 10 μL of the adducts sample was injected and separated using capillary LC with Luna C18-2 column (0.5 mm × 150 mm) with following gradient: 30% B for 20 mins, 30-60% B for 10 mins, 60% B for 10 mins, 60-100% B for 10 mins, 100-30% B for 10 mins and 30% B for 10 mins at a flow rate of 15 μL min−1. A 4000 QTRAP (AB Sciex, Foster City, CA) mass spectrometer with Analyst 1.4 software was operated in the positive ion mode was connected to the HPLC or capillary LC. Multiple reactions monitoring (MRM), and enhanced product ion (EPI) modes were done at 5000 V ion spray voltage, 60 V declustering potential, 15-35 eV collision energy (CE), and 0.15 s dwell time. From data on internal standards, a rough estimate of detection limit is ~0.3 fmol.
RESULTS
Characterization of ECL arrays and biocolloid reactor particles
Film compositions used in the array were characterized by making analogous films on 9 MHz gold-coated quartz crystal microbalance (QCM) resonators. Frequency shifts after deposition, washing and drying of each layer were used to estimate the weight of each component and nominal film thickness (SI, Fig. S1).39 QCM frequency decreased linearly with increasing layer number, demonstrating stable and reproducible film growth (SI, Fig. S1). The amount of each component adsorbed in the layers was 96 ± 10 ng for DNA, 310 ± 50 ng for RuPVP and 140 ± 20 ng for supersomes. Films had nominal thickness ~40 nm, suggesting that reactions will not be limited by mass transport.29
Amounts of biomolecules on the 1 μm magnetic particles (Table 1) were estimated by measuring the concentration remaining in solution after adsorption using UV absorbance and subtracting from the initial concentration. The total amount of supersomal protein on particles was estimated using a Bradford assay.40 The amount of DNA was obtained based on absorbance at 260 nm.41
Table 1.
The estimated amounts of immobilized supersomal proteins and DNA on magnetic beads.
| Composition (per mg particles) | Metabolic enzymes (μg of protein) | DNA (μg) |
|---|---|---|
| PDDA/1A1 | 513 ± 42 | – |
| PDDA/1A2 | 463 ± 20 | – |
| PDDA/1B1 | 482 ± 17 | – |
| PDDA/1A1/PDDA/DNA | 628 ± 47 | 32 ± 11 |
| PDDA/1A2/PDDA/DNA | 509 ± 10 | 37 ± 13 |
| PDDA/1B1/PDDA/DNA | 373 ± 38 | 27 ± 7 |
Activation of B[a]P and B[ghi]P by human liver supersomes
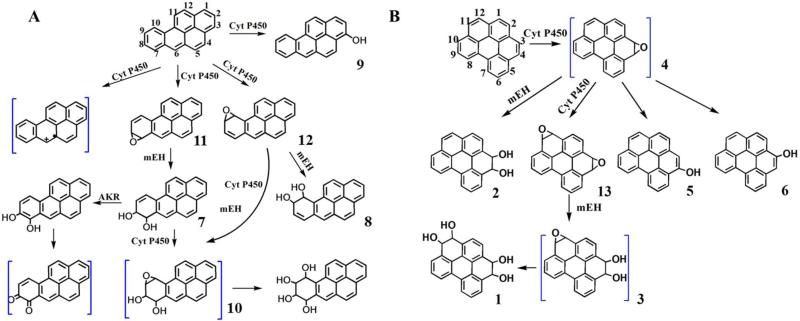
P450s 1A1, 1A2, and 1B1 are the major isoenzymes in the oxidation of PAHs,4 and were thus assembled in supersome films on biocolloid reactor particles to facilitate metabolic conversions of B[a]P and B[ghi]P. B[a]P is oxidized by cyt P4501A1 and 1B1 at 7, 8 and 9,10 positions, forming B[a]P 7,8-oxide (11, Scheme 2) and B[a]P 9,10-oxide (12).4 Both metabolites can be converted into BPDE (10), the ultimate carcinogen. The hydrolysis products of 11 and 12 by epoxy hydrolase are 7,8-dihydroxy-7,8-dihydro B[a]P (7, B[a]P 7, 8-diol), 9,10-dihydroxy-9,10-dihydro B[a]P (8, B[a]P 9, 10-diol). Also, cyt P450 1A1 and 1B1 can catalyze the formation of 3-hydroxy B[a]P (9, 3-OH B[a]P). Reaction of B[a]P with the supersomes-biocolloids produced LC peaks with characteristic UV spectra42 of 9, 7 or 11 and 8 or 12 (Supporting information Figure S2). Uncertainty arises due to similarities in the UV spectra of 11 and 12, and their hydrolysis products 7 and 8 that are the more likely final metabolites. They have relatively low retention times characteristic of more polar molecules, and both showed major ions of m/z 269 in positive MS mode, corresponded to molecular ions (m/z 287) losing a water molecule. Compound 9 gave m/z 269 ([M+H]+) in positive mode and m/z 267 ([M-H]−) in negative mode MS.
Scheme 2.
(A) Partial pathways of B[a]P metabolism.12,43,44 (B) Proposed partial pathways of B[ghi]P metabolism.22 Compounds in blue brackets are suspected metabolites with nucleosides reactivity. AKR, aldo-keto reductase, mEH, microsomal epoxide hydroloase.
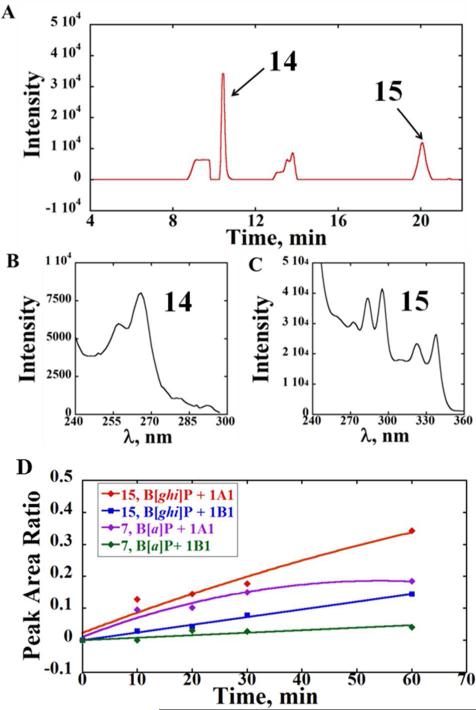
Biocolloid reactor particles containing individual cyt P450 1A1, 1B1 or 1A2 supersomes were also used to investigate the oxidation of B[ghi]P. Using an NADPH-regenerating system, similar chromatographic profiles were observed after metabolic conversions of B[ghi]P by P450 1A1 or 1B1 (Figure 2). Chromatographic peaks obtained with P450 1A2 were much smaller under the same conditions. Thus, we used cyt P450 1A1 and 1B1 for subsequent investigations. Of two major metabolites detected in the LC, the UV spectrum of metabolite we denote as 14 (tR ~11 min) was in agreement with oxidations at 3, 4 and 11, 12 positions of B[ghi]P,22 indicating formation of B[ghi]P 3,4,11,12-bisoxide. As above, it is uncertain whether 14 is B[ghi]P 3,4,11,12-bisoxide (13, Scheme 2) or the hydrolyzed product since both have the same UV spectrum.22 Compared with B[ghi]P 3,4,11,12-bisoxide, hydrolyzed product(s) are more polar with shorter retention times,22 and 14 eluted quite early (at 30% acetonitrile gradient). So it is quite possible that 14 (Figure 2) represents single or multiple diastereoisomers of 3,4,11,12-tetrahydroxy-3,4,11,12-tetrahydro-B[ghi]P (Scheme 2, 1, B[ghi]P 3,4,11,12-tetrol).
Figure 2.
Liquid chromatographic analysis of the reaction products from a 20-min incubation of 50 μM B[ghi]P with bioreactor particles containing P450 1A1 supersomes. (A) A partial photodiode array chromatogram at 260 nm showing two major metabolites 14 and 15, identified by their characteristic absorbance spectra. (B) UV spectrum of metabolite 14, and (C) UV spectrum of metabolite 15. (D) Relative formation rates of metabolites 15 and 7, obtained from peak area ratios against the internal standard 6-hydroxychrysene after incubations with magnetic biocolloids containing P450s 1A1 or 1B1 supersomes at different times.
The UV spectrum of the metabolite we denote as 15 (Figure 2, tR ~20 min) suggests 3,4 oxidation of B[ghi]P and formation of B[ghi]P 3,4-oxide.22 Due to the short retention time, we suspect this product represents diastereoisomers of 3,4-dihydroxy-3,4-dihydro-B[ghi]P (Scheme 2, 2, B[ghi]P 3,4-diol). MS gave m/z of 293, corresponded to molecular ions (m/z 311) losing a water molecule (Figure S3). Major fragmentations of 293 were m/z 275 and 265, corresponding to loss of a neutral water or CO from the parent. Overall, observation of metabolites 14 and 15 suggests the formation of B[ghi]P 3,4-oxide (4) and B[ghi]P 3,4,11,12-bisoxide (3).25
The relative formation rates of metabolites from B[a]P and B[ghi]P were characterized based on the peak area ratios relative to internal standard 6-hydroxychrysene (Figure 2D). Faster formation of metabolites 15 and 7 were found using P450 1A1 supersomes compared to those obtained with P450 1B1 supersomes. For both isozymes, metabolite 7 of B[a]P formed at a faster formation rate than metabolite 15 of B[ghi]P, i.e. 0.0037 μM•min−1 for 7 and 0.0060 μM•min−1 for 15 under P450 1A1 catalysis; 0.0008 μM•min−1 for 7 and 0.0024 μM•min−1 for 15 under P450 1B1 catalysis. These results also suggest that the reactions proceed for up to 60 min and do not provide indication of enzyme inhibition.
ECL Array studies
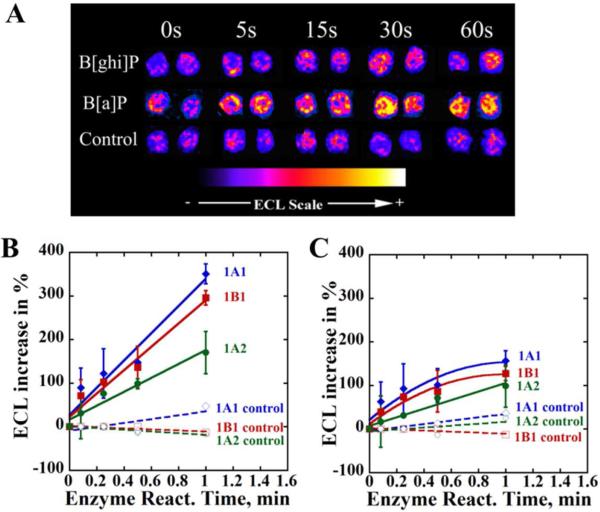
The ECL genotoxicity array (Fig. 1A) protocol features a two-step process: first, enzyme incubation, and second, DNA damage detection. The slope of the plot of % ECL increase vs. enzyme reaction time is proportional to the relative rate of DNA damage.29,30 P450s 1A1, 1A2 and 1B1 supersomes were incorporated into the array spots to facilitate the metabolic conversion. Three chips, each containing 36 array spots assembled with one type of enzyme along with RuPVP and DNA were employed to investigate B[a]P and B[ghi]P. Spots on each chip were exposed to incubation solutions containing B[a]P or B[ghi]P in the presence of an NADPH regenerating system.
Figure 3A shows a digitally reconstructed and recolored image from a DNA/RuPVP/1A1 array exposed to B[a]P and B[ghi]P together with an NADPH regeneration system. Controls are same spots exposed to B[a]P or B[ghi]P only, but no NADPH. ECL percentage increase normalized by dividing by the average amount of enzymes on the array spots (SI, Table S1) compared to 0 s versus enzyme reaction time are shown in Figures 3B and 3C. Figure 3A-3C show that both B[a]P and B[ghi]P produced increases in ECL light intensity with NADPH present, but not in the absence of NADPH. These results suggest that DNA damage occurred only when metabolic conversions of PAHs were activated. For both B[a]P and B[ghi]P, larger ECL signal increases were observed for supersomes 1A1 and 1B1 compared to 1A2. These results indicate that 1A1 and 1B1 are capable of catalyzing the formation of DNA-reactive metabolites at faster rates. As described in our earlier study,30 relative ECL turnover rates (R) for production of DNA-reactive metabolites were estimated as the slopes divided by concentration of the PAH in the study (Figs. 3B, 3C, and Table 2).30 Generally, B[a]P had much higher R-values than B[ghi]P for all three types of supersomes, suggesting a 3.4-fold faster overall rate (reactive metabolite and subsequence DNA adduct formation) of DNA damage by B[a]P compared to B[ghi]P.
Figure 3.
ECL array results for B[a]P and B[ghi]P (A) example of reconstructed ECL array spots containing DNA/RuPVP/1A1 exposed to either B[a]P and B[ghi]P using an NADPH regeneration system. Control spots were exposed to only B[ghi]P, but no NADPH. (B) and (C) ECL intensities increase (in percentage) versus enzyme reaction time for different enzymes, cyt P450 1A1(blue diamond), cyt P450 1A2 (green dot) and cyt P450 1B1 (red square) incubated with (B) 25 μM B[a]P and NADPH or (C) 50 μM B[ghi]P and NADPH. Controls (dashed lines) featured incubations with only 25 μM B[a]P or 50 μM B[ghi]P, but no NADPH.
Table 2.
Normalized ECL turnover rate (R) calculated from Fig. 3B and Fig. 3C.
| ECL Turnover R (μg−1•protein•min−1•mM) | 1A1 | 1B1 | 1A2 |
|---|---|---|---|
| B[a]P | 143 | 120 | 76.7 |
| B[ghi]P | 42.9 | 34.3 | 23.3 |
Nucleoside adducts of B[ghi]P 3,4-oxide
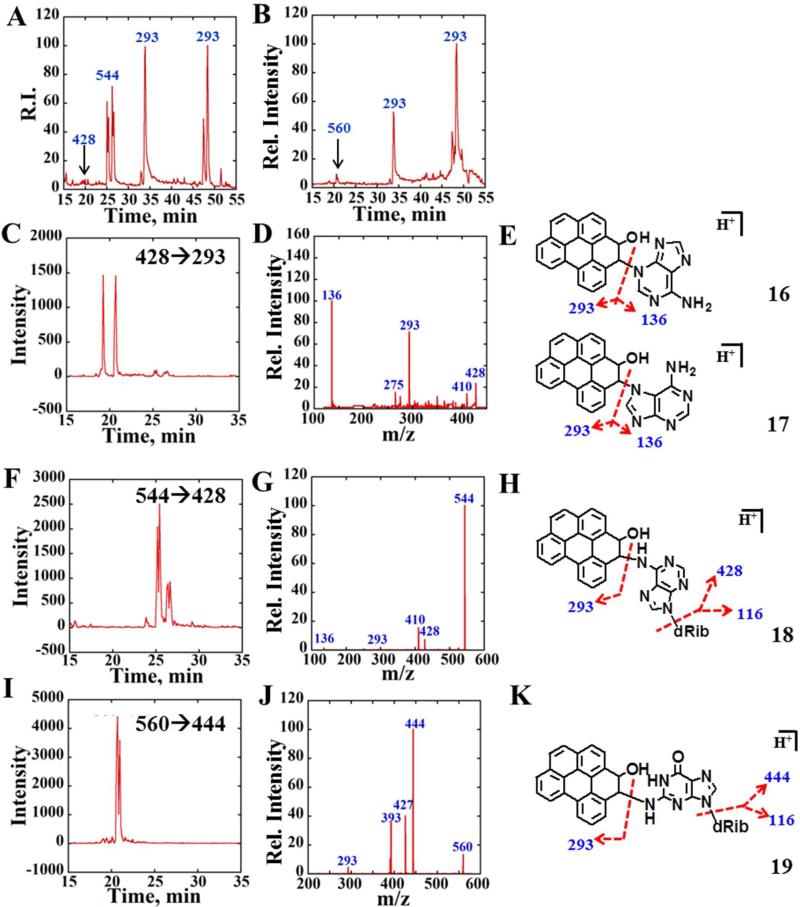
The detection of 15 as a metabolite of B[ghi]P, and the observation of reactive metabolite-derived DNA in ECL results (vide supra) as well as previous result with mouse microsomes25 suggested formation of 4, B[ghi]P 3,4-oxide. To help elucidate possible DNA adduct formation by 4, we synthesized it, obtaining a final product mixture of 40% B[ghi]P 3,4-oxide and 60% unreacted B[ghi]P as demonstrated by NMR (Supporting Information, Fig. S4). B[ghi]P 3,4-oxide was then reacted with dA or dG for 8 hrs to obtain the nucleoside adducts. A surrogate scan using total ion chromatogram was used to survey nucleoside adducts formed. Enhanced product ions (EPI) scan provided collision-induced dissociation (CID) spectra of selected ions, and was used for structural elucidation. Multiple reaction monitoring (MRM) revealed peaks representing mass transitions from precursor to product ion pairs.
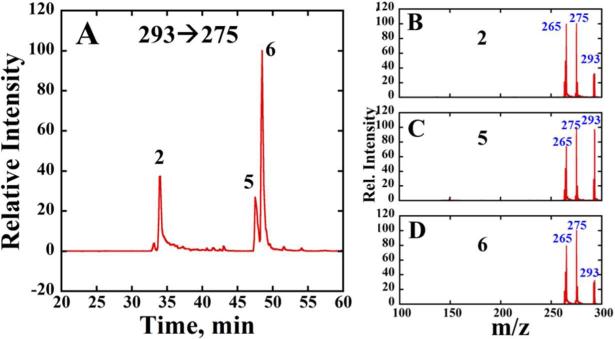
Total ion chromatograms (TIC) were obtained of all eluents containing molecular ions of m/z 544, 428, 293 for reaction of B[ghi]P 3,4-oxide with dA (Figure 4A), and for m/z 560, 293 for reaction of B[ghi]P 3,4-oxide with dG (Figure 4B). CID spectra of eluents at 33, 48, and 49 mins (Fig. 5B–D) presented major product ions, i.e. 275 and 265, of m/z 293. Therefore, these eluents most likely are transformation products of metabolite 2 derived from hydrolysis of 4. Previous work on benzo[e]pyrene 3,4-oxide revealed transformation into phenols 3-hydroxy and 4-hydroxy benzo[e]pyrene. 45,46 By analogy, we suspect that the two eluents with longer retention times (tR) were most likely the B[ghi]P monohydroxy phenols, 5 and 6 (m/z 293) (Scheme 2). Eluent with tR ~33 min is either 4 or hydrolyzed product 2. This observation suggest the formation of 5 and 6 from non-enzymatic hydrolysis of B[ghi]P 3,4-oxide. All these eluents have major mass transitions m/z 293→275 and extracted SRM chromatograms of m/z 293→275 (Figure 5A).
Figure 4.
LC-MS/MS analysis of reactions of synthesized B[ghi]P 3,4-oxide with dA and dG. (A) TIC containing ions of m/z 544, 428 and 293 for reaction between B[ghi]P 3,4-oxide and dA. (B) TIC containing ions of m/z 460, 444 and 293 for the reaction between B[ghi]P 3,4-oxide and dG. (C) Representative MRM chromatogram with a mass transition m/z 428→293. (D) Product ion spectrum of m/z 428 at CE 25 eV. (E) Possible structures of B[ghi]P 3,4-oxide dA adducts m/z 428 where reactions happen at N3 of dA (16) and N7 or dA (17). (F) Representative MRM chromatogram with mass transition m/z 544→428. (G) Product ion spectrum of m/z 544 at CE 15 eV. (H) Possible structure of B[ghi]P 3,4-oxide dA adduct m/z 544 where reaction happens at N6 of dA (18). (I) Representative MRM chromatogram with mass transition m/z 560→444. (J) Product ion spectrum of m/z 560 at CE 15 eV. (K) Possible structures of B[ghi]P 3,4-oxide dG adduct m/z 560 where reaction happens at N2 of dG (19).
Figure 5.
LC-MS/MS analysis of B[ghi]P 3,4-oxide after reaction with dG. (A) Representative MRM chromatogram with mass transition m/z 293→275 (B), (C) and (D) Product ion spectrum of m/z 293 at CE 35 eV for eluents 2, 5 and 6.
Total ion chromatogram (TIC) were obtained of all eluents containing molecular ions of m/z 544, 428, 293 for reaction of B[ghi]P 3,4-oxide with dA (Figure 4A), and for m/z 560, 293 for reaction of B[ghi]P 3,4-oxide with dG (Figure 4B). CID spectra of eluents at 33, 48, and 49 mins (Fig. 5B–D) presented major product ions, i.e. 275 and 265, of m/z 293. Therefore, these eluents most likely are transformation products of metabolite 2. Previous work on benzo[e]pyrene 3,4-oxide, with similar structure to B[ghi]P 3,4-oxide, revealed phenolic transformation into 3-hydroxy and 4-hydroxy benzo[e]pyrene.45,46 Therefore, we suspected that the two eluents with longer retentions were most likely the B[ghi]P monohydroxy phenols, 5 and 6 (m/z 293) (Scheme 2). Eluent with tR ~33 mins is either B[ghi]P 3,4-oxide or its hydrolyzed product 2, from unused B[ghi]P 3,4-oxide. This observation suggest the formation of 5 and 6 from non-enzymatic hydrolysis of B[ghi]P 3,4-oxide. All these eluents have major mass transitions m/z 293→275 and extracted SRM chromatograms of m/z 293→275 (Figure 5A).
Since m/z 293 ions were from either B[ghi]P 3,4-oxide or its derivatives, molecular ions of m/z 544, 428 (Fig. 4A) and 560 (Fig. 4B) were possible adducts produced by reaction of B[ghi]P 3,4-oxide with dA or dG. The CID spectrum in Fig. 4G shows product ions of m/z 544 (dA adducts), including m/z 428, 410, 293 and 136 (fragmentation pattern illustrated in Fig. 4H). The product ions of m/z 560 (dG adducts) were m/z 444, 427, 393 and 293, as shown in Fig. 4J. Both CID spectra show major product ions resulting from the parent ions losing 116, such as 428 from parent 544 and 444 from parent 560. A neutral loss of 116 is the fingerprint fragmentation of stable DNA adducts at low collision energy (CE).31,32 In addition, the MW of ions m/z 544 ([M+H]+) matches the sum up of B[ghi]P 3,4-oxide (m/z 292) and dA (m/z=251). Similarly, the MW of ions m/z 560 ([M+H]+) matches the sum of B[ghi]P 3,4-oxide (m/z 292) and dG (m/z=267). Therefore, it is highly likely that the m/z 544 or 560 ions are exocyclic DNA adducts derived from B[ghi]P 3,4-oxide attacking the N6 position of dA (Fig. 4H, 18) or N2 position of dG (Fig. 4K, 19). These positions are usually prone to reaction with bulky PAH metabolites.3'6 This assumption was strengthened by the common product ions m/z 293, which correspond to 3,4-dihydro B[ghi]P 3-ol, derived from loss of either dA or dG from their parent ions. (Fig. 4H and 4K). Considering products of m/z 544 ions, m/z 410 represents neutral water loss from m/z 428, and m/z 136 corresponds to an adenine. The product ion m/z 393, from m/z 560, was not a feature ion and the structure was not unidentified. The finger print MRM spectra for these adducts were m/z 544→428 for dA and m/z 560→444 for dG (Fig. 4F,I)
In addition to peaks for exocyclic dA and dG adducts of 4, two minor peaks m/z 428 with tR ~ 20 mins (black arrows, Fig. 4A) were also observed. Intensities are much lower than m/z 544, indicating relatively low abundance. Fragmentation of m/z 428 yielded major ions of m/z 293 and m/z 136, and minor ions of m/z 410 and m/z 275 (Fig. 4D, fragmentation illustrated in Fig. 4E). These ions match the m/z of moieties, 3,4-dihydro B[ghi]P 3-ol (m/z 293), adenine (m/z 136), B[ghi]P (m/z 275) and the water loss of parent 428 (m/z 410). Therefore, we concluded that the m/z 428 belongs to depurinated labile adducts of dA47 (Fig 4E, 16 and 17) that are likely formed at N3 and/or N7 positions of dA. No m/z 444 was detected correlating to the labile N7-dG adduct. The major transition for depurinated adducts was m/z 428→293 (Fig. 4C). The structures of compounds 16, 17, 18 and 19 (Fig. 4) are shown for nucleoside alkylating the 4-position of B[ghi]P 3,4-oxide (diastereoisomers not shown). Corresponding isomers with nucleosides alkylating the 3-position (structures not shown) may cause the split peaks in the chromatograms. Major product ions and mass transitions are summarized in Table 3, and were used to search for B[ghi]P-derived DNA adducts in subsequent reactions with DNA-enzyme biocolloid particles as described in the following section.
Table 3.
Mass spectrometry methods can used for detection of B[ghi]P 3,4-oxide nucleosides adducts.
| MS Methods (ESI+) | |||
|---|---|---|---|
| Targets | MRM | EPI | Major Product Ions |
| dA Adducts | 544→428, 428→293 | 544, 428 | 428, 293, 275 |
| dG Adducts | 560→444 | 560, 444 | 444, 293 |
Detection of DNA Adducts of PAH metabolites
As described above, cyt P450s 1A1 and 1B1 generate larger quantities of DNA-reactive B[a]P and B[ghi]P metabolites than cyt P450 1A2.34,48 Therefore, supersomes 1A1 and 1B1 were used in calf thymus DNA/enzyme biocolloid reactions to generate DNA adducts. B[a]P and B[ghi]P were reacted separately with magnetic PDDA/supersomes/PDDA/DNA biocolloids using NADPH regeneration in 96-well reaction/filtration plates, with enzyme hydrolysis to prepare the DNA for LC-MS/MS analysis.
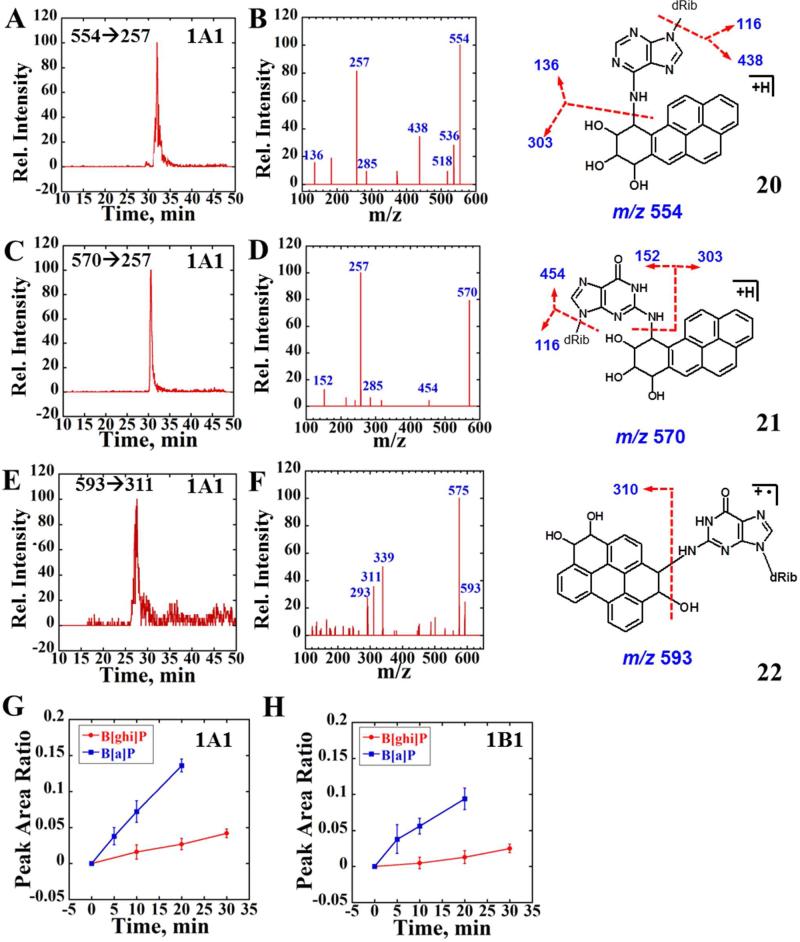
The active diol epoxide metabolite of B[a]P, BPDE, can react with both dA and dG in DNA forming stable adducts BPDE-dA and BPDE-dG.6 In ESI+ mode, molecular ions of BPDE-dA and BPDE-dG are m/z 554 and 570, structures 20 and 21 (Figure 6). Fragmentation of these adducts yielded high intensity of product ions m/z 257 other than deglycosylated ions (m/z 438 or m/z 454).49 The m/z 257 ions, corresponding to 7,9-dihydro-8H cyclopenta[2,1-b]pyren-8-one, derive from ions of m/z 285 that result from sequential loss of the nucleosides and two water molecules.49 Therefore, the dominant mass transitions m/z 554→257 and m/z 570→257 were used to monitor formation of BPDE-dA and BPDE-dG. EPI modes of product ions m/z 554 and m/z 570 were also employed to verify adduct structures.
Figure 6.
LC-MS/MS analysis of reactions of magnetic biocolloid reactors with B[a]P and B[ghi]P using 1A1 supersomes. (A) and (C) Representative SRM chromatogram with mass transition m/z 554→257 and m/z 570→257 indicating the formation of BPDE-dA and BPDE-dG adducts after 20 min reaction followed by enzyme hydrolysis using supersomes 1A1. (B) and (D) Product ion spectrum of m/z 554 and m/z 560 at CE 30 eV. (E) Representative SRM chromatogram with mass transition m/z 593→311 indicating the formation of B[ghi]P derived dG adduct, possibly formed from B[ghi]P 3,4,11,12-bisoxide after 20 min reaction followed by enzyme hydrolysis using supersomes 1A1. (F) Product ion spectrum of m/z 593 at CE 30 eV. (G) and (H) Blue curves reflect the peak area ratios to the internal standard versus time for the sum of both BPDE-dA and BPDE-dG formation (enzymes are as indicated), red curves represent the ratio for B[ghi]P 3,4,11,12-bisoixdes-dG formation. Possible structures of BPDE-dA (20), BPDE-dG (21) and B[ghi]P 3,4,11,12-biosoxides-dG (22) are also shown with red arrows indicating the major cleave sites.
Figures 6A-6D confirm two major DNA adducts with mass transitions m/z 554→257 and m/z 570→257 were observed when DNA/enzyme biocolloids reacted with B[a]P using supersomes 1A1 and NADPH. Similar data using supersomes 1B1 are shown in Supporting Information Fig. S5. The product profiles of molecular ions m/z 554 and m/z 570 in Fig. 6B, 6D demonstrate that the ions originated from reaction of BPDE and nucleosides since both deglycosylated products, m/z 438 and m/z 454 were observed along with adenine and guanine residues (m/z 136 and m/z 152). The same mass transition was not observed when biocolloid reactors were incubated with B[a]P alone, indicating bioactivation was necessary for the formation of BPDE nucleoside adducts. The blue curves in Figure 6G and 6H represent the total BPDE-DNA adducts peak area of m/z 554→257 and m/z 570→257 mass transition relative to internal standard (7-methylguanosine, m/z transition 298→166). B[ghi]P 3,4,11,12-bisoixdes-dG formation is also expressed as a ratio to internal standard.
For 20-mins reaction of B[ghi]P with biocolloid enzyme/DNA reactors, no peaks were found to match mass transitions listed in Table 3, suggesting little formation of DNA adducts of B[ghi]P 3,4-oxide. According to previous work,25 B[ghi]P 3,4,11,12-bioxides can hydrolyze to form compound 3 (diastereoisomers not shown). Experiments were done to investigate possible B[ghi]P 3,4,11,12-bisoxide DNA adducts, including (1) monitoring the signature neutral loss 116 of the sugar if exocyclic adducts are formed, and (2) scanning precursor ion (PIS) containing fragment of m/z 293, 275 (the feature B[ghi]P and B[ghi]P 3(or 4)-ol ions). Neutral loss produced no results, but PIS showed that m/z 593 ions produced m/z 293 and 275 fragments. Molecular ion m/z 593 correlates to the conjugated product of compound 3 (M.W. 326) with dG (M.W. 267), presumably derived from B[ghi]P 3,4,11,12-bisoxide (22, Fig. 6), although stereoisomers are possible. The CID spectrum of m/z 593 (Fig. 6F) showed major fragments contained B[ghi]P 3,4-diol ([M+H]+= 311) and B[ghi]P 3,4-oxide ([M+H]+= 293), indicating m/z 593 is a derivative of reaction product of B[ghi]P metabolite(s) with dG. The major SRM transitions 593→311 was used for following relative quantitation.
Relative formation rates of DNA adducts derived from B[a]P (570→257 and 554→257) and B[ghi]P (593→311) were estimated using SRM peak area ratios vs. the internal standard, 7-methylguanosine (298→166) (Figure 6G,H). Red curves showed increases in relative amount of B[ghi]P derived dG adduct with reaction time. Compared with BPDE-DNA adducts (Fig. 6G-H, blue curves), B[ghi]P metabolism produced only about 20% of the BPDE-DNA adducts in 20-min enzyme reactions for 1A1 and 1B1 supersomes. This observation is consistent with ECL array results, suggesting that DNA-reactive B[ghi]P metabolites are produced at lower levels than those of B[a]P.
DISCUSSION
Results described above demonstrate that ECL genotoxicity arrays with follow up biocolloid reactor metabolite-DNA adduct analysis by LC-MS/MS are a powerful combination to help elucidate complex genotoxicity-related chemical pathways. High throughput features facilitate comprehensive investigations of metabolite reactivity with DNA. Toxicity profiles of the PAHs were initially provided by ECL arrays, then LC-MS/MS analysis of products from enzyme- and DNA/enzyme-biocolloid reactor beads provided metabolite profiling, and structures and formation rates of important metabolites and DNA adducts. Results also revealed for the first time that a major human DNA adduct may result from reaction between B[ghi]P 3,4,11,12-bisoxide and deoxyguanosine.
ECL array results suggested 3.5-fold faster metabolic bioactivation of B[a]P toward DNA damage than B[ghi]P using human cyt P4501A1, 1B1 and 1A2 (Fig. 3). The bioreactor-LC-MS/MS approach validated more reactive B[a]P metabolites than B[ghi]P metabolites, and relative formation of major DNA adducts of B[a]P metabolite BPDE was ~5 times faster than DNA adducts of B[ghi]P metabolites.
The biotransformation of B[a]P involves three important metabolites ((Scheme 2, blue brackets) B[a]P radical, BPDE and B[a]P 7,8 quinone, which all react with DNA.15,16,44,49 B[ghi]P is presumably oxidized mainly at 3,4 or 11,12 positions to form K region epoxides and the active metabolites B[ghi]P 3,4-oxide (4) and B[ghi]P 3,4,11,12-bisoxide (13) that may react with DNA.22,25 Metabolites detected using enzyme biocolloid reactors and LC-MS/MS are consistent with K region epoxidation of B[ghi]P. Compound 1 and 2, the hydrolyzed products of 3 and 4, were the two major products found when B[ghi]P reacted with cyt P450 1A1 and 1B1 (Fig. 2). This is consistent with previous results using induced mouse liver microsomes.22 In addition to K-region epoxidized metabolites, Platt et al. also observed large quantities of phenol and quinone metabolites.22 The difference from ours is most like related to species differences, since human supersomes were used in our study and induced rat liver microsomes were used in Platt's work. We found that human cyt P450s 1A1 and 1B1 are mainly responsible for K-region epoxidation and catalyzed faster metabolism than cyt P450 1A2, similar to B[a]P.34 However, other enzymes that metabolize B[ghi]P cannot be ruled out.
Using 32P-labeling and chromatography, Platt's group revealed several DNA adducts including a major adduct generated from reaction of B[ghi]P 3,4-oxide and DNA when B[ghi]P was activated by induced rat liver microsomes.25 In our work, synthetic B[ghi]P 3,4-oxide was used to confirm generation of dG and dA adducts by LC-MS/MS (Fig. 4), which were consistent with Platt's observations. Our structural information from LC-MS/MS (Fig. 4) indicated that major stable B[ghi]P 3,4-oxide DNA adducts are most likely to form at positions N2 of dG or N6 of dA (Table 3). Each individual peak in the spectra may possibly represent the stereoisomers of the corresponded adduct as they would possess same m/z and fragmentation pattern. Possible structures of these adducts were presented.
In the ECL arrays, BPDE, the active metabolite of B[a]P (SI, Fig. S2) reacts with dA and dG in the DNA/microsome films to induce a significant ECL increase (Fig. 3B). Formation of the corresponding DNA adducts was confirmed by LC/MS-MS (Fig. 6A-D). In general, increases in ECL intensities correlate well with increasing amounts of BPDE dA and dG adducts observed by LC-MS/MS (Fig. 3B, Fig. 6G, H). Although B[ghi]P 3,4-oxide was able to attack DNA form stable dA and dG adducts (Fig. 4C-J), formation of those DNA adducts from B[ghi]P metabolites generated by human cyt P450 supersomes were not found in LC-MS/MS experiments. However, adducts derived from reaction between B[ghi]P derived bisoxides, possibly B[ghi]P-3,4,11,12-bisoxide, and dG were detected (Figure 6E-F). The observed B[ghi]P-3,4,11,12-bisoxide-dG adduct is likely to be the stable adduct according to its m/z. However, the LC-MS/MS method utilizing magnetic bioreactors may not detection of depurinate, unstable PAH-DNA adducts as these unstable adducts can detach from the DNA on the bioreactors and be lost during washing. Compared with the total dA and dG adducts formed from BPDE, B[ghi]P-3,4,11,12-bisoxide resulted in a lower rate of DNA adduct formation (Fig. 6G, H), indicating relative less reactivity and .or smaller formation rate of B[ghi]P-3,4,11,12-bisoxide. This result is consistent with in vivo experiments in which B[ghi]P was not found to initiate tumors in mouse models.23
Undetectable B[ghi]P 3,4-oxide adducts imply very low quantities of such adducts formed in the human enzyme/DNA bioreactor system. Reasons for this may include (a) the hydrophobicity and structurally hindered nature of B[ghi]P 3,4-oxide, (b) the lack of a route featuring carbenium ions (SI, Fig. S6) that can form with BPDE whereas B[ghi]P can only undergo epoxidation at the 3,4 position,50 and (c) rapid hydrolysis of B[ghi]P 3,4-oxide to 2 (Scheme 2) and conversion to phenols 5 and 6 (Fig. 5) in competing reactions to adduct formation.
In summary, ECL genotoxicity arrays and human enzyme/DNA bioreactor LC-MS/MS studies were used to rapidly elucidate differences in genotoxic chemistry featuring metabolite-induced DNA damage. Our findings confirm a considerably lower genotoxic profile of B[ghi]P than B[a]P and identified a new DNA adduct of human B[ghi]P metabolism. Clearly, improvements in LC-MS/MS sensitivity could provide detection of lower abundance metabolites and nucleoside adducts, and is currently being pursued in our laboratory.
Supplementary Material
ACKNOWLEDGMENT
The authors thank Yanke Liang, Kaddy Camara and Dr. Amy Howell for assistance with DMDO synthesis.
Funding
We gratefully acknowledge financial support from the National Institute of Environmental Health Sciences (NIEHS), NIH, USA, Grant No. ES03154.
ABBREVIATIONS
- AKR
aldo-keto reductase
- B[a]P
benzo[a]pyrene
- B[a]P 7, 8-diol
7,8-dihydroxy-7,8-dihydro benzo[a]pyrene
- B[a]P 9
10-diol, 9,10-dihydroxy-9,10-dihydro benzo[a]pyrene
- 3-OH B[a]P
3-hydroxy benzo[a]pyrene
- B[ghi]P
benzo[ghi]perylene
- B[ghi]P 3,4-oxide
3,4-epoxy-3,4-dihydro-B[ghi]P
- B[ghi]P 3,4-diol
3,4-dihydroxy-3,4-dihydro-benzo[ghi]perylene
- B[ghi]P 3,4,11,12-tetrol
3,4,11,12-tetrahydroxy-3,4,11,12-tetrahydro-benzo[ghi]perylene
- 3-OH B[ghi]P
3-hydroxy benzo[ghi]perylene
- 4-OH B[ghi]P
4-hydroxy benzo[ghi]perylene
- BPDE
benzo[a]pyrene 7,8-dihydrodiol 9,10-epoxide
- cyt P450 1A1
cytochrome P4501A1
- cyt P450 1A2
cytochrome P4501A2
- cyt P4501B1
cytochrome P4501B1
- dA
deoxyadenosine
- dG
deoxyguanosine
- DMDO
dimethyldioxirane
- ECL
electrochemiluminescence
- EPI
enhanced product ion
- ESI
electron spray ionization
- mEH
microsomal epoxide hydroloase
- MRM
multiple reactions monitoring
- PAHs
Polycyclic aromatic hydrocarbons
- PDDA
Polydiallyldimethylammonium chloride
- RuPVP
[Ru(bpy)2(PVP)10](ClO4)2
Footnotes
Supporting Information. Figures of metabolic transformation of B[a]P by supersomes, NMR characterization of B[ghi]P 3,4-oxide and QCM characterization of layer-by-layer assembly of supersomes in ECL arrays. This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interests.
REFERENCES
- 1.Neilson AH. PAHs and Related Compounds. Springer; Berlin: 1998. [Google Scholar]
- 2.Gelboin HV. Benzo[a]pyrene metabolism, activation and carcinogenesis: role and regulation of mixed-function oxidases and related enzymes. Physiol. Rev. 1980;60:1107–1166. doi: 10.1152/physrev.1980.60.4.1107. [DOI] [PubMed] [Google Scholar]
- 3.Harvey RG. Metabolic activation, DNA binding, and mechanisms of carcinogenesis. Polycyclic Aromatic Hydrocarbons: Chemistry and Carcinogenicity. Cambridge University Press; New York: 1991. pp. 50–78. [Google Scholar]
- 4.Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shimada T, Oda Y, Gillam EMJ, Guengerich FP, Inoue K. Metabolic Activation of Polycyclic Aromatic Hydrocarbons and Other Procarcinogens by Cytochromes P450 1A1 and P450 1B1 Allelic Variants and Other Human Cytochromes P450 in Salmonella typhimurium NM2009. Drug Metab. Disp. 2001;29:1176–1182. [PubMed] [Google Scholar]
- 6.Szeliga J, Dipple A. DNA Adduct Formation by Polycyclic Aromatic Hydrocarbon Dihydrodiol Epoxides. Chem. Res. Toxicol. 1998;11:1–11. doi: 10.1021/tx970142f. [DOI] [PubMed] [Google Scholar]
- 7.Arif JM, Smith WA, Gupta RC. Tissue distribution of DNA adducts in rats treated by intramammillary injection with dibenzo[a,l]pyrene, 7,12-dimethylbenz[a]anthracene and benzo[a]pyrene. Mut. Res. 1997;378:31–39. doi: 10.1016/s0027-5107(97)00095-x. [DOI] [PubMed] [Google Scholar]
- 8.Prahalad AK, Ross JA, Nelson GB, Roop BC, King LC, Nesnow S, Mass MJ. Dibenzo[a,l]pyrene-induced DNA adduction, tumorigenicity, and Ki-ras oncogene mutations in strain A/J mouse lung. Carcinogenesis. 1997;18:1955–1963. doi: 10.1093/carcin/18.10.1955. [DOI] [PubMed] [Google Scholar]
- 9.Ross JA, Nelson GB, Wilson KH, Rabinowitz JR, Galati A, Stoner GD, Nesnow S, Mass MJ. Adenomas Induced by Polycyclic Aromatic Hydrocarbons in Strain A/J Mouse Lung Correlate with Time.integrated DNA Adduct Levels. Cancer Res. 1995;55:1039–1044. [PubMed] [Google Scholar]
- 10.Poirier MC. Chemical-induced DNA damage and human cancer risk. Nat. Rev. Cancer. 2004;4:630–637. doi: 10.1038/nrc1410. [DOI] [PubMed] [Google Scholar]
- 11.Boysen G, Hecht SS. Analysis of DNA and protein adducts of benzo[a]pyrene in human tissues using structure.specific methods. Mut. Res. 2003;543:17–30. doi: 10.1016/s1383-5742(02)00068-6. [DOI] [PubMed] [Google Scholar]
- 12.Hall M, Grover PL. Polycyclic aromatic hydrocarbon: metabolism, activation and tumor initiation. In: Cooper CS, Grover PL, editors. In Chemical Carcinogenesis and Mutagenesis. Springer; Berlin: 1990. pp. 327–372. [Google Scholar]
- 13.Nebert DW, Dalton TP. The role of cytochrome P450 enzymes in endogenous signalling pathways and environmental carcinogenesis. Nat. Rev. Cancer. 2006;6:947–960. doi: 10.1038/nrc2015. [DOI] [PubMed] [Google Scholar]
- 14.Luch A. On the impact of the molecule structure in chemical carcinogenesis. In: Luch A, editor. Molecular, clinical and environmental toxicology, vol. 1: molecular toxicology. Birkhäuser Basel; Switzerland: 2009. pp. 159–179. [Google Scholar]
- 15.Conney AH, Chang RL, Jerina DM, Caroline Wei SJ. Studies on the Metabolism of Benzo[a]Pyrene and Dose-Dependent Differences in the Mutagenic Profile of Its Ultimate Carcinogenic Metabolite. Drug Metab. Rev. 1994;26:125–163. doi: 10.3109/03602539409029788. [DOI] [PubMed] [Google Scholar]
- 16.Rogan EG, Devanesan PD, RamaKrishna NVS, Higginbotham S, Padmavathi NS, Chapman K, Cavalieri EL, Jeong H, Jankowiak R, Small GJ. Identification and quantitation of benzo[a]pyrene.DNA adducts formed in mouse skin. Chem. Res. Toxicol. 1993;6:356–363. doi: 10.1021/tx00033a017. [DOI] [PubMed] [Google Scholar]
- 17.Chen L, Devanesan PD, Higginbotham S, Ariese F, Jankowiak R, Small GJ, Rogan EG, Cavalieri EL. Expanded analysis of benzo[a]pyrene.DNA adducts formed in vitro and in mouse skin: their significance in tumor initiation. Chem. Res. Toxicol. 1996;9:897–903. doi: 10.1021/tx960004a. [DOI] [PubMed] [Google Scholar]
- 18.Ross JA, Nesnow S. Polycyclic aromatic hydrocarbons: correlations between DNA adducts and ras oncogene mutations. Mutat. Res. 1999;424:155–166. doi: 10.1016/s0027-5107(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 19.Melendez-Colon VJ, Luch A, Seidel A, Baird WM. Cancer initiation by polycyclic aromatic hydrocarbons results from formation of stable DNA adducts rather than apurinic sites. Carcinogenesis. 1999;20:1885–1891. doi: 10.1093/carcin/20.10.1885. [DOI] [PubMed] [Google Scholar]
- 20.Andrews AW, Thibault LH, Lijinsky W. The relationship between carcinogenicity and mutagenicity of some polynuclear hydrocarbons. Mutat. Res. 1978;51:311–318. doi: 10.1016/0027-5107(78)90120-3. [DOI] [PubMed] [Google Scholar]
- 21.Grimmer G, Brune H, Deutsch-Wenzel R, Naujack KW, Misfeld J, Timm J. On the contribution of polycyclic aromatic hydrocarbons to the carcinogenic impact of automobile exhaust condensate evaluated by local application onto mouse skin. Cancer Lett. 1983;21:105–113. doi: 10.1016/0304-3835(83)90089-7. [DOI] [PubMed] [Google Scholar]
- 22.Platt KL, Grupe S. Microsomal biotransformation of benzo[ghi]perylene, a mutagenic polycyclic aromatic hydrocarbon without a “classic” bay region. Chem. Res. Toxicol. 2005;18:700–710. doi: 10.1021/tx049698a. [DOI] [PubMed] [Google Scholar]
- 23.a Sakai M, Yoshida D, Mizusaki S. Mutagenicity of polycyclic aromatic hydrocarbons and quinones on Salmonella typhimurium TA97. Mutat. Res. 1985;156:61–67. doi: 10.1016/0165-1218(85)90007-2. [DOI] [PubMed] [Google Scholar]; b Lijinsky W, Saffiotti U. Relationships between structure and skin tumorigenic activity among hydrogenated derivatives of several polycyclic aromatic hydrocarbons. Ann. Ital. Dermatol. Clin. Sper. 1965;19:34–41. [Google Scholar]
- 24.Hughes NC, Phillips DH. 32P-postlabelling analysis of the covalent binding of benzo[ghi]perylene to DNA in vivo and in vitro. Carcinogenesis. 1993;14:127–133. doi: 10.1093/carcin/14.1.127. [DOI] [PubMed] [Google Scholar]
- 25.Platt KL, Grupe S, Fickler M. The 3,4-oxide is responsible for the DNA binding of benzo[ghi]perylene, a polycyclic aromatic hydrocarbon without a “classic” bay.region. Chem-biol. Interact. 2008;176:179–187. doi: 10.1016/j.cbi.2008.07.010. [DOI] [PubMed] [Google Scholar]
- 26.Platt KL, Aderhold S, Kulpe K, Fickler M. Unexpected DNA damage caused by polycyclic aromatic hydrocarbons under standard laboratory conditions. Mutat. Res. 2008;650:96–103. doi: 10.1016/j.mrgentox.2007.09.011. [DOI] [PubMed] [Google Scholar]
- 27.Rusling JF, Hvastkovs EG, Schenkman JB. Screening of Reactive Metabolites using Genotoxicity Arrays and Enzyme/DNA Biocolloids. In: Nassar A, Hollenburg PF, Scatina J, editors. Drug Metabolism Handbook. J. Wiley; New Jersey: 2009. pp. 307–340. [Google Scholar]
- 28.Hvastkovs EG, Schenkman JB, Rusling JF. Metabolic Toxicity Screening Using Electrochemiluminescence Arrays Coupled with Enzyme-DNA Biocolloid Reactors and Liquid Chromatography-Mass Spectrometry. Annu. Rev. Anal. Chem. 2012;5:79–105. doi: 10.1146/annurev.anchem.111808.073659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hvastkovs EG, So M, Krishnan S, Bajrami B, Tarun M, Jansson I, Schenkman JB, Rusling JF. Electrochemiluminescent Arrays for Cytochrome P450-Activated Genotoxicity Screening. DNA Damage from Benzo[a]pyrene Metabolites. Anal. Chem. 2007;79:1897–1906. doi: 10.1021/ac061975q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan S, Zhao L, Schenkman JB, Rusling JF. Evaluation of Electrochemiluminescent Metabolic Toxicity Screening Arrays Using a Multiple Compound Set. Anal. Chem. 2011;83:2754–2760. doi: 10.1021/ac200050n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bajrami B, Zhao L, Schenkman JB, Rusling JF. Rapid LC-MS Drug Metabolite Profiling Using Microsomal Enzyme Bioreactors in a Parallel Processing Format. Anal. Chem. 2009;81:9921–9929. doi: 10.1021/ac9015853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao L, Schenkman JB, Rusling JF. High-Throughput Metabolic Toxicity Screening Using Magnetic Biocolloid Reactors and LC−MS/MS. Anal. Chem. 2010;82:10172–10178. doi: 10.1021/ac102317a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Forster RJ, Vos JG. Synthesis, characterization, and properties of a series of osmium- and ruthenium-containing metallopolymers. Macromolecules. 1990;23:4372–4377. [Google Scholar]
- 34.Kim JH, Stansbury KH, Walker NJ, Trush MA, Strickland PT, Sutter TR. Metabolism of benzo[a]pyrene and benzo[a]pyrene-7,8-diol by human cytochrome P450 1B1. Carcinogenesis. 1998;19:1847–1853. doi: 10.1093/carcin/19.10.1847. [DOI] [PubMed] [Google Scholar]
- 35.Zhao L, Schenkman JB, Rusling JF. Screening for reactive metabolites using electro-optical arrays featuring human liver cytosol and microsomal enzyme sources and DNA. Chem. Commun. 5386-5388. 2009 doi: 10.1039/b909372a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rusling JF, Hvastkovs EG, Hull DO, Schenkman JB. Biochemical applications of ultrathin films of enzymes, polyions and DNA. Chem. Commun. 2008:141–154. doi: 10.1039/b709121b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Murray RW. Chemistry of dioxiranes. 12. Dioxiranes. Chem. Rev. 1989;89:1187–1201. [Google Scholar]
- 38.Murray RW, Jeyaraman R. Dioxiranes: synthesis and reactions of methyldioxiranes. J. Org. Chem. 1985;50:2847–2853. [Google Scholar]
- 39.Lvov Y. Electrostatic Layer-by-layer Assembly of Proteins and Polyions. In: Lvov Y, Möhwald H, editors. Protein Architecture: Interfacing Molecular Assemblies and Immobilization Biotechnology. CRC Press; New York: 2000. pp. 125–167. [Google Scholar]
- 40.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 41.Vennison SJ. Laboratory manual of genetic engineering. PHI Learning; New Delhi: 2010. Estimation of nuclear acids. pp. 27–30. [Google Scholar]
- 42.Chemical, N. C. I. D. C. E., Branch, P. C., Repository, N.C. C. R. S., and Institute, M. R. Handbook of Analytical and Spectral Data for Polycyclic Aromatic Hydrocarbons: Benzo[a] pyrene and its metabolites. Midwest Research Institute; Kansas city: 1993. pp. 1–100. [Google Scholar]
- 43.Conney AH, Chang RL, Jerina DM, Caroline Wei S-J. Studies on the Metabolism of Benzo[a]Pyrene and Dose.Dependent Differences in the Mutagenic Profile of Its Ultimate Carcinogenic Metabolite. Drug Metab. Rev. 1994;26:125–163. doi: 10.3109/03602539409029788. [DOI] [PubMed] [Google Scholar]
- 44.Melendez-Colon VJ, Luch A, Seidel A, Baird WM. Cancer initiation by polycyclic aromatic hydrocarbons results from formation of stable DNA adducts rather than apurinic sites. Carcinogenesis. 1999;20:1885–1891. doi: 10.1093/carcin/20.10.1885. [DOI] [PubMed] [Google Scholar]
- 45.Lee H, Shyamasundar N, Harvey RG. Isomeric phenols of benzo[e]pyrene. J. Org. Chem. 1981;46:2889–2895. [Google Scholar]
- 46.Murray RW, Singh M, Rath NP. X-ray Crystal Structures of Some Arene Oxides. Struct. Chem. 1999;10:419–427. [Google Scholar]
- 47.Pullman A, Pullman B. Molecular electrostatic potential of the nucleic acids. Q. Rev. Biophys. 1981;14:289–380. doi: 10.1017/s0033583500002341. [DOI] [PubMed] [Google Scholar]
- 48.Shimada T, Gillam EMJ, Oda Y, Tsumura F, Sutter TR, Guengerich FP, Inoue K. Metabolism of Benzo[a]pyrene to trans-7,8-Dihydroxy.7,8.dihydrobenzo[a]pyrene by Recombinant Human Cytochrome P450 1B1 and Purified Liver Epoxide Hydrolase. Chem. Res. Toxicol. 1999;12:623–629. doi: 10.1021/tx990028s. [DOI] [PubMed] [Google Scholar]
- 49.Beland FA, Churchwell MI, Von Tungeln LS, Chen S, Fu PP, Culp SJ, Schoket B, Gyırffy E, Minárovits J, Poirier MC, Bowman ED, Weston A, Doerge DR. High-Performance Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry for the Detection and Quantitation of Benzo[a]pyrene−DNA Adducts. Chem. Res. Toxicol. 2005;18:1306–1315. doi: 10.1021/tx050068y. [DOI] [PubMed] [Google Scholar]
- 50.Enoch SJ, Cronin MT. A review of the electrophilic reaction chemistry involved in covalent DNA binding. Crit. Rev. Toxicol. 2010;40:728–748. doi: 10.3109/10408444.2010.494175. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.