Abstract
Cytoplasmic dynein is responsible for the transport and delivery of cargoes in organisms ranging from humans to fungi. Dysfunction of dynein motor machinery due to mutations in dynein or its activating complex dynactin can result in one of several neurological diseases in mammals. The mouse Legs at odd angles (Loa) mutation in the tail domain of the dynein heavy chain has been shown to lead to progressive neurodegeneration in mice. The mechanism by which the Loa mutation affects dynein function is just beginning to be understood. In this work, we generated the dynein tail mutation observed in Loa mice into the Neurospora crassa genome and utilized cell biological and complementing biochemical approaches to characterize how that tail mutation affected dynein function. We determined that the Loa mutation exhibits several subtle defects upon dynein function in N. crassa that were not seen in mice, including alterations in dynein localization, impaired velocity of vesicle transport, and in the biochemical properties of purified motors. Our work provides new information on the role of the tail domain on dynein function and points out areas of future research that will be of interest to pursue in mammalian systems.
Keywords: cytoplasmic dynein, dynactin, intracellular transport, Legs at odd angles (Loa) mutation, microtubules, Neurospora crassa
Introduction
Dynein is a megadalton-sized, multiprotein motor complex composed of four classes of gene products: the heavy chains (DHCs), intermediate chains (DICs), light intermediate chains, and light chains. The DHCs form the force producing motor entity of the dynein complex and the rest of the components are involved in linking dynein to cargoes [Bielli et al., 2001; Traer et al., 2007; Cai et al., 2010]. The heavy chain is comprised of a ~4300–4600 amino acid long polypeptide chain that can be divided into two regions, a motor head and a tail domain. The motor head of DHC is a member of the hexameric ATPase associated with various cellular activities (AAA+) family of ATPases [Neuwald et al., 1999; Iyer et al., 2004] that encode six AAA+ modules in a single polypeptide chain that folds into a ring-like structure [Samso et al., 1998; Burgess et al., 2003; Carter et al., 2011; Kon et al., 2011]. The motor head is responsible for the ATP hydrolysis as well as the microtubule interaction of the dynein complex [Gibbons et al., 1987; Gee et al., 1997; Koonce, 1997]. The tail domain is comprised of the N-terminal ~1500–1800 amino acids and is involved in the homodimerization of DHC as well as interaction with the other dynein subunits [Habura et al., 1999; King, 2000; Tynan et al., 2000]. Thus far the motor head and a part of the tail domain termed the linker have received much attention due to their involvement in dynein force production [Gee et al., 1997; Burgess et al., 2003; Mallik et al., 2004; Kon et al., 2005; Numata et al., 2008; Roberts et al., 2009]. In fact, several studies have reported recombinant dynein motors lacking the tail domain to be functional [Nishiura et al., 2004; Reck-Peterson et al., 2006; Shima et al., 2006; Mogami et al., 2007; Cho et al., 2008]. However, perturbation of the integrity of the dynein tail domain has been shown to alter motility properties of dynein [Pollock et al., 1998; Ori-McKenney et al., 2010; Markus and Lee, 2011].
Recent studies have reported mutations in the tail domain of DHC in mice [Hafezparast et al., 2003; Chen et al., 2007]. One such mutation is an ethylnitrosourea-induced missense mutation (F580Y) termed as the Legs at odd angles (Loa) mutation. Mice bearing a heterozygous Loa mutation (Loa+/−) exhibit abnormal posture; late-onset motor neuron degeneration as well as early-onset proprioceptive sensory neuropathy, and defects in muscle force, motor function, and neuronal migration [Hafezparast et al., 2003; Ilieva et al., 2008; Ori-McKenney and Vallee, 2011]. Mice homozygous for the Loa mutation (Loa−/−) exhibit perinatal mortality and neurons cultured from the homozygotes display retrograde axonal transport defects as well as Lewy body-like cytoplasmic aggregates [Hafezparast et al., 2003]. Collectively, these studies suggest a possible regulatory role of the tail domain on dynein function.
We decided to examine the role of the tail domain of dynein in Neurospora crassa, which has several significant advantages as a system to study dynein. DHCs show high structural conservation between N. crassa and higher eukaryotes (Figs. 1A and S1, Supporting Information). Furthermore, unlike other eukaryotes where dynein is essential for the survival of cells, N. crassa does not depend on dynein function for cellular viability. In addition, N. crassa has a haploid genome, which ensures that any mutation introduced in the single DHC locus will result in a homogenous pool of dynein complexes. In this work, we generated the dynein tail mutation observed in Loa mice into the N. crassa genome and characterized how that tail mutation affected dynein function. We determined that the Loa mutation exhibits several subtle defects upon dynein function in N. crassa including alterations in dynein localization and in the biochemical properties of purified motors.
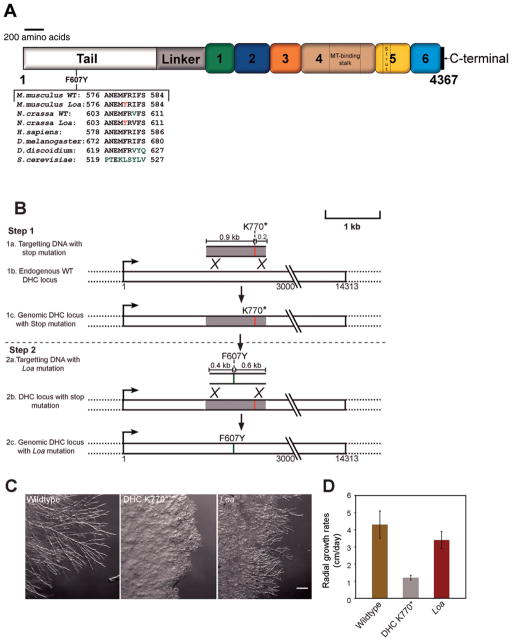
Fig. 1. Generation of an N. crassa Loa mutant strain.
A: Schematic representation of the cytoplasmic dynein heavy chain from N. crassa showing the position of the Loa mutation. Numbers 1–6 indicate appropriate AAA+ domains. MT and C represent the microtubule binding domain and C-terminal region respectively. The alignment of a subset of sequences shows the conservation of the DHC Loa mutation (F607Y) between N. crassa and other species. Black letters indicate conservation, green letters indicate altered amino acids, and red letters indicate the Loa mutation. B: A schematic representation of the two step strategy used to generate the Loa mutant strain. Step 1, introduction of a stop mutation to generate the ropy strain DHCK770*. Step 2, introduction of the Loa mutation and the rescue of the stop mutation. C: Comparison of colony morphologies between wildtype, mutant DHCK770* and Loa mutant strains. Note the Loa mutant strain exhibits mild ropy growth morphology in comparison to the wildtype strain. D: Bar graph showing radial growth rates of wildtype, mutant DHCK770* and Loa mutant strains. Data are shown as mean ± SD.
Results
Generation of a Fungal Strain Carrying a Mammalian Disease Mutation
Complete loss of function of the DHC gene in N. crassa leads to an altered hyphal growth phenotype termed as ropy [Plamann et al., 1994; Bruno et al., 1996]. Utilizing this phenomenon, we initially attempted to introduce the Loa mutation in the native DHC locus using a one-step homologous recombination based strategy. We reasoned that if the Loa mutation causes a complete loss of dynein function, transformed strains carrying the Loa mutation would display a ropy growth phenotype and therefore could be easily identified in a screen for strains exhibiting abnormal growth. However, after a series of transformation experiments, none of the resultant transformation progeny exhibited the ropy growth phenotype. Therefore, we considered the possibility that the Loa mutation may not lead to a complete loss of dynein function. We then devised a two step strategy to generate the Loa mutant strain (Fig. 1B). First, we introduced a stop codon into a wildtype DHC gene sequence at amino acid position 770 to generate a DHC mutant strain (DHCK770*) that encoded a truncated form of the DHC polypeptide. This mutant strain exhibited altered hyphal morphology resulting in a severe ropy growth phenotype and slow colony growth rates (Figs. 1C and 1D). The mutant growth morphology exhibited by DHCK770* strain was equivalent to DHC null N. crassa strains [Seiler et al., 1999; Riquelme et al., 2000] suggesting that there was a complete loss of dynein function in the DHCK770* strain. In the second step, the DHCK770* mutant strain was transformed with a DNA fragment that encoded the Loa mutation (F607Y) along with flanking native DHC DNA sequences that would rescue the K770* nonsense codon. Therefore, we screened this second round of transformants for improved growth phenotypes, which could indicate that homologous recombination occurred between the transforming DNA and the DHCK770* locus. We isolated 24 improved growth transformants and sequenced the ~15 kb DHC gene from six of those transformants. We determined that all six transformants carried the Loa mutation (Fig. 1A) and there were no other mutations within the DHC gene. The mutant Loa strains exhibited hyphal growth morphology and colony growth rates (Figs. 1C and 1D) that were intermediate between wildtype and DHCK770* strains. The intermediate effect of the Loa mutation on N. crassa growth phenotype is consistent with our hypothesis that the Loa mutation may not cause a complete loss of dynein function.
The Loa Mutation Causes Subtle Phenotypic Changes in Dynein Function
To examine the effect of the Loa mutation upon dynein localization in living cells, we crossed the Loa strain with a strain that expresses a dynein intermediate chain-mCherry chimeric fusion (DIC-mCherry). The DICmCherry chimera was the sole DIC in the haploid genome and was produced by homologous replacement of the endogenous DIC gene with a fully functional DIC-mCherry construct [Sivagurunathan et al., 2012]. As shown previously, expression of this chimeric DICmCherry had no effect on any dynein functions in N. crassa [Sivagurunathan et al., 2012]. The incorporation of the DIC-mCherry into Loa dynein allowed us to localize dynein in live cells.
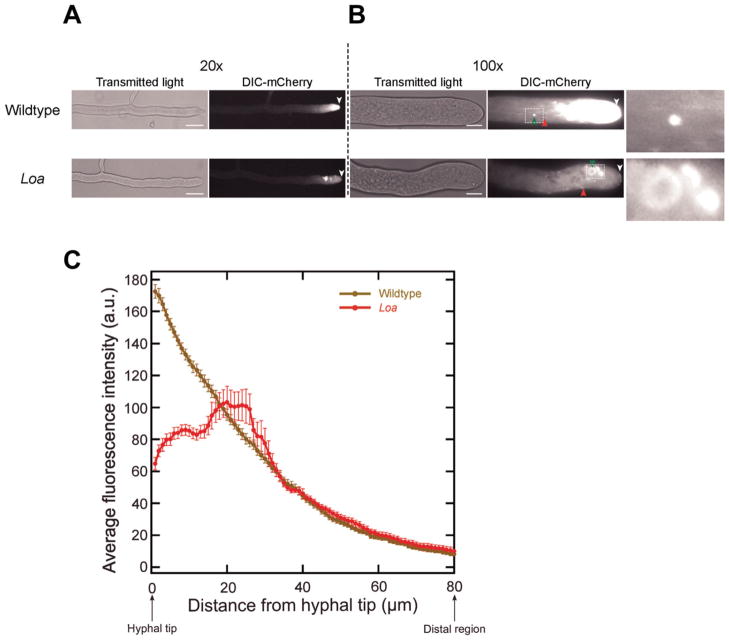
In wildtype strains, dynein accumulates as a bright cloud at the hyphal tips (Fig. 2A) [Sivagurunathan et al., 2012]. This fluorescent signal at the tip gradually tapered off as a function of distance from the hyphal tips in wildtype cells (Fig. 2C). Further analyses showed that wildtype dynein is localized to additional locations including microtubule end-binding comet tails and what appear to be spherical organelles [Fig. 2B; see also Sivagurunathan et al., 2012]. When we characterized the localization of mutant Loa dynein molecules in actively growing hyphae, we observed an approximately twofold reduction in the accumulation of dynein fluorescence to the apical 20 μm of hyphal tips (Figs. 2A and 2C). The Loa tip-based dynein fluorescence mirrored that of wildtype dynein in more distal regions; there was no detectable increase in the overall fluorescence of Loa dynein in the syncytial hyphal cytosol compared to wildtype dynein. Further examination revealed that the Loa dynein localized to comet tails and associated with spherical organelles similar to wildtype dynein behavior (Fig. 2B). However, we also saw that there were three subtle but significant variations in how mutant Loa dynein localized to spherical organelles in comparison to wildtype strains. First, fluorescence intensity measurements of the mutant Loa spherical structures indicated a greater than twofold increase in mCherry fluorescence in comparison to wildtype spherical structure fluorescence ((Fig. S2B, Supporting Information); wildtype =16 ± 6, Loa = 40 ± 8; n = 25; P < 0.005). Second, the Loa spherical structures were located much closer to the hyphal tips than their wildtype counterparts ((Fig. S2A, Supporting Information); wildtype = 47 ± 10 μm, Loa = 23 ± 5 μm; n = 20; P < 0.005). In addition to these differences, Loa spherical structures more often appeared to be clustered than the wildtype spherical structures. Previously, we examined 34 DHC mutations that resulted in the production of full-length DHC proteins with altered function, and determined that the defective dynein motors exhibited five distinct dynein localization phenotypes [Sivagurunathan et al., 2012]. The mild alterations observed with Loa dynein localization (reduced hyphal tip accumulation and altered vesicle organelle localization) are distinct from the more pronounced mutant dynein mislocalization phenotypes reported in those other DHC mutations [Sivagurunathan et al., 2012] and indicate that the Loa mutation alters dynein function by a distinct mechanism compared to those other mutations.
Fig. 2. Localization of dynein in mutant strains of N. crassa.
A: Hyphae from the edges of colonies of wildtype and Loa mutant strains were visualized at intermediate magnification to show the distance between dynein signal and the hyphal tip (arrowheads). Bars: 50 μm. B: Hyphae visualized using higher magnification show details of dynein localization. In wildtype strains dynein is accumulated at hyphal tips and localizes to comet tails (red arrows) and to spherical structures (green arrows). In the Loa mutant strains, dynein showed reduced accumulation to hyphal tips as well as enhanced localization to spherical structures. Left panels = transmitted light; right panels = mCherry fluorescence. Arrowheads indicate position of the hyphal tip. Bars: 10 μm. Enlarged views of the boxed regions are shown on the right. C: Fluorescence intensity profiles as a function of distance from the hyphal tips for wildtype (olive) and Loa strains (maroon). Data are shown as mean ± S.E. (n = 15 hyphal tips for each strain).
We examined the effect of the Loa mutation on vesicle transport in growing hyphae of N. crassa by utilizing the lipophilic styryl dye FM 4-64. Colonies of wildtype and Loa mutant strains were treated with FM 4-64 and hyphae at the colony edges were imaged with time-lapse microscopy to generate movies of endomembrane vesicle transport that could be analyzed in detail for motility characteristics (see videos and Fig. S3, Supporting Information). Our previously characterized DHC mutations exhibited drastic defects in endomembrane transport, including decreased numbers of motility events, decreased vesicle velocity and distance traveled, and an overall decrease in the motility index of the mutant strains [Sivagurunathan et al., 2012]. In contrast, we determined that the overall vesicle motility in Loa mutant strains was only slightly different from that of wildtype strains (Table I). Surprisingly, we observed that there was actually an increase in the number of motility events per micron of hyphal length in Loa hyphae compared to wildtype hyphae (Table I). This increase corresponds to a shift to more inward movements relative to outward movements. The additional motility events caused an increase in the motility index of Loa mutant hyphae relative to wildtype hyphae, opposite to the decrease observed in motility index in stronger DHC mutant alleles [Sivagurunathan et al., 2012]. The most statistically significant defect we found in Loa hyphae was a decrease in the velocity of vesicles that moved inward from the hyphal tip toward distal regions in the mutant colonies (Table I). At the extreme tip of growing hyphae, microtubules are arranged with their plus ends outward [Riquelme et al., 2002; Freitag et al., 2004; Sampson and Heath, 2005; Uchida et al., 2008]. Therefore, the in vivo defect of reduced velocity of inward directed motility is consistent with a modest defect in dynein function. Overall, the subtle differences in vesicle transport in Loa mutant strains and the intermediate colony morphology phenotypes these strains exhibit are consistent with a model in which the Loa mutation has slightly reduced activity compared to more severe DHC alleles.
Table I.
Summary of Vesicle Transport Dynamics
| Wildtype | Loa | |
|---|---|---|
| Motility index | 24.6 | 26.9 |
| Overall movements | ||
| Mean movements/μm hyphal length | 3.13 | 3.81 |
| N | 727 | 746 |
| Inward movements | ||
| Percent of total movements (%) | 50.8 | 54.7 |
| Mean velocity (μm/s ± SD) | 1.66 ± 0.62 | 1.52 ± 0.66** |
| Mean distance (μm ± SD) | 4.13 ± 2.24 | 3.88 ± 1.78* |
| Outward movements | ||
| Percent of total movements (%) | 49.2 | 45.3 |
| Mean velocity (μm/s ± SD) | 1.78 ± 0.68 | 1.70 ± 0.60* |
| Mean distance (μm ± SD) | 4.47 ± 2.50 | 4.54 ± 2.08 |
Statistically significant difference (P < 0.05) from wildtype value.
Statistically significant difference (P < 0.001) from wildtype value.
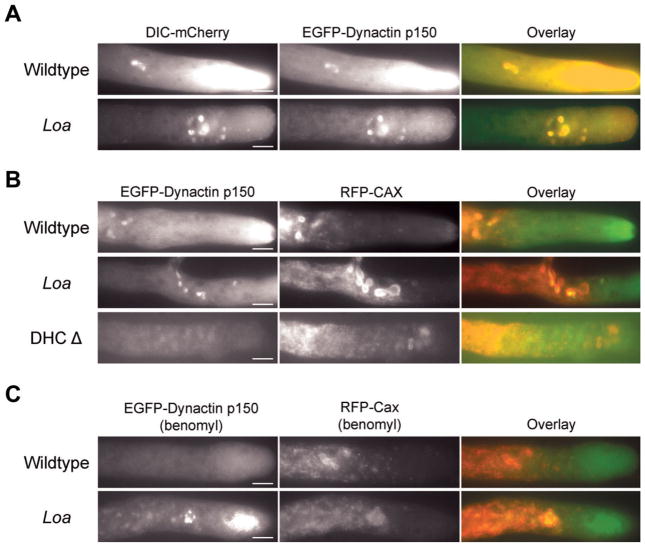
A previous report suggested that the Loa mutation may reduce the interaction between dynein and dynactin [Deng et al., 2010]. We examined the localization of dynactin in Loa mutant strains by replacing the endogenous dynactin p150 gene with an EGFP-p150 recombinant gene that had no effect on dynactin function or localization [Sivagurunathan et al., 2012]. We found that dynactin (as seen by EGFP-p150 signals) was colocalized with Loa dynein to hyphal tips, comet tails and the clustered spherical structures (Fig. 3A), as is expected in wild-type strains. This indicates that the Loa mutation does not significantly affect the interaction of Loa dynein with dynactin in cells.
Fig. 3. Colocalization of dynein, dynactin, and CAX in wildtype and Loa mutant strains.
A: Epifluorescence images of growing hyphae in strains expressing DIC-mCherry and EGFP- dynactin p150. Left panels: dynein; middle panels: dynactin; right panels: overlay. Dynein mCherry fluorescence colocalizes with dynactin GFP fluorescence. Bars: 10 μm. B: Colocalization of dynactin with CAX organelles. Left panels: EGFP-dynactin p150; middle panels: RFP-CAX; right panels: overlay. The majority of CAX positive organelles have dynactin p150 on their surface in both wildtype and Loa mutant heterokaryons. In DHC deletion strains, the CAX positive organelles are still present near the hyphal tip but do not have dynactin present on their surface. Bars: 10 μm. C: The Loa mutation induced colocalization of dynactin with CAX organelles does not require microtubules. Left panels: EGFP-dynactin p150; middle panels: RFP-CAX, right panels: overlay. The loss of microtubules does not alter the association of dynactin with CAX positive organelles in Loa strains but wildtype strains lose any CAX association. Bars: 10 μm.
Because there was an interesting difference in how Loa dynein interacted with spherical organelles, we further explored the identity of these spherical structures. To this end we utilized strains expressing RFP (tDimer2 (12)) labeled N. crassa endomembrane markers such as calcium/proton exchange protein (CAX), NCA-2, NCA-3, a subunit of the vacuolar ATPase (VMA-1), and VAM-3 [Bowman et al., 2009]. Since the membrane markers were labeled with RFP, we used EGFP-p150 dynactin as a marker of dynein. Due to the detrimental action of the N. crassa genetic defense mechanism with the repeated RFP tag, we could not generate a double mutant strain and instead utilized heterokaryon strains expressing GFP labeled dynactin and RFP labeled endomembrane markers (see “Materials and Methods” section). In both the wildtype DHC and Loa DHC heterokaryons, we saw no colocalization between dynactin and NCA-2, NCA-3, or VAM-3. However, the spherical structures visualized by dynactin localization did exhibit colocalization with either VMA-1 or CAX spherical structures in both wildtype and Loa strains (Fig. 3B). We also examined the localization of EGFP-p150 labeled dynactin in combination with RFP-CAX in a DHC deletion background (Fig. 3B). We found that the CAX-positive organelles were still present near the hyphal tip but that there was no colocalization of EGFP-p150 signal to the CAX organelle surface. According to recent reports, VMA-1 and CAX, a Ca2+/H+ exchanger, were both localized to a novel organelle implicated in calcium sequestration [Bowman et al., 2009, 2011].
In a previous study, we determined that DHC mutations may alter microtubule organization patterns and that the acute loss of microtubules can affect dynein localization phenotypes [Sivagurunathan et al., 2012]. To determine if the Loa mutation alters microtubule organization, we crossed the DIC-mCherry Loa mutant strain to a strain expressing β-tubulin GFP. In these strains, we observed that there were fewer microtubules closer to the tips than in control strains and these Loa microtubules were predominantly arranged parallel to the longitudinal axis of the hyphae (Fig. 4A). As expected, the comet tails in wildtype and Loa mutant strains showed colinearity with microtubules, suggesting that the comet tails and the hyphal tip accumulation of dynein in the wildtype and Loa mutant strains are dependent on microtubules.
Fig. 4. Localization of dynein and microtubules in wildtype and Loa mutant strains.
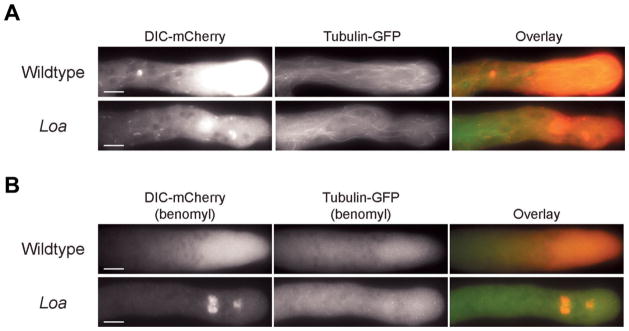
A: Epifluorescence images displaying hyphal localization of DIC-mCherry (left panels) and β-tubulin-GFP (middle panels) in wildtype and Loa mutant strains. Overlay images are shown in the right panels. Comet tail dynein fluorescence in wildtype and Loa mutant strains localize with microtubules. Bars: 10 μm. B: Dependence of dynein localization patterns on intact microtubule network. Benomyl treatment depolymerizes microtubules and alters comet tail localization and tip accumulation of dynein in wildtype and Loa mutant strains expressing DIC-mCherry and β-tubulin-GFP. Dynein is shown in left panels; microtubules are shown in the middle panels; overlay images are shown in the right panels. In Loa mutant strains, dynein localization to spherical structures was persistent even after benomyl treatment. Bars: 10 μm.
We performed two additional experiments to further examine the link between microtubules and dynein accumulation to vesicular organelles in Loa strains. First, we examined the effects of the microtubule depolymerizing drug benomyl upon Loa and wildtype strains carrying the β-tubulin-GFP and DIC-mCherry genes as markers for tubulin and dynein. In wildtype strains, benomyl-mediated microtubule disruption lowered dynein accumulation to hyphal tips and completely abolished dynein localization to comet tails and spherical structures (Fig. 4B). Benomyl treatment of Loa mutant strains led to the disruption of dynein comet tail and the hyphal tip fluorescence, similar to wildtype. However, dynein remained colocalized with large spherical structures even after the complete disruption of microtubules in the Loa mutant strains (Fig. 4B). Second, to definitively show that the dynein mislocalization in the previous experiment was to CAX/VMA-1 organelles, we utilized Loa and wildtype heterokaryon strains (described above) expressing GFP-labeled dynactin and RFP-CAX labeled endomembrane markers. When we treated those heterokaryon strains with benomyl, we saw extensive colocalization between the dynactin and CAX signals in the Loa heterokaryons but not in the wildtype heterokaryons (Fig. 3C). These data indicate that the accumulation of Loa dynein to CAX/VMA-1 vesicles occurs even in the absence of microtubules. One interpretation of these data is that wildtype dynein and dynactin complexes are transiently associated with the CAX/VMA-1 organelles and when benomyl is added, the dynein and dynactin complexes that leave the CAX/VMA-1 organelles are not replaced with new motor complexes. It is likely that the Loa mutation results in dynein and dynactin complexes that for some unknown reason associate more strongly with the CAX/VMA-1 organelles resulting in a strong signal in the presence or absence of benomyl.
Biochemical Analyses of Loa Dynein
We purified dynein motors from Loa mutant strains using an affinity based method [Sivagurunathan et al., 2012] to determine if the Loa mutation altered the biochemical properties of dynein. The purification profile of the Loa dynein was indistinguishable from that of the wildtype dynein, indicating that the mutation did not result in gross alterations to dynein composition (Fig. S4, Supporting Information). Next, we examined the effect of the Loa mutation on the nucleotide-sensitive microtubule binding behavior of dynein. Several studies have shown that the dynein–microtubule interaction can be modulated by different ATP and ATP analog nucleotide conditions [Imamula et al., 2007; Mizuno et al., 2007; Ori-McKenney et al., 2010]. We tested the microtubule binding properties of Loa dynein under varying nucleotide conditions using a microtubule cosedimentation assay. In the presence of 1 mM ATP, wildtype dynein showed reduced cosedimentation with microtubules, in contrast to the robust cosedimentation observed without nucleotides or in the presence of 1 mM ADP or AMPPNP (Figs. 5 and S5, Supporting Information). Loa dynein exhibited nucleotide- sensitive microtubule binding behavior similar to that of wildtype dynein (Fig. 5), which suggests that the Loa mutation does not significantly alter the microtubule interaction behavior of dynein in N. crassa.
Fig. 5. Quantification of the percentage of dynein in pellet fractions in comparison to total dynein.
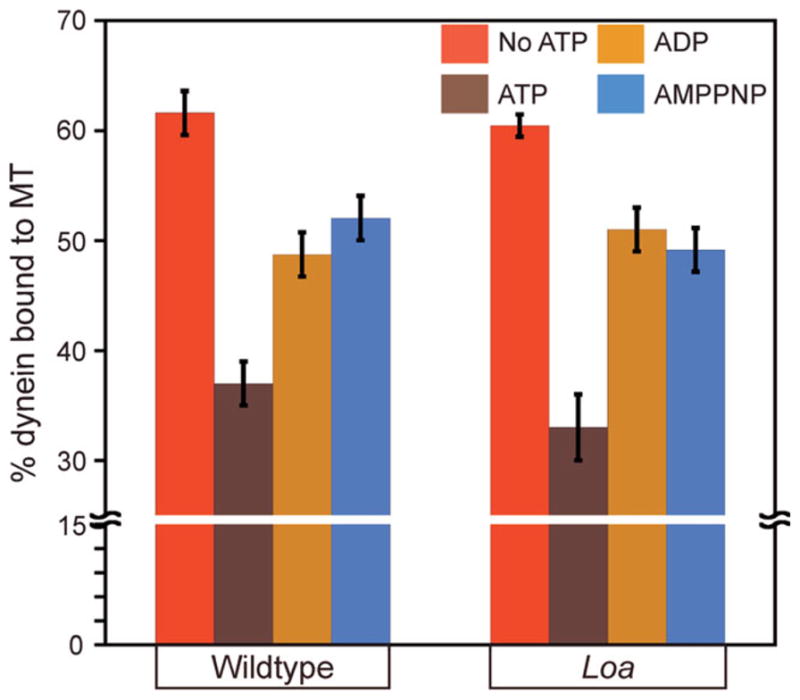
Cosedimentation was quantified from three independent experiments. Data shown as mean ± SEM. Wildtype and Loa dynein cosedimentation in the presence of 1 mM ATP was significantly less than dynein cosedimentation in the other conditions examined (t-test, P < 0.05). There was no significant difference between wildtype and Loa dynein at any single nucleotide condition (P > 0.05).
We examined the effect of the Loa mutation on the ATPase activity of dynein by measuring the basal and microtubule-stimulated ATPase rates of the mutant dynein molecules. The basal ATPase rate of Loa dynein was approximately 50% higher than the wildtype basal ATPase rate (Table II). In the presence of microtubules, the ATPase rate of both wildtype and Loa dynein doubled above their basal ATPase rate. These results indicate that the Loa mutation significantly alters the basal enzymatic activity of dynein relative to wildtype with no apparent effect on the mechanisms employed for sensing and responding to microtubules.
Table II.
ATPase Activities of Dynein Isolated from Wildtype and Loa Strains
| Dynein | Basal ATPase (nmol/min/ mg dynein) | MT-stimulated ATPase (nmol/min/ mg dynein) | Fold change |
|---|---|---|---|
| Wildtype | 115 ± 7 | 201 ± 3 | ~ 1.7 |
| Loa | 166 ± 20 | 350 ± 28 | ~ 2.1 |
Basal and microtubule (5 μM) stimulated ATPase activities are expressed as mean ± SD derived from three independent experiments. The Loa mutation significantly altered both the basal and microtubule-stimulated ATPase rates (P < 0.005).
Because the Loa dynein showed a microtubule binding behavior similar to wildtype dynein but displayed altered enzymatic properties, we examined Loa dynein in an in vitro microtubule-based bead motility assay to more precisely analyze its function. In this assay Loa or wildtype dynein molecules were coated onto polystyrene beads and these beads were added to a chamber with microtubules adhered to coverslip surface. The properties of N. crassa wildtype dynein are quite similar to mammalian dynein in terms of velocity and processivity Table III; Fig. 6 [Sivagurunathan et al., in press]. When we examined Loa dynein coated beads, we found that the beads bound to microtubules and moved along those microtubules as far as wildtype dynein (Table III). However, the average velocity of Loa dynein was reduced almost twofold in comparison to wildtype dynein (Table III; Fig. 6).
Table III.
In Vitro Motility Characteristics of Beads Coated with Dynein Isolated from Wildtype and Loa Strains
| Dynein | Velocity (μm/s) | P | Distance (μm) | n |
|---|---|---|---|---|
| Wildtype | 0.7 ± 0.3 | – | 2.6 ± 1.7 | 104 |
| Loa | 0.4 ± 0.3 | <0.001 | 2.3 ± 1.3 | 98 |
Velocities and distances are expressed as mean ± SD; P = probability from a Student’s t test in comparison to wildtype dynein velocities. n = number of motility events tracked.
Fig. 6. Histogram of velocity distributions of wildtype and Loa dynein in in vitro motility assay.
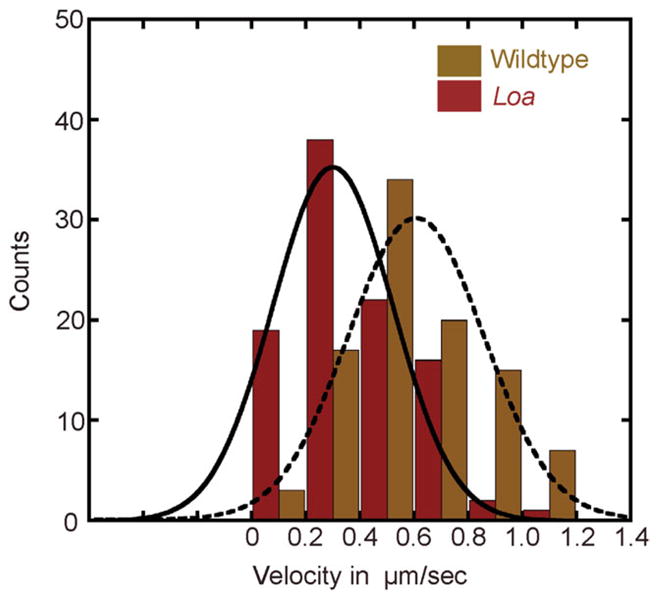
The dashed and solid curved lines correspond to a Gaussian fit of the wildtype and Loa dynein velocity distributions respectively.
Discussion
In our construction and initial characterizations of a N. crassa dynein strain with a tail mutation, it was clear that the Loa mutation caused a ropy colony morphology phenotype that was intermediate between that seen by wild-type and dynein null strains (Figs. 1C and 1D). When we explored the effect of the Loa mutation at the growing tips of N. crassa hyphae, we found that the dynein localization patterns in both wildtype and Loa strains initially appeared to be similar (dynein was found as a hyphal tip accumulation, in comet tails, and on spherical structures). However, there were subtle but reproducible differences in the overall pattern of Loa dynein distribution at the hyphal tip: the overall amount of Loa dynein at the extreme apex of the hyphae was reduced about twofold and the binding of Loa dynein to a specific subset of vesicle structures was altered (Figs. 2 and 3). In addition we observed that the Loa microtubule network was slightly disorganized compared to wildtype (Fig. 4). The studies performed to date on Loa+/− and Loa−/− mice have not examined the intracellular localization of dynein, or the potential interplay between Loa mutations and the cellular microtubule network. Based on our studies in N. crassa it will be very interesting for future work to determine if similar dynein mislocalization and organelle distribution phenotypes are present in mammalian systems.
When we explored the biochemical properties of Loa dynein motors from N. crassa, we observed both an increase in ATPase activity and a reduction in the velocity of the Loa dynein motors both in vivo and in vitro. The enzymatic and motility behaviors of Loa dynein from N. crassa are different from a previous report that saw no alteration in the ATPase rates from heterozygous Loa+/− mice and a reduction in the processivity of dynein isolated from either homozygous Loa−/− mice or heterozygous Loa+/− mice [Ori-McKenney et al., 2010]. We were surprised by the difference in the motility properties from processivity to velocity in the dynein preparations from the two organisms. However, it should be noted that the velocity reduction we observed with N. crassa Loa dynein is consistent with the observed reduction in the velocity of dynein driven intracellular retrograde movements in mouse neurons carrying homozygous Loa−/− mutations [Hafezparast et al., 2003; Ori-McKenney et al., 2010] and is further supported by reports that observed a similar effect on motor velocity upon perturbation of dynein tail domain integrity [Pollock et al., 1998; Markus and Lee, 2011]. The observed reduction in motor velocity and enhanced ATPase activity could be a consequence of the loss of dynein head–head coordination as demonstrated by a previous report [Ori-McKenney et al., 2010] that in turn may uncouple communication between ATP consumption and motility. Our data in conjunction with previous studies demonstrate the important role of an intact tail domain for proper dynein function in systems ranging from mammals to fungi.
In a recent study, we characterized the effects of 34 independent DHC mutations and found that these mutations entrapped dynein in one of five positions in an intracellular dynein cycle [Sivagurunathan et al., 2012]. Furthermore, we determined that all of these strains exhibited four common phenotypes: severe ropy colony growth morphology, complete loss of the hyphal tip accumulation of dynein, significant defects in the transport of FM 4-64 vesicles, and severe disorganization of the hyphal microtubule network [Sivagurunathan et al., 2012]. In comparison to these severe phenotypes, it is interesting that the N. crassa Loa mutation causes an intermediate ropy phenotype, a modest microtubule organization defect, a relatively subtle velocity defect in vesicle transport, and the Loa mutant strain actually exhibits hyphal tip accumulation of dynein, albeit reduced relative to wildtype strains. In short, the N. crassa Loa mutation does not appear to entrap dynein in a particular stage of the dynein cycle [Sivagurunathan et al., 2012] and instead the N. crassa Loa mutation appears to have partial function. These results are consistent with comparisons of homozygous dynein null and homozygous dynein Loa−/− mutations in mice. The mouse dynein null mutations only develop to late blastula stage [Harada et al., 1998] whereas mouse Loa−/− embryos survive until birth [Hafezparast et al., 2003], showing that some partial dynein activity is present.
By using N. crassa as our model system to study the effect of mutations on the function of dynein in cells, we have the advantages of (1) high sequence conservation with mammalian dynein genes, (2) dynein function being nonessential for viability of the organism, (3) a haploid genome so all mutations are homozygous, (4) an organism with well-defined genetics, and (5) the ability to purify enough motor for biochemical and motility assays. Our work in this model system provides new information on the role of the tail domain on dynein function and points out areas of future research that will be of interest to pursue in mammalian systems.
Materials and Methods
Strains, Media and Growth Conditions
Table SI, Supporting Information, lists the primary N. crassa strains used in this study. N. crassa strains were grown on Vogel’s minimal medium, supplemented with 2% sucrose (VSM) at 28°C. All reagents for media, supplements, and buffers used were purchased from Thermo Fisher Scientific (Hampton, NH) unless otherwise indicated. Radial growth rates were determined as described in Sivagurunathan et al. [2012].
Generation of the DHC Loa Mutant Strain
A DNA fragment encoding the N-terminal 840 amino acids of the N. crassa DHC (ro-1) was cloned into pBlue-script using standard PCR and cloning techniques to generate pDHC. This plasmid was used as a template for the introduction of the stop mutation and Loa mutation to generate the plasmids pDHCK770* and pDHCF607Y, respectively. The Loa mutant strain was generated in two steps. We showed previously that loss of dynein function leads to partial suppression of a cot-1ts mutation [Plamann et al., 1994]. Using the plasmid pDHCK770* as a template, we PCR amplified a 1.1 kb linear transformable DNA fragment encoding the stop mutation and flanking DNA sequences for integration into the native DHC locus. The DNA was transformed into the cot-1ts strain modified for efficient homologous recombination [Ninomiya et al., 2004; Kuck and Hoff, 2010]. Transformants carrying the DHC stop codon mutation were identified as partial revertants of cot-1ts and displayed the characteristic ropy growth phenotype. The presence of the stop codon at position 770 was confirmed by DNA sequencing. The DHCK770* strain was then transformed with a DNA fragment encoding the Loa mutation. Integration of the transformed DNA rescued the ropy growth phenotype but did not suppress cot-1ts indicating that the Loa mutation does not impart a complete loss of dynein function. Strains exhibiting rescued growth were subject to DNA sequencing, which confirmed the presence of the Loa mutation and the replacement of the K770 stop codon.
Generation of Heterokaryons
To study colocalization of EGFP-dynactin p150 with VMA-1/CAX-RFP fusions we generated heterokaryon strains. We used heterokaryons instead of generating strains that were doubly labeled since N. crassa employs several mechanisms to silence repetitive DNA sequences during specific stages of its sexual phase of life cycle [Selker, 1990; Cogoni and Macino, 1999; Shiu et al., 2001]. Because the endomembrane markers were labeled with tDimer2 (12), which is a direct tandem repeat of the dsRed fluorescent tag, the gene would have been silenced by the genomic-defense mechanisms of N. crassa during mating [Freitag and Selker, 2005]. To generate heterokaryons, conidia from the corresponding strains were collected and suspended in VSM media using standard techniques.
Imaging
Live-cell imaging was performed as described [Sivagurunathan et al., 2012]. The images were processed in Adobe Photoshop. Distance measurements were made using SPOT software (Sterling Heights, MI). Data are expressed as the mean ± SD. Fluorescence intensity profiles (expressed as mean gray values) were generated as described [Sivagurunathan et al., 2012] by using the line-scan tool in Metamorph software (Molecular Devices, Sunnyvale, CA). The mean and the standard error from analyzing 15 individual hyphae from each strain were plotted using Kaleidagraph (Synergy Software, Reading, PA). Fluorescence intensities of the spherical structures in wildtype and Loa mutant strains were determined using NIH ImageJ software. The fluorescence intensity (expressed as mean gray values) of individual spheres were determined by manually selecting spheres and measuring the mean gray value within the selection and normalizing the data with the gray value measurements of regions close to corresponding spheres. The final fluorescence intensity measurement from each strain was determined from the average of the gray values from measuring 25 individual spheres from each strain.
Vesicle transport imaging experiments utilized N-(3-Triethylammoniumpropyl)-4-(6-(4-(Diethylamino) Phenyl) Hexatrienyl) Pyridinium Dibromide (trade-name “FM 4-64,” Life Technologies Corporation, Carlsbad, CA), which has no appreciable fluorescence unless it is in a lipid membrane. Slide cultures of fungal strains were prepared the same as described earlier. VSM liquid media containing FM 4-64 was added to a final concentration of 5 μM onto a growing colony and a coverslip was placed on top. The hyphae were imaged in a time window between 5 and 15 min following dye treatment. Time-lapse image stacks (30 s duration) were generated with 40 ms exposures captured every 200 ms. The individual image stacks were analyzed using Metamorph software to generate movement kymographs. The velocities, distances traveled, and the total number of the movements were determined from those kymographs using custom designed software. Cytoplasmic streaming based transport was identified by the slower movement of vesicles (<0.5 μm/s) in unison. In this study, we did not further examine the movement cytoplasmic streaming based movement of vesicles. Individual vesicles were considered to have undergone a motility event if they exhibited a directed displacement at a velocity of at least 0.5 μm/s. Student t-tests were used to examine significance between various experimental data sets. The motility index was calculated by taking the sum of the velocity multiplied by the distance of all motility events for a strain divided by the total length of the hyphae examined. Representative live cell movies of FM 4-64 labeled vesicle trafficking in hyphal tips from Loa and wildtype strains are included in the online supplemental information. Each supplemental movie is 86 × 65 μm2 in size and is sped up sixfold for playback.
Benomyl Treatment
VSM liquid media containing 10 μM benomyl (DuPont, Wilmington, DE) was added to a colony on the slide cultures at room temperature. Benomyl treatment led to the disruption of microtubules in approximately 5 min after which the hyphae were imaged. As a control, media containing DMSO was added to the colony.
N. crassa Dynein Purification and Characterization
N. crassa dynein was purified using affinity chromatographic methods as described previously [Sivagurunathan et al., 2012]. A detailed description of microtubule cosedimentation assays, ATPase and motility experiments is described in our previous study [Sivagurunathan et al., 2012]. Briefly, dynein purified from wildtype and Loa strains was incubated at 32°C for 20 min’s with 5 μM paclitaxel stabilized microtubules under nucleotide-free or in the presence of 1 mM ATP, ADP, AMPPNP in PM buffer (100 mM PIPES–NaOH, 2 mMEGTA, 1 mMMgSO4, 5 mM GTP, and 100 μM paclitaxel). Samples were subject to high-speed spins (45,000 rpm) and the resultant supernatants were transferred to fresh tubes and the pellets were resuspended in PM buffer. Equal volumes of both fractions were resolved by SDS-PAGE, stained with Coomassie Brilliant Blue and digitized using a Hewlett Packard (Palo Alto, CA) Scanjet 7400c scanner. Digital images were analyzed using NIH ImageJ software and dynein heavy chain bands in the supernatant and pellet fractions were quantified densitometrically.
The ATPase activity of dynein was measured by a radio thin-layer chromatography (TLC) assay as described previously [Gilbert and Mackey, 2000; Mesngon et al., 2006; Sivagurunathan et al., 2012]. Briefly, dynein purified from wildtype and Loa strains were incubated with 1 mM ATP, and 10 μCi α-32P-ATP (Perkin Elmer, Boston, MA) in the absence or presence of 5 μM paclitaxel stabilized microtubules in BRB80 buffer (80 mM PIPES, 1 mM EGTA, 1 mM MgCl2, pH 6.9). Radiolabeled ADP generated after 60 min was resolved from ATP using PEI-Cellulose F TLC plates (EMD Chemicals, Gibbstown, NJ). The TLC plates were subject to phosphor imaging (Molecular Dynamics, Sunnyvale, CA). Densitometric quantification of ATP and ADP spots was performed using ImageQuant software (Molecular Dynamics, Sunnyvale, CA). The concentration of dynein isolated from wildtype and Loa strains relative to purified bovine dynein was measured by densitometry and was used to determine the specific activity.
In vitro bead based motility assays were performed as reported earlier [King and Schroer, 2000; Mallik et al., 2004; Culver-Hanlon et al., 2006; Sivagurunathan et al., 2012]. A flow-chamber coated with polymerized microtubules was generated. Purified dynein was bound to poly-bead carboxylate microspheres (0.21 μm diameter; Polysciences, Warrington, PA) by nonspecific adsorption at dynein: bead molar ratios between 20:1 and 40:1 in the presence of 10 μm ATP. The dynein bead mixture was added to the flow chamber and visualized by video-enhanced differential interference contrast microscopy. Bead motility characteristics were determined by tracking bead movement using custom-built image-processing software (Labview 6.1, National Instruments). The wildtype and Loa velocity histograms were generated using Kaleidagraph software and were fitted with a single Gaussian function.
Supplementary Material
Acknowledgments
Strains and plasmids were obtained from the Fungal Genetics Stock Center (Kansas City, Missouri). The authors thank Barry Bowman for the strains labeled with endomembrane markers. They thank Linda King for critical reading of the manuscript. This work was supported by grants to S.J.K. (NIH NS048501) and to M.P. (NSF MCB0235871) and (NIH PO1 GM069087).
Abbreviations used
- AAA+
ATPase associated with various cellular activities
- CAX
calcium/proton exchange protein
- DHC
dynein heavy chain
- DIC
dynein intermediate chain
- Loa
legs at odd angles
- VMA-1
a subunit of the vacuolar ATPase
Footnotes
Additional Supporting Information may be found in the online version of this article.
References
- Bielli A, Thornqvist PO, Hendrick AG, Finn R, Fitzgerald K, McCaffrey MW. The small GTPase Rab4A interacts with the central region of cytoplasmic dynein light intermediate chain-1. Biochem Biophys Res Commun. 2001;281(5):1141–1153. doi: 10.1006/bbrc.2001.4468. [DOI] [PubMed] [Google Scholar]
- Bowman BJ, Draskovic M, Freitag M, Bowman EJ. Structure and distribution of organelles and cellular location of calcium transporters in Neurospora crassa. Eukaryot Cell. 2009;8(12):1845–1855. doi: 10.1128/EC.00174-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman BJ, Abreu S, Margolles-Clark E, Draskovic M, Bowman EJ. Role of four calcium transport proteins, encoded by nca-1, nca-2, nca-3, and cax, in maintaining intracellular calcium levels in Neurospora crassa. Eukaryot Cell. 2011;10(5):654–661. doi: 10.1128/EC.00239-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno KS, Tinsley JH, Minke PF, Plamann M. Genetic interactions among cytoplasmic dynein, dynactin, and nuclear distribution mutants of Neurospora crassa. Proc Natl Acad Sci USA. 1996;93(10):4775–4780. doi: 10.1073/pnas.93.10.4775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgess SA, Walker ML, Sakakibara H, Knight PJ, Oiwa K. Dynein structure and power stroke. Nature. 2003;421(6924):715–718. doi: 10.1038/nature01377. [DOI] [PubMed] [Google Scholar]
- Cai Q, Lu L, Tian JH, Zhu YB, Qiao H, Sheng ZH. Snapin-regulated late endosomal transport is critical for efficient autophagy-lysosomal function in neurons. Neuron. 2010;68(1):73–86. doi: 10.1016/j.neuron.2010.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter AP, Cho C, Jin L, Vale RD. Crystal structure of the dynein motor domain. Science. 2011;331(6021):1159–1165. doi: 10.1126/science.1202393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen XJ, Levedakou EN, Millen KJ, Wollmann RL, Soliven B, Popko B. Proprioceptive sensory neuropathy in mice with a mutation in the cytoplasmic Dynein heavy chain 1 gene. J Neurosci. 2007;27(52):14515–14524. doi: 10.1523/JNEUROSCI.4338-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho C, Reck-Peterson SL, Vale RD. Regulatory ATPase sites of cytoplasmic dynein affect processivity and force generation. J Biol Chem. 2008;283(38):25839–25845. doi: 10.1074/jbc.M802951200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cogoni C, Macino G. Homology-dependent gene silencing in plants and fungi: a number of variations on the same theme. Curr Opin Microbiol. 1999;2(6):657–662. doi: 10.1016/s1369-5274(99)00041-7. [DOI] [PubMed] [Google Scholar]
- Culver-Hanlon TL, Lex SA, Stephens AD, Quintyne NJ, King SJ. A microtubule-binding domain in dynactin increases dynein processivity by skating along microtubules. Nat Cell Biol. 2006;8(3):264–270. doi: 10.1038/ncb1370. [DOI] [PubMed] [Google Scholar]
- Deng W, Garrett C, Dombert B, Soura V, Banks G, Fisher EM, van der Brug MP, Hafezparast M. Neurodegenerative mutation in cytoplasmic dynein alters its organization and dynein–dynactin and dynein–kinesin interactions. J Biol Chem. 2010;285(51):39922–39934. doi: 10.1074/jbc.M110.178087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freitag M, Selker EU. Expression and visualization of red fluorescent protein (RFP) in Neurospora. Fungal Genet Newsl. 2005;52:14–17. [Google Scholar]
- Freitag M, Hickey PC, Raju NB, Selker EU, Read ND. GFP as a tool to analyze the organization, dynamics and function of nuclei and microtubules in Neurospora crassa. Fungal Genet Biol. 2004;41(10):897–910. doi: 10.1016/j.fgb.2004.06.008. [DOI] [PubMed] [Google Scholar]
- Gee MA, Heuser JE, Vallee RB. An extended microtubule-binding structure within the dynein motor domain. Nature. 1997;390(6660):636–639. doi: 10.1038/37663. [DOI] [PubMed] [Google Scholar]
- Gibbons IR, Lee-Eiford A, Mocz G, Phillipson CA, Tang WJ, Gibbons BH. Photosensitized cleavage of dynein heavy chains. Cleavage at the “V1 site” by irradiation at 365 nm in the presence of ATP and vanadate. J Biol Chem. 1987;262(6):2780–2786. [PubMed] [Google Scholar]
- Gilbert SP, Mackey AT. Kinetics: a tool to study molecular motors. Methods. 2000;22(4):337–354. doi: 10.1006/meth.2000.1086. [DOI] [PubMed] [Google Scholar]
- Habura A, Tikhonenko I, Chisholm RL, Koonce MP. Interaction mapping of a dynein heavy chain. Identification of dimerization and intermediate-chain binding domains. J Biol Chem. 1999;274(22):15447–15453. doi: 10.1074/jbc.274.22.15447. [DOI] [PubMed] [Google Scholar]
- Hafezparast M, Klocke R, Ruhrberg C, Marquardt A, Ahmad-Annuar A, Bowen S, Lalli G, Witherden AS, Hummerich H, Nicholson S, et al. Mutations in dynein link motor neuron degeneration to defects in retrograde transport. Science. 2003;300(5620):808–812. doi: 10.1126/science.1083129. [DOI] [PubMed] [Google Scholar]
- Harada A, Takei Y, Kanai Y, Tanaka Y, Nonaka S, Hirokawa N. Golgi vesiculation and lysosome dispersion in cells lacking cytoplasmic dynein. J Cell Biol. 1998;141(1):51–59. doi: 10.1083/jcb.141.1.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilieva HS, Yamanaka K, Malkmus S, Kakinohana O, Yaksh T, Marsala M, Cleveland DW. Mutant dynein (Loa) triggers pro-prioceptive axon loss that extends survival only in the SOD1 ALS model with highest motor neuron death. Proc Natl Acad Sci USA. 2008;105(34):12599–12604. doi: 10.1073/pnas.0805422105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamula K, Kon T, Ohkura R, Sutoh K. The coordination of cyclic microtubule association/dissociation and tail swing of cytoplasmic dynein. Proc Natl Acad Sci USA. 2007;104(41):16134–16139. doi: 10.1073/pnas.0702370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Leipe DD, Koonin EV, Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol. 2004;146(1–2):11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- King SM. AAA domains and organization of the dynein motor unit. J Cell Sci. 2000;113(Pt 14):2521–2526. doi: 10.1242/jcs.113.14.2521. [DOI] [PubMed] [Google Scholar]
- King SJ, Schroer TA. Dynactin increases the processivity of the cytoplasmic dynein motor. Nat Cell Biol. 2000;2(1):20–24. doi: 10.1038/71338. [DOI] [PubMed] [Google Scholar]
- Kon T, Mogami T, Ohkura R, Nishiura M, Sutoh K. ATP hydrolysis cycle-dependent tail motions in cytoplasmic dynein. Nat Struct Mol Biol. 2005;12(6):513–519. doi: 10.1038/nsmb930. [DOI] [PubMed] [Google Scholar]
- Kon T, Sutoh K, Kurisu G. X-ray structure of a functional full-length dynein motor domain. Nat Struct Mol Biol. 2011;18(6):638–642. doi: 10.1038/nsmb.2074. [DOI] [PubMed] [Google Scholar]
- Koonce MP. Identification of a microtubule-binding domain in a cytoplasmic dynein heavy chain. J Biol Chem. 1997;272(32):19714–19718. doi: 10.1074/jbc.272.32.19714. [DOI] [PubMed] [Google Scholar]
- Kuck U, Hoff B. New tools for the genetic manipulation of filamentous fungi. Appl Microbiol Biotechnol. 2010;86(1):51–62. doi: 10.1007/s00253-009-2416-7. [DOI] [PubMed] [Google Scholar]
- Mallik R, Carter BC, Lex SA, King SJ, Gross SP. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427(6975):649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- Markus SM, Lee WL. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell. 2011;20(5):639–651. doi: 10.1016/j.devcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesngon MT, Tarricone C, Hebbar S, Guillotte AM, Schmitt EW, Lanier L, Musacchio A, King SJ, Smith DS. Regulation of cytoplasmic dynein ATPase by Lis1. J Neurosci. 2006;26(7):2132–2139. doi: 10.1523/JNEUROSCI.5095-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno N, Narita A, Kon T, Sutoh K, Kikkawa M. Three-dimensional structure of cytoplasmic dynein bound to microtubules. Proc Natl Acad Sci USA. 2007;104(52):20832–20837. doi: 10.1073/pnas.0710406105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogami T, Kon T, Ito K, Sutoh K. Kinetic characterization of tail swing steps in the ATPase cycle of Dictyostelium cytoplasmic dynein. J Biol Chem. 2007;282(30):21639–21644. doi: 10.1074/jbc.M701914200. [DOI] [PubMed] [Google Scholar]
- Neuwald AF, Aravind L, Spouge JL, Koonin EV. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9(1):27–43. [PubMed] [Google Scholar]
- Ninomiya Y, Suzuki K, Ishii C, Inoue H. Highly efficient gene replacements in Neurospora strains deficient for nonhomologous end-joining. Proc Natl Acad Sci USA. 2004;101(33):12248–12253. doi: 10.1073/pnas.0402780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiura M, Kon T, Shiroguchi K, Ohkura R, Shima T, Toyoshima YY, Sutoh K. A single-headed recombinant fragment of Dictyostelium cytoplasmic dynein can drive the robust sliding of microtubules. J Biol Chem. 2004;279(22):22799–22802. doi: 10.1074/jbc.M313362200. [DOI] [PubMed] [Google Scholar]
- Numata N, Kon T, Shima T, Imamula K, Mogami T, Ohkura R, Sutoh K. Molecular mechanism of force generation by dynein, a molecular motor belonging to the AAA+ family. Biochem Soc Trans. 2008;36(Pt 1):131–135. doi: 10.1042/BST0360131. [DOI] [PubMed] [Google Scholar]
- Ori-McKenney KM, Vallee RB. Neuronal migration defects in the Loa dynein mutant mouse. Neural Dev. 2011;6(1):26. doi: 10.1186/1749-8104-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori-McKenney KM, Xu J, Gross SP, Vallee RB. A cytoplasmic dynein tail mutation impairs motor processivity. Nat Cell Biol. 2010;12(12):1228–1234. doi: 10.1038/ncb2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plamann M, Minke PF, Tinsley JH, Bruno KS. Cytoplasmic dynein and actin-related protein Arp1 are required for normal nuclear distribution in filamentous fungi. J Cell Biol. 1994;127(1):139–149. doi: 10.1083/jcb.127.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollock N, Koonce MP, de Hostos EL, Vale RD. In vitro microtubule-based organelle transport in wild-type Dictyostelium and cells overexpressing a truncated dynein heavy chain. Cell Motil Cytoskeleton. 1998;40(3):304–314. doi: 10.1002/(SICI)1097-0169(1998)40:3<304::AID-CM8>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126(2):335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riquelme M, Gierz G, Bartnicki-Garcia S. Dynein and dynactin deficiencies affect the formation and function of the Spitzenkorper and distort hyphal morphogenesis of Neurospora crassa. Microbiology. 2000;146(Pt 7):1743–1752. doi: 10.1099/00221287-146-7-1743. [DOI] [PubMed] [Google Scholar]
- Riquelme M, Roberson RW, McDaniel DP, Bartnicki-Garcia S. The effects of ropy-1 mutation on cytoplasmic organization and intracellular motility in mature hyphae of Neurospora crassa. Fungal Genet Biol. 2002;37(2):171–179. doi: 10.1016/s1087-1845(02)00506-6. [DOI] [PubMed] [Google Scholar]
- Roberts AJ, Numata N, Walker ML, Kato YS, Malkova B, Kon T, Ohkura R, Arisaka F, Knight PJ, Sutoh K, et al. AAA+ ring and linker swing mechanism in the dynein motor. Cell. 2009;136(3):485–495. doi: 10.1016/j.cell.2008.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampson K, Heath IB. The dynamic behaviour of microtubules and their contributions to hyphal tip growth in Aspergillus nidulans. Microbiology. 2005;151(Pt 5):1543–1555. doi: 10.1099/mic.0.27750-0. [DOI] [PubMed] [Google Scholar]
- Samso M, Radermacher M, Frank J, Koonce MP. Structural characterization of a dynein motor domain. J Mol Biol. 1998;276(5):927–937. doi: 10.1006/jmbi.1997.1584. [DOI] [PubMed] [Google Scholar]
- Seiler S, Plamann M, Schliwa M. Kinesin and dynein mutants provide novel insights into the roles of vesicle traffic during cell morphogenesis in Neurospora. Curr Biol. 1999;9(15):779–785. doi: 10.1016/s0960-9822(99)80360-1. [DOI] [PubMed] [Google Scholar]
- Selker EU. Premeiotic instability of repeated sequences in Neurospora crassa. Annu Rev Genet. 1990;24:579–613. doi: 10.1146/annurev.ge.24.120190.003051. [DOI] [PubMed] [Google Scholar]
- Shima T, Imamula K, Kon T, Ohkura R, Sutoh K. Head-head coordination is required for the processive motion of cytoplasmic dynein, an AAA+ molecular motor. J Struct Biol. 2006;156(1):182–189. doi: 10.1016/j.jsb.2006.03.014. [DOI] [PubMed] [Google Scholar]
- Shiu PK, Raju NB, Zickler D, Metzenberg RL. Meiotic silencing by unpaired DNA. Cell. 2001;107(7):905–916. doi: 10.1016/s0092-8674(01)00609-2. [DOI] [PubMed] [Google Scholar]
- Sivagurunathan S, Schnittker RR, Razafsky DS, Nandini S, Plamann MD, King SJ. Analyses of dynein heavy chain mutations reveal complex interactions between dynein motor domains and cellular dynein functions. Genetics. 2012 doi: 10.1534/genetics.112.141580. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traer CJ, Rutherford AC, Palmer KJ, Wassmer T, Oakley J, Attar N, Carlton JG, Kremerskothen J, Stephens DJ, Cullen PJ. SNX4 coordinates endosomal sorting of TfnR with dynein-mediated transport into the endocytic recycling compartment. Nat Cell Biol. 2007;9(12):1370–1380. doi: 10.1038/ncb1656. [DOI] [PubMed] [Google Scholar]
- Tynan SH, Gee MA, Vallee RB. Distinct but overlapping sites within the cytoplasmic dynein heavy chain for dimerization and for intermediate chain and light intermediate chain binding. J Biol Chem. 2000;275(42):32769–32774. doi: 10.1074/jbc.M001537200. [DOI] [PubMed] [Google Scholar]
- Uchida M, Mourino-Perez RR, Freitag M, Bartnicki-Garcia S, Roberson RW. Microtubule dynamics and the role of molecular motors in Neurospora crassa. Fungal Genet Biol. 2008;45(5):683–692. doi: 10.1016/j.fgb.2007.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.