Abstract
DNA interstrand crosslinks (ICLs) are complex lesions that block essential DNA transactions including DNA replication, recombination, and RNA transcription. Naturally occurring ICLs are rare, yet these lesions are the major cause of toxicity following treatment with several classes of crosslinking cancer chemotherapeutic drugs. ICLs are repaired during and outside of S-phase by pathways with overlapping as well as distinct features. Here, we discuss some recent insights into the mechanisms of replication-dependent and replication–independent repair of ICLs with special emphasis on the differences between these repair pathways.
ICLs: definition and significance
Interstrand crosslinks (ICLs) are cytotoxic DNA lesions that are induced in DNA from exogenous sources and may also arise endogenously. It has been suggested that malondialdehyde, a product of lipid peroxidation, can generate adducts in DNA that get rearranged to form ICLs endogenously [1, 2]. ICLs are covalent linkages between the opposing stands of the DNA helix and are particularly deleterious to cells since they block the processes of transcription, replication and other DNA transactions that require unwinding of the DNA helix. Although they occur infrequently, they are thought to participate in cellular aging.
Importantly, several classes of crosslinking drugs widely used in cancer therapy induce ICLs much more frequently than the endogenous rate. ICL-forming drugs are bifunctional agents that react with bases on opposite strands of the DNA helix to form a covalent bond between the two strands. There are significant differences in the structure and chemical nature between different types of ICL. Notably, the chemistry of DNA adducts and the resulting structures of the lesions influence ICL repair pathways. In particular, repair in the absence of replication or transcription may rely more on ICL-induced distortion of the DNA molecule to initiate ICL repair [3]. Of the types of crosslinking agents used in studies discussed in this review, cisplatin is the most distorting to the DNA helix. Cisplatin reacts with the N7 of guanine in 5′-GC-3′ sequences to first form a monoadduct that then joins with the N7 of a guanine on the opposite strand to cause significant unwinding and bending of the DNA helix. Nitrogen mustards also cause distorting lesions; they create lesions between the N7 of guanines in a 5′-GNC-3 sequence, resulting in localized distortion of the DNA helix [4]. Mitomycin C, another commonly used reagent, reacts with guanine residues of 5′-CG-3′ sequences through the minor groove of DNA and only minimally perturbs the structure of the DNA at the site of the crosslink [5]. Psoralens intercalate into DNA and can be induced to form interstrand crosslinks with UV light. These lesions form preferentially between thymines in 5′-TA-3′ or 5′-AT-3′ sequences, resulting in small amounts of DNA unwinding but no bending of the DNA helix [6]. A more detailed description of how ICLs affect DNA structure can be found in [7, 8]. ICLs induced by these drugs are thought to be the primary cause of their cellular toxicity, even though ICLs only account for 1-15% of the lesions induced by these agents (reviewed in [9]).
ICLs can be repaired either in the presence or absence of DNA replication. Nonetheless, both pathways require several common key steps. The ICL must first be recognized to activate the ICL repair pathway, DNA damage signaling, and recruitment of downstream repair proteins. Among the recruited proteins are nucleases, which are thought to make incisions 5′ and 3′ to the lesion on the same strand of DNA, resulting in unhooking of the lesion. In replication-dependent pathways this leads to a double strand break (DSB) intermediate. Next, repair synthesis across the unhooked lesion by translesion (TLS) polymerases is thought to occur. The remnant, unhooked lesion is probably removed by general nucleotide incision repair (NER) and, in the case of replication-dependent repair, the DSB is repaired by homologous recombination [10]. There are significant differences in how these key steps are performed. In particular, replication-dependent repair relies on a blocked DNA polymerase to initiate ICL repair, whereas replication-independent repair has evolved alternative mechanisms to recognize ICL lesions. FA proteins, which regulate DNA transactions at replication forks, play a critical role in replication-dependent ICL repair whereas the FA pathway is mostly dispensable for replication-independent ICL repair.
ICL repair is critical for the survival of proliferating cells. The repair of ICLs has been most extensively studied in rapidly proliferating cells in culture. However, replication-independent ICL repair (RIR) is critical for cellular homeostasis, notably in post-mitotic cells, such as neurons, or rarely dividing cells, such as stem cells.
Experimental systems for the study of interstrand crosslinks
A significant part of our understanding of ICL repair comes from studies on the sensitivity of yeast strains or mammalian cell lines to crosslinking agents. In particular, cells from patients with the rare inherited disease Fanconi Anemia (FA) are abnormally susceptible to chromosomal breakage by bi-functional alkylating agents compared with other types of DNA damaging agents [11]. FA results from mutations in at least 15 genes. FA proteins can be grouped into three complexes. The core complex (FANCA, -B, -C, -E, -F, -G, -L, and -M, and associated proteins FAAP24 and FAAP100) is required for the mono-ubiquitylation of both FANCD2 and -I within the ID complex, whereas FANCD1, FANCJ, and FANCN act downstream of the ID complex. Of note, cells derived from patients with mutations in any of the FA genes are sensitive to crosslinking agents. Indeed, crosslinker-induced chromosomal breakage has become the standard cytogenetic test for the diagnosis of FA. Therefore, FA cell lines have provided multiple insights into the mechanisms of ICL repair. However, whereas the cellular hypersensitivity to crosslinking agents exhibited by FA cells indicates that the FA pathway plays an essential role in ICL repair, the mechanism by which FA proteins promote ICL repair is not fully elucidated.
A caveat with inferring ICL repair capacity from crosslinking sensitivity is that only 1-15% of the lesions induced in DNA in response to crosslinking agents are ICLs (reviewed in [9]). The remaining lesions are mono-adducts or intrastrand crosslinks. Unlike ICLs, these are thought to be repaired primarily via the NER pathway. This issue can be addressed by comparing mono- and bi-functional analogs of crosslinking drugs such as angelicin and psoralen [12] or HN1 and HN2 (nitrogen mustards) [13]. Host reactivation assays in cells have provided a more direct approach for studying ICL repair in the absence of other types of lesions [14-16]. In these assays plasmids harboring single site-specific ICLs in luciferase or GFP reporters are transfected into cell lines, and ICL repair is measured by the levels of luciferase or GFP expression.
In vitro biochemistry using purified proteins and DNA templates have also shed light on the mechanisms of ICL recognition and repair. Proteins implicated in ICL repair through cellular sensitivity studies have been assayed on DNA substrates with structures mimicking stalled replication forks or which contain site-specific ICLs, and their ability to bind to or process such substrates has been assessed [8].
Finally, cell-free extracts derived from mammalian cells [17] and Xenopus eggs have been instrumental in deciphering ICL repair mechanisms. Repair of DNA substrates with site-specific ICLs and no other DNA lesions can be directly studied these extracts [18-20]. In Xenopus cell-free extracts the repair of circular plasmids harboring a single ICL can be followed in a soluble system competent for DNA replication and activation of a DNA damage response. In this system, replication can also be switched off to monitor replication-independent repair events.
In summary, the mutations in genes which cause Fanconi Anemia have led to the discovery of many proteins which are involved in ICL repair. However there are still outstanding questions as to the precise functions of these proteins in ICL repair pathways. Whereas cells with defined DNA repair defects provide a physiological setting to study the effects of crosslinking drugs, cell-free extracts and in vitro biochemistry can directly lead to advances in unraveling the precise mechanisms of ICL repair.
Recognition
Collision of one or possibly two replication forks with an ICL lesion blocks helicase progression and initiates replication-dependent ICL repair. However, this stalled fork must be specifically directed into the ICL repair pathway. The FANCM/FAAP24 sub-complex is thought to act as a “molecular sensor”, recognizing a replication fork stalled at an ICL and recruiting downstream components of the ICL repair machinery. FANCM and its associated proteins FAAP24, MHF1 and MHF2 (FANCM-interacting histone fold proteins 1 and 2) bind preferentially to branched DNA structures in vitro [21-23] and FANCM-depleted cells fail to recruit the Fanconi core complex or to mono-ubiquitylate the FANCD2-FANCI heterodimer, key events that occur during normal ICL repair [24]. Furthermore, FANCM can promote reversal of model replication forks [25] (Figure 1). However, whether FANCM specifically interacts with forks stalled at an ICL is not established. The EUFA867 cell line used to study the roles of FANCM also harbors a biallelic mutation in FANCA. Correction of the FANCA defect in these cells rescues FANCD2 monoubiquitylation but not hypersensitivity to the crosslinking drug mitomycin C (MMC) [26]. Also, deletion of FANCM in DT40 cells does not completely abrogate FANCD2 ubiquitylation [27, 28]. This strongly suggests that FANCM is not the only sensor in the FA pathway. Indeed, FANCM and human MutS homologues (MSH), which recognize and bind mismatched bases during mismatch repair, could play redundant roles in activating the FA pathway and recognizing ICLs [29]. Cells depleted of MSH2, 3 or 6 are deficient in FANCD2 ubiquitylation and fail to properly recruit the Fanconi core complex in response to MMC. Importantly, co-depletion of both FANCM and MSH2 results in an even greater defect in FA pathway activation than depletion of either protein individually [29]. An additional study also points to physical and functional interactions between the FA pathway and mismatch repair (MMR) proteins [30]. Moreover, studies in in vitro mammalian cell extracts have shown that MutSβ is required for ICL-induced DNA synthesis [17]. This is consistent with the sensitivity of cells lacking MSH2 (a component of MutSβ to crosslinking drugs [31-33]. Genetic evidence in budding yeast also supports a role for MSH2 in ICL repair [34] and, interestingly, the yeast FANCM ortholog Mph1 physically and functionally interacts with the MutSβ mismatch repair factor (Msh2-Msh6) [35]. However, the role of MSH2 still remains contentious; three recent studies find no ICL repair defect in MSH2 deficient cells [12, 16, 19]. It is possible that MSH2 plays a redundant role in ICL repair for some types of ICL lesion, which would explain the lack of an ICL repair phenotype in some studies.
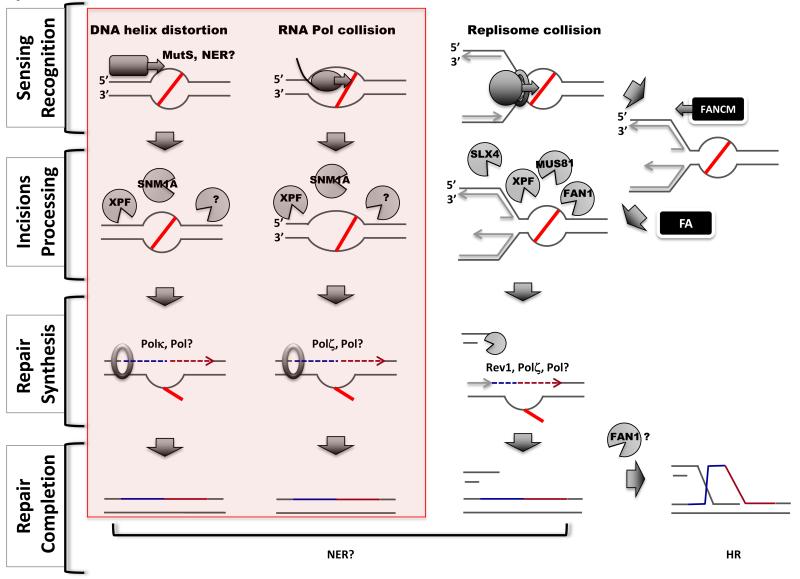
Figure 1.
Pathways of ICL repair. Replication-independent mechanisms are highlighted in the pink box (left). Four steps are shown (left). Three distinct mechanisms can sense ICL lesions (indicated by a red line) (top): repair proteins recognizing distortion in the DNA helix (left column), a transcribing polymerase (middle column) or a replisome blocked by the lesion (right column). Recognition can occur through dedicated repair enzymes (NER or MMR) or following collision during DNA transactions. Several nucleases are thought to participate in the incision steps (second row), however, their exact sites of action are not defined. XPR/ERCC1, by analogy with NER, is thought to act 5′ of the ICLs, whereas the 3′ nuclease acting in RIR is not known (?). The FA pathway regulates the activity of nucleases in the replication-dependent pathway. FANCM facilitates fork regression whereas other FA proteins could recruit nucleases. Specific translesion synthesis nucleases participate in distinct repair pathways (third row): Polκ and Polζ in replication-independent repair and Rev1 and Polζ in the replication-dependent pathway. More than one DNA polymerase could be involved in the repair of an ICL. Replication-dependent ICL repair generates a DSB intermediate (bottom left), which is ultimately repaired via homologous recombination.
How are ICLs sensed in the absence of a stalled replication fork? One potential mechanism could be a collision between a transcribing RNA polymerase and the ICL (Figure 1). NER of lesions on only one strand of DNA occurs preferentially on the transcribed strand. Similarly, ICLs generated by psoralen or cisplatin are also repaired preferentially in actively transcribed genes, as determined by renaturing agarose gel electrophoresis [36, 37]. Furthermore, the transcription-coupled repair (TCR) genes CSA and CSB (Cockaine Syndrome A and B) are required for efficient repair of an MMC ICL positioned in an actively transcribing region of a luciferase reporter plasmid [15]. Cells harboring XPC (Xeroderma Pigmentosum complementation group C) mutations (XPC is an NER protein that is only required for global genome NER [GG-NER] in the absence of transcription) were also significantly impaired in repair of this ICL, suggesting that GG-NER likewise plays a significant role in the repair of MMC ICLs. By contrast, cisplatin-generated ICLs require XPA, XPF and XPG (Xeroderma Pigmentosum complementation group A, F and G) for repair but not XPC [16]. This suggests that the less distorting MMC ICLs might rely more on the XPC lesion-sensing protein for repair, than the more distorting cisplatin ICLs. Structurally distinct ICLs are repaired with varying efficiencies, indicating that the extent of DNA distortion induced by the ICL might define the repair pathway and explain some of the variation in protein requirements for the repair of different types of ICL lesion [19]. However, one limitation of the above studies is that they monitor repair of a crosslink placed in actively transcribed regions of the reporter plasmids, thus they are biased towards TCR. By contrast, a study using a laser to randomly induce psoralen crosslinks in G1 cells found that there was a dependence on the global NER protein, XPC. This suggests that in cells, in the absence of replication, global repair mechanisms also contribute to RIR [12].
ICL repair has also been demonstrated in Xenopus cell-free extracts that do not support replication or transcription. Thus, ICLs are repaired in the absence of collision between the ICL and a replication fork or a transcription bubble [38, 39]. As yet there is no protein that has been unequivocally shown to recognize ICLs in the absence of transcription or replication. However, good candidates include MMR or NER proteins. In vitro studies using purified components show that proteins in the MMR and NER pathways are able to bind specifically to ICL-containing duplexes that do not mimic a replication fork. As mentioned above, MMR complexes are implicated in ICL repair in both replicating cells and non-replicating mammalian cell extracts. Furthermore, MutSβ was shown to recognize ICLs in in vitro gel shift assays, and bind to psoralen-crosslinked substrates in a reaction stimulated by PCNA (Proliferating Cell Nuclear Antigen) [17, 40]. NER protein complexes XPA-RPA (Replication Protein A) and XPC-hR23B (specific for GG-NER) can also bind to psoralen ICLs in vitro, either individually or together to form a larger complex [41, 42]. Additionally, the XPA-RPA complex, but not the XPC-RAD32 complex, interacted with psoralen ICLs simultaneously with MutS. Therefore, it is possible that NER or MMR pathway proteins – together or independently - might act to recognize ICLs in the absence of replication in a manner which is dependent on the type or the chemical or stuctural nature of the ICL.
The stucture of the crosslinked DNA is likely to play an important role in ICL recognition. For example, the NER protein XPC binds to photoproducts through interaction with non-hydrogen bonded bases opposite the lesion [43, 44]. This suggests that ICLs that induce partial unwinding of the DNA helix, such as cisplatin [45] and psoralen [46], could be recognised by XPC in this fashion. XPA has been postulated to act as a damage verification protein, showing a preference for kinked DNA structures [47]. In NER, these kinked structures are induced by upstream NER proteins, but it is possible that ICL lesions that generate significant bending of the DNA helix, such as cisplatin [45] and nitrogen mustards [4], might be sufficient for XPA binding in the the absence of XPC and other proteins. Experiments using mammalian cell extracts have shown that the efficiency of an incision 5′ to the ICL is dependent on the extent of helical distortion induced by the crosslink, with less-distorting lesions showing ~3-fold less incision [3].
In summary, the recognition of ICL lesions in the presence of replication or transcription is a straightforward process which occurs when either a replication fork or transcription bubble collides with the ICL. These collisions are likely then further recognised by other proteins such as FANCM in the case of replication or CSB in the case of transcription, which funnels the lesions into downstream repair processes. In the absence of replication or transcription, the recognition of an ICL is probably a slower process which may require the action of multiple proteins from different pathways such as NER, mismatch repair or some as yet unidentified pathway.
Incision
Following ICL recognition, it is thought that dual incisions are made 5′ and 3′ to the lesion. This “unhooking” of the ICL is considered to be a prerequisite for downstream processing of the damage by other repair proteins (Figure 1). The nucleases responsible for these incisions are not known. In the context of replication-dependent repair, incision of the DNA on both sides of the ICL will result in collapse of the stalled fork and repair by DSB-dependent recombination. In the replication-independent pathway, so long as incision only occurs on one strand or on both strands but widely separated, a DSB will not form.
The acute sensitivity of XPF- or ERCC1-deficient cells to crosslinking agents makes the XPF-ERCC1 heterodimer a prime candidate for being a nuclease involved in the incision step of ICL repair [13, 48-50]. XPF- and ERCC1-deficient CHO cells appear to be impaired in unhooking, as determined by a modified comet assay [13]. DNA double-strand breaks (DSBs) induced by irradiation are routinely measured by embedding cells in agar and monitoring DNA fragmentation following electrophoresis by the appearance of a “tail” of genomic DNA. In this standard comet assay, the tail moment is proportional to the level of DSBs. In the modified comet assay the level of interstrand cross-linking is proportional to the decrease in the tail moment in the irradiated sample treated with a crosslinking agent compared to the irradiated untreated control. Unhooking is expected to generate a DSB in replication-dependent ICL repair, therefore, it is expected that cells lacking XPF-ERCC1 would have less DSBs in response to crosslinking agents. However, similar numbers of DSBs are formed in response to crosslinking drug treatment in XPF- and ERCC1-null cells as compared to their wild-type counterparts [13, 51]. DSBs generated in response to MMC persisted for much longer in ERCC1−/− than in WT cells, suggesting that XPF-ERCC1 may play a role downstream in the resolution of ICL-induced DSBs through recombinational repair [51]. Thus, it is possible that an alternative interpretation of the comet assays is that loss of XPF or ERCC1 results in failed recombination substrates that migrate with similar mobility to crosslinked DNA [52]. Of note, persistent DSBs observed in the absence of XPF might be pathological structures generated by Mus81-Eme1 [53].
SLX4, a structure-specific endonuclease, has recently been identified as an FA protein (FANCP) [54] and is suggested to be a scaffold for the recruitment of incision nucleases because it interacts with both XPF-ERCC1 and MUS81-EME1 (see below). SLX4 forms a dimeric nuclease with SLX1 in which SLX1 provides the catalytic activity and SLX4 is the regulatory subunit. SLX4-null cells are sensitive to crosslinking agents and PARP inhibitors [54, 55]. Interestingly, sensitivity to PARP inhibitors (a surrogate marker of homologous recombination defects) of SLX4-null cells can be complemented with SLX4 mutants that do not interact with XPF. However, the sensitivity of these cells to MMC cannot be rescued with XPF interaction-deficient mutants [56]. The sensitivity of SLX4-defective DT40 cells to crosslinking agents is also not rescued by a mutant lacking the ubiquitin-interacting domain responsible for interacting with ubiquitylated FANCD2 [57]. Together the data suggest that SLX4, XPF and FANCD2 cooperate in the resolution of intermediates generated during the processing of ICLs.
The MUS81-EME1 structure-specific nuclease has also been implicated in ICL repair because mouse embryonic stem cells lacking this complex are sensitive to MMC. In vitro, MUS81-EME1 cleaves DNA structures such as 3′ flaps or replication fork-like stuctures suggesting that a stalled replication fork at an ICL might make a good in vivo substrate for this complex [58]. However, recent work suggests a more downstream role for MUS81-EME1 where it acts as a late responder that promotes repair when other pathways fail [53]. Finally, FAN1 nuclease may be involved in incision, because the FAN1 endonuclease prefers 5′ flap structures [59-61]. However, although FAN1-depleted cells are sensitive to crosslinking agents, ICL-dependent DSBs arise in these cells with normal frequency, but persist longer than in wild-type cells [59, 60]. This suggests that FAN1 is not involved in incision, but rather might act downstream in the repair pathway to resolve DSBs (Figure 1).
It is not clear whether the nucleases required for unhooking are the same for replication-dependent and -independent ICL repair. In some in vitro studies, the XPF-ERCC1 complex was only able to cleave 5′ and 3′ to ICLs in the context of Y-shaped junctions [62, 63]. This implies that XPF can only make incisions when a replication fork is stalled at an ICL. However, in more recent studies XPF-ERCC1 was active on completely duplex DNA, making an incision 5′ to the ICL [53]. Furthermore, linear substrates are cleaved 5′ to the ICL in non-replicating mammalian cell-free extracts in an XPF-ERCC1- and XPG-dependent manner [3]. This latter study also observed NER-independent unhooking in these extracts, suggesting an as yet unidentified protein activity that is able to unhook ICLs in a replication-independent manner.
In the replication-dependent ICL repair pathway, the ubiquitylated FANCD2-FANCI complex promotes incision [64]. It has been suggested, but not yet clearly demonstrated, that ubiquitylation of FANCD2-FANCI might recruit the scaffold protein SLX4 via its ubiquitin binding domains, which in turn might recruit nucleases such as XPF to sites of ICL damage. However, work from our laboratories [38, 39] and others [16] show that RIR does not require the FA pathway. This highlights a major difference between the two repair modes. It suggests that in the absence of replication, nucleases are recruited independently of FANCD2-FANCI. However, it should be noted that although FA proteins may not be required in RIR, the proteins are loaded onto episomal plasmids containing site-specific ICLs even in the absence of replication [65]. Shen et al. used an episomal chromatin immunoprecipitation system containing an Epstein-Barr virus (EBV) replication start site and a site-specific ICL. Replication could be induced with Epstein-Barr nuclear antigen-1 (EBNA). These authors found that even in the absence of EBNA, FANCD2 and FA core proteins were recruited to the ICL-containing plasmid. It is plausible that whilst FA proteins are not specifically required to promote RIR, they might activate a checkpoint. Indeed, Chk1 is phosphorylated in response to ICL damage in a FANCD2 and FA core complex-dependent manner [38].
Finally, cells deficient for the exonuclease SNM1A are sensitive to crosslinking agents. SNM1A has been proposed to act after the incision to digest DNA surrounding an unhooked crosslink to create a better substrate for the translesion synthesis step. In support of this, in vitro assays showed that SNM1A can load from a single strand nick and digest past an ICL [53].
In summary, several nucleases, XPF-ERCC1, MUS81-EME1, SLX1-SLX4, FAN1, FANCD2 and SNM1A, have been associated with ICL repair. Loss of any of these proteins is associated with sensitivity to crosslinking drugs, however, the precise role of these enzymes in ICL repair remains to be elucidated. During replication-dependent ICL repair, incisions are required not only for ICL unhooking but also for the resolution of repair intermediates downstream of the ICL incision, making the task of assigning each nuclease to a defined repair step more difficult than in RIR.
Translesion synthesis
Another common step in both replication-dependent and -independent pathways is translesion DNA synthesis (TLS) (Figure 1). Studies in E. coli have clearly demonstrated HR-independent and -dependent pathways for ICL repair. Efficient repair of a plasmid-borne ICL was detected in the absence of HR in a pathway that was entirely NER-dependent, and partially dependent on the TLS polymerase, PolB. An NER-dependent HR repair pathway was also demonstrated [66]. In eukyaryotes, several TLS polymerases involved in lesion bypass are thought to participate in ICL repair. Pol ζ- and Rev1-deficient cells are sensitive to ICL-forming agents [67-71]. These two proteins are tight interaction partners [72] and in yeast have an epistatic relationship with respect to ICL repair [73]. In replicating Xenopus laevis extracts, depletion of Rev7, the non-catalytic but essential subunit of Pol ζ, blocks ICL repair subsequent to nucleotide insertion opposite the unhooked lesion [20]. Genetic studies in yeast also demonstrate a role for Pol ζ and Rev1 in repair of ICLs in the G1 phase of the cell cycle [73]. Further evidence for an RIR role for Pol ζ and Rev1 comes from work using reporter plasmids in mammalian cells. The plasmids carry either a psoralen or cisplatin lesion in the transcribed region of a GFP or luciferase reporter plasmid. These plasmids do not replicate and are biased towards transcription-coupled repair. Both Rev1 and Pol ζ are required for efficient ICL repair in the plasmid-based systems [16, 74].
By contrast, work in Xenopus cell-free extracts has shown that Pol ζ is not required for repair of ICL lesions in the absence of both transcription and replication [39]. Instead, repair of ICLs in these extracts requires Polκ, a Y family TLS polymerase. Mammalian cells lacking Polκ are also moderately defective in ICL repair [16, 39]. Taken together, these results suggest that in the absence of transcription, Polκ may be preferentially used to bypass unhooked ICLs. Consistent with a role for Polκ in RIR, Polκ−/− MEFs in G0 are exquisitely sensitive to MMC, suggesting that ICL repair requires Polκ in the absence of DNA replication, and when transcription levels are low. Similarly, Polκ is specifically required for NER in G0 cells [75].
A role for Polη in ICL repair has also been suggested by sensitivity assays. Polη-deficient XP-V cells are hypersensitive to crosslinking agents. XP-V cells have greater levels of γH2AX than wild-type cells in response to psoralen crosslinks but not to mono-adducts generated by angelicin [76]. DSBs form only during replication-dependent ICL repair, and γH2AX is a marker for DSBs, which argues for a role for Polη in replication-dependent ICL repair. Polη is not required for transcription-coupled repair of cisplatin lesions [16], but it plays a minor role in repair of psoralen lesions [14] and a more significant role in repair of MMC lesions [15]. These conclusions are based on host reactivation assays that are recombination-independent, and therefore presumably replication-independent. These differences could reflect the fact that different unhooked adducts have varying structures which may fit more or less efficiently into the active site of Polη.
Many of the TLS polymerases mentioned above are able to bypass ICL lesions in vitro [77]. Additionally the Y-family TLS polymerase Polι, which has not yet been shown genetically to have a role in ICL repair, can also insert a nucleotide opposite a guanine in a cisplatin lesion and can bypass a nitrogen mustard lesion in a similar fashion to Polκ and Polη [77]. Polι could therefore be redundant with other Y-family TLS.
Another potential difference between the replication-dependent and -independent pathways is the role of FA proteins in the TLS step. As mentioned above, RIR pathways are independent of FA proteins. By contrast, the FA pathway and TLS are functionally related in ICL repair in the replication-dependent pathway. FA has long been linked to TLS; cells from FA patients and CHO cells deficient for Fanconi proteins are hypomutable, meaning that fewer spontaneous mutations, and fewer mutations in response to DNA damage are observed in these cells [78, 79]. Moreover, chicken DT40 cells lacking FANCC are hypomutable at the IgM locus [80] and FANCC is also epistatic with REV1 and REV3 in terms of cellular sensitivity to crosslinking agents [80]. More recently, a specific association between the TLS polymerase Rev1 and FANCA via the Fanconi anemia-associated protein 20 (FAAP20) has been observed [81]. FAAP20 interacts with REV1, stabilizes REV1 foci and appears to promote TLS as shown using a shuttle-vector-based mutagenesis assay. However, the precise mechanism by which FAAP20 promotes TLS is still not clear. Although this work was related to UV lesions rather than to ICLs, this is nevertheless the first evidence for a physical linkage between FA proteins and a TLS polymerase. It is thus possible that Fanconi proteins play a similar role in regulating TLS in replication-dependent ICL repair.
Homologous recombination
Repair of ICLs in S phase involves the generation of DSBs [13]. The availability of a homologous template makes homologous recombination (HR) a likely candidate to repair these DSBs. This hypothesis is strongly supported by genetic evidence that cells deficient in the HR proteins BRCA1 (Breast and ovarian cancer susceptibility 1), BRCA2 and Rad51 are sensitive to crosslinking agents [82-85], whereas those deficient in non-homologous end-joining (NHEJ) proteins are not [13, 86], indicating that DSB intermediates generated during ICL repair are not substrates for end-joining repair. Interestingly, an HR-independent role for BRCA1 in ICL repair was recently reported. The sensitivity of BRCA1-null MEFs to PARP inhibitors (a measure of HR efficiency) was suppressed by simultaneously knocking down 53BP1, a protein that antagonizes the function of BRCA1 in the early steps of HR. However, these cells were still hypersensitive to cisplatin, suggesting a role for BRCA1 upstream of the HR step in ICL repair [87].
Nakanishi et al. recently described a sophisticated system using a modified DR-GFP (direct repeat GFP) reporter to monitor ICL-induced HR specifically. The reporter has an EBNA binding site and a site-specific psoralen ICL introduced into an incomplete copy of the GFP gene. GFP positive cells can thus only derive from ICL-induced HR. This study found that ICL-induced HR was dependent on induction of replication with EBNA and reduced in FANCA-deficient cells. This suggests a role for the FA pathway in promoting repair of DSBs created by replication fork collisions with ICLs [88].
Recently, direct evidence for HR in replication-dependent ICL repair was demonstrated in Xenopus leavis extracts. Inhibition of Rad51 in replicating extracts ablated repair of a single site-specific ICL. Moreover, Rad51 appeared to bind to replication forks stalled at ICLs prior to induction of a DSB [89]. This suggests that Rad51 may help to stabilize stalled forks and promote stand invasion as soon as a DSB is formed. It should be noted that in this study, Rad51 binding to ICLs was not reduced in the absence of FANCD2, suggesting that FANCD2 is not required for the early accumulation of Rad51.
Since it is generally accepted that a DSB is not generated in RIR, it follows that this pathway does not require HR proteins. Indeed, it has been shown that RIR does not require Rad51 [39] or the Rad51 paralogs XRCC2/3 [15, 16].
Concluding remarks
Understanding all the mechanisms of ICL repair should lead to improvement of crosslinking chemotherapeutic drugs and help to understand why cancers become resistant to such treatment. There is now solid evidence that higher eukaryotes repair ICLs both by a replication- and HR-dependent pathway and by a replication- and/or recombination-independent pathway. It is likely that replication-dependent ICL repair is dominant in rapidly dividing cells. This might explain why RIR has until recently been overlooked. However, RIR is the sole repair mechanism by which non- or slowly-dividing cells maintain genomic integrity and transcriptional activity after ICL generation. The physiological relevance of this pathway is emphasized by several studies, including our own, which have shown that cells lacking proteins such as Polκ [39], Polζ [73] and CSB [16], which are involved in RIR, are more sensitive to crosslinking drugs when treated in G0/G1. Moreover, Enoui et al. showed that inactivation of genes involved in replication-dependent and -independent ICL repair have an additive effect on cellular sensitivity to cisplatin and MMC, suggesting that the two pathways are not mutually exclusive and can act together. This might be particularly important when cells are exposed to especially high doses of crosslinking drugs. In G1 or quiescent cells, RIR would be the only means of defense against crosslinking chemotherapeutic drugs. Therefore, elucidating the distinction between replication-independent and replication-dependent ICL repair should guide the rational design of drugs to target slowly-cycling tumor cells or to help alleviate the side effects of crosslinking drugs on post-mitotic cells. In addition to the implications of RIR for chemotherapy, there is evidence that exposure to ICL-forming agents promotes aging (reviewed in [90]). RIR therefore, might be important in non-replicating cells to slow the process of cellular aging.
Highlights.
Interstrand crosslinks are repaired by distinct pathways during and outside of S-phase.
ICL repair outside of S-phase is critical for cellular homeostasis, particularly in post-mitotic or rarely-dividing cells.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Summerfield FW, Tappel AL. Detection and measurement by high-performance liquid chromatography of malondialdehyde crosslinks in DNA. Anal Biochem. 1984;143(2):265–71. doi: 10.1016/0003-2697(84)90662-6. [DOI] [PubMed] [Google Scholar]
- 2.Summerfield FW, Tappel AL. Cross-linking of DNA in liver and testes of rats fed 1,3-propanediol. Chem Biol Interact. 1984;50(1):87–96. doi: 10.1016/0009-2797(84)90134-0. [DOI] [PubMed] [Google Scholar]
- 3.Smeaton MB, et al. Distortion-dependent unhooking of interstrand cross-links in mammalian cell extracts. Biochemistry. 2008;47(37):9920–30. doi: 10.1021/bi800925e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rink SM, Hopkins PB. A mechlorethamine-induced DNA interstrand cross-link bends duplex DNA. Biochemistry. 1995;34(4):1439–45. doi: 10.1021/bi00004a039. [DOI] [PubMed] [Google Scholar]
- 5.Tomasz M, et al. Isolation and structure of a covalent cross-link adduct between mitomycin C and DNA. Science. 1987;235(4793):1204–8. doi: 10.1126/science.3103215. [DOI] [PubMed] [Google Scholar]
- 6.Roberts JJ, Friedlos F. Quantitative estimation of cisplatin-induced DNA interstrand cross-links and their repair in mammalian cells: relationship to toxicity. Pharmacology & therapeutics. 1987;34(2):215–46. doi: 10.1016/0163-7258(87)90012-x. [DOI] [PubMed] [Google Scholar]
- 7.Noll DM, Mason TM, Miller PS. Formation and repair of interstrand cross-links in DNA. Chem Rev. 2006;106(2):277–301. doi: 10.1021/cr040478b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guainazzi A, Scharer OD. Using synthetic DNA interstrand crosslinks to elucidate repair pathways and identify new therapeutic targets for cancer chemotherapy. Cell Mol Life Sci. 2010;67(21):3683–97. doi: 10.1007/s00018-010-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang LC, Gautier J. The Fanconi anemia pathway and ICL repair: implications for cancer therapy. Critical reviews in biochemistry and molecular biology. 2010;45(5):424–39. doi: 10.3109/10409238.2010.502166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Molecular cell. 2010;40(2):179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasaki MS, Tonomura A. A high susceptibility of Fanconi’s anemia to chromosome breakage by DNA cross-linking agents. Cancer research. 1973;33(8):1829–36. [PubMed] [Google Scholar]
- 12.Muniandy PA, et al. Repair of laser-localized DNA interstrand cross-links in G1 phase mammalian cells. The Journal of biological chemistry. 2009;284(41):27908–17. doi: 10.1074/jbc.M109.029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Silva IU, et al. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Molecular and cellular biology. 2000;20(21):7980–90. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. Involvement of nucleotide excision repair in a recombination-independent and error-prone pathway of DNA interstrand cross-link repair. Molecular and cellular biology. 2001;21(3):713–20. doi: 10.1128/MCB.21.3.713-720.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng H, et al. Nucleotide excision repair- and polymerase eta-mediated error-prone removal of mitomycin C interstrand cross-links. Molecular and cellular biology. 2003;23(2):754–61. doi: 10.1128/MCB.23.2.754-761.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Enoiu M, Jiricny J, Scharer OD. Repair of cisplatin-induced DNA interstrand crosslinks by a replication-independent pathway involving transcription-coupled repair and translesion synthesis. Nucleic acids research. 2012;40(18):8953–64. doi: 10.1093/nar/gks670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang N, et al. hMutSbeta is required for the recognition and uncoupling of psoralen interstrand cross-links in vitro. Molecular and cellular biology. 2002;22(7):2388–97. doi: 10.1128/MCB.22.7.2388-2397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Yehoyada M, et al. Checkpoint signaling from a single DNA interstrand crosslink. Molecular cell. 2009;35(5):704–15. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hlavin EM, et al. Cross-link structure affects replication-independent DNA interstrand cross-link repair in mammalian cells. Biochemistry. 2010;49(18):3977–88. doi: 10.1021/bi902169q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Räschle M, et al. Mechanism of Replication-Coupled DNA Interstrand Crosslink Repair. Cell. 2008;134(6):969–980. doi: 10.1016/j.cell.2008.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ciccia A, et al. Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Molecular cell. 2007;25(3):331–43. doi: 10.1016/j.molcel.2007.01.003. [DOI] [PubMed] [Google Scholar]
- 22.Yan Z, et al. A histone-fold complex and FANCM form a conserved DNA-remodeling complex to maintain genome stability. Molecular cell. 2010;37(6):865–78. doi: 10.1016/j.molcel.2010.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Singh TR, et al. MHF1-MHF2, a histone-fold-containing protein complex, participates in the Fanconi anemia pathway via FANCM. Molecular cell. 2010;37(6):879–86. doi: 10.1016/j.molcel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim JM, et al. Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood. 2008;111(10):5215–22. doi: 10.1182/blood-2007-09-113092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gari K, et al. Remodeling of DNA replication structures by the branch point translocase FANCM. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(42):16107–12. doi: 10.1073/pnas.0804777105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Singh TR, et al. Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood. 2009;114(1):174–80. doi: 10.1182/blood-2009-02-207811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosado IV, et al. The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA crosslink repair. Nucleic Acids Res. 2009;37(13):4360–70. doi: 10.1093/nar/gkp365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang Y, et al. FANCM and FAAP24 maintain genome stability via cooperative as well as unique functions. Mol Cell. 2013;49(5):997–1009. doi: 10.1016/j.molcel.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huang M, et al. Human MutS and FANCM complexes function as redundant DNA damage sensors in the Fanconi Anemia pathway. DNA repair. 2011;10(12):1203–12. doi: 10.1016/j.dnarep.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 30.Williams SA, et al. Functional and physical interaction between the mismatch repair and FA-BRCA pathways. Human molecular genetics. 2011;20(22):4395–410. doi: 10.1093/hmg/ddr366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Aquilina G, et al. N-(2-chloroethyl)-N’-cyclohexyl-N-nitrosourea sensitivity in mismatch repair-defective human cells. Cancer research. 1998;58(1):135–41. [PubMed] [Google Scholar]
- 32.Fiumicino S, et al. Sensitivity to DNA cross-linking chemotherapeutic agents in mismatch repair-defective cells in vitro and in xenografts. International journal of cancer. Journal international du cancer. 2000;85(4):590–6. doi: 10.1002/(sici)1097-0215(20000215)85:4<590::aid-ijc23>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 33.Wu Q, et al. Mismatch repair participates in error-free processing of DNA interstrand crosslinks in human cells. EMBO reports. 2005;6(6):551–7. doi: 10.1038/sj.embor.7400418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barber LJ, et al. DNA interstrand cross-link repair in the Saccharomyces cerevisiae cell cycle: overlapping roles for PSO2 (SNM1) with MutS factors and EXO1 during S phase. Molecular and cellular biology. 2005;25(6):2297–309. doi: 10.1128/MCB.25.6.2297-2309.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ward TA, et al. Components of a Fanconi-like pathway control Pso2-independent DNA interstrand crosslink repair in yeast. PLoS genetics. 2012;8(8):e1002884. doi: 10.1371/journal.pgen.1002884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Larminat F, Zhen W, Bohr VA. Gene-specific DNA repair of interstrand cross-links induced by chemotherapeutic agents can be preferential. The Journal of biological chemistry. 1993;268(4):2649–54. [PubMed] [Google Scholar]
- 37.Islas AL, Vos JM, Hanawalt PC. Differential introduction and repair of psoralen photoadducts to DNA in specific human genes. Cancer research. 1991;51(11):2867–73. [PubMed] [Google Scholar]
- 38.Ben-Yehoyada M, et al. Checkpoint Signaling from a Single DNA Interstrand Crosslink. Molecular Cell. 2009;35(5):704–715. doi: 10.1016/j.molcel.2009.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams HL, Gottesman ME, Gautier J. Replication-independent repair of DNA interstrand crosslinks. Molecular cell. 2012;47(1):140–7. doi: 10.1016/j.molcel.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhao J, et al. Mismatch repair and nucleotide excision repair proteins cooperate in the recognition of DNA interstrand crosslinks. Nucleic acids research. 2009;37(13):4420–9. doi: 10.1093/nar/gkp399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thoma BS, et al. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic acids research. 2005;33(9):2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vasquez KM, et al. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(9):5848–53. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Camenisch U, et al. Two-stage dynamic DNA quality check by xeroderma pigmentosum group C protein. The EMBO journal. 2009;28(16):2387–99. doi: 10.1038/emboj.2009.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maillard O, Solyom S, Naegeli H. An aromatic sensor with aversion to damaged strands confers versatility to DNA repair. PLoS biology. 2007;5(4):e79. doi: 10.1371/journal.pbio.0050079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coste F, et al. Crystal structure of a double-stranded DNA containing a cisplatin interstrand cross-link at 1.63 A resolution: hydration at the platinated site. Nucleic acids research. 1999;27(8):1837–46. doi: 10.1093/nar/27.8.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spielmann HP, et al. Solution structures of psoralen monoadducted and cross-linked DNA oligomers by NMR spectroscopy and restrained molecular dynamics. Biochemistry. 1995;34(40):12937–53. [PubMed] [Google Scholar]
- 47.Camenisch U, et al. Recognition of helical kinks by xeroderma pigmentosum group A protein triggers DNA excision repair. Nature structural & molecular biology. 2006;13(3):278–84. doi: 10.1038/nsmb1061. [DOI] [PubMed] [Google Scholar]
- 48.Hoy CA, et al. Defective DNA cross-link removal in Chinese hamster cell mutants hypersensitive to bifunctional alkylating agents. Cancer research. 1985;45(4):1737–43. [PubMed] [Google Scholar]
- 49.Andersson BS, et al. Nucleotide excision repair genes as determinants of cellular sensitivity to cyclophosphamide analogs. Cancer chemotherapy and pharmacology. 1996;38(5):406–16. doi: 10.1007/s002800050504. [DOI] [PubMed] [Google Scholar]
- 50.De Silva IU, et al. Defects in interstrand cross-link uncoupling do not account for the extreme sensitivity of ERCC1 and XPF cells to cisplatin. Nucleic acids research. 2002;30(17):3848–56. doi: 10.1093/nar/gkf479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niedernhofer LJ, et al. The structure-specific endonuclease Ercc1-Xpf is required to resolve DNA interstrand cross-link-induced double-strand breaks. Mol Cell Biol. 2004;24(13):5776–87. doi: 10.1128/MCB.24.13.5776-5787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bergstralh DT, Sekelsky J. Interstrand crosslink repair: can XPF-ERCC1 be let off the hook? Trends in Genetics. 2008;24(2):70–76. doi: 10.1016/j.tig.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 53.Wang AT, et al. Human SNM1A and XPF-ERCC1 collaborate to initiate DNA interstrand cross-link repair. Genes & development. 2011;25(17):1859–70. doi: 10.1101/gad.15699211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim Y, et al. Mutations of the SLX4 gene in Fanconi anemia. Nature genetics. 2011;43(2):142–6. doi: 10.1038/ng.750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fekairi S, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138(1):78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, et al. Regulation of multiple DNA repair pathways by the Fanconi anemia protein SLX4. Blood. 2013;121(1):54–63. doi: 10.1182/blood-2012-07-441212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamamoto KN, et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011;108(16):6492–6. doi: 10.1073/pnas.1018487108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hanada K, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. The EMBO journal. 2006;25(20):4921–32. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142(1):77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 60.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142(1):65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Molecular cell. 2010;39(1):36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fisher LA, Bessho M, Bessho T. Processing of a psoralen DNA interstrand cross-link by XPF-ERCC1 complex in vitro. The Journal of biological chemistry. 2008;283(3):1275–81. doi: 10.1074/jbc.M708072200. [DOI] [PubMed] [Google Scholar]
- 63.Kuraoka I, et al. Repair of an interstrand DNA cross-link initiated by ERCC1-XPF repair/recombination nuclease. The Journal of biological chemistry. 2000;275(34):26632–6. doi: 10.1074/jbc.C000337200. [DOI] [PubMed] [Google Scholar]
- 64.Knipscheer P, et al. The Fanconi anemia pathway promotes replication-dependent DNA interstrand cross-link repair. Science. 2009;326(5960):1698–701. doi: 10.1126/science.1182372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shen X, et al. Recruitment of fanconi anemia and breast cancer proteins to DNA damage sites is differentially governed by replication. Mol Cell. 2009;35(5):716–23. doi: 10.1016/j.molcel.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Berardini M, Foster PL, Loechler EL. DNA polymerase II (polB) is involved in a new DNA repair pathway for DNA interstrand cross-links in Escherichia coli. J Bacteriol. 1999;181(9):2878–82. doi: 10.1093/gao/9781884446054.article.t031385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu F, et al. DNA polymerase zeta regulates cisplatin cytotoxicity, mutagenicity, and the rate of development of cisplatin resistance. Cancer research. 2004;64(21):8029–35. doi: 10.1158/0008-5472.CAN-03-3942. [DOI] [PubMed] [Google Scholar]
- 68.Okada T, et al. Multiple roles of vertebrate REV genes in DNA repair and recombination. Molecular and cellular biology. 2005;25(14):6103–11. doi: 10.1128/MCB.25.14.6103-6111.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sonoda E, et al. Multiple roles of Rev3, the catalytic subunit of polzeta in maintaining genome stability in vertebrates. The EMBO journal. 2003;22(12):3188–97. doi: 10.1093/emboj/cdg308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wittschieben JP, et al. Loss of DNA polymerase zeta causes chromosomal instability in mammalian cells. Cancer research. 2006;66(1):134–42. doi: 10.1158/0008-5472.CAN-05-2982. [DOI] [PubMed] [Google Scholar]
- 71.Simpson LJ, Sale JE. Rev1 is essential for DNA damage tolerance and non-templated immunoglobulin gene mutation in a vertebrate cell line. The EMBO journal. 2003;22(7):1654–64. doi: 10.1093/emboj/cdg161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Murakumo Y, et al. Interactions in the error-prone postreplication repair proteins hREV1, hREV3, and hREV7. The Journal of biological chemistry. 2001;276(38):35644–51. doi: 10.1074/jbc.M102051200. [DOI] [PubMed] [Google Scholar]
- 73.Sarkar S, et al. DNA interstrand crosslink repair during G1 involves nucleotide excision repair and DNA polymerase zeta. EMBO J. 2006;25(6):1285–94. doi: 10.1038/sj.emboj.7600993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shen X, et al. REV3 and REV1 play major roles in recombination-independent repair of DNA interstrand cross-links mediated by monoubiquitinated proliferating cell nuclear antigen (PCNA) J Biol Chem. 2006;281(20):13869–72. doi: 10.1074/jbc.C600071200. [DOI] [PubMed] [Google Scholar]
- 75.Ogi T, et al. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Molecular cell. 2010;37(5):714–27. doi: 10.1016/j.molcel.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 76.Mogi S, Butcher CE, Oh DH. DNA polymerase eta reduces the gamma-H2AX response to psoralen interstrand crosslinks in human cells. Experimental cell research. 2008;314(4):887–95. doi: 10.1016/j.yexcr.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ho TV, et al. Structure-dependent bypass of DNA interstrand crosslinks by translesion synthesis polymerases. Nucleic acids research. 2011;39(17):7455–64. doi: 10.1093/nar/gkr448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hinz JM, et al. The Fanconi anemia pathway limits the severity of mutagenesis. DNA repair. 2006;5(8):875–84. doi: 10.1016/j.dnarep.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 79.Papadopoulo D, et al. Decreased mutagenicity in Fanconi’s anemia lymphoblasts following treatment with photoactivated psoralens. Progress in clinical and biological research. 1990;340A:241–8. [PubMed] [Google Scholar]
- 80.Niedzwiedz W, et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Molecular cell. 2004;15(4):607–20. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 81.Kim H, et al. Regulation of Rev1 by the Fanconi anemia core complex. Nature structural & molecular biology. 2012;19(2):164–70. doi: 10.1038/nsmb.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bhattacharyya A, et al. The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. The Journal of biological chemistry. 2000;275(31):23899–903. doi: 10.1074/jbc.C000276200. [DOI] [PubMed] [Google Scholar]
- 83.Grossmann KF, et al. S. cerevisiae has three pathways for DNA interstrand crosslink repair. Mutation research. 2001;487(3-4):73–83. doi: 10.1016/s0921-8777(01)00106-9. [DOI] [PubMed] [Google Scholar]
- 84.Moynahan ME, Cui TY, Jasin M. Homology-directed dna repair, mitomycin-c resistance, and chromosome stability is restored with correction of a Brca1 mutation. Cancer research. 2001;61(12):4842–50. [PubMed] [Google Scholar]
- 85.Kraakman-van der Zwet M, et al. Brca2 (XRCC11) deficiency results in radioresistant DNA synthesis and a higher frequency of spontaneous deletions. Molecular and cellular biology. 2002;22(2):669–79. doi: 10.1128/MCB.22.2.669-679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pace P, et al. Ku70 corrupts DNA repair in the absence of the Fanconi anemia pathway. Science. 2010;329(5988):219–23. doi: 10.1126/science.1192277. [DOI] [PubMed] [Google Scholar]
- 87.Bunting SF, et al. BRCA1 functions independently of homologous recombination in DNA interstrand crosslink repair. Molecular cell. 2012;46(2):125–35. doi: 10.1016/j.molcel.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nakanishi K, et al. Homology-directed Fanconi anemia pathway cross-link repair is dependent on DNA replication. Nature structural & molecular biology. 2011;18(4):500–3. doi: 10.1038/nsmb.2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Long DT, et al. Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science. 2011;333(6038):84–7. doi: 10.1126/science.1204258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Grillari J, Katinger H, Voglauer R. Contributions of DNA interstrand cross-links to aging of cells and organisms. Nucleic acids research. 2007;35(22):7566–76. doi: 10.1093/nar/gkm1065. [DOI] [PMC free article] [PubMed] [Google Scholar]