Summary
Autophagy, an intrinsically nonselective process, can also target selective cargo for degradation. The mechanism of selective peroxisome turnover by autophagy-related processes (pexophagy), termed micropexophagy and macropexophagy,is unknown. We show how a Pichia pastoris protein, PpAtg30, mediates peroxisome selection during pexophagy. It is necessary for pexophagy, but not for other selective and nonselective autophagy-related processes. It localizes at the peroxisome membrane via interaction with peroxins, and during pexophagy it colocalizes transiently at the preautophagosomal structure (PAS) and interacts with the autophagy machinery. PpAtg30 is required for formation of pexophagy intermediates, such as the micropexophagy apparatus (MIPA) and the pexophagosome (Ppg). During pexophagy, PpAtg30 undergoes multiple phosphorylations, at least one of which is required for pexophagy. PpAtg30 overexpression stimulates pexophagy even under peroxisome-induction conditions, impairing peroxisome biogenesis. Therefore, PpAtg30 is a key player in the selection of peroxisomes as cargo and in their delivery to the autophagy machinery for pexophagy.
Introduction
Autophagy involves several pathways culminating in the lysosomal or vacuolar degradation of intracellular proteins and organelles in all eukaryotic cells (Klionsky and Ohsumi, 1999). These pathways deliver either specific or nonspecific cargos to the vacuole in yeasts, or to lysosomes in higher eukaryotes (Abeliovich and Klionsky, 2001).
Eukaryotic cells exhibit two general modes of autophagy. In macroautophagy (also referred to as autophagy), an inducible pathway required for cell survival under starvation conditions in yeasts (Yorimitsu and Klionsky, 2005), organelles and cytosolic proteins are nonselectively sequestered into double-membrane vesicles (autophagosomes), which fuse with the vacuole or lysosome. In higher eukaryotes, macroautophagy is implicated in diverse functions, including innate immunity, development, clearance of protein aggregates, and tumor suppression (Levine, 2005). Microautophagy also targets cytosolic proteins and organelles nonspecifically, but is distinct from macroautophagy in that proteins and organelles are captured directly by alysosome or vacuole, without prior sequestration in a separate vesicle.
Many proteins required for macroautophagy in yeast are shared with those necessary for selective cargo degradation, such as the cytosol-to-vacuole (Cvt) pathway in Saccharomyces cerevisiae (Scott et al., 1996), which transports specific cargos (aminopeptidase I/ScApe1 and α-mannosidase) for delivery to the vacuole, where they are processed to their functional forms. Given this overlap of the machinery components, it is not surprising that in S. cerevisiae many proteins required for the Cvt and/or autophagic pathways localize at least transiently to the PAS (Kim et al., 2001a), a site of cargo sequestration and vesicle formation for the Cvt and autophagy pathways.
Pexophagy, the specific degradation of peroxisomes by autophagy-related pathways, occurs in a range of organisms from unicellular eukaryotes to mammals but has been studied the most in methylotrophic yeasts (Dunn et al., 2005; Farré and Subramani, 2004; Kiel et al., 2003; Kim and Klionsky, 2000). Growing Pichia pastoris or Hansenula polymorpha cells on methanol as a carbon and energy source induces peroxisomes and the synthesis of peroxisomal alcohol oxidase (AOX) and other enzymes required for methanol metabolism. When peroxisomes are no longer required, two morphologically and genetically distinct, selective autophagic pathways are exploited for pexophagy. In P. pastoris, adaptation from methanol to glucose medium induces micropexophagy, whereas a switch from methanol to ethanol medium induces macropexophagy (Farré and Subramani, 2004; Tuttle and Dunn, 1995).
The micro- and macropexophagy pathways are morphologically similar to the micro- and macroautophagy pathways, but the primary distinction in pexophagy is that peroxisomes are selectively chosen and delivered to the autophagy machinery for degradation. During micropexophagy, peroxisomes are sequestered by arm-like extensions from the vacuole. These sequestering membrane arms emanate from perivacuolar structures (PVS) (Chang et al., 2005) and septate to form sequestering vacuolar arms that almost completely surround the peroxisomes (Mukaiyama et al., 2002). A second membrane structure, the MIPA, forms between the membrane tips of an engulfing vacuole and completes the engulfment and sequestration of peroxisomes (Mukaiyama et al., 2004). Membrane fusion events occur, releasing a single-membrane vesicle containing peroxisomes (micropexophagic body) into the lumen of the vacuole for degradation.In contrast, during macropexophagy, single peroxisomes are sequestered by a newly formed membrane, which enwraps the peroxisome to form a double-membrane vesicle, the Ppg (Ano et al., 2005). The outer membrane of the Ppg fuses with the vacuole, delivering a macropexophagic body into the vacuolar lumen.
Not surprisingly, most proteins required for macroautophagy are also necessary for pexophagy. However, pexophagy requires the specific recognition of peroxisomes, as well as signaling proteins to up- and downregulate the process. In P. pastoris, the α subunit of phosphofructokinase (PpPfk1) is involved in signaling events during micropexophagy (Yuan et al., 1997), and PpAtg11, PpAtg26, and PpAtg28 may be required in the early events following peroxisome recognition. In H. polymorpha, HpPex3 and HpPex14, two of the main components of the peroxisome biogenesis machinery required for membrane and matrix protein import, also have roles in macropexophagy (Bellu et al., 2001, 2002; Zutphen et al., 2008).
Although current progress has defined certain peroxisomal membrane and autophagic machinery components necessary for pexophagy, two central unanswered questions are how selectivity is achieved and how peroxisomes and the autophagic machinery are coupled during pexophagy. We describe a novel P. pastoris protein (PpAtg30) involved in peroxisome recognition and perhaps signaling during both micro- and macropexophagy. PpAtg30 interacts with both autophagic and peroxisomal proteins, but its activation of the autophagic machinery is dependent upon its phosphorylation, which occurs only under peroxisome turnover conditions. We present a model to explain how peroxisomes are selected by PpAtg30 and delivered to the autophagic machinery for pexophagy.
Results
Identification of PpATG30
An unknown ORF, called PpATG30 hereafter (nucleotide sequences have been deposited in the Entrez database with accession number:AY310405),was identified from a collection of micropexophagy mutants obtained by restriction enzyme mediated integration (REMI; Farré et al., 2007). It encodes a 44 kDa protein with two coiled-coil domains and is not well conserved. A few putative homologs of PpATG30 were found in H. polymorpha (Jan Kiel, personal communication), P. guilliermondii (accession number: XP_001485854), P. stipitis (accession number: XP_001387397), C. albicans (accession number: XM_710803), L. elongisporus (accession number: XM_001526852), and D. hansenii (accession number: XM_458648).
PpATG30 Is Essential for Pexophagy but Not for the Cvt or Autophagy Pathways
Fluorescence microscopy confirmed the pexophagy defect in Ppatg30Δ cells and indicated the stage at which micropexophagy is blocked (Figure 1A). Using Ppatg30Δ and wild-type cells expressing BFP-SKL (BFP fused to Peroxisome Targeting Signal 1), peroxisomes were induced in methanol medium (in the presence of FM4-64 to stain the vacuole membrane) and then shifted either to glucose or ethanol medium to induce micropexophagy or macropexophagy, respectively. After 2 hr of glucose adaptation, diffuse BFP fluorescence, a hallmark of peroxisome degradation, appeared in the vacuole lumen of wild-type cells (Figure 1A). Micropexophagy in the Ppatg30Δ strain was blocked at a late stage (referred to elsewhere as stage 1c; Mukaiyama et al., 2002), just prior to the final stage of peroxisome sequestration by septated vacuoles. After 3.5 hr of ethanol adaptation, the wild-type cells had degraded most peroxisomes as the BFP fluorescence was inside the vacuole (Figure 1A). In Ppatg30Δ cells, by contrast, the BFP fluorescence remained peroxisomal and outside the vacuole.
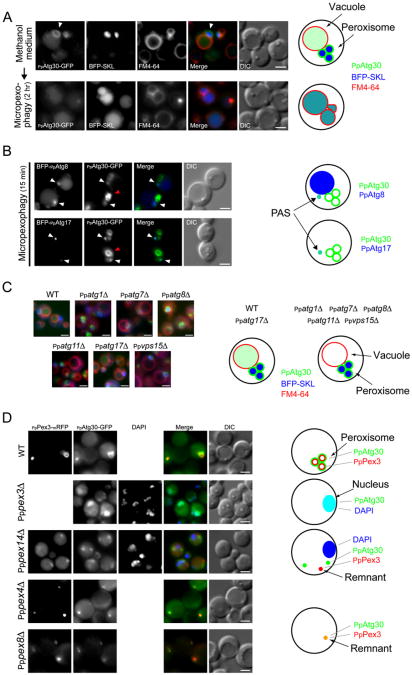
Figure 1. PpAtg30 Is Essential for Micropexophagy and Macropexophagy, but Not for the Cvt or Autophagy Pathway.
(A) Wild-type (SJCF247) and Ppatg30Δ (SJCF332) cells expressing BFP-SKL were transferred from methanol to glucose or ethanol medium to induce micropexophagy or macropexophagy, respectively. Merged images show vacuoles stained by FM4-64 (red) and peroxisomes labeled with BFP-SKL (blue). Bars = 2 μm.
(B) Visualization of AOX activity after glucose adaptation for 10 hr (micropexophagy) and ethanol adaptation for 30 hr (macropexophagy) in wild-type (PPY12), Ppatg8Δ (SJCF257), and Ppatg30Δ (SJCF44) cells.
(C) Ppatg30Δ (SJCF44) and Ppatg30Δ cells expressing PpAtg30-HA (SJCF590) were grown in methanol medium overnight and shifted to glucose medium. Five OD (optical density at 600 nm) of cells were lysed and analyzed as described in the Experimental Procedures. AOX, PpPex3, and PpPex17 were used to follow peroxisome degradation. The β subunit of F0/F1 ATPase served as loading control. Filled and open arrowheads show unmodified and modified PpAtg30, respectively.
(D) Processing of PpApe1-CFP by the Cvt and autophagy pathways. Wild-type (SJCF483) and Ppatg30Δ (SJCF651) cells expressing PpApe1-CFP were grown in glucose medium (+N, Cvt pathway) and switched to nitrogen starvation medium for 4 hr (−N, autophagy pathway). PpApe1-CFP was detected in cell lysates. Precursor (pr) and mature (m) forms, respectively, of PpApe1 are shown.
(E) Wild-type (SJCF547), Ppatg30Δ (SJCF826), Ppatg9Δ (SJCF843), Ppatg11Δ (SJCF777), and pep4 prb1(SJCF557) cells expressing GFP-PpAtg8 were grown in glucose medium (+N) and shifted to SD-N for 6 hr (−N). Protein extracts were immunoblotted with anti-GFP antibody.
(F) Ppatg30Δ cells remain viable in nitrogen starvation conditions. Wild-type (PPY12), Ppatg30Δ (SJCF44), and Ppvps15Δ cells were grown toapproximately1 OD/ml in YPD medium, washed, and resuspended in nitrogen starvation medium at 0.5 OD/ml. Viability was assessed as described in the Experimental Procedures.
The inability of the Ppatg30Δ cells to perform pexophagy was also followed by disappearance of several peroxisomal markers. In wild-type cells, but not in the Ppatg8Δ strain (deficient in all autophagy-related processes) and Ppatg30Δ cells, upon 10 hr of glucose adaptation (or 30 hr of ethanol adaptation), the peroxisomal AOX activity was barely detected (Figure 1B). Furthermore, in Ppatg30Δ cells complemented with PpAtg30-HA, a significant loss of AOX, PpPex3, and PpPex17 proteins was seen during pexophagy (Figure 1C). Similar results were obtained when cells were grown in oleate or methanol medium and pexophagy was induced by a shift to nitrogen starvation medium (Figure S1A, see the Supplemental Data available with this article online).
Recently, we found a Cvt pathway in P. pastoris (Farré et al., 2007). As in S. cerevisiae, the PpApe1 is delivered to the vacuole by this pathway that requires several autophagy proteins and is cleaved to a lower molecular weight mature form by a vacuolar protease. PpApe1 processing was normal in Ppatg30Δ cells (Figure 1D) resembling that in wild-type cells both in rich medium (+N, Cvt pathway) and in nitrogen starvation conditions (−N, macroautophagy pathway).
Atg8, an ubiquitin-like protein required for autophagy-related pathways, is normally also degraded in the vacuole (Shintani and Klionsky, 2004). Recently, a biochemical assay was used to monitor autophagy using GFP-Atg8 exploiting the resistance of GFP to vacuolar protease digestion, causing the appearance of free GFP in autophagy-competent cells (Shintani and Klionsky, 2004). As expected, the formation of free GFP was detected during autophagy conditions (–N) in WT cells and in the Cvt mutant Ppatg11Δ, but not in the autophagy mutant Ppatg9Δ, or in the protease-deficient strain pep4 prb1 (Figure 1E). During autophagy conditions, Ppatg30Δ cells were similar to wild-type and Ppatg11Δ cells.
Cell viability assays showed that as in wild-type cells, Ppatg30Δ cells survived long periods in nitrogen starvation medium, whereas autophagy mutants, such as Ppvps15Δ cells, did not (Figure 1F).
Therefore PpATG30 is required for both micropexophagy and macropexophagy, but not for the Cvt or autophagy pathways. It is, to our knowledge, the first gene required specifically for both pexophagy pathways.
PpAtg30 Localization
We examined PpAtg30 localization by fluorescence microscopy. PpAtg30-GFP surrounded the peroxisome cluster (Figure 2A, arrow), but a small amount of PpAtg30-GFP was also inside the vacuole in methanol-grown cells. Upon induction of micropexophagy, the PpAtg30-GFP and the peroxisome cluster (marked by BFP-SKL) were inside the vacuolar lumen.
Figure 2. PpAtg30 Colocalizes with Peroxisomes, and with the PAS, as well as with Vacuoles, during Pexophagy.
(A) PpAtg30 colocalizes with the peroxisomes. Ppatg30Δ cells coexpressing PpAtg30-GFP and BFP-SKL (SJCF385) were grown overnight in methanol medium with FM4-64, shifted to glucose medium for 2 hr, and examined by fluorescence microscopy. Arrowhead indicates peroxisomes.
(B) PpAtg30 colocalizes with PpAtg8 and PpAtg17. Ppatg8Δ cells coexpressing PpAtg30-GFP and BFP-PpAtg8 (SJCF764), and wild-type cells coexpressing PpAtg30-GFP and BFP-PpAtg17 (SJCF409), were grown overnight in methanol medium, adapted to glucose medium for 15 min, and examined by fluorescence microscopy. White arrow, colocalization of PpAtg30-GFP with BFP-PpAtg8 and/or BFP-PpAtg17, probably at the PAS; red arrow, PpAtg30 around the peroxisome.
(C) Fluorescence microscopy of FM4-64-stained wild-type (SJCF393), Ppatg1Δ (SJCF624), Ppatg7Δ (SJCF387), Ppatg8Δ (SJCF389), Ppatg11Δ (SJCF623), Ppatg17Δ (SJCF390), and Ppvps15Δ (SJCF391) cells expressing BFP-SKL and PpAtg30-GFP. Cells were grown in methanol medium for 6 hr.
(D) Fluorescence and DIC microscopy of wild-type (SJCF768), Pppex14Δ (SJCF587), Pppex4Δ (SJCF769), and Pppex8Δ (SJCF770) cells coexpressing PpPex3-mRFP and PpAtg30-GFP; and Pppex3Δ (SJCF330) cells expressing PpAtg30-GFP. Nuclei were stained with DAPI. Cells were grown in methanol medium for 6 hr. PpAtg30 was expressed from its endogenous promoter in (A), (B), (C), and (D). Bars = 2 μm.
Some of the PpAtg30 also localized near peroxisomes during early stages of micropexophagy. To identify if this localization was at the PAS or MIPA, we used two autophagy proteins, PpAtg8 and PpAtg17. In S. cerevisiae, ScAtg8 and ScAtg17 localize to the PAS (Suzuki et al., 2007). In P. pastoris, PpAtg8 also localizes transiently to the MIPA and the Ppg during micropexophagy and macropexophagy, respectively (Mukaiyama et al., 2004; Oku et al., 2003). Functional N-terminal fusions of BFP-PpAtg8 or BFP-PpAtg17 colocalized transiently with the nonperoxisomal PpAtg30-GFP soon after glucose adaptation (15 min) (Figure 2B and Figure S2). BFP-PpAtg8 localized in a perivacuolar dot (the PAS), and also at the MIPA in 5%–10% of cells examined during the early stages of micropexophagy, and later in the vacuolar lumen of most cells. In cells where BFP-PpAtg8 formed a dot near the peroxisome cluster, it colocalized with PpAtg30-GFP. The proximity of large peroxisomes, where most of the PpAtg30 localizes, to the MIPA, precluded the unambiguous localization of PpAtg30 to the MIPA.
The location of PpAtg30 was also examined by subcellular fractionation and immunoelectron microscopy (Figure S3). PpAtg30 was in the membrane pellet fraction (P200), as was PpPex17 (Figure S3A). Treatment of the pellet fraction with Na2CO3 (to strip off peripheral membrane proteins) or 1% Triton X-100 did not solubilize PpAtg30 (Figure S3B). The absence of predicted transmembrane segments in PpAtg30 suggests it is a peripheral membrane protein, but it associates strongly with the membrane fraction and it may form a detergent-resistant complex as described for ScAtg11 (Kim et al., 2001b).The peroxisomal membrane localization of PpAtg30 was also confirmed by immunoeletron microscopy (Figure S3C). Examination of many such micrographs showed that 89% of the immunogold label for PpAtg30 localized close to or on the peroxisomal membrane.
We then localized PpAtg30-GFP in autophagy and autophagy-related mutants (Figure 2C). As described above, PpAtg30-GFP in wild-type cells localized mostly around the peroxisomes, with a small fraction in the vacuolar lumen. In most pexophagy mutants tested (Ppatg1Δ, Ppatg7Δ, Ppatg8Δ, Ppatg11Δ, and Ppvps15Δ), PpAtg30-GFP expressed from its endogenous promoter accumulated even more densely around the peroxisomes and was absent from the vacuole in methanol-grown cells and during pexophagy conditions. The brighter signal probably reflects a block in delivery to the vacuole for degradation. PpAtg17 has a minor effect during pexophagy (unpublished data), and unlike the other autophagy-related mutants tested, Ppatg17Δ cells did not accumulate PpAtg30-GFP around the peroxisomes. Instead, as in wild-type cells, a small fraction of the PpAtg30-GFP was inside the vacuole.
In H. polymorpha, HpPex3 and HpPex14 are directly or indirectly involved in macropexophagy (Bellu et al., 2001, 2002; Zutphen et al., 2008). The localization of PpAtg30 was affected in corresponding pex mutants of P. pastoris (Figure 2D). In wild-type cells, PpAtg30-GFP expressed from the endogenous promoter localized at the peroxisomes and colocalized with PpPex3-mRFP. In Pppex3Δ cells, PpAtg30-GFP mislocalized to the DAPI-stained nucleus. In Pppex14Δ cells, PpPex3-mRFP localized to aborted peroxisomal structures called “remnants.” PpAtg30-GFP did not localize to remnants or to the nucleus in Pppex14Δ cells, but instead mislocalized to an unknown structure in the cytosol. In other pex mutants, such as Pppex4Δ or Pppex8Δ, PpAtg30-GFP localization was not affected and it colocalized with PpPex3-mRFP at the remnants.
The colocalization of PpAtg30 with peroxisomes, and transiently with the autophagy proteins PpAtg8 and PpAtg17, supports its role in pexophagy.
PpAtg30 Phosphorylation and Its Requirement for Pexophagy
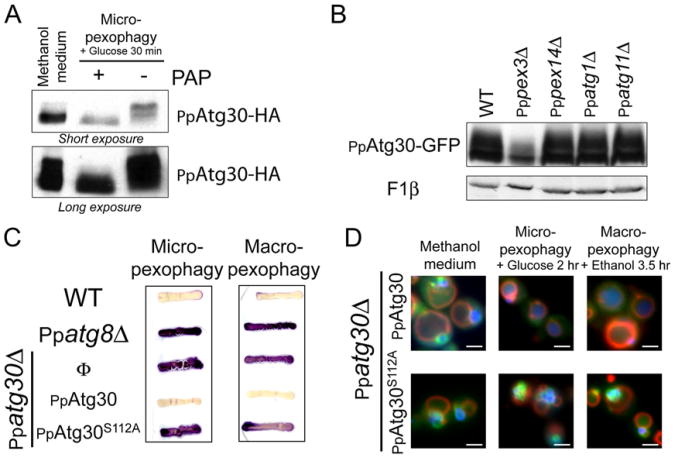
In glucose medium, PpAtg30-HA was barely discernible (data not shown), but its expression increased when cells were shifted to methanol medium (time 0 hr during micropexophagy) and declined when cells were shifted back to glucose medium (time 1.5–6 hr during micropexophagy; Figure 1C), coincident with its vacuolar delivery (Figure 2A). The shift from methanol to either glucose or ethanol also resulted in increased prominence of modified forms of PpAtg30 (with an additional apparent molecular weight up to 5 kDa). These bands were in low abundance in cells grown on methanol medium (Figure 1C at 0 hr), but dramatically increased to become the most abundant species during adaptation to glucose (Figure 1C at 1.5 hr and Figure S1B) orethanol medium (data not shown). The density of higher bands increased concomitantly with a decrease in density of the lower band. Both the unmodified and modified forms were undetectable after 6 hr of adaptation to glucose or ethanol medium, suggesting that PpAtg30 is also degraded during pexophagy (Figure 1C). The slower-migrating bands from wild-type cells were either absent or present at greatly reduced levels in phosphatase-treated samples, confirming that the modification was phosphorylation (Figure 3A). The phosphorylation status of PpAtg30 was analyzed in two strains deficient in peroxisome biogenesis (Pppex3Δ and Pppex14Δ) and in pexophagy (Ppatg1Δ and Ppatg11Δ). Only PpPex3 was required for the phosphorylation of PpAtg30 (Figure 3B), suggesting that PpAtg30 phosphorylation follows its recruitment to peroxisomes.
Figure 3. PpAtg30 Phosphorylation.
(A) PpAtg30 is a phosphoprotein. Ppatg30Δ cells expressing PpAtg30-HA (SJCF590) were grown 6 hr in methanol and micropexophagy was induced for 30 min. Three OD of cells were incubated for 20 min with and without PAP at 30° C.
(B) PpAtg30 phosphorylation depends on PpPex3. PpAtg30 phosphorylation was analyzed by immunoblotting of cell lysates expressing PpAtg30-GFP: wild-type (SJCF600), Pppex3Δ (SJCF330), Pppex14Δ (SJCF366), Ppatg1 D (SJCF473), and Ppatg11Δ (SJCF420).
(C) S112A mutation in PpAtg30 blocks pexophagy. Strains used for the AOX test are: wild-type (PPY12), Ppatg8Δ (SJCF257), Ppatg30Δ (SJCF44), Ppatg30Δ expressing PpAtg30-GFP (SJCF632), and Ppatg30Δ expressing PpAtg30S112A-GFP (SJCF736).
(D) The mutation S112A does not affect PpAtg30 localization. Ppatg30Δ cells coexpressing PpAtg30-GFP or PpAtg30S112A-GFP and BFP-SKL (SJCF385 andSJCF757) were analyzed by fluorescence microscopy.
Using different short deletions, the region from amino acid 95–119 of PpAtg30 was found to be essential for pexophagy, but not its localization. This region of 25 amino acids contains six serines, one threonine, and four tyrosines. Several point mutations in this region were constructed and expressed in the Ppatg30Δ strain (data not shown). PpAtg30S112A failed to complement either micro- or macropexophagy in Ppatg30Δ cells (Figure 3C). Upon induction of pexophagy, AOX activity persisted as in the Ppatg8Δ cells, but not in wild-type cells. By fluorescence microscopy, although PpAtg30S112A-GFP localized normally around peroxisomes in methanol-grown cells (Figure 3D), both the GFP and BFP-SKL remained in clusters outside the vacuole after 2 hr of glucose adaptation or 3.5 hr of ethanol adaptation. Ppatg30Δ cells complemented with the wild-type protein completely degraded the peroxisomes under the same conditions. To verify that S112 is phosphorylated, affinity-purified PpAtg30-ProteinA was subjected to mass spectrometry (MS). The MS data revealed that PpAtg30 was phosphorylated on several residues representing at least nine phosphorylation sites. Upon testing of the single point mutations of each of these sites, only one, S112, blocked pexophagy completely. The MS-MS chromatogram of the peptide 95–116 showed phosphorylation of serine 112 (Figure S4B).
Overexpression of Atg30 Induces Pexophagy during Peroxisome Proliferation Conditions
We analyzed the effects of altering the PpAtg30 expression level on cells growing in methanol. Moderate overexpression of PpAtg30 from one additional locus (driven by its own promoter) did not affect peroxisome biogenesis as judged by the similar levels of PpPex3 and PpPex17 in strains expressing either one or two copies of PpAtg30 (Figure 4A). When, however, PpAtg30 was overexpressed from the strong GAPDH promoter, levels of PpPex3 and PpPex17 were severely reduced (Figure 4A and Figure S5C) and cell growth slowed significantly (Figure 4B). The growth defect caused by strong PpAtg30 overexpression mimics the phenotype of peroxisome biogenesis mutants to a degree, although strains overexpressing PpAtg30 did grow after 48 hr and eventually reached the same cell numbers at stationary phase as wild-type cells.
Figure 4. PpAtg30 Induces Pexophagy.
(A) Overexpression of PpAtg30 induces pexophagy in methanol medium. Strains expressed PpAtg30-GFP at three different levels: (1, SJCF137) PpAtg30 expressed from the endogenous promoter (endogenous expression), (2, SJCF600) endogenous expression supplemented by one copy of PpAtg30 driven by its own promoter (double expression), and (3, SJCF20) PpAtg30 expressed from the strong and constitutive GAPDH promoter (high-level expression). Cells were grown in methanol medium for 6 hr.
(B) Growth curves in methanol medium of wild-type strains expressing normal (SJCF600) or high levels of PpAtg30 (SJCF20). Cells were precultured to mid-log phase in YPD medium, washed, and resuspended at a density of 0.2 OD/ml in methanol medium.
(C) These same strains were transformed with a plasmid expressing BFP-SKL (normal expression [SJCF393] and overexpression [SJCF314]) and examined by fluorescence microscopy.
(D) Fluorescence microscopy of methanol-grown cells (wild-type [SJCF247 and SJCF578], Ppatg30Δ [SJCF332 and SJCF909], Ppatg1Δ [SJCF907], and Ppatg11Δ [SJCF908]) expressing BFP-SKL and PpAtg30-GFP under the control of the copper-inducible CUP1 promoter. Cells were grown for 14 hr in methanol medium and for an additional 2 hr in the presence of 200 μM CuSO4. White arrowhead shows colocalization between BFP-SKL (peroxisomes) and PpAtg30-GFP at the vacuole lumen. Bars = 2 μm.
(E) Immunoblotting of cell lysates expressing different levels of PpAtg30 from its own promoter or from the GAPDH promoter in wild-type (SJCF600 and SJCF20),Ppatg30Δ transformed with PpAtg30S112A (SJCF910 and SJCF758), Ppatg1Δ (SJCF473 and SJCF471), Ppatg2Δ (SJCF837 and SJCF827), Ppatg8Δ (SJCF823and SJCF279), Ppatg11Δ (SJCF420 and SJCF829), and Ppatg28Δ (SJCF848 and SJCF839) cells. (F) Immunoblotting of cell lysates from wild-type (SJCF600 and SJCF20), Pppex3Δ (SJCF330 and SJCF893), and Pppex14Δ (SJCF366 and SJCF892) cells expressing PpAtg30-GFP from its own promoter or the GAPDH promoter. The wedge in (E) and (F) indicates increasing PpAtg30 expression levels. Cells in (A), (C), (E), and (F) were grown in methanol for 6 hr. Bars = 2 μm.
Strong PpAtg30 overexpression also inhibited the peroxisomal import of BFP-SKL in cells growing in methanol medium. After 6 hr, no peroxisomes were seen by differential interference contrast (DIC) or fluorescence microscopy (Figure 4C). Most of the BFP-SKL was mislocalized to the cytosol and the PpAtg30-GFP was in the vacuolar lumen.
To further confirm that the mislocalization of BFP-SKL under these conditions was caused by peroxisome degradation in the vacuole and not due to direct or indirect effects of PpAtg30 overexpression on biogenesis, we induced PpAtg30-GFP after the peroxisomes had been formed and confirmed their degradation by fluorescence microscopy (Figure 4D). BFP-SKL expressed from the AOX promoter was used to label peroxisomes and we overexpressed PpAtg30 using a copper-inducible promoter CUP1 (Koller et al., 2000). Cells were grown in methanol medium for 14 hr prior to inducing PpAtg30 with 200 mM CuSO4 for 2 hr. We used this protocol to analyze four different strains (WT, Ppatg1Δ, Ppatg11Δ, and Ppatg30Δ). In wild-type and Ppatg30Δ cells examined without or before the addition of copper, peroxisomes proliferated normally and PpAtg30 was only faintly visible. After copper addition, most of PpAtg30-GFP and BFP-SKL colocalized as a diffuse haze inside the vacuole. Cells deficient in Ppatg1Δ and Ppatg11Δ resembled the wild-type before the addition of copper. After induction, PpAtg30-GFP accumulated around the peroxisomes and pexophagy failed to occur. Furthermore, in Ppatg1Δ, cells, PpAtg30-GFP accumulated more strongly than in Ppatg11Δ cells, probably due to the fact that all autophagy-related pathways are defunct in the Ppatg1Δ cells as compared to Ppatg11Δ. Similar results were obtained with cells grown on oleate. Overexpression of PpAtg30 led to delivery of BFP-SKL to the vacuole and a decrease in the number of peroxisomes, while overexpression of PpAtg30S112A in oleate did not induce pexophagy (Figure S5D).
In methanol-grown cells, pexophagy induced by PpAtg30 overexpression did not occur in either the autophagy/pexophagy/Cvt mutants (Ppatg1Δ, Ppatg2Δ, and Ppatg8Δ) or the pexophagy/Cvt mutants (Ppatg11Δ and Ppatg28Δ). In each case, the levels of PpPex3 and PpPex17 were not reduced by overexpression of PpAtg30 (Figure 4E). As with the autophagy mutants, both Pppex3Δ and Pppex14Δ cells failed to degrade PpPex3 or/and PpPex17 (Figure 4F). When PpAtg30S112A-GFP was overexpressed in Ppatg30Δ cells, PpPex3 and PpPex17 degradation were drastically reduced (Figure 4E) and delivery of BFP-SKL to the vacuole was impaired in oleate grown cells (Figure S5D).
PpAtg30 Is Required for Functional MIPA and Ppg Formation and for Relocation of Atg11 to the Peroxisome-Sequestering Arms of the Vacuole
The MIPA and Ppg contain at least PpAtg8 and PpAtg26 (Mukaiyama et al., 2004). In wild-type cells, the MIPA or Ppg were found in 5% of the cells during glucose and ethanol adaptation, respectively (Figure 5A and Figure S6A). In Ppatg30Δ cells, these structures were not formed and GFP-PpAtg8 remained primarily cytosolic and at the PAS (Figures S6A and S6B). These data demonstrate that PpAtg30 is essential for the relocation of GFP-PpAtg8 to the MIPA during micropexophagy, and to the Ppg during macropexophagy. Thus PpAtg30 is required for the formation of functional MIPA and Ppg compartments.
Figure 5. PpAtg30 Is Required for MIPA and Pexophagosome Formation and PpAtg11 Localization.
(A) Fluorescence and DIC microscopy of wild-type (SJCF320) and Ppatg30Δ (SJCF376) cells expressing BFP-SKL and GFP-PpAtg8 under the control of the ATG8 promoter. Cells were grown overnight in methanol medium and shifted to glucose medium or ethanol for 20 min to induce micropexophagy or macropexophagy, respectively. The MIPA and Ppg are indicated by arrowheads. Bars = 2 μm.
(B) PpAtg8 lipidation status is not affected in Ppatg30Δ cells. Wild-type (SJCF911), Ppatg7Δ (SJCF912), and Ppatg30Δ (SJCF913) cells expressing HA-PpAtg8 under its own promoter were grown in methanol medium (SM) to mid-log phase and shifted for 60 min on glucose medium (SD). HA-PpAtg8-PE was separated from HA-PpAtg8 by 15% SDS page gels containing 6% urea and analyzed by immunoblotting.
(C) Fluorescence and DIC microscopy of wild-type (SJCF594) and Ppatg30Δ (SJCF432) cells expressing GFP-PpAtg11 and BFP-SKL. Cells were grown overnight in methanol and shifted to glucose for 10 min to induce micropexophagy. Bars = 2 μm.
Atg8 undergoes a series of posttranslational modifications resulting in conjugation to phosphatidylethanolamine (PE), thereby anchoring Atg8 to membranes (Ichimura et al., 2000). We extended our analysis by examining lipidation of PpAtg8 in cells grown in methanol medium (SM) and undergoing micropexophagy (60 min in glucose medium [SD]; Figure 5B). As a control for lipidation we used Ppatg7Δ cells, which lack the E1-like protein involved in Atg8 activation (Ichimura et al., 2000). Wild-type and Ppatg30Δ cells, but not the Ppatg7Δ cells, displayed significant amounts of lipidated HA-PpAtg8 (HA-PpAtg8-PE).
ScAtg11 is involved in cargo recognition in the Cvt pathway and is also required for pexophagy (Kim et al., 2001b; Yorimitsu and Klionsky, 2005). In wild-type cells, GFP-PpAtg11 localized mostly at the vacuolar membrane and in several dots on the membrane, some of which are on the sequestering membranes that surround the peroxisomes (BFP-SKL; Figure 5C). In Ppatg30Δ cells, GFP-PpAtg11 remained localized to the vacuolar membrane, but the puncta at the sequestering arms were absent. Thus, during micropexophagy in the absence of PpAtg30, events downstream of peroxisome recognition, such as proper formation of the MIPA and the Ppg, as well as the relocalization of PpAtg11 to the peroxisome-sequestering arms of the vacuole, do not proceed normally.
PpAtg30 Interacts with Pex and Atg Proteins
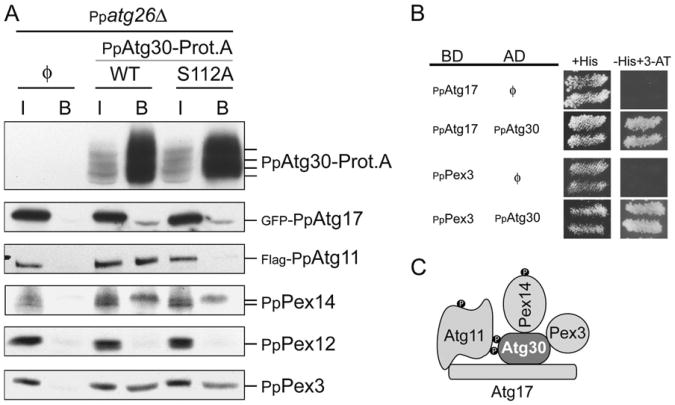
Detecting the interactions of PpAtg30 with other proteins is complicated by its low expression level, its strong membrane association, and its degradation when pexophagy is induced. To circumvent these problems, we affinity-purified PpAtg30-ProteinA fusion overexpressed from the GAPDH promoter in Ppatg26Δ cells to block pexophagy and to enrich its complexes. PpAtg30 interacted in vivo with two peroxins, PpPex3 and PpPex14, and two Atg proteins, PpAtg17 and PpAtg11 (Figure 6A). In P. pastoris, PpPex14 is present in two forms (Johnson et al., 2001), unmodified and phosphorylated. Only the phosphorylated form of PpPex14 appeared to interact with PpAtg30.
Figure 6. PpAtg30 Interacts with PpPex3, PpPex14, PpAtg11, and PpAtg17.
(A) ProteinA affinity isolation was used to purify PpAtg30-Prot.A. The background strain isPpatg26Δ. The strains used are: without PpAtg30-Prot.A (ø) [SJCF765], with PpAtg30-Prot.A (WT)[SJCF767], and with PpAtg30S112A-Prot.A (S112)[SJCF766]. The antibodies used are indicated and the PpAtg30-Prot.A has an extra tag (CBP), used for detection on immunoblots. I, Input; B, bound.
(B) The interaction by yeast two-hybrid assays was determined by growth on medium lacking histidine supplemented with 15-100 mM 3-aminotriazole(3-AT). Strain AH109 was transformed with plasmids containing the binding domain (BD)-fused to PpAtg17 or PpPex3 proteins and the activation domain (AD)-fused to PpAtg30 as indicated.
(C) Summary of the interaction domains of PpAtg30, PpPex3, PpPex14, PpAtg11, and PpAtg17. • indicates phosphorylation.
To understand the function of PpAtg30 phosphorylation, we affinity-isolated the PpAtg30S112A-ProteinA fusion and analyzed the same interactions. PpPex3, PpPex14, and PpAtg17 interaction with PpAtg30 were unaffected by the S112A mutation, but the PpAtg11/PpAtg30 interaction was inhibited suggesting that phosphorylation of PpAtg30 at S112 may be necessary for its interaction with PpAtg11 (Figure 6A).
Potential PpAtg30 interactions with peroxins and autophagy proteins were also investigated by yeast two-hybrid analyses (Figure 6B). PpAtg30 interacted with PpPex3 and PpAtg17. However, PpAtg30 did not interact with PpPex14 and PpAtg11, perhaps because PpAtg30 and PpPex14 are not modified. Also PpAtg30 did not interact with the peroxins PpPex7, PpPex8, PpPex11, PpPex22, PpPex25, PpPex28, PpPex29, PpPex30, and PpPex31, and autophagy proteins PpAtg1, PpAtg6, PpAtg8, PpAtg20, PpAtg24, PpAtg28, and PpVps34 (data not shown). A summary of these interactions is in Figure 6C, and mapping of the domains of interaction between PpAtg30 and PpAtg17 is presented in Figures S7A–S7D.
Discussion
PpAtg30 Is the Peroxisome Receptor for Pexophagy
Since many genes required for pexophagy overlap with those required for other autophagy-related pathways, a central question in understanding pexophagy is how the nonselective autophagic machinery is recruited and constrained to achieve selective peroxisome degradation.
Our data provide several strong lines of evidence in support of a role for PpAtg30 as the peroxisome receptor during pexophagy. PpAtg30 is required for both forms of pexophagy, independent of the peroxisome proliferation medium (oleate or methanol), but not for the Cvt and autophagy pathways (Figure 1 and Figure S1A); its expression level and turnover in various media mirror those of peroxisomal proteins (Figure 1C and Figure S1B); it localizes to peroxisomes upon their induction (interacting with at least two peroxins, PpPex3 and PpPex14; Figures 2 and 6, and Figure S5A) and is transported concomitantly with them to the vacuole (Figure 2A); and it also interacts with two proteins integral to the autophagy machinery (PpAtg11 and PpAtg17, Figure 6). The observation that upon induction of micropexophagy, vacuolar invagination and septation occur normally in Ppatg30Δ cells (Figure 1A), but peroxisomes fail to enter the vacuole, suggests that the signaling event at the vacuolar membrane is not disrupted in the absence of PpAtg30, but that the autophagy machinery fails either to assemble correctly or to recognize peroxisomes. Finally, even when both pexophagy and autophagy are induced simultaneously (methanol- or oleate-grown cells shifted to glucose medium without nitrogen), peroxisome degradation still requires PpAtg30 (Figure S1).
Our experiments also address why and how PpAtg30 is required for pexophagy. In addition to localizing mainly around peroxisomes when they are induced, PpAtg30-GFP localized transiently atthe PAS, colocalizing with PpAtg8 and PpAtg17, after pexophagy was induced (Figure 2B). The PAS localization of PpAtg30 may be a consequence of its interactions with PpAtg11 and PpAtg17 and appears to be independent of peroxisomal association (Figure S2). The subsequent degradation of PpAtg30 occurs (as with peroxisomes) in the vacuole (Figure 2A) and depends on several components of the autophagy machinery (data not shown).
PpAtg30 is, to our knowledge, the first protein described to interact with both peroxisomal proteins (PpPex3 and PpPex14) and autophagy proteins (PpAtg11 and PpAtg17; Figure 6). It is interesting that pexophagy targets two key peroxins of the peroxisome biogenesis machinery required for membrane and matrix protein import. Pex3 is an integral membrane protein involved in the docking of most peroxisomal-membrane proteins (Rayapuram and Subramani, 2006). Pex3 thus acts at an early stage of peroxisome development (Hoepfner et al., 2005). Its absence in yeast or mammalian cells affects peroxisome formation. Pex14 is involved in the docking of the PTS receptors, Pex5 and Pex7, required for peroxisomal matrix protein import (Rayapuram and Subramani, 2006). It is currently unclear if PpAtg30/PpPex3/PpPex14 interactions are direct, but the two-hybrid experiments performed in heterologous S. cerevisiae cells and the mislocalization of PpAtg30 in Pppex3Δ and Pppex14Δ suggest direct binding between PpAtg30 and PpPex3. These interactions of PpAtg30 strongly indicate a coupling between the biogenesis and degradative arms of the peroxisome homeostasis machineries.
PpAtg30 Regulates Pexophagy
PpAtg30 is a phosphoprotein. During peroxisome proliferation, 5%–10% of the total PpAtg30 is modified. The proportion increases to 80%–90% when pexophagy is induced (Figure 1C and Figure S4A). PpAtg30 phosphorylation (at least of serine 112) is likely to be required for pexophagy because the PpAtg30S112A mutant does not interact with PpAtg11, but does associate with peroxisomes and peroxins (Figures 3D and 6A), suggesting that PpAtg30 might first tag the peroxisomes prior to its activation by phosphorylation at this site, which would then deliver peroxisomes to the autophagic machinery via PpAtg30 interactions with PpAtg11. Low levels of PpAtg30 phosphorylation in cells growing on methanol or oleate may allow the cells to remove deficient peroxisomes and could explain the presence of some PpAtg30 in the vacuolar lumen in cells grown in methanol. Although PpAtg30 is phosphorylated at many sites, at present only S112 is known to be physiologically essential for pexophagy.
PpAtg17 interacts with PpAtg30 independent of its S112 phosphorylation. The significance of this association remains to be elucidated. In P. pastoris, PpAtg17 is not essential for pexophagy, when tested in glucose medium with nitrogen (data not shown). As seen for the Cvt pathway (Suzuki et al., 2007), PpAtg17 function could also be required for pexophagy under starvation conditions. Its importance as a scaffold protein for the pexophagy machinery in rich medium may be substituted by PpAtg11.
The levels of modified PpAtg30 are critical, and imbalances severely affect pexophagy (Figure 4). When PpAtg30 is overexpressed, the proportion that is phosphorylated remains the same (5%–10%), but the absolute amount of phosphorylated PpAtg30 increases (Figure 4A). This induces pexophagy even in conditions otherwise favoring peroxisome proliferation, without any requirement for other signaling events (Figure 4 and Figure S5A). A high level of phosphorylated PpAtg30 might “switch on” pexophagy, whereas its degradation (with the peroxisomes) “switches off” the pathway. Gross PpAtg30 phosphorylation does not depend on autophagy proteins, like PpAtg1, PpAtg11 or PpVps15, or the peroxin PpPex14, but does depend on PpPex3 (Figure 3B) whose absence causes the mislocalization of PpAtg30 to the nucleus, which may insulate it from the kinase(s) (Figure 2D). PpAtg30 is polyphosphorylated and we have not yet determined whether one or several kinasesphosphorylate PpAtg30. Even if a kinase, such as PpAtg1, did phosphorylate PpAtg30 on a few sites, its effect might be masked by phosphorylation at different sites by other kinase(s) (Figure 3B).
PpAtg30 Is Required for Functional MIPA and Ppg Formation and for PpAtg11 Localization at the Perivacuolar Structures and Vacuolar-Sequestering Arms
During micropexophagy, sequestering membranes emanating from the vacuole engulf peroxisomes for subsequent degradation. As these structures form, PpAtg11 accumulates at the PVS, close to the site of emergence of the sequestering membranes (Chang et al., 2005). The MIPA forms between the tips of the sequestering membranes, probably facilitating fusion of the sequestering membranes around peroxisomes (Mukaiyama et al., 2004; Oku et al., 2003). Assembly of the MIPA requires the conjugation of PpAtg8 to PE. PpAtg8 lipidation (mediated by PpAtg4, PpAtg7, and PpAtg3; Ichimura et al., 2000) is required for MIPA and Ppg formation during pexophagy. In the absence of PpAtg30, neither a functional MIPA nor the Ppg is formed, although Atg8 is lipidated (Figures 5A and 5B and Figure S6). In the absence of PpAtg30, PpAtg11 fails to localize to the PVS and the peroxisome-sequestering arms of the vacuolar membrane during micropexophagy, although its localization at the vacuolar membrane is preserved (Figure 5C). Despite PpAtg11 mislocalization, the sequestering membranes form normally (Figure 1A).
Our data suggest that late peroxisome sequestration events and proper MIPA assembly require the recognition of the peroxisomes by its receptor PpAtg30. A likely model is that the MIPA and Ppg are formed using the peroxisome surface as a template until the elongating structure reaches the tips of the sequestering membranes (for the MIPA) or the vacuolar membrane (for the Ppg).
Model for Peroxisome Recognition during Pexophagy
In our current model (Figure 7), the autophagy machinery required for pexophagy is primed to function in any condition and (at least for pexophagy induced by PpAtg30 overexpression) needs no extra signals other than PpAtg30 phosphorylation. This is not surprising because most of the proteins involved in pexophagy are required for autophagy (during starvation conditions) and/or constitutively for the Cvt pathway. An hitherto unknown kinase phosphorylates PpAtg30 (which is localized at the peroxisome surface via interactions with PpPex3 and PpPex14) normally when the peroxisomes are not required. It is interesting to note that the phosphorylation of PpAtg30 requires the presence of PpPex3. When PpAtg30 is modified, it interacts with PpAtg11 and PpAtg17 and triggers pexophagy.
Figure 7. Model for Peroxisome Recognition during Pexophagy.
(A) Cells growing in rich medium have few small peroxisomes per cell and peroxins and PpAtg30 are either weakly, or not, expressed.
(B) During peroxisome proliferation, the expression of peroxins and PpAtg30 is induced. PpAtg30 localizes at the peroxisome membrane by interacting with PpPex3 and PpPex14.
(C) Pexophagy conditions activate an unknown kinase that phosphorylates peroxisome-associated PpAtg30. PpAtg30 is not required for the formation of the peroxisome-sequestering arms, but is needed for movement of PpAtg11 to the arms. PpAtg30 delivers peroxisomes to the autophagy machinery via interaction with PpAtg11 and PpAtg17.
(D) The peroxisome cluster tagged with phosphorylated PpAtg30 is fully surrounded by the vacuole, sequestering arms, and MIPA.
Experimental Procedures
Yeast Strains, Plasmids, and Media
The P. pastoris strains and plasmids used are listed in Table S1 in the Supplemental Data. Growth media components are described in the Supplemental Data.
Isolation of the Ppatg30 Mutant
The Ppatg30 mutant was isolated from among Zeocin-resistant mutants by screening for AOX activity after a glucose shift from the methanol condition (Farre et al., 2007; Schroder et al., 2007).
Cell Viability Assay
Wild-type (PPY12), Ppatg30Δ (SJCF44), and Ppvps15Δ cells were grown to approximately 1 OD/ml in YPD medium, washed twice in water, resuspended in nitrogen starvation medium at 0.5 OD/ml, and incubated on a shaker at 30°C. Appropriate dilutions of aliquots were spread onto YPD plates in triplicate. Colonies were counted after 2-3 days and the mean values were plotted.
Phosphatase Treatment
Cell lysates were prepared by the postalkaline extraction procedure. Briefly, cells were centrifuged, washed once with sterile distilled water, and resuspended in 0.2 M NaOH, 1/100th volume of b-mercaptoethanol, and 1 mM PMSF. After 10 min of incubation on ice, proteins were precipitated by ice-cold acetone extraction and centrifugation at 15,000 × g for 10 min at 4°C. The pellet was air-dried and was resuspended in phosphatase buffer (50 mM MES [pH 6.0], 1 mM DTT) containing protease inhibitor cocktail (Sigma) and 1mM PMSF. For potato acid phosphatase (PAP) treatment, lysates corresponding to approximately 3 OD equivalents were treated with 3 U of PAP (Sigma) at 30°C for 20 min. The PAP was not added to untreated samples. The reaction was stopped by addition of 6× sample loading buffer and boiling at 100°Cfor 10 min.
Yeast Two-Hybrid Analysis
The GAL4-based Matchmaker yeast two-hybrid system (Clontech Laboratories, Inc.) was used according to the manufacturer's specifications. A full-length clone of PpPex3, PpAtg17, and PpAtg30 was amplified by PCR and inserted into pGAD-GH (Activation Domain, AD) and/or pGBT9 (Binding Domain, BD).
ProteinA Affinity Isolation Analyses
Cells were grown overnight on methanol and transferred for 30 min on glucose medium without ammonium sulfate before extraction. Fifty OD equivalents of yeast cells were washed in phosphate-buffered saline (pH 7.4) and lysed with glass beads (vortexed 5 times for 2 min at 4°C) in 1 ml IP lysis buffer (50 mM HEPES-KOH [pH 7.5], 0.15 M NaCl, 1% CHAPS, 10% glycerol, 1 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitor cocktail) and centrifuged (500 × g, 10 min). The membrane protein solubilization was performed by incubation the supernatant at 4°C for 30 min with rotation and was then centrifuged (21,000 × g, 10 min). Human IgG-Agarose (Sigma) was added to the supernatant (100 μl/ml of lysate) and incubated for 1 hr. Beads were washed five times (5 ml) with the IP lysis buffer for 5 min. Bound protein was eluted with 100 μl of sample buffer, resolved by SDS-PAGE, and visualized by immunoblotting. Loading was as follows: input (I), 0.2 OD equivalent and immunopurified proteins (B), 7.5 OD equivalent.
Protein Extracts of Cells Expressing Different Levels of PpAtg30
Cells were precultured to mid-log phase in YPD medium, washed, resuspended at a density of 0.5 OD/ml in methanol medium, and grown at 30°C for 6 hr unless otherwise indicated. Cells were disrupted with glass beads as for ProteinA affinity-isolation analyses and the lysate was centrifuged at 21,000 × g for 10 min at 4°C. The supernatant was considered the raw extract. Proteins were assayed and 20 μg were loaded per lane.
Supplementary Material
Acknowledgments
We are very grateful to E. Peters (GNF) and D. Mason (GNF) for demonstrating the phosphorylation of S112 and other sites, R. Tsien (UCSD) for the mRFP construct, and K. Booher and A. Sekhar for technical support. This work was supported by an EMBO fellowship to J.C.F and by NIH grants (DK41737 and GM069373) to S.S. R.D.M. was supported by a CMG training grant.
Footnotes
Accession Numbers: Nucleotide sequences have been deposited in the Entrez database with accession code AY310405.
Supplemental Data: Supplemental Data include seven figures, one table, Supplemental Experimental Procedures, and Supplemental References and can be found with this article online at http://www.developmentalcell.com/cgi/content/full/14/3/365/DC1/.
References
- Abeliovich H, Klionsky DJ. Autophagy in yeast: mechanistic insights and physiological function. Microbiol Mol Biol Rev. 2001;65:463–479. doi: 10.1128/MMBR.65.3.463-479.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ano Y, Hattori T, Oku M, Mukaiyama H, Baba M, Ohsumi Y, Kato N, Sakai Y. A sorting nexin PpAtg24 regulates vacuolar membrane dynamics during pexophagy via binding to phosphatidylinositol-3-phosphate. Mol Biol Cell. 2005;16:446–457. doi: 10.1091/mbc.E04-09-0842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellu AR, Komori M, van der Klei IJ, Kiel JA, Veenhuis M. Peroxisome biogenesis and selective degradation converge at Pex14p. J Biol Chem. 2001;276:44570–44574. doi: 10.1074/jbc.M107599200. [DOI] [PubMed] [Google Scholar]
- Bellu AR, Salomons FA, Kiel JA, Veenhuis M, Van Der Klei IJ. Removal of Pex3p is an important initial stage in selective peroxisome degradation in Hansenula polymorpha. J Biol Chem. 2002;277:42875–42880. doi: 10.1074/jbc.M205437200. [DOI] [PubMed] [Google Scholar]
- Chang T, Schroder LA, Thomson JM, Klocman AS, Tomasini AJ, Stromhaug PE, Dunn WA., Jr PpATG9 encodes a novel membrane protein that traffics to vacuolar membranes, which sequester peroxisomes during pexophagy in Pichia pastoris. Mol Biol Cell. 2005;16:4941–4953. doi: 10.1091/mbc.E05-02-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn WA, Jr, Cregg JM, Kiel JA, van der Klei IJ, Oku M, Sakai Y, Sibirny AA, Stasyk OV, Veenhuis M. Pexophagy, the selective autophagy of peroxisomes. Autophagy. 2005;1:75–83. doi: 10.4161/auto.1.2.1737. [DOI] [PubMed] [Google Scholar]
- Farré JC, Subramani S. Peroxisome turnover by micropexophagy: an autophagy-related process. Trends Cell Biol. 2004;14:515–523. doi: 10.1016/j.tcb.2004.07.014. [DOI] [PubMed] [Google Scholar]
- Farré JC, Vidal J, Subramani S. A cytoplasm to vacuole targeting pathway in P. pastoris. Autophagy. 2007;3:230–234. doi: 10.4161/auto.3905. [DOI] [PubMed] [Google Scholar]
- Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- Johnson MA, Snyder WB, Cereghino JL, Veenhuis M, Subramani S, Cregg JM. Pichia pastoris Pex14p, a phosphorylated peroxisomal membrane protein, is part of a PTS-receptor docking complex and interacts with many peroxins. Yeast. 2001;18:621–641. doi: 10.1002/yea.711. [DOI] [PubMed] [Google Scholar]
- Kiel JA, Komduur JA, van der Klei IJ, Veenhuis M. Micropexophagy in Hansenula polymorpha: facts and views. FEBS Lett. 2003;549:1–6. doi: 10.1016/s0014-5793(03)00794-4. [DOI] [PubMed] [Google Scholar]
- Kim J, Klionsky DJ. Autophagy, cytoplasm-to-vacuole targeting pathway, and pexophagy in yeast and mammalian cells. Annu Rev Biochem. 2000;69:303–342. doi: 10.1146/annurev.biochem.69.1.303. [DOI] [PubMed] [Google Scholar]
- Kim J, Huang WP, Klionsky DJ. Membrane recruitment of Aut7p in the autophagy and cytoplasm to vacuole targeting pathways requires Aut1p, Aut2p, and the autophagy conjugation complex. J Cell Biol. 2001a;152:51–64. doi: 10.1083/jcb.152.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, Scott SV, Ohsumi Y, Dunn WA, Jr, Klionsky DJ. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001b;153:381–396. doi: 10.1083/jcb.153.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky DJ, Ohsumi Y. Vacuolar import of proteins and organelles from the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:1–32. doi: 10.1146/annurev.cellbio.15.1.1. [DOI] [PubMed] [Google Scholar]
- Koller A, Valesco J, Subramani S. The CUP1 promoter of Saccharomyces cerevisiae is inducible by copper in Pichia pastoris. Yeast (Chichester, England) 2000;16:651–656. doi: 10.1002/(SICI)1097-0061(200005)16:7<651::AID-YEA580>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Levine B. Eating oneself and uninvited guests: autophagy-related pathways in cellular defense. Cell. 2005;120:159–162. doi: 10.1016/j.cell.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Mukaiyama H, Baba M, Osumi M, Aoyagi S, Kato N, Ohsumi Y, Sakai Y. Modification of a ubiquitin-like protein Paz2 conducted micropexophagy through formation of a novel membrane structure. Mol Biol Cell. 2004;15:58–70. doi: 10.1091/mbc.E03-05-0340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaiyama H, Oku M, Baba M, Samizo T, Hammond AT, Glick BS, Kato N, Sakai Y. Paz2 and 13 other PAZ gene products regulate vacuolar engulfment of peroxisomes during micropexophagy. Genes Cells. 2002;7:75–90. doi: 10.1046/j.1356-9597.2001.00499.x. [DOI] [PubMed] [Google Scholar]
- Oku M, Warnecke D, Noda T, Muller F, Heinz E, Mukaiyama H, Kato N, Sakai Y. Peroxisome degradation requires catalytically active sterol glucosyltransferase with a GRAM domain. EMBO J. 2003;22:3231–3241. doi: 10.1093/emboj/cdg331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayapuram N, Subramani S. The importomer–a peroxisomal membrane complex involved in protein translocation into the peroxisome matrix. Biochim Biophys Acta. 2006;1763:1613–1619. doi: 10.1016/j.bbamcr.2006.08.035. [DOI] [PubMed] [Google Scholar]
- Schroder LA, Glick BS, Dunn WA. Identification of pexophagy genes by restriction enzyme-mediated integration. Methods Mol Biol. 2007;389:203–218. doi: 10.1007/978-1-59745-456-8_15. [DOI] [PubMed] [Google Scholar]
- Scott SV, Hefner-Gravink A, Morano KA, Noda T, Ohsumi Y, Klionsky DJ. Cytoplasm-to-vacuole targeting and autophagy employ the same machinery to deliver proteins to the yeast vacuole. Proc Natl Acad Sci USA. 1996;93:12304–12308. doi: 10.1073/pnas.93.22.12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, Klionsky DJ. Cargo proteins facilitate the formation of transport vesicles in the cytoplasm to vacuole targeting pathway. J Biol Chem. 2004;279:29889–29894. doi: 10.1074/jbc.M404399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–218. doi: 10.1111/j.1365-2443.2007.01050.x. [DOI] [PubMed] [Google Scholar]
- Tuttle DL, Dunn WA., Jr Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Klionsky DJ. Autophagy: molecular machinery for self-eating. Cell Death Differ. 2005;12(Suppl 2):1542–1552. doi: 10.1038/sj.cdd.4401765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan W, Tuttle DL, Shi YJ, Ralph GS, Dunn WA., Jr Glucose-induced microautophagy in Pichia pastoris requires the alpha-subunit of phosphofructokinase. J Cell Sci. 1997;110:1935–1945. doi: 10.1242/jcs.110.16.1935. [DOI] [PubMed] [Google Scholar]
- Zutphen T, Veenhuis M, van der Klei IJ. Pex14 is the sole component of the peroxisomal translocon that is required for pexophagy. Autophagy. 2008;4:63–66. doi: 10.4161/auto.5076. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.