Abstract
Mantle cell lymphoma (MCL) is a heterogeneous disease with most patients following an aggressive clinical course while others have an indolent behavior. We performed an integrative and multidisciplinary analysis of 177 MCL to determine whether the immunogenetic features of the clonotypic B cell receptors may identify different subsets of tumors. ‘Truly unmutated’ (100% identity) IGHV genes were found in 24% cases, 40% were ‘minimally/borderline mutated’ (99.9-97%), 19% ‘significantly mutated’ (96.9-95%) and 17% ‘hypermutated’ (<95%). Tumors with high (≥97%) or low (<97%) mutational load used different IGHV genes and their gene expression profiles were also different for several gene pathways. A gene set enrichment analysis showed that MCL with high and low IGHV mutations were enriched in memory and naïve B-cell signatures, respectively. Furthermore, the highly mutated tumors displayed less genomic complexity, were preferentially SOX11 negative, and showed more frequently non-nodal disease. The best cut-off of germline identity of IGHV genes to predict survival was 3%. Patients with high and low mutational load had significant different outcome with 5-year overall survival of 59% and 40%, respectively (P=0.004). Nodal presentation and SOX11 expression also predicted for poor overall survival. In a multivariate analysis, IGHV gene status and SOX11 expression were independent risk factors. In conclusion, these observations suggest the idea that MCL with mutated IGHV, SOX11 negativity, and non-nodal presentation correspond to a subtype of the disease with more indolent behavior.
Keywords: mantle cell lymphoma, immunoglobulin rearrangement, SOX11, expression profiling, prognosis
Introduction
Mantle cell lymphoma (MCL) is a mature B-cell neoplasm clinically characterized by a generalized lymphadenopathy, disseminated disease at diagnosis and a poor clinical evolution (1). The translocation t(11;14)(q13;q32) deregulating cyclin D1 expression is considered the primary oncogenic event (2,3). Additionally, MCL usually carry a high number of secondary chromosomal alterations targeting different oncogenic pathways that contribute to the progression of the disease (4–8).
The immunogenetic analysis of the clonogenic B cell receptors (BcRs) in B-cell neoplasms has made significant contributions towards understanding their ontogenetic derivation, obtaining evidence for the possible involvement of antigen selection in their pathogenesis, and identifying biological subtypes with clinical implications (9–12). These studies have also proven to be of clinical relevance as exemplified by chronic lymphocytic leukemia (CLL), where molecular analysis of the immunoglobulin (IG) genes expressed by the clonogenic BcRs identifies subsets of tumors with different biological features, clinical presentation and outcome, indicating that the functional antigen reactivity of the clonogenic BcRs is critically implicated in shaping the biological behavior of the malignant clones (9,10,13–16).
MCL shares certain phenotypic and biological analogies with CLL (1). Several studies have shown that 15–40% of MCL have a somatically hypermutated BcR and a strong restriction in IGHV gene usage (17,18). Similar to CLL (19), closely homologous (stereotyped) VH CDR3s have been recognized in MCL, albeit with molecular features clearly distinct from those described in CLL (17,19). Altogether, these immunogenetic findings strongly argue for antigen-driven selection in the clonogenic expansion of tumor cells in MCL (17). However, contrary to CLL, the clinical implications of immunogenetic analysis in MCL remain controversial (18,20–26). Most studies have found no relationship between the mutational status of the clonogenic IGHV genes and the evolution of the disease (23,27). Although a tendency to longer survival has been reported for patients with a high number of somatic mutations or carrying specific IGHV genes (18,21,22,28). Moreover, a subset of patients with a very indolent clinical course and SOX11 negative expression seem to express IGs with a high load of somatic hypermutation (SHM) (20). A potential confounding issue in most relevant studies has been the application of a 2% identity cut-off value for assigning cases to the mutated or unmutated subgroup. This cut-off has been utilized widely for prognostication in CLL but may not be appropriate outside this context. In particular, it may mask the biological (and, perhaps, clinical) heterogeneity of MCL as also indicated by the recent finding that subsets of MCL cases with different mutational load display marked immunogenetic differences, even when comparing cases with limited mutations (98–99.9% identity) to those with no mutations at all (100% identity) (17). In the present study, we performed an integrative and multidisciplinary analysis of a large series of MCL in order to determine whether the specific molecular features of the clonotypic BcR may identify subsets of tumors with biological differences potentially underlying a different clinical behavior.
Materials and Methods
Study population
177 patients with MCL were selected based on the availability of tumor samples. All cases had the t(11;14)(q13;q32) and/or cyclin D1 overexpression. Samples were obtained from peripheral blood (n=99), lymph nodes (n=41), spleen (n=15), bone marrow (n=7), and other tissues (n=15). The IGHV gene mutational status of 41 out of 177 cases was reported previously (17). Clinical information is summarized in Table 1. The patients were managed heterogeneously: thirteen patients did not receive chemotherapy and fifteen patients received front-line intensive treatment with high-dose araC and/or autologous stem cell transplant. Ninety patients were treated with different regiments of conventional chemotherapy. Rituximab was administered to 43 patients at some stage during the course of their disease. Tissue sections for evaluation of the histological variants and additional pathological features were available in 95 cases (Table 1). The study was approved by the Institutional Review Board and informed consent was obtained from each patient.
Table 1.
Clinico-biological characteristics of MCL patients according to the mutational status of IGHV.
| Total | HM (<95%) | SM (95%–96.9) | MBM (97%–99.9%) | TU (100%) | P | |
|---|---|---|---|---|---|---|
| No. of cases (%) | 177 | 30 (17%) | 33 (19%) | 71 (40%) | 43 (24%) | |
| Pathological and molecular data | ||||||
| Morphology | 0.045 | |||||
| Classical/small cell (%) | 71/95 (75) | 6/6 (100) | 8/13 (66) | 29/44 (66) | 28/32 (87) | |
| Blastoid/pleomorphic (%) | 24/95 (25) | 0/6 | 5/13 (38) | 15/44 (34) | 4/32 (13) | |
| Ki-67 (≥35%) | 24/75 (32) | 1/8 (12) | 4/10 (40) | 14/34 (41) | 5/23 (22) | 0.249 |
| CD5-positive | 72/83 (87) | 9/16 (56) | 11/13 (85) | 32/34 (94) | 20/20 (100) | <0.001 |
| SOX11-positive | 111/161 (69) | 5/26 (19) | 11/26 (42) | 57/67 (85) | 38/42 (90) | <0.001 |
| CNA, mean number (range) | 12.4 (0–50) | 2.8 (0–26) | 6.3 (0–33) | 9.5 (0–50) | 8.6 (0–34) | <0.001 |
| High genomic complexity (≥4 CNA) | 62/101 (61) | 4/18 (22) | 6/16 (37) | 30/39 (77) | 22/28 (79) | <0.001 |
| TP53 mutated | 21/85 (25) | 3/12 (25) | 3/13 (23) | 12/35 (37) | 2/25 (8) | 0.083 |
| 17p alteration/TP53 mutated | 33/109 (30) | 7/20 (35) | 4/17 (23) | 15/43 (35) | 7/29 (24) | 0.676 |
| Clinical data | ||||||
| Median age (range) | 66 (29–90) | 68 (45–88) | 70 (44–85) | 66 (29–89) | 63 (33–90) | |
| Ratio Male/Female | 112/39 | 11/11 | 17/8 | 51/13 | 33/7 | 0.027 |
| Lymph node (>1 cm) (%) | 84/122 (69) | 5/19 (26) | 9/18 (50) | 42/50 (84) | 28/35 (80) | <0.001 |
| Palpable splenomegaly (%) | 67/110 (60) | 7/16 (44) | 7/16 (44) | 30/47 (63) | 23/31 (74) | 0.094 |
| High serum LDH (>450) (%) | 30/77 (39) | 2/14 (14) | 3/11 (27) | 14/33 (42) | 11/18 (61) | 0.044 |
| Stage IV (%) | 70/79 (89) | 7/8 (88) | 7/10 (70) | 32/35 (91) | 24/26 (92) | 0.246 |
| Lymphocytosis (median, L/mm3) | 8800 | 10812 | 11368 | 18374 | 38633 | 0.396 |
| Follow up data | ||||||
| Median follow-up, years (range) | 3.25 (0.23–21) | 4.08 (0.33–8) | 3.50 (0.50–21) | 3.63 (0.23–12) | 2.65 (0.68–6.58) | 0.748 |
| Chemotherapy at any time (%) | 105/118 (89) | 13/19 (68) | 13/18 (72) | 47/49 (96) | 32/32 (100) | <0.001 |
| Median time to treatment (months) | 1 | 4.4 | 2 | 1 | 1 | 0.157 |
| Complete response rate (%) | 30 | 15 | 15 | 52 | 19 | 0.02 |
| 5-year overall survival (%) | 46 | 62 | 55 | 49 | 25 | 0.023 |
Amplification of IGHV-IGHD-IGHJ rearrangements, sequence analysis and interpretation
PCR amplification was performed using either complementary or genomic DNA extracted from cryopreserved blood cells, frozen tissues and formalin-fixed paraffin embedded (FFPE) tissues (17 cases). RNA and DNA were extracted by using the TRIZOL reagent (Invitrogen Life Technologies, Inc., Gaithersburg, MD), QIAamp DNA mini-kit, AllPrep DNA/RNA Mini Kit (for tissue samples), and FFPE RNeasy minikit (for FFPE tissues)(QIAGEN, Germantown, MA, USA).
IGHV-IGHD-IGHJ rearrangements were amplified and analyzed as reported previously (10,29). The sequences were analyzed using IMGT® databases and tools (30,31) (32). Only productive rearrangements were evaluated. Output data from IMGT/V-QUEST was used to obtain: IGHV gene usage, percentage of identity to germ line, length and composition of the VH CDR3.
Gene expression profiling
We studied the gene expression profiling (GEP) of 38 cases using highly purified leukemic cells (>95% by flow cytometry) from untreated MCL patients. RNA was hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays (Affymetrix, Santa Clara, CA), as described previously (20). The analysis of the scanned images and the determination of the signal value were obtained with GeneChip® Command Console® Software (Affymetrix). Raw data were imported to R Package. The data was normalized using the Robust Multichip Analysis algorithm of the BioConductor affy Package and the 25% of genes with lower interquartile range were excluded. Differential gene expression between the subgroups of cases was performed using moderated t-statistics with empirical Bayes shrinkage of the standard errors, implemented in the BioConductor limma Package (33). The false discovery rate (FDR) method of Benjamini and Hochberg was used to adjust the P-value for each gene based on a significance level of 0.05. Functional enrichment analysis of the differentially expressed genes was performed using the DAVID and IPA applications. The primary data of the microarrays are available from the Gene Expression Omnibus (GEO) of the National Center for Biotechnology Information, accession number GSE36000.
Twenty differentially expressed genes were selected and validated by quantitative PCR (qPCR) using the Fluidigm BioMark Real Time System (Fluidigm, San Francisco, CA). Total RNA was retrotranscribed to cDNA using the high capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA) and pre-amplified with 14 cycles according to the manufacturer’s recommendations. The GUSB and B2M genes were used as endogenous controls and Universal Human Reference RNA (Stratagene, La Jolla, CA, US) as the calibrator sample.
Gene set enrichment analysis (GSEA)
We performed an enrichment pathway analysis using the GSEA desktop application (GSEA, Broad Institute at MIT, Cambridge, MA). We used a pre-ranked gene list based on the limma’s statistic obtained in the differential expression analysis using the curated collection of canonical pathways (3276 gene sets). We added four additional gene sets generated in-house to capture gene expression signatures associated with different B-cell origin, using data available at the GEO (accession number GDS3516) (Supplemental Table S1). The B-cell origin signatures were defined by those genes that were more significantly expressed, exhibiting at least 20% higher levels compared to the second highest expressing group (for naïve and memory B-cell signatures) and 50% (for germinal center cell and plasma cell signatures) in order to obtain approximately 100–200 genes in each signature.
Analysis of SOX11 expression
SOX11 expression was evaluated in 161/177 (90%) cases and categorized as positive or negative according to previous defined criteria (20,29,34). One hundred forty-three cases were analyzed by qPCR using the cut-off of 9 relative units (29), 64 cases were analyzed by immunohistochemisty (34) and 50 cases by GEP using as cutoff a signal value of 120 for the probe set 204913_s_at following a MAS5 normalization with a target intensity of 150. Overall, 51% of the cases were evaluated with at least two different techniques, with fully concordant results (Supplemental Figure S1).
Molecular analysis
TP53 mutational analysis was performed as previously described (29) and the genomic profile of 101 cases was investigated using the Affymetrix Genome-Wide Human SNP Array 6.0 in 73 cases and the 100K SNP-array in 28 cases, as previously described (Affymetrix, Santa Clara, CA) (35,36).
Statistical analysis
The independence between categorical clinical parameters and the MCL subgroups was evaluated using Fisher’s exact test and continuous variables were compared by Mann-Whitney or Kruskall-Wallis tests. To find the best IGHV gene identity % cut-off related to survival we used a maximally selected log-rank statistic (maxstat package). Overall survival (OS) was measured from date of diagnosis to date of death or last follow-up. Survival was estimated using the Kaplan-Meier method and survival curves were compared using the log-rank test. The association between different variables and outcome was estimated using univariate Cox regression analysis, whereas the independence of IGHV gene mutational status was estimated by multivariate Cox regression analysis. Covariates included in the multivariate were age, nodal presentation, IGHV gene mutational status and SOX11 expression. The variables selected for the multivariate analysis were the ones statistically significant in the univariate analysis and which had information available in more than 100 patients. The covariates used in the model did not show co-linearity and did not violate the proportional hazard assumption (by assessing the plots of smoothed martingale residuals). P values ≤0.05 were considered statistically significant. All analyses were performed with SPSS software v.18.0 (Chicago, IL) and R Package.
Results
IG gene repertoires and SHM
A total of 177 productive IGHV-IGHD-IGHJ rearrangements from 177 patients were analyzed. Based on the approach introduced by Hadzidimitriou et al (17) we subdivided the cohort in three subsets: i) ‘truly unmutated’ (TU; 100% identity), ii) ‘minimally/borderline mutated’ (MBM; 99.9-97% identity), and iii) ‘significantly mutated’ (SM; 96.9-95% identity). We included an additional subset defined as ‘hypermutated’ (HM; <95% identity). Overall, 24% of the cases were classified as TU, 40% as MBM, 19% as SM and 17% as HM IGHV (Table 1).
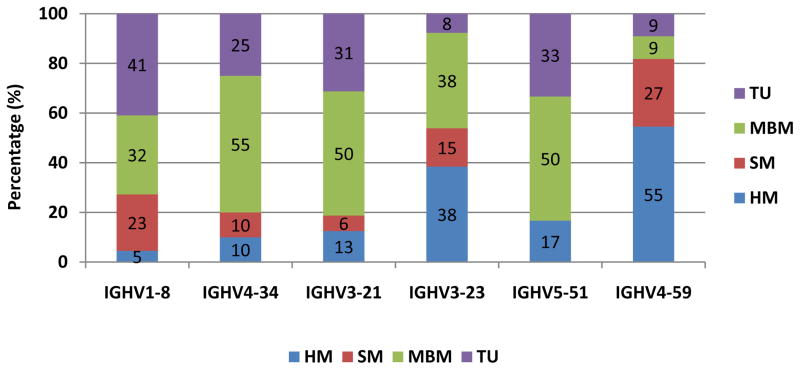
As expected, IGHV3 and IGHV4 were predominant (39% and 28% of cases, respectively). The IG repertoire is skewed, since 53% of the gene usage was represented by only six genes, IGHV1-8 (12%), IGHV4-34 (11%), IGHV3-21 (9%), IGHV3-23 (7%), IGHV5-51 (7%), and IGHV4-59 (6%) (Supplemental Table S2), this bias in the repertoire is similar to the previously reported in a large cohort of MCL (17). The IGHV gene repertoire of the four mutational subsets was different: IGHV1-8 was preferentially used by TU cases (41%) and MBM cases (32%); IGHV4-34 and IGHV3-21 were more common in MBM (55 and 50%, respectively); IGHV4-59 predominated in the HM subset (55%); and IGHV3-23 was found with similar frequencies in the HM and MBM subsets (38%) (Figure 1 and Supplemental Table S2).
Figure 1.
Distribution of rearrangements of the six more frequent IGHV genes in subgroups of MCL with different mutational status.
We identified twenty-four IGHD genes among which the IGHD2-2 (13%), IGHD3-3 (10%), IGHD2-15 (8%) and IGHD1-26 (7%) predominated (Supplementary Table S2). Interestingly, IGHD1-26 was more frequently associated with the TU group (16%) and IGHD2-2 was mainly found in HM and SM (17% and 24%, respectively). Most of the cases utilized IGHJ4 (39%) and IGHJ6 (28%) with no differences among the four IGHV mutational subsets (Supplemental Table S2). The median VH CDR3 length was 16 amino acids (aa) (range 7–30). IGHV5-51 and IGHV3-23 displaying shorter VH CDR3 compared to IGHV3-34 and IGHV3-21 rearrangements (11 and 13 aa vs. 18 and 19 aa, respectively). Differential patterns of IGHV-IGHD and IGHV-IGHJ associations were identified that were concordant with our previous findings (17).
Recurrent aa changes introduced by SHM in conserved positions of the VH domain are considered as suggestive of antigen selection (9,12,17,19). We found such changes in mainly in IGHV4-59, IGHV3-23 and IGHV1-8 rearrangements (Supplemental Figure S2). As an example, codon 92 (FR3) of the IGHV4-59 rearrangements was mutated in 55% of the cases, with the substitution S-to-T in this codon being detected in 36% of cases (Supplementary Figure S2).
MCL subsets exhibit distinct pathological and molecular features
We explored potential associations between IGHV gene mutational status and several features of the MCL tumors. HM-MCL was conspicuous for the complete absence of blastoid/pleomorphic variants, whereas their proportion was similar in the other subgroups (P=0.045) (Table 1). Only half of the HM-MCL cases expressed CD5 whereas almost all tumors of the other mutational subsets were positive (P<0.001) (Table 1).
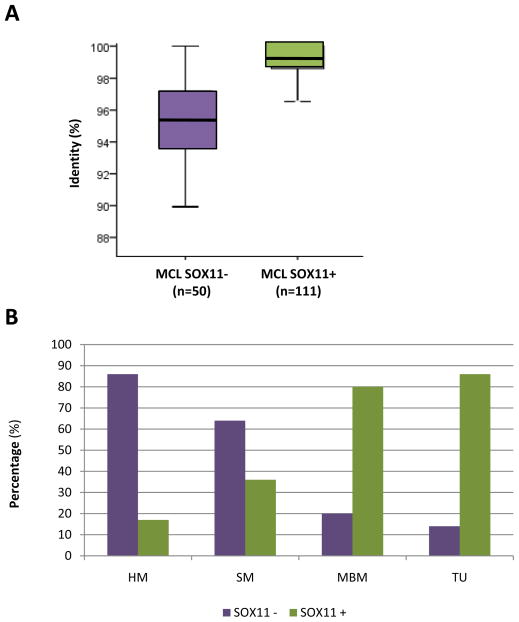
SOX11 expression was positive in 69% of evaluated (Table 1). SOX11-positive tumors had significantly lower IGHV gene mutations than SOX11-negative tumors (mean % identity: 98.7% and 95.1%, respectively; P<0.001) (Figure 2A). Notably, SOX11 expression was predominantly found in tumors with no or low IGHV mutations (TU 86%; MBM 80%) whereas only a small number of MCL with high number of IGHV mutations expressed SOX11 (HM 17%; SM 36%) (P<0.001) (Figure 2B; Table 1).
Figure 2.
IGHV mutational load according to SOX11 expression. A, Box plot of the percentage of IGHV identity in MCL cases negative and positive for SOX11 expression. B, Distribution of the cases according to four subgroups of IGHV gene mutational status and SOX11 expression.
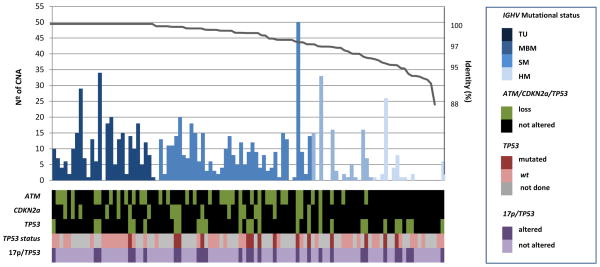
The profile of gains and losses of 101 cases analyzed by SNP-arrays was concordant with the pattern previously defined (5). The main losses were located at 11q (35%), 8p (33%), 13q (30%), 17p (26%) and 9p (21%) whereas regions with gains were 3q (30%) and 8q (23%). The number of copy number alterations (CNA) was inversely related to the level of SHM and varied from 2.8 (range 0–26) in HM tumors to 8.6 (range 0–34) in TU MCL (P<0.001) (Table 1). The number of cases with high genomic complexity (≥4 alterations) was significantly lower in HM and SM (22% and 37%) than in MBM and TU tumors (77% and 79%) (P<0.001) (Figure 3, Table 1). Interestingly, no deletions of the ATM and CDKN2A were observed in HM MCL compared to the remaining three groups whereas TP53 gene mutations and 17p alterations/TP53 mutations were similarly distributed among the four subsets of MCL (Table 1 and Figure 3).
Figure 3.
Characterization of MCL cases according to their distinct genetic and molecular features. The 101 cases in which SNP-array analyses were performed are represented. In the upper panel the number of genomic alterations (bar plots) and the % of identity of IGHV genes (from left to right in decreasing order).
GEP reveals distinct signatures and putative cell of origin in MCL subsets with different IGHV gene mutational status
To identify biological features that could distinguish the different subsets of MCL according to SHM status, we performed a genome-wide GEP of 38 purified peripheral blood untreated tumor samples. We grouped the HM and SM cases as mutated MCL (M-MCL) and the MBM and TU as unmutated MCL (U-MCL) and found 518 genes differentially expressed: 395 genes were upregulated in U-MCL and 123 in M-MCL (Supplemental Table S3). The signature of 13 genes, including SOX11, that we found previously underexpressed in indolent MCL (20), was also downregulated in M-MCL in this independent series. The previously described GEP proliferation signature (37) and survival predictor signature (38) were not significantly different between the two subgroups, although there was a trend to lower proliferation (mean±standard deviation; 0.14±0.25 vs 0.51±1.04) and better prognosis score (−0.09±0.41 vs. 0.32±1.03) in M-MCL compared to U-MCL, respectively.
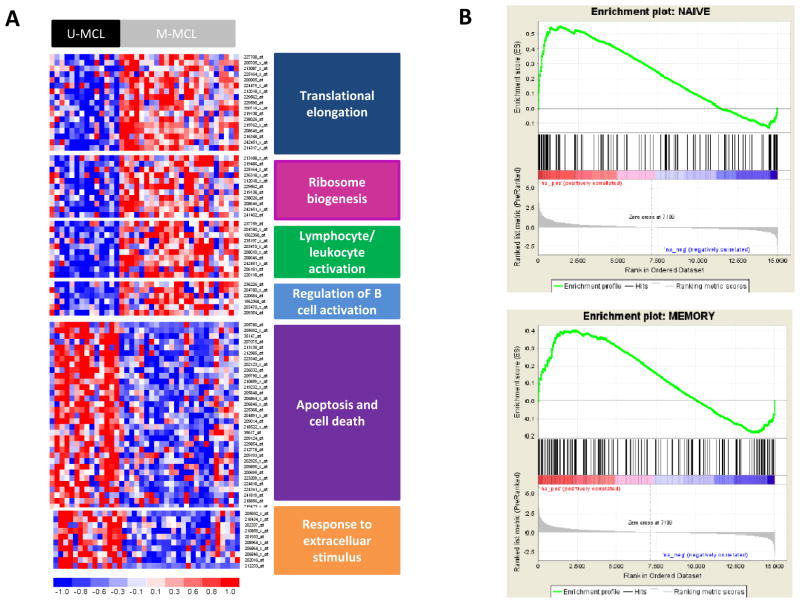
To gain insights into the biological meaning of the differential expression profile between M-MCL and U-MCL, we performed a functional enrichment analysis using the DAVID application. The most significant biological processes enriched among the up-regulated genes in M-MCL were related to translational elongation, ribosome biogenesis, regulation of B-cell activation and leukocyte/lymphocyte activation. On the other hand, the regulation of transcription, apoptosis/cell death and response to extracellular stimulus were the functional terms enriched among the genes significantly upregulated in U-MCL group (Figure 4A). Very similar results were also obtained using IPA (data not shown).
Figure 4.
A, Differences in the gene expression profile between U-MCL and M-MCL. Heatmap displaying the main pathways enriched among the genes differentially expressed. B, Enrichment plots obtained from GSEA. Significant enrichment of the naïve B-cell gene set in the U-MCL/SOX11-possitive and significant enrichment of the memory B-cell gene set in M-MCL/SOX11-negative.
To validate the GEP results we performed qPCR of 20 selected genes in 38 cases, 21 M-MCL and 17 U-MCL, six tumors studied by GEP and 32 independent cases. The results were concordant in 18/20 (90%) (Supplemental Table S4).
To investigate whether the different GEP of the U-MCL and M-MCL could be related to a particular subtype of normal B-cell counterpart we performed a GSEA using four specific gene sets related to different normal B-cell subtypes, this analysis indicated that the SOX11-positive U-MCL expressed a signature enriched in genes related to naïve B-cells (FDR: 0.001 and NES: 1.7) whereas SOX11-negative M-MCL had a signature related to memory B-cells (FDR: 0.086 and NES: 1.28) (Figure 4B).
IGHV gene mutational status defines MCL subsets with distinct clinical presentation and outcome
The male/female ratio was significantly different in the four MCL subsets, varying from 1 in HM MCL to 4.7 in TU tumors (P=0.027) (Table 1). The major clinical difference concerned nodal presentation, which was less common in HM (26%) and SM (50%) than in MBM (84%) and TU (80%) MCL (P<0.001). Blood lymphocytosis was higher in MBM- and TU-MCL but the differences were not statistically significant (Table 1).
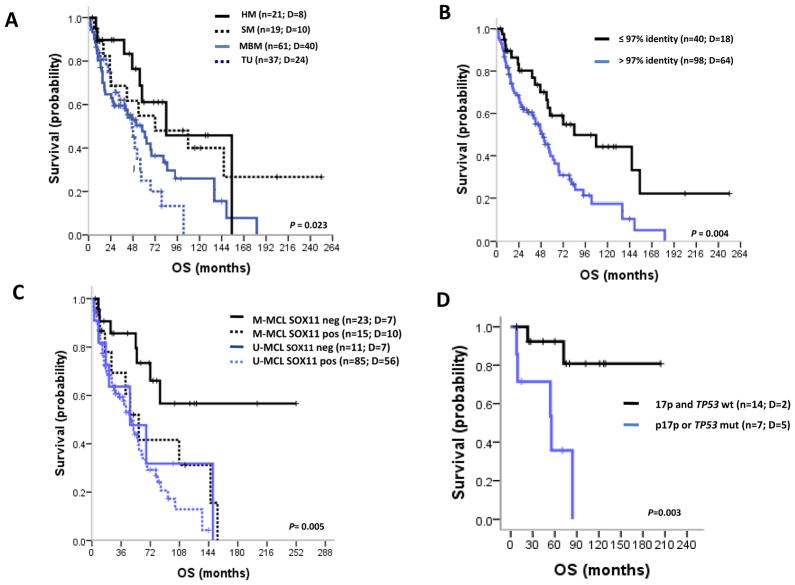
The four MCL subsets did not differ regarding the use of different treatments, including either high dose AraC and/or autologous stem cell transplantation or Rituximab at any time during the course of the disease. Overall, 68 and 71% of HM and SM MCL were treated during the clinical course compared to 96 and 100% of MBM and TU, respectively (P<0.001). M-MCL patients had a significantly better OS than U-MCL patients (P=0.023) (Table 1 and Figure 5A). The IGHV gene identity cut-offs that best discriminates the patient’s outcome were 96.6% and 97% (Supplementary Figure S3). For simplicity we selected the 97% cut-off and found that M-MCL had a significantly better outcome than U-MCL with 5-year OS rates of 59% and 40%, respectively (P=0.004) (Figure 5B).
Figure 5.
Kaplan-Meier estimates of OS for MCL patients according to IGHV gene mutational status and SOX11 expression. A, OS for the four different subgroups of IGHV identity. HM and SM have better OS (5-year OS 61%, 95% confidence interval [CI], 35–87 and 5-year OS 55%, 95% CI, 29–81, respectively) compared to MBM (5-year OS 14%, 95% CI, 34–62) and TU (5-year OS 18%, 95% CI, 7–43). B, OS subgroups of MCL with <97% (Unmutated U-MCL) and ≥97% (Mutated M-MCL) identity. M-MCL showed better OS (5-year OS 59%, 95% CI, 41–77) than U-MCL (5-year OS 40%, 95% CI, 28–52). C, OS according to IGHV gene mutational status and SOX11 expression. M-MCL SOX11-negative showed a better OS (5-year OS 73%, 95% CI, 52–94) than the other groups (M-MCL SOX11-pos: 5-year OS 42%, 95% CI, 12–72; U-MCL SOX11-neg: 5-year OS 48%, 95% CI, 13–83; U-MCL SOX11-pos: 5-year OS 38%, 95% CI, 26–50). D, OS of the M-MCL SOX11-negative patients with 17p/TP53 alterations showed a worse OS (5-year OS 36%, 95% CI, 0–76) than patients without 17p/TP53 alterations (5-year OS 92%, 95% CI, 77–100).
In addition to SHM, variables predicting for poor OS on univariate analysis were advanced age, high serum LDH, nodal presentation, blastoid morphology, high Ki67 expression, positive SOX11 expression and 17p/TP53 alteration (Table 2). Because both, high IGHV gene mutational load and SOX11-negativity have been associated with a more indolent evolution of MCL, we examined which of the two was the most important variable for OS (Figure 5C). Interestingly, M-MCL patients negative for SOX11 (n=23, 5-year OS, 73%) had a significantly better prognosis than the other subgroups of patients (P=0.005). In addition, 17p/TP53 alterations in this subgroup of M-MCL recognized a subset of patients with a significant worst outcome (Figure 5D).
Table 2.
Analysis for overall survival in MCL patients.
| Variable | N | Events | OS (months) 5-year OS (95% CI) | P | RR | Multivariate analysis 95% CI | P |
|---|---|---|---|---|---|---|---|
| Agea | 0.019 | 1.03 | 1.01–1.06 | 0.001 | |||
| Gender | 0.514 | ||||||
| Female | 25 | 20 | 61 (43–79) | ||||
| Male | 98 | 58 | 56 (41–70) | ||||
| LDH | <0.001 | ||||||
| Low | 45 | 23 | 72 (40–104) | ||||
| High | 29 | 23 | 17 (12–22) | ||||
| Lymphocytes (log10 L/mm3) | 0.22 | ||||||
| ≥ 4 | 42 | 22 | 46 (44–48) | ||||
| < 4 | 40 | 23 | 38 (36–40) | ||||
| Nodal presentation | 0.004 | ||||||
| No | 38 | 17 | 81 (59–103) | ||||
| Yes | 80 | 53 | 47 (34–59) | ||||
| Morphology | 0.001 | ||||||
| Classical | 64 | 42 | 57 (50–64) | ||||
| Blastoid | 22 | 18 | 23 (3–43) | ||||
| Ki67 | <0.001 | ||||||
| High (≥35%) | 45 | 30 | 53 (37–69) | ||||
| Low (<35%) | 22 | 18 | 18 (0–36) | ||||
| IGHV gene | 0.004 | 2.01 | 1.08–3.74 | 0.029 | |||
| Mutated (<97%) | 40 | 18 | 84 (20–148) | ||||
| Unmutated (≥97%) | 80 | 64 | 50 (38–62) | ||||
| SOX11 expression | 0.003 | 2.44 | 1.22–4.9 | 0.012 | |||
| Negative | 34 | 14 | 84 (12–156) | ||||
| Positive | 100 | 66 | 48 (38–58) | ||||
| TP53 statusb | <0.001 | ||||||
| Wild type | 31 | 14 | 59 (38–80) | ||||
| Mutated | 19 | 17 | 19 (0–39) | ||||
| 17p/TP53b | <0.001 | ||||||
| Not altered | 60 | 24 | 53 (17–53) | ||||
| Altered | 28 | 22 | 25 (6–44) |
Age was treated as a continuous variable.
Survival for TP53 and 17p/TP53 alterations was calculated from the time of sample assessment instead of from time of diagnosis.
Finally, we performed a multivariate analysis with: age, nodal presentation, IGHV gene mutational status, and SOX11 expression. In the final model with 112 cases, age (relative risk [RR]: 1.03; 95% confidence interval [CI]: 1.01–1.06; P=0.001), IGHV gene mutational status (RR: 2.01; 95% CI: 1.08–3.74; P=0.029), and SOX11 expression (RR: 2.44; 95% CI: 1.22–4.9; P=0.012) were identified as independent risk factors for OS, whereas the nodal presentation no longer retained any prognostic value.
Discussion
The immunogenetic analysis of the BcR in B-cell neoplasms has helped to identify biological subtypes with clinical implications (9–12,12,39–42). A recent study of the IGHV genes in a large series of MCL provided strong molecular evidence for antigen-driven selection in the pathogenesis of at least a subset of cases and also led to the identification of distinct molecular subsets of MCL defined by the repertoire and mutational status of the clonogenic IGs (17). In the present study, we report that these molecular subsets exhibit distinct genetic, molecular and clinical characteristics suggesting that they may correspond to different subtypes of the disease.
The clinical relevance of the IGHV gene mutational status in MCL has been controversial. Almost all previous studies have discriminated subsets of MCL following the 98% IGHV gene germline identity cut-off established for CLL (10,41). However, as highlighted by Hadzidimitriou et al (17), this approach may have overlooked the particular biological characteristics of MCL. Following the IGHV mutational subset definition of this study, we stratified our MCL series in truly unmutated, minimally/borderline mutated and significantly mutated subsets, an included an additional group of hypermutated MCL (17).
Our series was enriched in tumors with SM and HM IGHV genes, similar to the study of Orchard et al (18) who reported 29% of cases in their cohort exhibiting <97% identity. Interestingly, this immunogenetic bias in these two cohorts likely reflects the higher frequency of the non-nodal clinical presentation (present study: 31%; Orchard et al: 46%) compared to most other MCL studies (17). These differences are probably due to the characteristics of the collaborative centers in our study that include reference laboratories for leukemic patients in addition to surgical pathology groups. In contrast, most previous studies of the BcR in MCL have been performed in patients from clinical trials that required histological confirmation of the tumor (22) or in tumor samples recruited from surgical pathology departments (21,23–25,43) and, therefore, may have underestimated the subset of patients presenting with leukemic non-nodal disease.
The major clinical and biological differences among patients were observed between the HM and TU-MCL, whereas SM and MBM tumors had intermediate features with a tendency to resemble the HM and TU tumors, respectively (Table 1). The most significant differences among the mutational subsets concerned IGHV gene usage, CD5 expression, genomic complexity, gene expression profiles, including SOX11 expression, gender distribution, and nodal presentation All these findings support the concept that the mutational status of the IGHV genes identifies biologically and clinically distinct subsets of MCL.
The relationship between IGHV gene mutational status and the clinical course of MCL has been addressed in different studies but the results have been inconclusive with only some tendencies to favorable outcome in tumors with high mutational load. In our study, we found that 97% identity was the best cut-off for predicting survival. This cut-off enabled the delimitation of HM/SM vs. MBM/TU MCL and defined a significant difference in the overall survival of these two subgroups of patients. On these grounds, it is not unreasonable to claim that the inability of previous studies to detect clinical implications for SHM in MCL may be due, at least in part, to the application of the “CLL-relevant” 2% cut-off and the relatively lower number of cases with high SHM that reduced the statistical power of the analysis.
In keeping with previous studies (18,20,26), we found that nodal presentation and SOX11 expression were also predictors of poor outcome but, interestingly, in the multivariate analysis only the IGHV gene mutational status and SOX11 expression remained as independent variables. A recent study by Nygren et al (43) has shown an apparently conflicting result with SOX11-negative MCL having a worse prognosis than SOX11-positive tumors. However, 9/13 (69%) of the SOX11-negative cases in that study were strongly positive for p53 by immunohistochemistry, suggesting that these tumors carried TP53 gene mutations. These findings are concordant with our recent observation indicating that TP53 mutations and 17p deletions in SOX11-negative MCL are associated with complex genomic karyotypes and a rapid clinical evolution, whereas SOX11-negative MCL with wild type TP53/17p have a very stable disease and a long survival (29). Altogether, these observations support the idea that MCL with mutated IGHV, SOX11-negativity and non-nodal clinical presentation may correspond to a subtype of the disease with more indolent behavior. However, the inactivation of TP53, similar to other small B-cell lymphomas, confers a more aggressive behavior with development of nodal dissemination and more rapid clinical evolution.
The comprehensive analysis of the BcR in MCL is modifying our views about its potential ontogeny and the role of antigen selection in the pathogenesis of the disease (17,44). The definition of a cell of origin for this tumor is facing the same challenges already encountered in CLL (15). The spectrum of SHM, the molecular evidence for antigen selection, and the differences in the GEP of the two major subsets of MCL according to SHM load raise the possibility, as suggested for CLL (15), of a scenario in which the different subsets of MCL may be related to different normal counterparts. The postulated cells may include in addition to naïve B-cells for some TU tumors, intermediate cells between naïve and germinal center cells (45), transitional B-cells (46), or even a post-germinal center memory B-cells for the tumors with high load of SHM. Our GEP analysis of the two major subset of MCL supports this hypothesis with an enrichment of the naïve B-cell signature in MCL with unmutated IGHV and the memory B-cell signature in the mutated IGHV tumors.
In conclusion, our results suggest the idea that the mutational status of the IGHV genes and SOX11 expression recognize two major subsets of MCL with distinct molecular and genetic features and also distinct clinical presentation and evolution. The gene expression profile analysis supports relationship of MCL with unmutated and mutated IGHV with naïve and memory B-cells, respectively, reinforcing the hypothesis that they may correspond to different subtypes of the disease.
Supplementary Material
Acknowledgments
Grant Support: Spanish Ministry of Science SAF 2008-03630 (E.C.), “Instituto de Salud Carlos III, Fondo Investigaciones Sanitarias” (PI08/0077 and PI11/01177) (S.B.), “Red Temática de Investigación Cooperativa de Cáncer” RD06/0020/0039, (RD06/0020/0014, RD06/0020/0051, “Formación de Personal Investigador” (BES-2007-16330) (A.N.), Fondo Europeo de Desarrollo Regional. Unión Europea. “Una manera de hacer Europa”, European Mantle Cell Lymphoma Network, Generalitat de Catalunya (2009-SGR-992; 2009-SGR-967), European Science Foundation, ‘Frontiers of Functional Genomics’ (L.S.), Cariplo Foundation and Associazione Italiana per la Ricerca sul Cancro (Italy), ENosAI project (09SYN-13-880), co-funded by the EU and the Hellenic General Secretariat for Research and Technology.
Footnotes
Conflict of interests: no conflicts of interest to be disclosed.
References
- 1.Swerdlow S, Campo E, Harris N, Jaffe E, Pileri S, Stein H, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues. 4. Lyon: 2008. [Google Scholar]
- 2.Welzel N, Le T, Marculescu R, Mitterbauer G, Chott A, Pott C, et al. Templated nucleotide addition and immunoglobulin JH-gene utilization in t(11;14) junctions: implications for the mechanism of translocation and the origin of mantle cell lymphoma. Cancer Res. 2001;61:1629–36. [PubMed] [Google Scholar]
- 3.Jares P, Colomer D, Campo E. Genetic and molecular pathogenesis of mantle cell lymphoma: perspectives for new targeted therapeutics. Nat Rev Cancer. 2007;7:750–62. doi: 10.1038/nrc2230. [DOI] [PubMed] [Google Scholar]
- 4.Bea S, Campo E. Secondary genomic alterations in non-Hodgkin’s lymphomas: tumor-specific profiles with impact on clinical behavior. Haematologica. 2008;93:641–5. doi: 10.3324/haematol.13030. [DOI] [PubMed] [Google Scholar]
- 5.Royo C, Salaverria I, Hartmann E, Rosenwald A, Campo E, Bea S. The complex landscape of genetic alterations in mantle cell lymphoma. Semin Cancer Biol. 2011;5:322–34. doi: 10.1016/j.semcancer.2011.09.007. [DOI] [PubMed] [Google Scholar]
- 6.Bea S, Ribas M, Hernandez JM, Bosch F, Pinyol M, Hernandez L, et al. Increased number of chromosomal imbalances and high-level DNA amplifications in mantle cell lymphoma are associated with blastoid variants. Blood. 1999;93:4365–74. [PubMed] [Google Scholar]
- 7.Perez-Galan P, Dreyling M, Wiestner A. Mantle cell lymphoma: biology, pathogenesis, and the molecular basis of treatment in the genomic era. Blood. 2011;117:26–38. doi: 10.1182/blood-2010-04-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hartmann EM, Campo E, Wright G, Lenz G, Salaverria I, Jares P, et al. Pathway discovery in mantle cell lymphoma by integrated analysis of high-resolution gene expression and copy number profiling. Blood. 2010;116:953–61. doi: 10.1182/blood-2010-01-263806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murray F, Darzentas N, Hadzidimitriou A, Tobin G, Boudjogra M, Scielzo C, et al. Stereotyped patterns of somatic hypermutation in subsets of patients with chronic lymphocytic leukemia: implications for the role of antigen selection in leukemogenesis. Blood. 2008;111:1524–33. doi: 10.1182/blood-2007-07-099564. [DOI] [PubMed] [Google Scholar]
- 10.Hamblin TJ, Davis Z, Gardiner A, Oscier DG, Stevenson FK. Unmutated Ig V(H) genes are associated with a more aggressive form of chronic lymphocytic leukemia. Blood. 1999;94:1848–54. [PubMed] [Google Scholar]
- 11.Zibellini S, Capello D, Forconi F, Marcatili P, Rossi D, Rattotti S, et al. Stereotyped patterns of B-cell receptor in splenic marginal zone lymphoma. Haematologica. 2010;95:1792–6. doi: 10.3324/haematol.2010.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bikos V, Darzentas N, Hadzidimitriou A, Davis Z, Hockley S, Traverse-Glehen A, et al. Over 30% of patients with splenic marginal zone lymphoma express the same immunoglobulin heavy variable gene: ontogenetic implications. Leukemia. 2012 doi: 10.1038/leu.2012.3. [DOI] [PubMed] [Google Scholar]
- 13.Maura F, Cutrona G, Fabris S, Colombo M, Tuana G, Agnelli L, et al. Relevance of stereotyped B-cell receptors in the context of the molecular, cytogenetic and clinical features of chronic lymphocytic leukemia. PLoS One. 2011;6:e24313. doi: 10.1371/journal.pone.0024313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Szankasi P, Bahler DW. Clinical laboratory analysis of immunoglobulin heavy chain variable region genes for chronic lymphocytic leukemia prognosis. J Mol Diagn. 2010;12:244–9. doi: 10.2353/jmoldx.2010.090091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiorazzi N, Ferrarini M. Cellular origin(s) of chronic lymphocytic leukemia: cautionary notes and additional considerations and possibilities. Blood. 2011;117:1781–91. doi: 10.1182/blood-2010-07-155663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stamatopoulos K. Antigens in CLL: themes and variations. Blood. 2010;115:3855–6. doi: 10.1182/blood-2010-02-270249. [DOI] [PubMed] [Google Scholar]
- 17.Hadzidimitriou A, Agathangelidis A, Darzentas N, Murray F, fau-Larue MH, Bredo PL, et al. Is there a role for antigen selection in mantle cell lymphoma? Immunogenetic support from a series of 807 cases. Blood. 2011;118:3088–95. doi: 10.1182/blood-2011-03-343434. [DOI] [PubMed] [Google Scholar]
- 18.Orchard J, Garand R, Davis Z, Babbage G, Sahota S, Matutes E, et al. A subset of t(11;14) lymphoma with mantle cell features displays mutated IgVH genes and includes patients with good prognosis, nonnodal disease. Blood. 2003;101:4975–81. doi: 10.1182/blood-2002-06-1864. [DOI] [PubMed] [Google Scholar]
- 19.Agathangelidis A, Darzentas N, Hadzidimitriou A, Brochet X, Murray F, Yan XJ, et al. Stereotyped B-cell receptors in one third of chronic lymphocytic leukemia: towards a molecular classification with implications for targeted therapeutic interventions. Blood. 2012;119:4467–75. doi: 10.1182/blood-2011-11-393694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fernandez V, Salamero O, Espinet B, Sole F, Royo C, Navarro A, et al. Genomic and gene expression profiling defines indolent forms of mantle cell lymphoma. Cancer Res. 2010;70:1408–18. doi: 10.1158/0008-5472.CAN-09-3419. [DOI] [PubMed] [Google Scholar]
- 21.Camacho FI, Algara P, Rodriguez A, Ruiz-Ballesteros E, Mollejo M, Martinez N, et al. Molecular heterogeneity in MCL defined by the use of specific VH genes and the frequency of somatic mutations. Blood. 2003;101:4042–6. doi: 10.1182/blood-2002-11-3456. [DOI] [PubMed] [Google Scholar]
- 22.Kienle D, Krober A, Katzenberger T, Ott G, Leupolt E, Barth TF, et al. VH mutation status and VDJ rearrangement structure in mantle cell lymphoma: correlation with genomic aberrations, clinical characteristics, and outcome. Blood. 2003;102:3003–9. doi: 10.1182/blood-2003-05-1383. [DOI] [PubMed] [Google Scholar]
- 23.Schraders M, Oeschger S, Kluin PM, Hebeda K, Schuuring E, Groenen PJ, et al. Hypermutation in mantle cell lymphoma does not indicate a clinical or biological subentity. Mod Pathol. 2009;22:416–25. doi: 10.1038/modpathol.2008.199. [DOI] [PubMed] [Google Scholar]
- 24.Walsh SH, Thorselius M, Johnson A, Soderberg O, Jerkeman M, Bjorck E, et al. Mutated VH genes and preferential VH3-21 use define new subsets of mantle cell lymphoma. Blood. 2003;101:4047–54. doi: 10.1182/blood-2002-11-3479. [DOI] [PubMed] [Google Scholar]
- 25.Thorselius M, Walsh S, Eriksson I, Thunberg U, Johnson A, Backlin C, et al. Somatic hypermutation and V(H) gene usage in mantle cell lymphoma. Eur J Haematol. 2002;68:217–24. doi: 10.1034/j.1600-0609.2002.01662.x. [DOI] [PubMed] [Google Scholar]
- 26.Ondrejka SL, Lai R, Kumar N, Smith SD, Hsi ED. Indolent mantle cell leukemia: clinicopathologic variant characterized by isolated lymphocytosis, interstitial bone marrow involvement, kappa light chain restriction, and good prognosis. Haematologica. 2011;96:1121–7. doi: 10.3324/haematol.2010.036277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cogliatti SB, Bertoni F, Zimmermann DR, Henz S, Diss TC, Ghielmini M, et al. IgV H mutations in blastoid mantle cell lymphoma characterize a subgroup with a tendency to more favourable clinical outcome. J Pathol. 2005;206:320–7. doi: 10.1002/path.1781. [DOI] [PubMed] [Google Scholar]
- 28.Thelander EF, Walsh SH, Thorselius M, Laurell A, Landgren O, Larsson C, et al. Mantle cell lymphomas with clonal immunoglobulin V(H)3–21 gene rearrangements exhibit fewer genomic imbalances than mantle cell lymphomas utilizing other immunoglobulin V(H) genes. Mod Pathol. 2005;18:331–9. doi: 10.1038/modpathol.3800237. [DOI] [PubMed] [Google Scholar]
- 29.Royo C, Navarro A, Clot G, Salaverria I, Giné E, Jares P, et al. Non-nodal type of mantle cell lymphoma is a specific biological and clinical subgroup of the disease. Leukemia. 2012 doi: 10.1038/leu.2012.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Giudicelli V, Duroux P, Ginestoux C, Folch G, Jabado-Michaloud J, Chaume D, et al. IMGT/LIGM-DB, the IMGT comprehensive database of immunoglobulin and T cell receptor nucleotide sequences. Nucleic Acids Res. 2006;34:D781–D784. doi: 10.1093/nar/gkj088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lefranc MP, Giudicelli V, Ginestoux C, Jabado-Michaloud J, Folch G, Bellahcene F, et al. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 2009;37:D1006–D1012. doi: 10.1093/nar/gkn838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brochet X, Lefranc MP, Giudicelli V. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 2008;36:W503–W508. doi: 10.1093/nar/gkn316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. (3) 2004:Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 34.Mozos A, Royo C, Hartmann E, De JD, Baro C, Valera A, et al. SOX11 expression is highly specific for mantle cell lymphoma and identifies the cyclin D1-negative subtype. Haematologica. 2009;94:1555–62. doi: 10.3324/haematol.2009.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Puente XS, Pinyol M, Quesada V, Conde L, Ordonez GR, Villamor N, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475:101–5. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bea S, Salaverria I, Armengol L, Pinyol M, Fernandez V, Hartmann EM, et al. Uniparental disomies, homozygous deletions, amplifications, and target genes in mantle cell lymphoma revealed by integrative high-resolution whole-genome profiling. Blood. 2009;113:3059–69. doi: 10.1182/blood-2008-07-170183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenwald A, Wright G, Wiestner A, Chan WC, Connors JM, Campo E, et al. The proliferation gene expression signature is a quantitative integrator of oncogenic events that predicts survival in mantle cell lymphoma. Cancer Cell. 2003;3:185–97. doi: 10.1016/s1535-6108(03)00028-x. [DOI] [PubMed] [Google Scholar]
- 38.Blenk S, Engelmann JC, Pinkert S, Weniger M, Schultz J, Rosenwald A, et al. Explorative data analysis of MCL reveals gene expression networks implicated in survival and prognosis supported by explorative CGH analysis. BMC Cancer. 2008;8:106. doi: 10.1186/1471-2407-8-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stamatopoulos K, Belessi C, Moreno C, Boudjograh M, Guida G, Smilevska T, et al. Over 20% of patients with chronic lymphocytic leukemia carry stereotyped receptors: Pathogenetic implications and clinical correlations. Blood. 2007;109:259–70. doi: 10.1182/blood-2006-03-012948. [DOI] [PubMed] [Google Scholar]
- 40.Arons E, Suntum T, Stetler-Stevenson M, Kreitman RJ. VH4-34+ hairy cell leukemia, a new variant with poor prognosis despite standard therapy. Blood. 2009;114:4687–95. doi: 10.1182/blood-2009-01-201731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Damle RN, Wasil T, Fais F, Ghiotto F, Valetto A, Allen SL, et al. Ig V gene mutation status and CD38 expression as novel prognostic indicators in chronic lymphocytic leukemia. Blood. 1999;94:1840–7. [PubMed] [Google Scholar]
- 42.Forconi F, Sozzi E, Cencini E, Zaja F, Intermesoli T, Stelitano C, et al. Hairy cell leukemias with unmutated IGHV genes define the minor subset refractory to single-agent cladribine and with more aggressive behavior. Blood. 2009;114:4696–702. doi: 10.1182/blood-2009-03-212449. [DOI] [PubMed] [Google Scholar]
- 43.Nygren L, Baumgartner WS, Klimkowska M, Christensson B, Kimby E, Sander B. Prognostic role of SOX11 in a population-based cohort of mantle cell lymphoma. Blood. 2012;119:4215–23. doi: 10.1182/blood-2011-12-400580. [DOI] [PubMed] [Google Scholar]
- 44.Agathangelidis A, Hadzidimitriou A, Rosenquist R, Stamatopoulos K. Unlocking the secrets of immunoglobulin receptors in mantle cell lymphoma: implications for the origin and selection of the malignant cells. Semin Cancer Biol. 2011;21:299–307. doi: 10.1016/j.semcancer.2011.09.009. [DOI] [PubMed] [Google Scholar]
- 45.Kolar GR, Mehta D, Pelayo R, Capra JD. A novel human B cell subpopulation representing the initial germinal center population to express AID. Blood. 2007;109:2545–52. doi: 10.1182/blood-2006-07-037150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sims GP, Ettinger R, Shirota Y, Yarboro CH, Illei GG, Lipsky PE. Identification and characterization of circulating human transitional B cells. Blood. 2005;105:4390–8. doi: 10.1182/blood-2004-11-4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.