Abstract
UDP-galactose 4′-epimerase (GALE) catalyzes the interconversion of UDP-galactose and UDP-glucose, an important step in galactose catabolism. Type III galactosemia, an inherited metabolic disease, is associated with mutations in human GALE. The V94M mutation has been associated with a very severe form of type III galactosemia. While a variety of structural and biochemical studies have been reported that elucidate differences between the wildtype and this mutant form of human GALE, little is known about the dynamics of the protein and how mutations influence structure and function. We performed molecular dynamics simulations on the wildtype and V94M enzyme in different states of substrate and cofactor binding. In the mutant, the average distance between the substrate and both a key catalytic residue (Tyr-157) and the enzyme-bound NAD+ cofactor and the active site dynamics are altered making substrate binding slightly less stable. However, overall stability or dynamics of the protein is not altered. This is consistent with experimental findings that the impact is largely on the turnover number (kcat), with less substantial effects on Km. Active site fluctuations were found to be correlated in enzyme with substrate bound to just one of the subunits in the homodimer suggesting inter-subunit communication. Greater active site loop mobility in human GALE compared to the equivalent loop in Escherichia coli GALE explains why the former can catalyze the interconversion of UDP-N-acetylgalactosemine and UDP-N-acetylglucosamine while the bacterial enzyme cannot. This work illuminates molecular mechanisms of disease and may inform the design of small molecule therapies for type III galactosemia.
Introduction
UDP-galactose 4′-epimerase (GALE, EC 5.1.3.2) catalyzes the reversible interconversion of UDP-galactose and UDP-glucose (1) . In many organisms, including humans, the enzyme can also catalyze the interconversion of the N-acetylated forms of these UDP-sugars (2). The reaction with UDP-galactose is important because it plays a critical role in the Leloir pathway of galactose catabolism in which galactose is converted to the glycolytic intermediate glucose 6-phosphate (3, 4). Both UDP-sugar interconversions are important in the synthesis of glycoproteins and glycolipids. The majority of GALE enzymes characterized to date are homodimeric with each subunit containing one active site. These active sites contain an NAD+ molecule whose function is to transiently oxidize the UDP-sugar substrate at the C4-OH position. This oxidation is followed by reorientation of the substrate in the active site and re-reduction of the ketone to an alcohol (5, 6). The structure of human GALE (HsGALE) revealed that a key tyrosine residue (Tyr157) assists in this oxidation/re-reduction reaction cycle (7).
Mutations in enzymes of the Leloir pathway are associated with the inherited metabolic disease, galactosemia. This disease has highly varied symptoms which range from altered blood chemistry to death in childhood. The most commonly detected form, type I galactosemia (galactose 1-phosphate uridylyltransferase deficiency, OMIM #230400) generally has serious clinical manifestations which get progressively worse through childhood (8, 9). Currently the only treatment is the restriction of dietary galactose (and its precursors such as lactose). While this slows the appearance of symptoms and can lessen their severity, it is rarely completely effective and it is often difficult to impose a strict diet onto young children (10). In contrast, galactokinase deficiency which results in type II galactosemia (OMIM #230200) is a relatively mild disease in which the only verified symptom is early onset cataracts (11). Mutations in the gene encoding human GALE (HsGALE) are associated with type III galactosemia (OMIM #230350) (12-14). Historically, this disease was divided into two forms. The more severe, or “generalized” form was associated with severe symptoms similar to those of type I galactosemia and the recommended treatment is similar. The milder, or “peripheral”, form was associated with increased levels of galactose metabolites in the blood and no intervention was considered necessary. However, it has now been conclusively shown that this is an over-simplification. The disease presents a continuum of severities of symptoms which are influenced by the mutations(s) present in the patient’s GALE genes and on their environment (in which diet is a particularly important factor) (14, 15). A summary of mutations of hGALE and their effect on the protein can be found in (16). While the focus of this work is V94M, a substrate binding site mutation that causes the most severe form of type III galactosemia, there are other known mutations with severe impact on the enzyme. The G90E mutant is very severely impaired kinetically and, along with L183P, also is highly destabilized compared to wildtype (17). K161N is also very severely impaired kinetically, with the likely loss of the NAD cofactor. Interestingly, this mutant is thermally more stable than wildtype (18).
It is not clear why aberrant galactose metabolism can result in such severe consequences. Several studies have suggested that a build-up of galactose 1-phosphate is toxic to eukaryotic cells, but the exact, molecular cause of this toxicity is unknown (19). Altered UDP-sugar metabolism in type III galactosemia affects the pools of these precursors for glycoprotein and glycolipid synthesis (20, 21). In addition, aggregation of HsGALE has been observed in cells expressing disease-associated variants (22). As yet, there are no small molecule therapies for any form of galactosemia, although it has been suggested that a “small molecule chaperone” approach may be viable since many of the mutations result in decreased protein stability (1, 15, 17, 18, 23-25). In order to implement such therapies, however, it is important to understand more about not just the structures of the proteins and the disease-associated variants but also the flexibility and dynamics of these structures. Molecular Dynamics (MD) has been established as one of the prime methods to computationally probe the dynamics of biomolecular systems (26). It is thus ideally suited to compare the dynamics of different mutant forms of the same protein. In the realm of computer-aided drug discovery, MD can be used to account for receptor flexibility (27). Therefore, we undertook a detailed molecular dynamics investigation of HsGALE and the V94M variant which is the one most commonly associated with the most severe form of type III galactosemia (24).
Material and Methods
System preparation
Systems based on two different crystal structures were prepared for simulations: PDB entries 1EK6 (7) and 1I3L (28) for wildtype and V94M human UDP-galactose 4-epimerase respectively. These structures contain the substrate uridine-5′-diphosphate glucose (UDP-glc) and uridine-5′-diphosphate galactose (UDP-gal) respectively and were modeled as dimers. The cofactor was modeled as NAD+ in all structures. Missing residues (347,348 in 1EK6A, 1,347,348 in 1EK6B, 1 in 1I3LA and 1,347,348 in 1I3LB) were built in using Prime (29, 30). For each of the starting structures, four different systems were built: apo (no substrates, no NAD+ cofactors), NAD+-bound (no substrates, with two NAD+ cofactors), single-substrate-NAD+-bound (with one substrate and two NAD+ cofactors) and substrate-NAD+-bound (with two substrates and two NAD+ cofactors). Tleap (31) was used to neutralize the systems by adding Na+ counter ions (6,8,10, and 12 Na+ for the apo, NAD+-bound, single-substrate-NAD+-bound and substrate-NAD+- bound systems respectively) and solvating using a TIP3P water box. The fully solvated substrate-NAD+-bound systems contained 109037 (1EK6), and 115573 (1I3L) atoms, respectively. Simulations were performed on each of the eight different systems. Minimization using SANDER (31) was carried out in two stages: 1000 steps of minimization of solvent and ions with the protein, substrate, and cofactor restrained using a force constant of 500 kcal/mol/Å2, followed by a 2500 step minimization of the entire system. A short initial 20 ps MD simulation with weak restraints (10 kcal/mol/Å2) on the protein, substrate, and cofactor residues was used to heat the system to a temperature of 300K. Subsequently, 150 ns of MD simulations were performed on each of the eight systems under investigation.
Molecular Dynamics simulations
All MD simulations were performed under the NPT ensemble at 300 K using AMBER (31) and the ff99SBildn force field (32, 33). Periodic boundary conditions were used, along with a non-bonded interaction cutoff of 10 Å. Bonds involving hydrogen atoms were constrained using the SHAKE algorithm (34), allowing for a time step of 2 fs. For each system, 150 ns MD trajectories were generated totaling a simulation time of 1.2 μs.
Distance measurements
Assuming the proposed catalysis mechanism is correct, NAD+ needs to oxidize the sugar at the C4-OH group of the substrate. Similarly the Tyrosine 157 residue is required for the mechanism. The proximity of the nicotinamide group, the substrate C4-OH, as well as the Tyr 157 were monitored for wildtype and mutant substrate-NAD+-bound simulations. For this the pairwise distances between the C13 substrate atom, the C18 atom on the cofactor nicotinamide group and the OH on Tyr 157 were calculated.
Additionally, stabilizing hydrogen bonds between protein active site residues and the substrate or cofactor have been monitored for wildtype and mutant substrate-NAD+-bound simulations. This way changes in dynamics caused by the mutation that might affect cofactor or substrate binding stability can be investigated. A total of 12 H-bonds that are present both in the wildtype and mutant crystal structures were investigated: N224 O – substrate H1, F226 H – substrate O1, R300 2HH1 – substrate O8, R300 2HH2 – substrate O8, N187 1HD2 – substrate O11, R239 HE – substrate O11, D66 OD1 – cofactor H11, N34 H – cofactor N5, N37 H – cofactor O6, D33 OD1 – cofactor H5, K92 HZ1 – cofactor O8, I14 H – cofactor O9. Distances between 2.0 and 3.5 Å are representative of hydrogen bonds (35). All distance measurements were carried out at 200 ps intervals.
Secondary structure, RMSF, and cross correlation analysis
In order to investigate a possible change in stability upon the V94M mutation, the secondary structure content over the course of the simulations were calculated for all eight systems. Frames every 20 ps were extracted from the trajectories. The STRIDE algorithm (36) was used to calculate helical and strand content for each of the frames. Average secondary structure contents for the entire simulation were also calculated. The Root Mean Square Fluctuations (RMSF) were calculated using ptraj (AmberTools 12). For the comparison between human and E. coli loop fluctuations, the average RMSF of residues 298 through 311 (human) and their equivalent counterparts in the bacterial protein was calculated. To compare the active site dynamics between the two subunits of HsGALE, all residues that are within 7 Å of the substrate in the original substrate-NAD+-bound complex were determined and average RMSF values reported. The Dynamical Cross-Correlation Matrix (DCCM) function in R was used to calculate the cross correlation of atomic displacements within the simulations.
Results and Discussion
The V94M mutation has no effect on overall protein stability
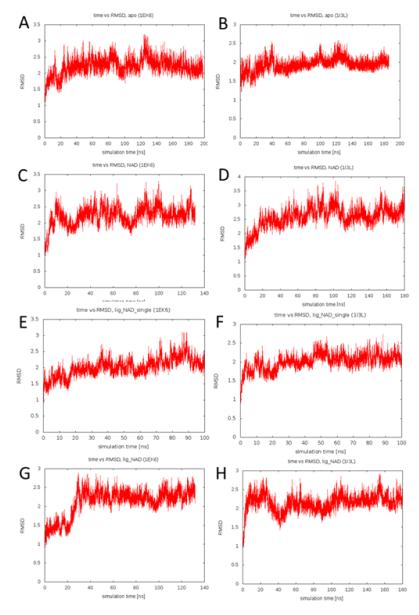
To assess the effect of the V94M mutation on protein stability we monitored the root mean square distance (RMSD) to starting structure over the course of the simulations. Figure 1 shows the RMSD vs simulation time plots for all eight simulations. All simulations converged to RMSDs of 2-3 Å with respect to the starting structure. There is no significant difference between wildtype and mutant protein systems. This suggests that the V94M alteration does not have a measurable impact on the stability of UDP-galactose 4′-epimerase on the timescale probed by the molecular dynamics simulations. We also calculated the average secondary structure content of the simulated systems in order to investigate whether partial unfolding of secondary structure elements occurs. This could be seen as an early indication of loss of protein stability. However, all simulations are characterized by a helical content of 38-40% and a strand content of 15-16%. Again, there is no difference between the wildtype and mutant forms.
Figure 1.
RMSD vs. simulation time plot for the A) apo wildtype, B) apo V94M, C) NAD+-bound wildtype, D) NAD+-bound V94M, E) single-substrate-NAD+-bound wildtype, F) single-substrate-NAD+- bound V94M, G) substrate-NAD+-bound wildtype and H) substrate-NAD+-bound V94M simulations.
These findings are in agreement with experimental data which suggest that, while V94M has a profound impact on the turnover number (kcat), the mutant protein form is not less stable than the wildtype (17, 24, 25). Indeed, limited proteolysis studies suggest that this variant may be slightly more stable than the wildtype (24).
The V94M mutation slows down catalysis
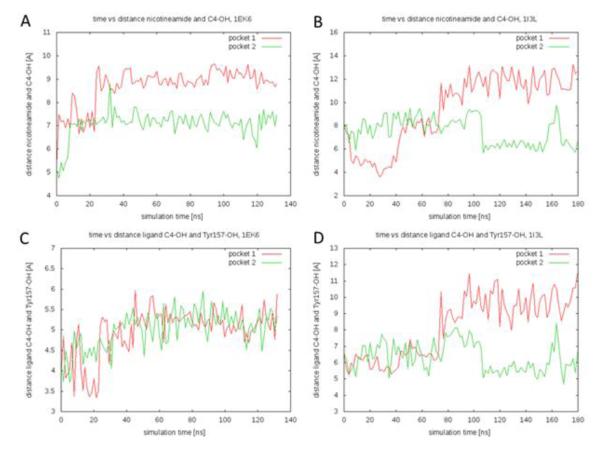
While there is no discernible influence of the V94M mutation on overall protein stability, there may well be an effect on the protein’s catalytic mechanism. According to the proposed mechanism (7) the NAD+ oxidizes the sugar at the C4 hydroxyl group. Additionally the hydroxyl group on Y157 is involved in catalysis. For this oxidation to happen, the nicotinamide group and the Y157 hydroxyl group have to be close to the C4-OH. We thus monitored the distances from the C4 substrate atom to the C18 atom on the cofactor nicotinamide group and the OH on Tyr157, respectively. Figure 2 shows these distances as a function of time for the substrate-NAD+-bound simulations for each protein subunit. The average distance between the substrate C4 and the cofactor nicotinamide group increases from 7.9 Å in the wildtype protein to 8.5 Å in the mutant simulations. The average distance between the substrate C4 and the Tyr157 hydroxyl group increases from 5.0 Å in the wildtype protein to 7.3 Å in the mutant simulations. Thus the average distances between the substrate C4 and both the cofactor nicotinamide group and the hydroxyl on Tyr157 are smaller in the wildtype simulations suggesting that the catalysis partners are closer together. This finding presents an interesting perspective on the effect of the mutation on the catalytic mechanism. While no overall difference in protein dynamics can be observed for the mutation, it seems that V94M alters the dynamics of the active site in a way that moves the substrate further from its interaction partners. We speculate that the greater distances would slow down proton transfer and, most likely, affect kcat. This is computational confirmation of the previously reported experimental hypothesis that V94M permits the substrate to adopt more non-productive conformations in the active site (1) and is in agreement with experiments which demonstrated that the main effect of the V94M alteration is a reduction in kcat (17, 25).
Figure 2.
Interatomic distances as a function of simulation time. A) Distances from the C4 substrate atom to the C18 atom on the cofactor nicotinamide group for the wildtype simulation. B) Distances from the C4 substrate atom to the C18 atom on the cofactor nicotinamide group for the V94M simulation. C) Distances from the C4 substrate atom to the OH on Tyr157 for the wildtype simulation. D) Distances from the C4 substrate atom to the OH on Tyr157 for the V94M simulation.
Changes that might affect cofactor or substrate binding
Another interesting aspect is whether there is an effect of the V94M mutation on either cofactor or substrate binding. None of the cofactors or substrates dissociated from the protein during the course of our simulations. We thus used the time evolution of the interactions stabilizing the cofactor or substrate in the active site (such as hydrogen bonds or salt bridges) as an indication of overall binding stability. The rationale behind this approach is that rapid or frequent breaking of these stabilizing interactions leads to an overall less stable binding. For the analysis we identified 12 representative hydrogen bonds that are present both in the wildtype and mutant crystal structures. Six of these bonds were between the substrate and protein, while six were between the cofactor and protein. The average fraction that these bonds are present in the substrate-NAD+-bound simulations was calculated. The hydrogen bonds between the substrate and protein were formed in 36% of the simulation time for the wildtype protein, compared with 30% for the V94M protein. On the other hand, the hydrogen bonds between the cofactor and protein were formed in 51% of the simulation time for the wildtype protein, compared with 50% for the V94M protein. This shows that the cofactor is more tightly bound than the substrate. These results also suggest that there is more substrate binding stability in the active site for the wildtype system compared to the mutant system, while the mutation does not seem to influence cofactor binding. These differences are not large however. This is in agreement with experimental findings that suggest that the V94M mutation only changes Km slightly (17, 25).
We also calculated the root mean square fluctuations (RMSF) of the active site residues for the wildtype and mutant substrate-NAD+-bound simulations. The average active site residues fluctuations in the wildtype simulation are 0.83 Å, compared to 0.96 Å for the V94M simulations. This increase in active site dynamics is another indication for a possible loss of binding stability caused by the mutation.
Hints of local inter-subunit communication upon substrate binding
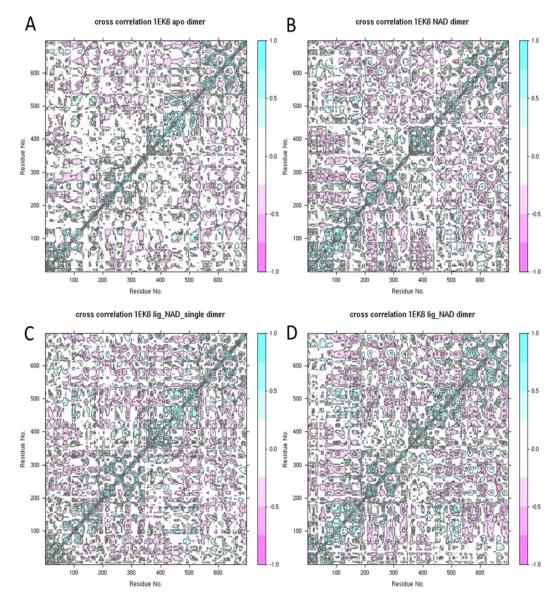
The physiologically functional form of HsGALE is a dimer (7, 17, 37), implying that the individual subunits may not function independently of each other. Indeed, evidence of inter-subunit communication has been observed in some fungal GALEs (38-41) and in wildtype HsGALE at reduced temperatures (24 °C) and for the M284K variant at 24 °C and 37 °C (23). Here we investigated the level of inter-subunit correlations, as well as local, active site inter-subunit communication in HsGALE. Molecular dynamics offers the opportunity to probe the degree to which the dynamics in one subunit are correlated with the dynamics in the other subunit. The cross-correlations of atomic displacements of all residue pairs within the dimer were calculated for the four wildtype simulations (Figure 3). The upper left square shows the inter-subunit correlations. The least amount of correlations is seen in the apo simulations, suggesting that the subunits dynamics are largely independent of each other. Upon cofactor binding, there is a marked increase in inter-subunit correlations. This effect is intensified when the substrate binds as well. Interestingly, the binding of a single substrate to one active site did not cause the same increase in inter-subunit correlations as binding of two substrates have. This implies that binding of the cofactors has a much bigger impact on inter-subunit communication than binding of a single substrate, and there is no hint of global communication upon substrate binding. We also investigated the dynamical fluctuations (average RMSF values) of the active site residues in contact with the substrate. The active site residues in the apo simulation have RMSF values of 1.21 Å and 1.12 Å in the first and second subunit respectively. The corresponding values for the NAD+-bound simulations are 1.31 Å and 1.14 Å. In both these simulations, the two subunits display differently strong fluctuations of active site residues, suggesting that the active site dynamics are independent in each subunit. In contrast to this, the fluctuations in the single-substrate-NAD+-bound are 0.93 Å in both subunits, while the fluctuations in the substrate-NAD+-bound are 0.97 Å in both subunits. This suggests that substrate binding evokes some communication between the subunits that influences the local active site dynamics. Interestingly, binding of a substrate to just one subunit is sufficient to yield the same effect even in the subunit that does not have substrate bound. This is a local sign of inter-subunit communication.
Figure 3.
Cross-correlations of atomic displacements of all residue pairs within the dimer for the wildtype A) apo, B) NAD+-bound, C) single-substrate-NAD+-bound and D) substrate-NAD+-bound simulations.
Differences between human and E. coli forms
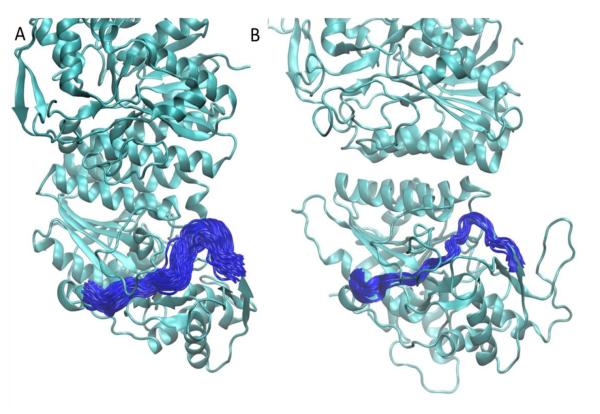
One of the most profound differences between the human and E. coli GALE enzymes is a difference in specificity for UDP-GalNAc / UDP-GlcNAc. HsGALE has been shown to be able to bind UDP-GalNAc / UDP-GlcNAc, while the bacterial enzyme does not have that ability. Careful previous studies established that the difference in specificity is due to key amino acid substitutions as well as differences in active site pocket volume between the proteins of the different species (42, 43). Here we investigated whether the dynamics of the protein loop containing the key C307 (human) / Y299 (E. coli) residues also has an effect on specificity. For this we calculated the RMSFs of the loop containing residues 298 through 311 (in the human protein) and the equivalent loop in the E. coli protein. Calculations for E. coli GALE were based on simulations reported previously (44). The average per residue fluctuation for the HsGALE loop in the substrate-NAD+-bound simulations was 1.40 Å, compared to 1.38 Å for the E. coli loop. The interactions between key substrate binding residues in the loop and the substrate restrain the loop dynamics in both species. Differences however emerge when comparing the simulations without substrate. The average per residue fluctuation for the HsGALE loop in the apo simulations was 1.48 Å, compared to only 1.07 Å for the E. coli simulations. Additionally, the average per residue fluctuation for the HsGALE loop in the NAD+-bound simulations was 1.70 Å. No similar simulations were performed for E. coli GALE. These results suggest that in addition to sequence differences that clearly impact specificity, the dynamics of the loop bordering the active site clearly also play a role in specificity. When the substrate is not bound, the fluctuations in the loop are larger, allowing the active site pocket to open up more, thus enabling the binding of larger substrates, such as those containing a NAc group. Figure 4 shows a cartoon representation of the loop conformations during the simulations for human and E. coli GALE. The loop samples a much wider variety of conformations in the human protein.
Figure 4.
Cartoon representation of dynamics of active site loop in apo A) human GALE and B) E. coli GALE. The protein is shown in green ribbon representation. The loop containing the C307 (human) / Y299 (E. coli) residues is shown in blue every 500 time frames. More substantial fluctuations are observed in the human enzyme.
Conclusions
A wide array of published experimental data comparing the wildtype HsGALE and the V94M mutant form (associated with the most severe form of type III galactosemia) explains their differences on a structural and biochemical level. Here we reported a set of molecular dynamics simulations aiming to compare the dynamics of wildtype and V94M HsGALE. Our results corroborate previously reported experimental findings, often times allowing us to add a dynamics-based explanation to the arguments based on structural data. We found that the mutation does not change the overall stability or dynamics of the protein. It does however change the active site dynamics in a way that make substrate, and probably also cofactor, binding less stable. Our simulations confirmed that the mutation also changes the active site dynamics which increases the average distance between the substrate, cofactor and Tyr-157 thus lowering the turnover number, kcat. Hints of local, active site inter-subunit communication were observed in simulations with a single substrate molecule bound to the dimeric protein. An increase in active site loop dynamics impacts specificity in the human GALE: a comparison between the human and E. coli GALE enzymes indicated that the dynamics of a crucial active site loop can help explain the differences in specificity for substrates containing a NAc group. Thus, the previously reported structural changes seen in the mutant crystal structure are only part of the reason for the difference between the two proteins. While beyond the scope of the current work, future studies will focus on simulating the effects of other severe mutations in hGALE and contrasting them to both wildtype and V94M. In addition to explaining vital protein dynamics, our simulations will also serve as a basis to account for receptor flexibility in future drug discovery studies. Recent work on other single gene disorders has demonstrated that small molecule therapies are viable where compounds can be identified which modulate the flexibility of the affected protein (for recent examples, see (45, 46)). Therefore, the work reported here will be important in the development of any such therapies for type III galactosemia.
Highlights.
We present molecular dynamics simulations of HsGALE and the V94M variant.
We find substrate binding to be slightly less stable in the mutant.
The overall stability or dynamics of the protein is not altered by the mutation.
Results may inform design of small molecule therapies for type III galactosemia.
Acknowledgements
We would like to thank Prof. Dr. J. Andrew McCammon, Dr. Jacob Durrant, Dr. Thomas McCorvie, Aaron Friedman and Samantha Banford for helpful discussions and Dr. Jacob Durrant and Aaron Friedman for allowing us to use their E. coli GALE simulations for the loop fluctuations analysis. This work was supported in part by grants to the McCammon lab from the National Institutes of Health, the National Science Foundation, the Howard Hughes Medical Institute, the National Biomedical Computation Resource, and the NSF Supercomputer Centers. Computational resources were supported, in part, by the National Science Foundation grant PHY-0822283, the Center for Theoretical Biological Physics. S. L. was supported by the American Heart Association and the Center for Theoretical Biological Physics.
Abbreviation List
- (GALE)
UDP-galactose 4′-epimerase
- (HsGALE)
Human UDP-galactose 4′-epimerase
- (kcat)
turnover number
- (galactose 1-phosphate uridylyltransferase deficiency, OMIM #230400)
type I galactosemia
- (OMIM #230200)
type II galactosemia
- (OMIM #230350)
type III galactosemia
- (MD)
Molecular Dynamics
- (UDP-glc)
uridine-5′-diphosphate glucose
- (UDP-gal)
uridine-5′-diphosphate galactose
- (RMSF)
Root Mean Square Fluctuations
- (DCCM)
Dynamical Cross-Correlation Matrix
- (RMSD)
root mean square distance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Timson DJ. The structural and molecular biology of type III galactosemia. IUBMB Life. 2006;58:83–9. doi: 10.1080/15216540600644846. [DOI] [PubMed] [Google Scholar]
- 2.McCorvie TJ, Timson DJ. UDP-galactose 4-epimerase (GALE) In: Taniguchi N, Honke K, Fukuda M, Narimatsu H, Yamaguchi Y, Angata T, editors. Handbook of Glycosyltransferases and Related Genes. Springer; 2013. [Google Scholar]
- 3.Frey PA. The Leloir pathway: a mechanistic imperative for three enzymes to change the stereochemical configuration of a single carbon in galactose. Faseb J. 1996;10:461–70. [PubMed] [Google Scholar]
- 4.Leloir LF. The enzymatic transformation of uridine diphosphate glucose into a galactose derivative. Arch Biochem Biophys. 1951;33:186–90. doi: 10.1016/0003-9861(51)90096-3. [DOI] [PubMed] [Google Scholar]
- 5.Maitra US, Ankel H. Uridine diphosphate-4-keto-glucose, an intermediate in the uridine diphosphate-galactose-4-epimerase reaction. Proc Natl Acad Sci U S A. 1971;68:2660–3. doi: 10.1073/pnas.68.11.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong SS, Frey PA. Fluorescence and nucleotide binding properties of Escherichia coli uridine diphosphate galactose 4-epimerase: support for a model for nonstereospedific action. Biochemistry. 1977;16:298–305. doi: 10.1021/bi00621a022. [DOI] [PubMed] [Google Scholar]
- 7.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 8.McCorvie TJ, Timson DJ. Structural and molecular biology of type I galactosemia: disease-associated mutations. IUBMB Life. 2011;63:949–54. doi: 10.1002/iub.510. [DOI] [PubMed] [Google Scholar]
- 9.McCorvie TJ, Timson DJ. The structural and molecular biology of type I galactosemia: Enzymology of galactose 1-phosphate uridylyltransferase. IUBMB Life. 2011;63:694–700. doi: 10.1002/iub.511. [DOI] [PubMed] [Google Scholar]
- 10.Fridovich-Keil JL. Galactosemia: the good, the bad, and the unknown. Journal of cellular physiology. 2006;209:701–5. doi: 10.1002/jcp.20820. [DOI] [PubMed] [Google Scholar]
- 11.Bosch AM, Bakker HD, van Gennip AH, van Kempen JV, Wanders RJ, Wijburg FA. Clinical features of galactokinase deficiency: a review of the literature. Journal of inherited metabolic disease. 2002;25:629–34. doi: 10.1023/a:1022875629436. [DOI] [PubMed] [Google Scholar]
- 12.Fridovich-Keil J, Bean L, He M, Schroer R. Epimerase Deficiency Galactosemia. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adam MP, editors. Epimerase Deficiency Galactosemia. Seattle (WA): 1993. [Google Scholar]
- 13.Holton JB, Gillett MG, MacFaul R, Young R. Galactosaemia: a new severe variant due to uridine diphosphate galactose-4-epimerase deficiency. Archives of disease in childhood. 1981;56:885–7. doi: 10.1136/adc.56.11.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Openo KK, Schulz JM, Vargas CA, Orton CS, Epstein MP, Schnur RE, et al. Epimerase-deficiency galactosemia is not a binary condition. Am J Hum Genet. 2006;78:89–102. doi: 10.1086/498985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chhay JS, Vargas CA, McCorvie TJ, Fridovich-Keil JL, Timson DJ. Analysis of UDP-galactose 4′-epimerase mutations associated with the intermediate form of type III galactosaemia. Journal of inherited metabolic disease. 2008;31:108–16. doi: 10.1007/s10545-007-0790-9. [DOI] [PubMed] [Google Scholar]
- 16.McCorvie TJ, Timson DJ. In silico prediction of the effects of mutations in the human UDP-galactose 4′-epimerase gene: Towards a predictive framework for type III galactosemia. Gene. 2013 doi: 10.1016/j.gene.2013.04.061. [DOI] [PubMed] [Google Scholar]
- 17.Timson DJ. Functional analysis of disease-causing mutations in human UDP-galactose 4- epimerase. Febs J. 2005;272:6170–7. doi: 10.1111/j.1742-4658.2005.05017.x. [DOI] [PubMed] [Google Scholar]
- 18.McCorvie TJ, Liu Y, Frazer A, Gleason TJ, Fridovich-Keil JL, Timson DJ. Altered cofactor binding affects stability and activity of human UDP-galactose 4′-epimerase: implications for type III galactosemia. Biochim Biophys Acta. 2012;1822:1516–26. doi: 10.1016/j.bbadis.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai K, Elsas LJ, Wierenga KJ. Galactose toxicity in animals. IUBMB Life. 2009;61:1063–74. doi: 10.1002/iub.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daenzer JM, Sanders RD, Hang D, Fridovich-Keil JL. UDP-galactose 4′-epimerase activities toward UDP-Gal and UDP-GalNAc play different roles in the development of Drosophila melanogaster. Plos Genet. 2012;8:e1002721. doi: 10.1371/journal.pgen.1002721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kingsley DM, Kozarsky KF, Hobbie L, Krieger M. Reversible defects in O-linked glycosylation and LDL receptor expression in a UDP-Gal/UDP-GalNAc 4-epimerase deficient mutant. Cell. 1986;44:749–59. doi: 10.1016/0092-8674(86)90841-x. [DOI] [PubMed] [Google Scholar]
- 22.Bang YL, Nguyen TT, Trinh TT, Kim YJ, Song J, Song YH. Functional analysis of mutations in UDP-galactose-4-epimerase (GALE) associated with galactosemia in Korean patients using mammalian GALE-null cells. Febs J. 2009;276:1952–61. doi: 10.1111/j.1742-4658.2009.06922.x. [DOI] [PubMed] [Google Scholar]
- 23.McCorvie TJ, Wasilenko J, Liu Y, Fridovich-Keil JL, Timson DJ. In vivo and in vitro function of human UDP-galactose 4′-epimerase variants. Biochimie. 2011;93:1747–54. doi: 10.1016/j.biochi.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wohlers TM, Christacos NC, Harreman MT, Fridovich-Keil JL. Identification and characterization of a mutation, in the human UDP-galactose-4-epimerase gene, associated with generalized epimerase-deficiency galactosemia. Am J Hum Genet. 1999;64:462–70. doi: 10.1086/302263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wohlers TM, Fridovich-Keil JL. Studies of the V94M-substituted human UDPgalactose-4- epimerase enzyme associated with generalized epimerase-deficiency galactosaemia. Journal of inherited metabolic disease. 2000;23:713–29. doi: 10.1023/a:1005682913784. [DOI] [PubMed] [Google Scholar]
- 26.Adcock SA, McCammon JA. Molecular dynamics: survey of methods for simulating the activity of proteins. Chemical reviews. 2006;106:1589–615. doi: 10.1021/cr040426m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sinko W, Lindert S, McCammon JA. Accounting for Receptor Flexibility and Enhanced Sampling Methods in Computer-Aided Drug Design. Chem Biol Drug Des. 2013;81:41–9. doi: 10.1111/cbdd.12051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Molecular basis for severe epimerase deficiency galactosemia. X-ray structure of the human V94m-substituted UDP-galactose 4- epimerase. J Biol Chem. 2001;276:20617–23. doi: 10.1074/jbc.M101304200. [DOI] [PubMed] [Google Scholar]
- 29.Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- 30.Jacobson MP, Pincus DL, Rapp CS, Day TJ, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–67. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- 31.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr., Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. Journal of computational chemistry. 2005;26:1668–88. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–25. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78:1950–8. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryckaert J-P, Ciccotti G, Berendsen HJC. Numerical integration of the cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. Journal of Computational Physics. 1977;23:327–41. [Google Scholar]
- 35.Wallwork SC. Hydrogen-Bond Radii. Acta Crystallogr. 1962;15:758. &. [Google Scholar]
- 36.Frishman D, Argos P. Knowledge-based protein secondary structure assignment. Proteins. 1995;23:566–79. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 37.Dutta SK, Ray M, Bhaduri A. Uridine 5′-Diphosphate Glucose 4-Epimerase from Ehrlich Ascites-Carcinoma Cells. J Bioscience. 1985;9:59–70. [Google Scholar]
- 38.Brahma A, Banerjee N, Bhattacharyya D. UDP-galactose 4-epimerase from Kluyveromyces fragilis--catalytic sites of the homodimeric enzyme are functional and regulated. Febs J. 2009;276:6725–40. doi: 10.1111/j.1742-4658.2009.07386.x. [DOI] [PubMed] [Google Scholar]
- 39.Hay M, Bhaduri A. UDPGlucose 4-epimerase from Saccharomyces fragilis. Allosteric kinetics with UDP-glucose as substrate. J Biol Chem. 1975;250:4373–5. [PubMed] [Google Scholar]
- 40.Nayar S, Brahma A, Barat B, Bhattacharyya D. UDP-galactose 4-epimerase from Kluyveromyces fragilis: analysis of its hysteretic behavior during catalysis. Biochemistry. 2004;43:10212–23. doi: 10.1021/bi049569t. [DOI] [PubMed] [Google Scholar]
- 41.Ray M, Bhaduri A. UDPglucose 4-epimerase from Saccharomyces fragilis: asymmetry in allosteric properties leads to unidirectional catalysis. Biochem Biophys Res Commun. 1978;85:242–8. doi: 10.1016/s0006-291x(78)80035-7. [DOI] [PubMed] [Google Scholar]
- 42.Thoden JB, Henderson JM, Fridovich-Keil JL, Holden HM. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J Biol Chem. 2002;277:27528–34. doi: 10.1074/jbc.M204413200. [DOI] [PubMed] [Google Scholar]
- 43.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4- epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J Biol Chem. 2001;276:15131–6. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- 44.Friedman AJ, Durrant JD, Pierce LC, McCorvie TJ, Timson DJ, McCammon JA. The molecular dynamics of Trypanosoma brucei UDP-galactose 4′-epimerase: a drug target for African sleeping sickness. Chem Biol Drug Des. 2012;80:173–81. doi: 10.1111/j.1747-0285.2012.01392.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bulawa CE, Connelly S, Devit M, Wang L, Weigel C, Fleming JA, Packman J, Powers ET, Wiseman RL, Foss TR, Wilson IA, Kelly JW, Labaudiniere R. Tafamidis, a potent and selective transthyretin kinetic stabilizer that inhibits the amyloid cascade. Proc Natl Acad Sci U S A. 2012;109:9629–34. doi: 10.1073/pnas.1121005109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Santos-Sierra S, Kirchmair J, Perna AM, Reiss D, Kemter K, Roschinger W, Glossmann H, Gersting SW, Muntau AC, Wolber G, Lagler FB. Novel pharmacological chaperones that correct phenylketonuria in mice. Hum Mol Genet. 2012;21:1877–87. doi: 10.1093/hmg/dds001. [DOI] [PubMed] [Google Scholar]