Abstract
Glioblastomas of children and young adults have a median survival of only 12-15months and are clinically and biologically distinct from histologically similar cancers in older adults1. They are defined by highly specific mutations in the gene encoding the histone H3.3 variant H3F3A2, occurring either at or close to key residues marked by methylation for regulation of transcription – K27 and G34. Here we show that the cerebral hemispheric-specific G34 mutation drives a distinct expression signature through differential genomic binding of the K36 trimethylation mark (H3K36me3). The transcriptional program induced recapitulates that of the developing forebrain, and involves numerous markers of stem cell maintenance, cell fate decisions and self-renewal. Critically, H3F3A G34 mutations cause profound upregulation of MYCN, a potent oncogene which is causative of glioblastomas when expressed in the correct developmental context. This driving aberration is selectively targetable in this patient population by inhibiting kinases responsible for stabilisation of the protein.
Keywords: glioma, ChIP-Seq, forebrain, H3F3A, G34R/V
INTRODUCTION
The clinical and molecular differences observed in glioblastoma of children and young adults compared with the more common, histologically similar lesions in older adults is strongly suggestive of a distinct underlying biology1. The identification of unique and highly specific mutations in the gene encoding the histone H3.3 variant H3F3A in glioblastoma of children and young adults has recently provided definitive proof of this hypothesis2. However, a mechanism for how mutations at or close to key residues associated with post-translational modification of the histone tail led to tumorigenesis was lacking.
We have sought to address this by examining how the differences in clinical presentation, anatomical location and gene expression associated with the different H3F3A mutations are manifested. By exploiting the only know G34 mutant model system we show that differential binding of the H3K36 trimethyl mark to underpin these processes and identify MYCN as the oncogenic driver during forebrain development, providing a novel avenue for targeted therapy in children with these tumours.
RESULTS
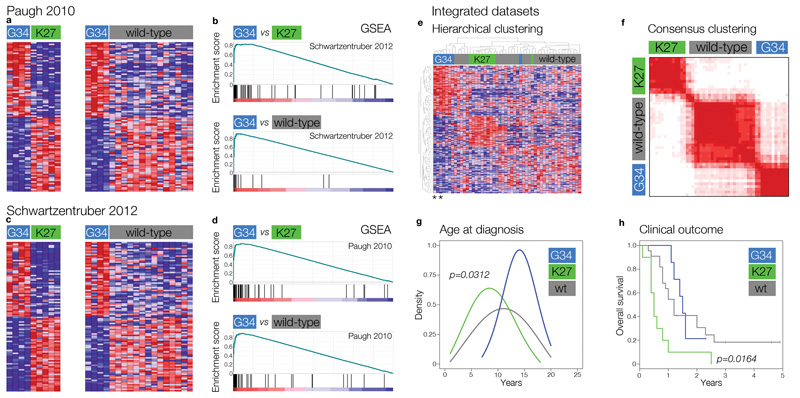
Initial evidence suggested a distinct gene expression signature associated with mutations at the K27 (lysine to methionine, K27M) versus G34 (glycine to either arginine, G34R, or valine, G34V) residues2. We validated these data by identifying differential expression patterns for mutations with G34 versus K27 mutations in two independent datasets for which mutation data were either publicly available or were ascertained in our laboratory2,3 (Figure 1). In both instances, highly significant differential gene expression was noted between G34 mutant tumours and K27 or wild-type cases (Figure 1A,C), which was consistent across the datasets as assessed by gene set enrichment analysis (GSEA) (Figures 1B,D) with enrichment scores (ES) of 0.833 – 0.943 and p (family-wise error rate (FWER)) and q (false discovery rate (FDR)) values of 0.0 – 0.04. Given the considerable overlap in gene expression signatures between studies, we subsequently utilised an integrated dataset (Supplementary Table S1), where hierarchical clustering resolved G34 and K27 mutant tumours from a more heterogeneous wild-type subgroup (Figure 1E), confirmed by k-means consensus clustering (Figure 1F). These subgroups also showed important clinical differences, as previously described2, with K27 mutant tumours arising in younger children (peak age 7 years, p=0.0312, t-test) (Figure 1G) and having a worse clinical outcome (p=0.0164, log-rank test) (Figure 1H) compared with G34 tumours (peak age 14 years) and H3F3A wild-type. There were no significant transcriptional or clinicopathological differences between G34R and G34V tumours, although a lack of the latter samples (n=2) precludes robust analyses.
Figure 1. Distinct molecular and clinical correlates of H3F3A mutation subgroups.
(a) Heatmap representing differential gene expression signatures between G34 versus K27, and G34 versus wild-type, paediatric GBM specimens from Paugh 2010. Top 100 differentially expressed genes are shown for each comparison. (b) Gene set enrichment analysis (GSEA) for differential gene expression signatures identified in Schwartzentruber 2012 versus those in Paugh 2010. Top – G34 versus K27: enrichment score (ES)=0.833, p (family-wise error rate (FWER))=0.0, q (false discovery rate (FDR))=0.0; Bottom – G34 versus wild-type: ES=0.94, FWER p=0.0, FDR q=0.0. (c) Heatmap representing differential gene expression signatures between G34 versus K27, and G34 versus wild-type, paediatric GBM specimens from Schwartzentruber 2012. Top 100 differentially expressed genes are shown for each comparison. (d) GSEA for differential gene expression signatures identified in Paugh 2010 versus those in Schwartzentruber 2012. Top – G34 versus K27: ES=0.88, FWER p=0.03, FDR q=0.04.; Bottom – G34 versus wild-type: ES=0.90, FWER p=0.0, FDR q=0.0. (e) Hierarchical clustering of the integrated gene expression datasets, highlighting specific clusters of G34 and K27 mutant tumours, distinct from a more heterogeneous group of wild-type cases. G34V tumours are represented by asterisks. (f) K-means consensus clustering finds the most stable number of subgroups to be three, marked by H3F3A mutation status. (g) K27 and G34 mutant paediatric GBM in our integrated dataset have distinct age incidence profiles, with K27 tumours peaking at 7 years in contrast to G34 at age 14. The two G34V tumours were diagnosed at age 14 and 20. (h) Kaplan-Meier plot for overall survival of paediatric GBM patients stratified by H3F3A status. K27 mutant tumours have significantly shorter survival than G34 (p=0.0164, log-rank test). A single G34V case for which data was available had an overall survival of 1.4 years.
To understand the functional significance of H3F3A mutations in cerebral hemispheric tumours, we turned to a well-characterised4 model of paediatric GBM, the KNS42 cell line, which was derived from a 16 year-old patient and harbours the G34V mutation (Figure 2A). In contrast to the reported data in a single paediatric GBM sample with G34R2, KNS42 cells did not show increased levels of total histone H3K36 trimethylation compared with a panel of H3F3A wild-type paediatric glioma cells (Figure 2B, Supplementary Figure S1). KNS42 cells harbour a non-synonymous coding change of ATRX (Q891E) that appears in the SNP databases (rs3088074), and Western blot analysis shows no diminution of protein levels. As ATRX is a known chaperone of histone H3.3 to the telomeres, a wild-type protein would not be expected to convey the alternative lengthening of telomeres (ALT) phenotype, as observed (Supplementary Figure S2), however ought not to play a significant role in gene transcription as deposition of H3.3 in euchromatin is carried out by alternative chaperones such as HIRA.
Figure 2. Differential binding of H3K36me3 in G34 mutant KNS42 cells drives paediatric GBM expression signatures.
(a) Sanger sequencing trace for KNS42 paediatric GBM cells reveals a heterozygous c.104G>T p.(Gly34Val) H3F3A mutation. (b) Western blot for mono-(me1), di-(me2) and tri-(me3) methylated histone H3 in G34 mutant KNS42 and wild-type paediatric glioma cell lines. Total H3 is used as an extracted histone loading control. (c) Circos plot representing the KNS42 genome, aligned with chromosomes 1 to X running clockwise from 12 o’clock. Outer ring - H3K36me3 ChIP-Seq binding. Grey: all binding; blue: differential binding in KNS42 versus SF188. Selected differentially bound developmental transcription factors and pluripotency genes are labelled. Inner ring – DNA copy number. Green points: copy number gain; black points: normal copy number; red points: copy number loss. Single base mutations in selected genes (H3F3A:G34V and TP53:R342*) are labelled inside the circle. (d) Correlation plot of RNA polymerase II versus H3K36me3 for 65 differentially trimethyl-bound regions by ChIP-Seq in KNS42 cells. R2=0.66, p<0.0001. (e) Heatmap representing a ranked list of differentially bound H3K36me3 and RNA polymerase II in G34V KNS42 versus wild-type SF188 cells, with top 20 genes listed. (f) GSEA for pre-ranked differentially bound genes identified in ChIP-Seq versus those in the integrated gene expression datasets. Top – G34 versus K27: ES=0.86, FWER p=0.03, FDR q=0.03.; Bottom – G34 versus wild-type: ES=0.84, FWER p=0.02, FDR q=0.04. (g) DAVID gene ontology analysis for pre-ranked list of differentially bound genes identified in ChIP-Seq. Fold enrichment of processes are plotted, and coloured by FDR q value. (h) Top: Mean expression of the G34 core enrichment signature in a temporal gene expression dataset of human brain development. Period 1, embryonal; periods 2-7, foetal; periods 8-12, post-natal; periods 13-15, adulthood. Bottom: Heatmap representing spatial differences in G34 core enrichment signature expression in structures within embryonic and early foetal development, with highest levels mapping to the ganglionic eminences and amygdala.
We performed chromatin immunoprecipitation linked to next-generation whole genome sequencing (ChIP-Seq) for H3K36me3 in order to test the hypothesis that, rather than total H3K36me3, the G34V mutation may instead result in differential binding of the trimethyl mark throughout the genome. Compared with H3F3A wild-type SF188 paediatric GBM cells, H3K36me3 was found to be significantly differentially bound in KNS42 cells at 5130 distinct regions of the genome corresponding to 156 genes (DESeq p<0.05, overall fold change >2, contiguous median coverage >2) (Supplementary Table S2). These observations were not due to differential gene amplification, as concurrent whole genome DNA sequencing demonstrated that these bound genes were not found in regions on cell line-specific copy number alterations (Figure 2C, Supplementary Figure S3, Supplementary Table S2). As the H3K36 residue is regarded as an activating mark for gene expression5, we concurrently performed ChIP-Seq for RNA polymerase II in order to produce a read-out of transcriptional activity, and observed a significant correlation between H3K36me3 and RNA polymerase II binding for the 156 differentially bound genes (R2=0.923, p<0.0001) (Figure 2D). By integrating the H3K36me3 and RNA polymerase II data, a ranked list of differentially trimethyl-bound and expressed genes was derived (Figure 2E). Interrogating this ranked list using our integrated paediatric GBM expression dataset showed highly significant enrichment for G34-associated gene signatures in the differentially bound and expressed genes in G34 mutant KNS42 cells (ES= 0.84-0.86, FWER p = 0.02-0.03; FDR q = 0.03-0.04) (Figure 2F).
To investigate the functions of the transcriptional programs targeted by this novel mechanism, we performed gene ontology analysis of the differentially bound and expressed genes. These data revealed highly significant enrichment of processes involved in forebrain and cortex development, as well as differentiation of neurons and regulation of cell proliferation (Figure 2G). We identified a subset of 16 genes to be part of the core enrichment group showing significant overlap between G34 mutant paediatric GBM specimens and transcription driven by differential binding of H3K36me3 in KNS42 cells (Supplementary Table S3). By mapping the expression of these genes to published signatures of restricted spatio-temporal areas of brain development6, we noted highly elevated levels at embryonic and early foetal time-points, which rapidly tailed off through mid-late foetal development and postnatal and adult periods (Figure 2H). Expression of the G34 core enrichment genes was particularly pronounced in the early foetal amygdala, inferior temporal cortex, and the caudal, medial and lateral ganglionic eminences. (Figure 2H). Developmental expression patterns of G34 mutation associated genes were in contrast to those observed with K27 mutation signatures derived from paediatric GBM specimens, which correlated with those of the embryonic upper rhombic lip, early-mid foetal thalamic and cerebellar structures, and peaked during the mid-late foetal period (Supplementary Figure S4).
Specifically, the G34 mutation drives expression of numerous highly developmentally regulated transcription factors, including as an exemplar DLX6 (distal-less homeobox 6), a homeobox transcription factor which plays a role in neuronal differentiation in the developing forebrain7. The highly significant differential H3K36me3 and RNA polymerase II binding observed by ChIP-Seq (Figure 3A) was validated by ChIP-qPCR (Figure 3B), and expression of DLX6 was noted to be significantly higher in G34 paediatric GBM samples than K27 mutant or wild-type tumours in the integrated gene expression datasets at the mRNA level (Figure 3C), and at the protein level in a tissue microarray comprising 46 paediatric and young adult GBM cases (Figure 3D and Supplementary Table S4). Other similarly validated forebrain development-associated transcription factors included ARX8, DLX57, FOXA19, NR2E110, POU3F211, and SP812 (Supplementary Figures S5-S10). Moreover, a number of key determinants of cell fate were also found to be differentially bound by H3K36me3 and expressed in G34 mutant cells. These included MSI1 (Musashi-1)13 (Supplementary Figure S11); EYA4 (eyes absent homolog 4)14 (Supplementary Figure S12); and SOX2, which is required for stem cell maintenance (Figure 3E-H).
Figure 3. G34 induces a transcriptional program linked to forebrain development and self-renewal.
DLX6: (a) ChIP-Seq of H3K36me3 and RNA polymerase II binding for G34 mutant KNS42 (blue) and wild-type SF188 cells (grey) for the DLX6 locus, which also encompasses the transcripts DLX5, DLX6-AS1 and DLX6-AS2. (b) Validation of ChIP-Seq data by ChIP-qPCR using specific primers targeting DLX6. Blue bars: KNS42, grey: SF188. *** p<0.0001, t-test. (c) Boxplot of DLX6 expression in the integrated paediatric GBM samples stratified by H3F3A status. Blue box: G34, green: K27, grey: wild-type. *** p<0.001, ANOVA. (d) Top – immunohistochemistry for DLX6 protein in a G34 mutant paediatric GBM sample RMH2465. Bottom – Barplot of DLX6 expression in a paediatric GBM tissue microarray stratified by H3F3A status. Blue bars: G34, green: K27, grey: wild-type. ++ strong expression, + moderate expression, - negative. SOX2: (e) ChIP-Seq of H3K36me3 and RNA polymerase II binding for G34 mutant KNS42 (blue) and wild-type SF188 cells (grey) for the SOX2 locus, which also encompasses the SOX2-OT transcript. (f) Validation of ChIP-Seq data by ChIP-qPCR using specific primers targeting SOX2. Blue bars: KNS42, grey: SF188. *** p<0.0001, t-test. (g) Boxplot of SOX2 expression in the integrated paediatric GBM samples stratified by H3F3A status. Blue box: G34, green: K27, grey: wild-type. * p<0.05, ANOVA. (h) Top – immunohistochemistry for SOX2 protein in a G34 mutant paediatric GBM sample RMH2465. Bottom – Barplot of SOX2 expression in a paediatric GBM tissue microarray stratified by H3F3A status. Blue bars: G34, green: K27, grey: wild-type. ++ strong expression, + moderate expression, - negative.
Strikingly, the most significant differentially bound and expressed gene in our G34 mutant KNS42 cells was MYCN (33-fold H3K36me3 compared with SF188, DESeq p=7.94×10−8; 60-fold RNA Pol II, DESeq p=1.59×10−9) (Figure 4A-D). Of note, a small number of H3F3A wild-type tumours also expressed high levels of MYCN, and were found to be MYCN gene amplified (Figure 4C). However, amplification was not seen in G34 mutant tumours, which parallels observations in diffuse intrinsic pontine glioma where MYCN amplification was found in wild-type, but not K27 mutant tumours15. Transduction of the G34V mutation into normal human astrocytes (NHA) and transformed human fetal glial cells (SVG) conferred an approximately 2-3 fold increase in MYCN transcript levels over wild-type-transduced controls, validating these observations (Supplementary Figure S13). H3F3A G34 mutation may therefore represent an alternative mechanism of enhancing expression levels of MYCN in paediatric glioblastoma.
Figure 4. G34 H3K36me3 upregulates MYCN which is selectively targetable by kinases that destabilise the protein.
MYCN: (a) ChIP-Seq of H3K36me3 and RNA polymerase II binding for G34 mutant KNS42 (blue) and wild-type SF188 cells (grey) for the MYCN locus, which also encompasses the MYCNOS transcript. (b) Validation of ChIP-Seq data by ChIP-qPCR using specific primers targeting MYCN. Blue bars: KNS42, grey: SF188. *** p<0.0001, t-test. (c) Boxplot of MYCN expression in the integrated paediatric GBM samples stratified by H3F3A status. Blue box: G34, green: K27, grey: wild-type. ** p<0.01, ANOVA. Wild-type tumours with high mRNA expression were frequently amplified (“Amp”). (d) Top – immunohistochemistry for MYCN protein in a G34 mutant paediatric GBM sample. RMH2465 Bottom – Barplot of MYCN expression in a paediatric GBM tissue microarray stratified by H3F3A status. Blue bars: G34, green: K27, grey: wild-type. ++ strong expression, + moderate expression, - negative. (e) Effects on cell viability of MYCN-knockdown in KNS42 cells. Western blot demonstrating efficiency of reduction of MYCN by three individual siRNAs targeting MYCN (named 6, 12 and W) and a pool of all three after 48 and 96 hours. Barplot demonstrating effects on KNS42 cell viability after siRNA knockdown at 7 days. *** p<0.001, t-test vs control. (f) siRNA screen for 714 human kinases in KNS42 cells. Western blot demonstrating expression of MYCN protein in G34 mutant KNS42 cells in contrast to a panel of wild-type paediatric glioma lines. GAPDH is used as a loading control. Kinase targets are plotted in plate well order along the x axis, and Z scores along the y axis. PLK1 is used as a positive control and is plotted in red. Negative controls are coloured light grey, and kinases with Z scores greater than -2.0 (no effect on cell viability) are coloured grey. ‘Hits’ (Z score less than -2.0) are coloured dark grey or blue, the latter if the effect on cell viability is specific to KNS42 cells and not in a panel of four H3F3A wild type paediatric glioma cell lines. The most significant, and selective hits were for CHK1 and AURKA. (g) Effect of knockdown of AURKA on MYCN levels in KNS42 cells. Western blot for AURKA and MYCN in KNS42 cells treated with individual oligonucleotides directed against AURKA for 48 and 96 hours. GAPDH is used as a loading control. (h) Effect of a selective small molecule inhibitor of AURKA on MYCN protein levels and cell viability. Left: Western blot for MYCN protein in KNS42 cells after exposure to 0.1, 0.5 and 2-5μM VX-689 (triangle). GAPDH is used as a loading control. Right: Barplot showing effects on cell viability of KNS42 cells exposed to 0.1, 0.5 and 2-5μM VX-689. ** p<0.01, t-test vs control.
Targeting MYCN is an attractive therapeutic intervention in tumours harbouring gene mutation such as neuroblastoma16, and direct inhibition by siRNA knockdown in KNS42 cells reduced cell viability in proportion to the reduction of protein levels observed (Figure 4E). Pharmacological agents which directly inhibit Myc transcription factors, however, remain elusive. We therefore carried out a synthetic lethal screen to ascertain how we might target these H3F3A G34 mutant, MYCN-driven tumours in the clinic. We utilised a series of siRNAs directed against 714 human kinases against our panel of paediatric glioma cell lines to identify those which conferred selective cell death to the MYCN-expressing KNS42 cells versus wild-type, non-MYCN-expressing controls (Figure 4F). The most significant synthetically lethal hits in the G34 mutant cells compared to H3F3A wild-type were kinases that have been previously associated with stabilisation of MYCN protein, specifically CHK1 (checkpoint kinase 1)17 and AURKA (aurora kinase A)18. Knockdown of AURKA by an independent set of four individual oligonucleotides targeting the gene led to a concurrent reduction of MYCN protein in KNS42 cells (Figure 4G). This destabilisation of MYCN was also observed in a dose-dependent manner using a highly selective small molecule inhibitor of AURKA, VX-689 (also known as MK-510819), at concentrations which in addition led to a significant reduction in viability of the G34 mutant cells (Figure 4H). Together, these data demonstrate the utility of targeting MYCN stability in H3F3A G34 mutant paediatric glioblastoma as a means of treating this subgroup of patients.
DISCUSSION
Emerging evidence is strongly suggestive that paediatric glioblastomas with H3F3A mutations can be subclassified into distinct entities. Our data indicate key molecular and clinical differences between G34 and K27 mutant tumours, reflecting the anatomical specificity (K27 tumours restricted to the pons and thalamus, G34 the cerebral hemispheres15) (Supplementary Table S4) and likely distinct developmental origins of these disease subgroups. Utilising the only known model of H3F3A mutant cells to date, we propose that the gene expression signature associated with G34 mutation in paediatric GBM patient samples is likely driven by a genomic differential binding of the transcriptionally activating H3K36me3 mark.
Mapping these gene expression signatures to publicly available datasets of human brain development shows a strong overlap with the ganglionic eminences of the embryonic and early foetal periods. These structures represent a transiently proliferating cell mass of the fetal subventricular zone, are the source of distinct neuroglial progenitors20, and are therefore strong candidates for the location of the cells of origin of cerebral hemispheric G34-driven paediatric GBM. As with other paediatric brain tumours21,22, mutation-driven subgroups of glioblastoma retain gene expression signatures related to discrete cell populations from which these distinct tumours may arise. In addition, this mutation-driven differential H3K36me3 binding leads to a significant upregulation of numerous genes associated with cell fate decisions. Thus we have identified a transcriptional readout of the likely developmental origin of G34 mutant glioblastoma coupled with a self-renewal signature we previously identified in KNS42 cells23 driven by mutation-induced differential binding of H3K36me3.
Significantly, the G34 mutation additionally upregulates MYCN through H3K36me3 binding. It was recently reported that forced overexpression of stabilised MYCN protein in neural stem cells of the developing mouse forebrain gave rise to glioblastomas24, and thus we provide the mechanism by which the initiating tumorigenic insult is delivered at the correct time and place25 during neurogenesis. Targeting stabilisation of MYCN protein via synthetic lethality approaches in H3F3A G34 mutant paediatric glioblastoma provides a potential novel means treating this subgroup of patients.
MATERIALS and METHODS
Primary paediatric glioblastoma expression profiling
Expression data from the Schwartzentruber2 (GSE34824) and Paugh3 (GSE19578) studies were retrieved from the Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/) and analysed in GenePattern using a signal-to-noise metric, and GSEA implemented for testing of enrichment of gene lists. Paediatric GBM expression signatures were mapped to specific developmental stages and anatomic locations using on a spatio-temporal gene expression dataset of human brain development in Kang et al.6 (GSE25219).
Tissue microarrays
Immunohistochemistry for DLX6 (NBP1-85929, Novus Biologicals, Littleton, CO, USA), SOX2 (EPR3131, Epitomics, Burlingame, CA, USA) and MYCN (#9405, Cell Signalling, Danvers, MA, USA) was carried out on tissue microarrays consisting of 46 cases of paediatric and young adult glioblastoma ascertained for H3F3A mutation by Sanger sequencing.
Cell line analysis
Paediatric glioblastoma KNS42 cells were obtained from the JCRB (Japan Cancer Research Resources) cell bank. Paediatric SF188 cells were kindly provided by Dr Daphne Haas-Kogan (UCSF, San Francisco, CA, USA), whilst UW479, Res259 and Res186 were kindly provided by Dr Michael Bobola (University of Washington, Seattle, WA, USA). All cells have been extensively characterised previously4, and were authenticated by short tandem repeat (STR) profiling. Western blot analysis was carried out for total histone H3 (ab97968, Abcam, Cambridge, UK), as well as H3K36 trimethylation (ab9050, Abcam), dimethylation (ab9049, Abcam) and monomethylation (ab9050, Abcam) after histone extraction using a histone purification minikit (ActiveMotif, Carlsbad, CA, USA), and quantitated by scanning on the Storm 860 Molecular Imager (GE Healthcare, Amersham, UK) and analyzed using ImageQuant software (GE Healthcare). Additional Western blots for MYCN (#9405, Cell Signaling, Danvers, MA, USA), ATRX (sc-15408, Santa Cruz) and GAPDH (#2118, Cell Signaling) were carried out according to standard procedures.
Chromatin immunoprecipitation
ChIP was carried out using antibodies against H3K36me3 and RNA polymerase II using the HistonePath™ and TranscriptionPath™ assays by ActiveMotif. Whole genome sequencing was carried out using an Illumina HiSeq2000 instrument with a fold coverage of >30 fold. Validation of active regions was carried out by ChIP-qPCR.
siRNA screening and validation
siRNA screening was carried out on a library of 714 human kinases using Dharmacon SMARTpools™ (Dharmacon, Lafayette, CO, USA), with cell viability estimated via a highly sensitive luminescent assay measuring cellular ATP levels (CellTiter-Glo™; Promega, Madison, WI, USA). Z-scores were calculated using the median absolute deviation of all effects in each cell line. Individual ON-TARGETplus™ oligonucleotides for validation were obtained from Dharmacon and knockdown validated by Western blot for AURKA (#4718, Cell Signaling) according to standard procedures for up to 96 hours. The AURKA-selective small molecule inhibitor VX-689 (MK-5108) was obtained from Selleckchem (Houston, TX) and assayed for up to five days. Effects on cell viability assessed by CellTiter-Glo™ (Promega). siRNAs targeting human MYCN were custom designs and kindly provided by Janet Shipley (ICR, London UK).
Supplementary Material
SIGNIFICANCE.
We provide the mechanistic explanation for how the first histone gene mutation in human disease biology acts to deliver MYCN, a potent tumorigenic initiator, into a stem cell compartment of the developing forebrain, selectively giving rise to incurable cerebral hemispheric glioblastoma. Employing synthetic lethal approaches to these mutant tumour cells provides a rational way to develop novel and highly selective treatment strategies.
ACKNOWLEDGEMENTS
This work is supported by Cancer Research UK, the Wellcome Trust, the Samantha Dickson Brain Tumour Trust and The Stravros Niarchos Foundation. LB, MN, AM, DAB, DC, KT, AB, AJ, SP, IB, CJL, AA, PW and CJ acknowledge NHS funding to the Biomedical Research Centre.
Footnotes
Conflict of interest statement: LB, AM, MN, AB, AJ, SP, DAB, KT, MV, IB, CJL, AA, PW and CJ are employees of the Institute of Cancer Research, which has a commercial interest in AURKA and CHK1 inhibitors
REFERENCES
- 1.Jones C, Perryman L, Hargrave D. Paediatric and adult malignant glioma: close relatives or distant cousins? Nature reviews. Clinical oncology. 2012;9:400–413. doi: 10.1038/nrclinonc.2012.87. [DOI] [PubMed] [Google Scholar]
- 2.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482:226–231. doi: 10.1038/nature10833. [DOI] [PubMed] [Google Scholar]
- 3.Paugh BS, Qu C, Jones C, Liu Z, Adamowicz-Brice M, Zhang J, et al. Integrated molecular genetic profiling of pediatric high-grade gliomas reveals key differences with the adult disease. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2010;28:3061–3068. doi: 10.1200/JCO.2009.26.7252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bax DA, Mackay A, Little SE, Carvalho D, Viana-Pereira M, Tamber N, et al. Molecular and phenotypic characterisation of paediatric glioma cell lines as models for preclinical drug development. PloS one. 2009;4:e5209. doi: 10.1371/journal.pone.0005209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nature reviews. Molecular cell biology. 2012;13:115–126. doi: 10.1038/nrm3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang HJ, Kawasawa YI, Cheng F, Zhu Y, Xu X, Li M, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Panganiban G, Rubenstein JL. Developmental functions of the Distal-less/Dlx homeobox genes. Development. 2002;129:4371–4386. doi: 10.1242/dev.129.19.4371. [DOI] [PubMed] [Google Scholar]
- 8.Kitamura K, Yanazawa M, Sugiyama N, Miura H, Iizuka-Kogo A, Kuaka M, et al. Mutation of ARX causes abnormal development of forebrain and testes in mice and X-linked lissencephaly with abnormal genitalia in humans. Nature genetics. 2002;32:359–369. doi: 10.1038/ng1009. [DOI] [PubMed] [Google Scholar]
- 9.Chatterjee S, Bourque G, Lufkin T. Conserved and non-conserved enhancers direct tissue specific transcription in ancient germ layer specific developmental control genes. BMC developmental biology. 2011;11:63. doi: 10.1186/1471-213X-11-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monaghan AP, Bock D, Gass P, Schwaeger A, Wolfer DP, Lipp HP, et al. Defective limbic system in mice lacking the tailless gene. Nature. 1997;390:515–517. doi: 10.1038/37364. [DOI] [PubMed] [Google Scholar]
- 11.McEvilly RJ, de Diaz MO, Schonemann MD, Hooshmand F, Rosenfeld MG. Transcriptional regulation of cortical neuron migration by POU domain factors. Science. 2002;295:1528–1532. doi: 10.1126/science.1067132. [DOI] [PubMed] [Google Scholar]
- 12.Zembrzycki A, Griesel G, Stoykova A, Mansouri A. Genetic interplay between the transcription factors Sp8 and Emx2 in the patterning of the forebrain. Neural development. 2007;2:8. doi: 10.1186/1749-8104-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Experimental cell research. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 14.Li X, Oghi KA, Zhang J, Krones A, Bush KT, Glass CK, et al. Eya protein phosphatase activity regulates Six1-Dach-Eya transcriptional effects in mammalian organogenesis. Nature. 2003;426:247–254. doi: 10.1038/nature02083. [DOI] [PubMed] [Google Scholar]
- 15.Khuong-Quang DA, Buczkowicz P, Rakopoulos P, Liu XY, Fontebasso AM, Bouffet E, et al. K27M mutation in histone H3.3 defines clinically and biologically distinct subgroups of pediatric diffuse intrinsic pontine gliomas. Acta neuropathologica. 2012;124:439–437. doi: 10.1007/s00401-012-0998-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gustafson WC, Weiss WA. Myc proteins as therapeutic targets. Oncogene. 2010;29:1249–1259. doi: 10.1038/onc.2009.512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cole KA, Huggins J, Laquaglia M, Hulderman CE, Russell MR, Bosse K, et al. RNAi screen of the protein kinome identifies checkpoint kinase 1 (CHK1) as a therapeutic target in neuroblastoma. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3336–3341. doi: 10.1073/pnas.1012351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto T, Horn S, Brockmann M, Eilers U, Schuettrumpf L, Popov N, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 19.Shimomura T, Hasako S, Nakatsuru Y, Mita T, Ichikawa K, Kodera T, et al. MK-5108, a highly selective Aurora-A kinase inhibitor, shows antitumor activity alone and in combination with docetaxel. Molecular cancer therapeutics. 2010;9:157–166. doi: 10.1158/1535-7163.MCT-09-0609. [DOI] [PubMed] [Google Scholar]
- 20.Miyoshi G, Hjerling-Leffler J, Karayannis T, Sousa VH, Butt SJ, Battiste J, et al. Genetic fate mapping reveals that the caudal ganglionic eminence produces a large and diverse population of superficial cortical interneurons. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2010;30:1582–1594. doi: 10.1523/JNEUROSCI.4515-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RA, Wright KD, Poppleton H, Mohankumar KM, Finkelstein D, Pounds SB, et al. Cross-species genomics matches driver mutations and cell compartments to model ependymoma. Nature. 2010;466:632–636. doi: 10.1038/nature09173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gibson P, Tong Y, Robinson G, Thompson MC, Currle DS, Eden C, et al. Subtypes of medulloblastoma have distinct developmental origins. Nature. 2010;468:1095–1099. doi: 10.1038/nature09587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gaspar N, Marshall L, Perryman L, Bax DA, Little SE, Viana-Pereira M, et al. MGMT-independent temozolomide resistance in pediatric glioblastoma cells associated with a PI3-kinase-mediated HOX/stem cell gene signature. Cancer research. 2010;70:9243–9252. doi: 10.1158/0008-5472.CAN-10-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swartling FJ, Savov V, Persson AI, Chen J, Hackett CS, Northcott PA, et al. Distinct Neural Stem Cell Populations Give Rise to Disparate Brain Tumors in Response to N-MYC. Cancer cell. 2012;21:601–613. doi: 10.1016/j.ccr.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Phoenix TN, Gilbertson RJ. There’s a time and a place for MYCN. Cancer cell. 2012;21:593–595. doi: 10.1016/j.ccr.2012.05.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.