Abstract
Androgens regulate body composition by interacting with the androgen receptor (AR) to control gene expression in a tissue-specific manner. To identify novel regulatory roles for AR in preadipocytes, we created a 3T3-L1 cell line stably expressing human AR. We found AR expression is required for androgen-mediated inhibition of 3T3-L1 adipogenesis. This inhibition is characterized by decreased lipid accumulation, reduced expression of adipogenic genes, and induction of genes associated with osteoblast differentiation. Collectively, our results suggest androgens promote an osteogenic gene program at the expense of adipocyte differentiation.
Keywords: Adipocyte, Osteoblast, Androgen receptor, Gene regulation
1. Introduction
The mechanisms by which androgens modulate fat distribution and bone density are clinically important, especially in an aging population where declining testosterone levels are prevalent [1]. Although a risk factor for type 2 diabetes mellitus, obesity is demonstrated to protect against bone loss [2–5]. Recent studies, however, have demonstrated a negative correlation between visceral fat accumulation and bone density, suggesting fat distribution may predict bone density and insulin resistance [6–9]. The interaction between visceral fat accumulation and bone mineral density remains poorly understood; yet, the mechanisms are important to improve healthspan in an aging world population.
Adipocytes and osteoblasts share a common mesenchymal stem cell (MSC) origin [10] with increasing evidence of transdifferentiation between cell types [11,12]. Accordingly, any factor controlling the balance between adipogenesis and osteogenesis in MSCs represents a potential regulator of body composition.
An early event in human adipocyte differentiation is the upregulation of AR expression, which opposes fat cell differentiation in an androgen-dependent manner [13]. In order to investigate androgen regulation of preadipocyte commitment more comprehensively, we created a 3T3-L1 cell line stably expressing full-length AR and analyzed the gene programs regulated by the androgen/AR axis.
2. Materials and methods
2.1. Cell culture, differentiation, and preparation of stable cell lines
3T3-L1 cells were maintained at 5% CO2/37 °C in DMEM/F12 (Invitrogen) with 10% fetal bovine serum (FBS; Gemini Bio-Products), 100 U/ml penicillin, and 100 μg/ml streptomycin. Postconfluent cells were differentiated with 5 μg/ml insulin, 1 μM dexamethasone, and 0.5 mM 3-isobutyl-1-methylxanthine in DMEM/F12 medium containing 10% FBS (DMI). After 48 h, the medium was changed to DMEM/F12 containing 10% FBS and 5 μg/ml insulin. Subsequently, the culture medium was replaced with DMEM/F12 containing 10% FBS every 48 h.
Flag-tagged human AR (fAR) was stably expressed at physiologically relevant levels in 3T3-L1 preadipocyte cells using lentivirus, as previously described [13]. Stable clones were selected in puromycin after single cell dilution.
2.2. Oil Red O staining
After differentiation, media was removed and 10% formalin was added for 5 min. Formalin was removed and a second volume of 10% formalin was added to wells for 1 h. Wells were then washed with 60% isopropanol and allowed to dry. Oil Red O (2 g/L) was applied 10 min, followed by extensive washing with distilled water. All steps were performed at room temperature. Images were acquired using a digital camera.
2.3. Antibodies and western blotting
Western blot analysis was performed with whole cell lysates run on 4–12% Bis-Tris NuPage® (Invitrogen) gels and transferred onto Immobilon-P Transfer Membranes (Millipore). After membrane blocking (SuperBlock, Pierce), primary antibodies (anti-AR rabbit polyclonal, Santa Cruz Biotechnology) were incubated overnight at 4 °C, followed by secondary antibodies for 1 h at room temperature. Immunoreactive bands were visualized by chemiluminescence. β-actin (mouse monoclonal, Sigma Chemical Co.) was used as the invariant control.
2.4. RNA extraction and qPCR analysis
RNA was extracted from cells using the RNeasy kit (Qiagen) following manufacturer instructions. To measure relative mRNA expression, qPCR was performed using the Taqman RT-PCR one-step master mix in conjunction with an ABI 7500 real-time PCR system (Applied Biosystems). Each sample was tested in duplicate in two independent experiments. β-actin was used as the invariant control. The following primer and probes (Roche Universal Probe Library) were used:
AR: 5′-tgtcaactccaggatgctctact-3′; 5′-tggctgtacatccgagacttg-3′. Probe: 5′-6-FAM-ttcaatgagtaccgcatgc-BHQ-3′.
FABP4: 5′-ggatggaaagtcgaccacaa-3′; 5′-tggaagtcacgcctttcata-3′. Roche probe #77.
CEBPB: 5′-aagatgcgcaacctggag-3′; 5′-cagggtgctgagctctcg-3′, Roche probe #67.
Pref1: 5′-cgggaaattctgcgaaatag-3′; 5′-tgtgcaggagcattcgtact-3′, Roche probe #80.
PPARG: 5′-gaaagacaacggacaaatcacc-3′; 5′-gggggtgatatgtttgaacttg-3′, Roche probe #7.
CEBPA: 5′-aaacaacgcaacgtggaga-3′; 5′-gcggtcattgtcactggtc-3′, Roche probe #67.
Ank: 5′-tcaccaacatagccatcgac-3′; 5′-actgcatcctccttgactgc-3′; Roche probe #32.
ENPP1: 5′-cggacgctatgattccttaga-3′; 5′-agcacaatgaagaagtgagtcg-3′; Roche probe #72.
Notch: 5′-ctggaccccatggacatc-3′; 5′-aggatgactgcacacattgc-3′; Roche probe #80.
SPP1: 5′-cccggtgaaagtgactgatt-3′; 5′-ttcttcagaggacacagcattc-3′; Roche probe #82.
Chrd: 5′-tcactgcccacctccttg-3′; 5′-atcttttaccacgccctgag-3′; Roche probe #66.
Cyr61: 5′-ggatctgtgaagtgcgtcct-3′; 5′-ctgcatttcttgcccttttt-3′; Roche probe #66.
2.5. DNA microarrays
Microarrays were performed by the Baylor College of Medicine Microarray Core Facility using Affymetrix Mouse Gene 1.0 ST arrays (Affymetrix). 3T3-L1 cells stably expressing human AR were differentiated in the presence or absence of 10 nM R1881 for 24 h, followed by total RNA isolation. All RNA samples were analyzed with a Bioanalyser 2100 (Agilent Technologies) before microarray hybridization.
2.6. Microarray analysis
Normalized gene expression values were calculated by the Robust Multiarray Averaging method using R.2.10 and Limma 2.19 analysis package, as previously described [13]. The false discovery rate was controlled by Benjamini and Hochberg correction to account for multiple testing error.
2.7. Statistical analysis
Student’s t-test was used for statistical analyses of qPCR data. A p-value cutoff of 0.05 was used to determine significance.
3. Results
3.1. Exogenous expression of AR is required for androgen signaling in 3T3-L1
AR mRNA is latently expressed during 3T3-L1 adipogenesis, leading to limited androgen responsiveness in these cells [13–15]. We investigated the relationship between AR and 3T3-L1 adipogenesis by measuring relative mRNA levels of AR and aP2 between days 0 and 8 after dexamethasone/IBMX/insulin (DMI) induction, representing preadipocytes and mature adipocytes, respectively. AR was weakly expressed in days 1, 2, and 3, reaching highest levels in days 6 through 8. As a reference, aP2 was upregulated 900-fold at day 6 while AR exhibited 6-fold induction (Fig. 1A), consistent with reported expression patterns [14]. To test the effect of androgens on 3T3-L1 adipogenesis, we added DHT or R1881 to 3T3-L1 cells beginning on day 0 of induction. In contrast to a previous report [16] and consistent with our recent findings [13], these ligands did not inhibit 3T3-L1 adipogenesis (Fig. 1A). We also evaluated AR expression levels in 3T3-L1 cells and mouse primary fat tissues (Fig. 1C). AR was expressed at very low levels in differentiated 3T3-L1 cells (day 8) compared to subcutaneous fat, epididymal fat, retroperitoneal fat and brown fat. Based on these results, we reasoned low levels of AR expression in 3T3-L1 prevented an inhibitory effect of androgens on adipogenesis.
Fig. 1.
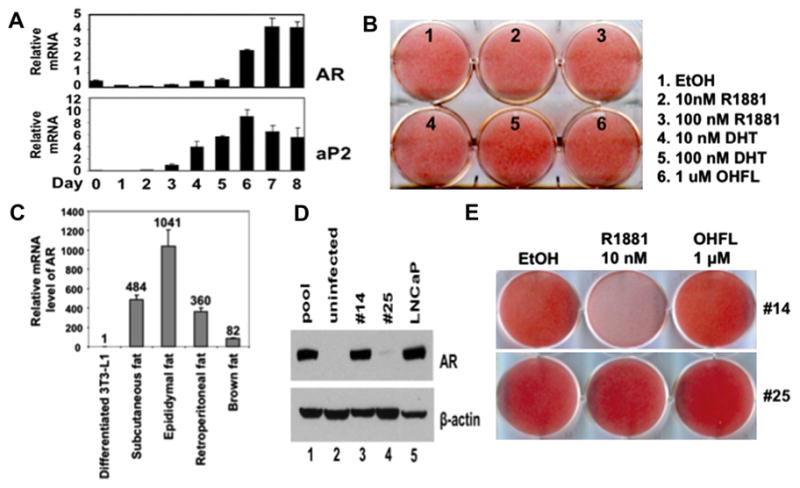
AR expression and action in mouse adipocytes. (A) Induced AR and aP2 mRNA levels in wild-type 3T3-L1 cells during adipogenic differentiation. (B) Wild type 3T3-L1 cells were treated with adipocyte differentiation cocktail in the presence of androgens for 8 d followed by Oil Red O staining. (C) Relative AR mRNA for in vitro differentiated 3T3-L1 cells and murine adipose tissues. (D) 3T3-L1-AR stable cell lines were created by infecting 3T3-L1 cells with lentivirus expressing flag-tagged AR. Western blot comparison of AR protein levels in pooled 3T3-L1 infected with fAR lentivirus and uninfected 3T3-L1 with 3T3-L1-fAR stable clones #14 and #25. LNCaP, a prostate cancer cell line with endogenous AR expression, was used for comparison of relative AR expression levels. (E) 3T3-L1 fAR stable clones #14 and #25 were differentiated in the presence of ethanol (EtOH), 10 nM R1881, or O-hydroxyflutamide (O-HFL) for 8 d. Oil-Red-O was used to determine effects of hormones on adipogenesis in both clones.
Next, we generated 3T3-L1 stable cell lines constitutively expressing Flag-tagged human AR (fAR). Briefly, 3T3-L1 cells were infected with lentivirus encoding fAR with verification of AR protein levels by Western blotting (Fig. 1D). AR was detected in pooled 3T3-L1 stable cells, single stable 3T3-L1 clone #14, and LNCaP cells. Clone #25 showed a weak band, indicating a low level of AR expression. AR protein in parental 3T3-L1 cells was undetectable, in agreement with the androgen-refractory nature of these cells. Subsequently, Clones #14 and #25 were induced to differentiate for 8 days in the presence of vehicle (EtOH), 10 nM R1881 or 1 μM hydroxyflutamide (OHFL), an AR antagonist, to test for androgen responsiveness (Fig. 1E). Conventional adipocyte differentiation conditions (EtOH) promoted adipogenesis in both clones. However, differentiation of Clone #14 cells was completely blocked by R1881, but not by OHFL. In contrast, adipogenesis in Clone #25 cells was not affected by either androgen or anti-androgen (OHFL). These results indicated adipogenesis of Clone #14 cells was inhibited by AR activation.
3.2. Androgen alters adipogenic genes when AR is expressed in 3T3-L1 cells
Adipogenic stimuli activate C/EBPβ, C/EBPδ and glucocorticoid receptor (GR) to induce genes encoding PPARγ and C/EBPα [17–20]. Marking the commitment phase, early activation of C/EBPβ, C/EBPδ and GR during 3T3-L1 differentiation has been shown to repress expression of the preadipocyte marker Pref-1 [21–23] Subsequently, PPARγ mRNA and protein expression are robustly induced in a feed-forward loop with C/EBPα to induce or repress adipose-specific genes [24,25].
We measured the expression of several adipogenic marker genes by qPCR to determine whether reduced Oil-Red-O staining in R1881-treated cells (Fig. 1E) resulted from androgen/AR-mediated alterations in the 3T3-L1 differentiation program. Stable expression of fAR in 3T3-L1 (Clone #14: fAR cells) followed by differentiation in the presence of 10 nM R1881 significantly reduced the induction of adipogenic marker genes, including PPARγ (Fig. 2A), C/EBPα (Fig. 2B), and aP2 (Fig. 2C).
Fig. 2.
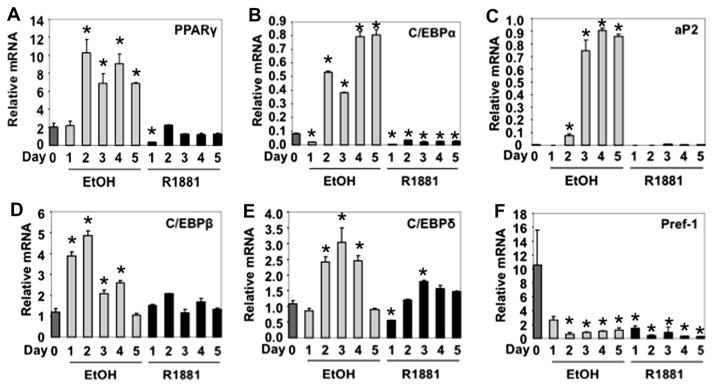
Effect of androgen treatment on the expression of key genes in adipogenesis using 3T3-L1-fAR stable cell line (#14). 3T3-L1 fAR cells were differentiated for up to 5 days in the presence or absence of 10 nM R1881. qPCR was used to measure mRNA expression for (A) PPARγ, (B) C/EBPα, (C) aP2, (D) C/EBPβ (E) C/EBPδ, and (F) Pref-1. All results are n = 2 ± S.E.M. Asterisks indicate mRNA statistically different from the vehicle control (*p < 0.05).
Upregulation of C/EBPβ (Fig. 2D) and C/EBPδ (Fig. 2E) by dexamethasone in the early stages of adipocyte differentiation [17,18] was abrogated by R1881 treatment as early as 2 h post DMI treatment. However, the kinetics of Pref-1 downregulation (Fig. 3F) were unaffected under these conditions, suggesting androgen/AR signaling did not block the differentiation program through maintenance of the preadipocyte state [21–23].
Fig. 3.
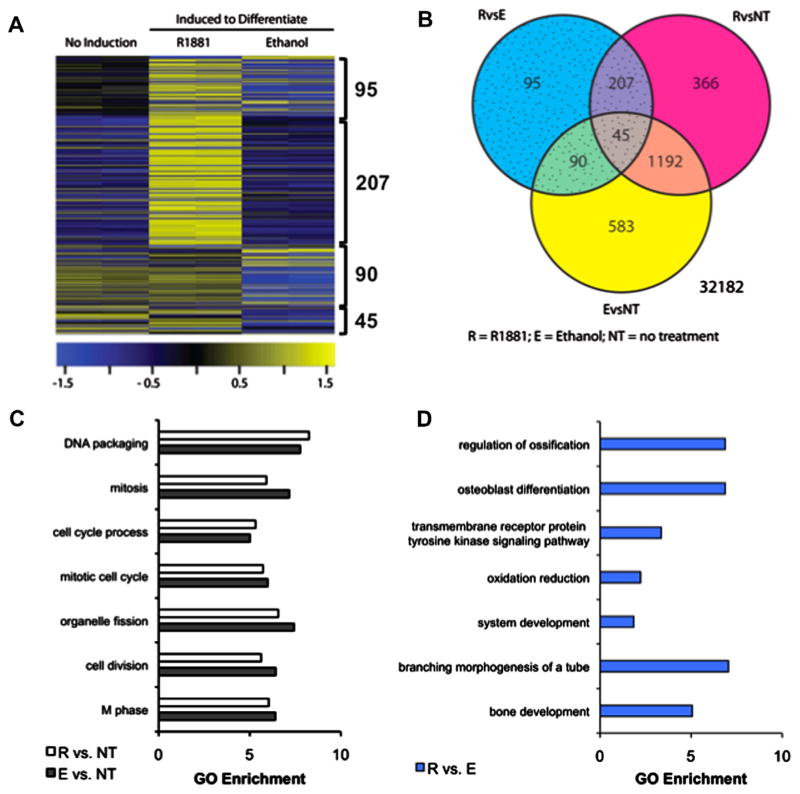
Androgen regulation of genome-wide gene expression during adipogenesis in 3T3-L1-fAR cells. Confluent 3T3-L1-fAR cells (clone #14) were induced to differentiate for 24 h in the presence of 10 nM R1881 (R), EtOH (E) or were left undifferentiated (no treatment; NT). (A) Shown are array probes significantly different between cells differentiated in the presence or absence of androgen; FC > 2; q-value < 0.05. Significantly changed probe sets in the R v. E contrast are highlighted. (B) Genes shared and unique to each treatment group represented by Venn diagram. (C) Gene Ontology (GO) analysis of R1881 (R) and ethanol (E) treatments versus NT controls showed no pathways were differentially enriched. (D) Further GO analysis of genes between R and E showed pathways associated with bone differentiation significantly enriched. Genes with FC > 2 and FDR < 0.05 were used for GO analysis.
3.3. Androgens promote an osteogenic gene signature in 3T3-L1 expressing AR
Our data suggested stable expression of AR in 3T3-L1 cells (fAR cells) inhibited adipocyte differentiation in an androgen-dependent manner. We next characterized the androgen-dependent transcriptome in fAR cells. Three treatment groups were used to determine the effect of androgen on gene expression: vehicle (no induction); DMI+EtOH (ethanol, E); and 10 nM R1881+DMI (R1881). fAR cells were treated for 24 h, followed by RNA isolation, microarray, and follow-up qPCR analysis. We detected 2578 probe sets (Supplemental File 1) differentially expressed (2-fold; q < 0.05) in three contrast groups (Fig. 3A): probe sets changed after 24 h in response to DMI (E v. NT), 10 nM R1881+DMI (R v. NT), or the difference between E v. NT and R v. NT (R v. E). We hypothesized the last contrast (R v. E) identified a subset of genes uniquely regulated by androgen during adipogenesis (Fig. 3B).
We next used gene ontology (GO) analysis to identify biological pathways enriched in each treatment group. Comparison of biological pathways altered by DMI+EtOH (E) or DMI+R1881 (R) relative to undifferentiated fAR cells demonstrated enrichment of mitosis-related biological processes (Fig. 3C), reflecting the requirementin 3T3-L1 cells of mitotic clonal expansion prior to terminal differentiation [26]. However, when we compared DMI+EtOH (E) to DMI+R1881 (R), three GO biological process categories associated with bone development were significantly enriched (Fig. 3D): regulation of ossification, osteoblast differentiation, bone development.
To study the bone phenotype at the gene expression level, we selected genes for qPCR analysis which exhibited the most significant fold change between DMI+EtOH (E) and DMI+R1881 (R) treatments: Ank (regulation of ossification), Cyr61 (osteoblast differentiation), Chrd (regulation of ossification, osteoblast differentiation), Enpp1 (regulation of ossification), Notch1 (osteoblast differentiation), and Spp1 (osteoblast differentiation). For qPCR validation, fAR cells were treated for 48 h with vehicle (EtOH), DMI, 10 nM R1881, or 10 nM R1881+DMI. DMI treatment alone repressed expression of Ank (Fig. 4A), Cyr61 (Fig. 4B), Chrd (Fig. 4C) and Spp1 (Fig. 4F). Enpp1 (Fig. 4D) and Notch1 (Fig. 4E), genes associated with ‘regulation of ossification’ and/or ‘osteoblast differentiation’ were unchanged by DMI treatment alone. In the presence of R1881 alone, or in combination with DMI, three patterns of response were observed when compared to the vehicle control. Genes were either (i) not repressed by androgen treatment (Ank); (ii) induced by androgen independently of DMI (Cyr61, Chrd, Enpp1, Notch1); or (iii) more significantly repressed by R1881 (Spp1). In summary, our gene expression analysis established androgen treatment in fAR cells promoted an osteoblast phenotype at the expense of adipogenesis.
Fig. 4.
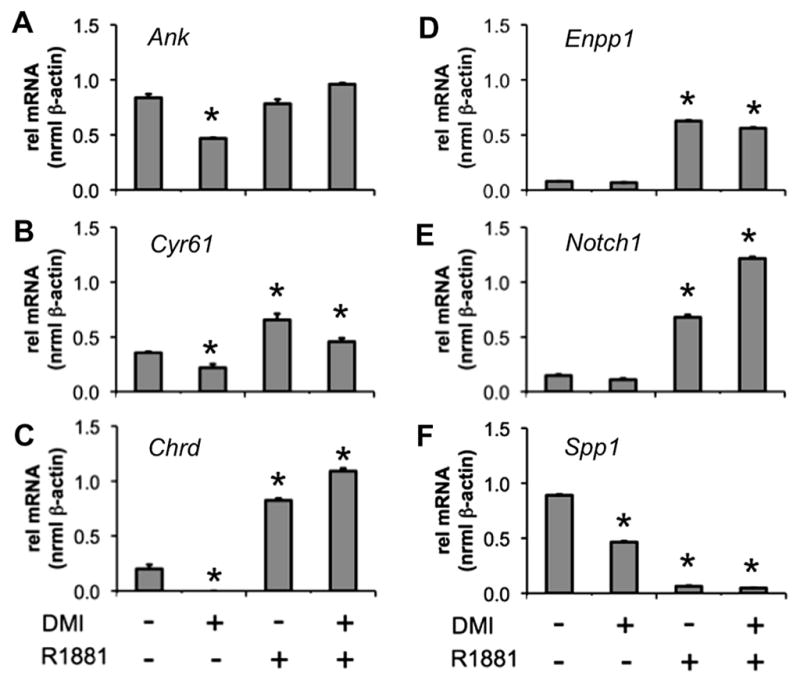
Androgen promotes expression of genes involved in osteogenesis. 3T3-L1 fAR cells were differentiated for 48 h in the presence or absence of 10 nM R1881. qPCR was used to measure mRNA expression for (A) Ank, (B) Cyr61, (C) Chrd, (D) Enpp1, (E) Notch, and (F) Spp1. All results are n = 2 ± S.E.M. Asterisks indicate mRNA statistically different from the vehicle control (*p < 0.05).
4. Discussion
The role of androgens and AR has expanded from reproductive tissues to multiple endocrine targets, including bone and adipose tissue [27]. Clinically, testosterone deficiency in aging men is associated with accumulation of central adiposity, decreased bone mineral density and type 2 diabetes mellitus [28]. Indeed, testosterone therapy has been shown to improve insulin sensitivity, reduce visceral fat mass, plasma triglycerides, cholesterol and fasting blood glucose levels, and increase bone density [29–31]. These facts underscore the importance of androgens in modulating energy balance and body composition.
We initially sought to characterize the roles of AR and androgen during 3T3-L1 preadipocyte differentiation. Consistent with previous data [14], we observed upregulation of AR late in the differentiation process, correlating with induction of the mature adipocyte marker, aP2. However adipogenesis was not affected by R1881. While AR modulates mature adipocyte function [32], our findings collectively suggested a limited role of AR and androgens in 3T3-L1 differentiation. Supporting this notion, AR mRNA levels in 3T3-L1 cells were extremely low compared to primary fat depots, consistent with time-resolved expression of nuclear receptors during 3T3-L1 adipogenesis [14,15], microarray profiling [33], and our recent work [13].
C/EBPβ, C/EBPδ, and GR co-operatively induce C/EBPα and PPARγ mRNA to initiate adipogenesis [17–20]. Then, C/EBPα and PPARγ coordinately activate the transcription of adipocyte-specific genes [25,26,34]. 3T3-L1 cells stably expressing fAR (clone #14) showed reduced mRNA levels for key adipogenic transcription factors, PPARγ and C/EBPα, and concomitant reduction of the adipocyte marker aP2 (Fig. 2). Other adipocyte-determining transcription factors, including C/EBPδ and C/EBPβ, were less affected.
Previous studies have shown dexamethasone drives preadipocytes toward a novel intermediate cellular state through transient activation of C/EBPβ and C/EBPδ, leading to repression of Pref-1 [21–23]. In this study, downregulation of Pref-1 was not affected by androgen treatment, suggesting activation of C/EBPβ and C/EBPδ was intact. Collectively, these results indicate androgens block adipogenesis prior to the induction of PPARγ and C/EBPα, but downstream of Pref-1 repression by dexamethasone.
Transcriptomic profiling of short-term (24 h) treatment with the synthetic androgen R1881 treatment on differentiating 3T3-L1 cells stably expressing fAR (Fig. 4) showed unique pathways enriched upon gene ontology analysis. When we compared R1881 and differentiation (EtOH) treatments to undifferentiated cells, expected pathways associated with mitotic clonal expansion [26] were enriched: M phase, mitotic cell cycle, cell cycle process. However, ontology analysis of genes differentially expressed between fAR cells differentiated in the presence of R1881 or differentiated in the presence of vehicle, showed enrichment of processes associated with bone development and osteogenesis.
Validation of key genes in each of these ontology categories substantiated the positive effect of androgens on bone differentiation. Ank [35,36], Chrd [37,38], Cyr61 [39,40], Enpp1 [35,41,42], Notch1 [43,44], and Spp1 [45] represent genes marking osteoblast function and bone remodeling, each showing regulation by R1881 alone or in combination with DMI. Collectively these findings position androgen signaling in preadipocytes as a mediator of osteogenic gene transcription.
Osteoblasts and adipocytes arise from a common mesenchymal stem cell (MSC) origin, as illustrated by the ability of MSCs to differentiate into either lineage [10]. Thus, we propose AR modulation may mediate the negative relationship between visceral fat and bone [7] to improve body composition.
Supplementary Material
Acknowledgments
We thank Lisa White for expert assistance with microarray experiments. This work was funded by NIH 1F32DK85979 (S.M.H.), NIH 1K01DK096093 (S.M.H.), Caroline Weiss Law Foundation (S.E.M.), NIH 3U19DK062434 NURSA (N.J.M.), NIH 5K01DK084209 (Q.F.), and NIH 5K01DK081446 (B.H.). The Genomic and RNA Profiling Core at Baylor College of Medicine is supported by P30-CA125123. This work was also partly supported by the Diabetes and Endocrinology Research Center (P30-DK079638) at Baylor College of Medicine.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.bbrc.2013.03.078.
References
- 1.Callewaert F, Boonen S, Vanderschueren D. Sex steroids and the male skeleton: a tale of two hormones. Trends Endocrinol Metab. 2010;21:89–95. doi: 10.1016/j.tem.2009.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Reid IR, Ames R, Evans MC, et al. Determinants of total body and regional bone mineral density in normal postmenopausal women-a key role for fat mass. J Clin Endocrinol Metab. 1992;75:45–51. doi: 10.1210/jcem.75.1.1619030. [DOI] [PubMed] [Google Scholar]
- 3.Reid IR, Plank LD, Evans MC. Fat mass is an important determinant of whole body bone density in premenopausal women but not in men. J Clin Endocrinol Metab. 1992;75:779–782. doi: 10.1210/jcem.75.3.1517366. [DOI] [PubMed] [Google Scholar]
- 4.Albala C, Yanez M, Devoto E, et al. Obesity as a protective factor postmenopausal osteoporosis. Int J Obes Relat Metab Disord. 1996;20:1027–1032. [PubMed] [Google Scholar]
- 5.Khosla S, Atkinson EJ, Riggs BL, et al. Relationship between body composition and bone mass in women. J Bone Miner Res. 1996;11:857–863. doi: 10.1002/jbmr.5650110618. [DOI] [PubMed] [Google Scholar]
- 6.Janicka A, Wren TA, Sanchez MM, et al. Fat mass is not beneficial to bone in adolescents and young adults. J Clin Endocrinol Metab. 2007;92:143–147. doi: 10.1210/jc.2006-0794. [DOI] [PubMed] [Google Scholar]
- 7.Gilsanz V, Chalfant J, Mo AO, et al. Reciprocal relations of subcutaneous and visceral fat to bone structure and strength. J Clin Endocrinol Metab. 2009;94:3387–3393. doi: 10.1210/jc.2008-2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russell M, Mendes N, Miller KK, et al. Visceral fat is a negative predictor of bone density measures in obese adolescent girls. J Clin Endocrinol Metab. 2010;95:1247–1255. doi: 10.1210/jc.2009-1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bredella MA, Torriani M, Ghomi RH, et al. Determinants of bone mineral density in obese premenopausal women. Bone. 2011;48:748–754. doi: 10.1016/j.bone.2010.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittenger MF, Mackay AM, Beck SC, et al. Multilineage potential of human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 11.Schiller PC, D’Ippolito G, Brambilla R, et al. Inhibition of gap-junctional communication induces the trans-differentiation of osteoblasts to an adipocytic phenotype in vitro. J Biol Chem. 2001;276:14133–14138. doi: 10.1074/jbc.M011055200. [DOI] [PubMed] [Google Scholar]
- 12.Wolf NS, Penn PE, Rao D, et al. Intraclonal plasticity for bone, smooth muscle, and adipocyte lineage in bone marrow stroma fibroblastoid cells. Exp Cell Res. 2003;290:346–357. doi: 10.1016/s0014-4827(03)00321-5. [DOI] [PubMed] [Google Scholar]
- 13.Hartig SM, He B, Newberg JY, et al. Feed-forward inhibition of androgen receptor activity by gluococorticoid action in human adipocytes. Chem Biol. 2012;19:1126–1141. doi: 10.1016/j.chembiol.2012.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fu M, Sun T, Bookout AL, et al. A nuclear receptor atlas: 3T3-L1 adipogenesis. Mol Endocinol. 2005;19:2437–2450. doi: 10.1210/me.2004-0539. [DOI] [PubMed] [Google Scholar]
- 15.Lahnalampi M, Heinaniemi M, Sinkkonen L, et al. Time-resolved expression profiling of the nuclear receptor superfamily in human adipogenesis. PLOS One. 2010;5:e12991. doi: 10.1371/journal.pone.0012991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh R, Artaza JN, Taylor WE, M, et al. Testosterone inhibits adipogenic differentiation in 3T3-L1 cells: nuclear translocation of androgen receptor complex with beta-catenin and T-cell factor 4 may bypass canonical Wnt signaling to down-regulate adipogenic transcription factors. Endocrinology. 2006;147:141–154. doi: 10.1210/en.2004-1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Christy RJ, Kaestner KH, Geiman DE, et al. CCAAT/enhancer binding protein gene promoter: binding of nuclear factors during differentiation of 3T3-L1 preadipocytes. Proc Natl Acad Sci USA. 1991;88:2593–2597. doi: 10.1073/pnas.88.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang QQ, Lane MD. Activation and centromeric localization of CCAAT/enhancer-binding proteins during the mitotic clonal expansion of adipocyte differentiation. Genes Dev. 1999;13:2231–2241. doi: 10.1101/gad.13.17.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steger DJ, Grant GR, Schupp M, et al. Propagation of adipogenic signals through an epigenomic transition state. Genes Dev. 2010;24:1035–1044. doi: 10.1101/gad.1907110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siersbaek R, Nielsen R, John S, et al. Extensive chromatin remodeling and establisment of transcription factor ‘hotspots’ during early adipogenesis. EMBO J. 2011;30:1459–1472. doi: 10.1038/emboj.2011.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smas CM, Hui HS. Pref-1, a protein containing EGF-like repeats, inhibits adipocyte differentiation. Cell. 1993;73:725–734. doi: 10.1016/0092-8674(93)90252-l. [DOI] [PubMed] [Google Scholar]
- 22.Smas CM, Kachinskas D, Liu CM, et al. Transcriptional control of the pref-1 gene in 3T3-L1 adipocyte differentiation. Sequence requirement for differentiation-dependent suppression. J Biol Chem. 1998;273:31751–31758. doi: 10.1074/jbc.273.48.31751. [DOI] [PubMed] [Google Scholar]
- 23.Pantoja C, Huff JT, Yamamoto KR. Glucocorticoid signaling defines a novel commitment state during adipogenesis in vitro. Mol Biol Cell. 2008;19:4032–4041. doi: 10.1091/mbc.E08-04-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu ZD, Rosen ED, Brun R, et al. Cross-regulation of C/EBP alpha and PPAR gamma controls the transcriptional pathway of adipogenesis and insulin sensitivity. Mol Cell. 1999;3:151–158. doi: 10.1016/s1097-2765(00)80306-8. [DOI] [PubMed] [Google Scholar]
- 25.Rosen ED, Hsu CH, Wang XZ, et al. C/EBP alpha induces adipogenesis through PPAR gamma: a unified pathway. Genes Dev. 2002;16:22–26. doi: 10.1101/gad.948702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tang QQ, Otto TC, Lane MD. Mitotic clonal expansion: a synchronous process required for adipogenesis. Proc Natl Acad Sci USA. 2003;100:44–49. doi: 10.1073/pnas.0137044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zitzmann M. Testosterone deficiency, insulin resistance and the metabolic syndrome. Nat Rev Endocrinol. 2009;5(2009):673–681. doi: 10.1038/nrendo.2009.212. [DOI] [PubMed] [Google Scholar]
- 28.Wang C, Nieschlag E, Swerdloff R, Behre HM, Hellstrom WJ, Gooren LJ, Kaufman JM, Legros JJ, Lunenfeld B, Morales A, Morley JE, Schulman C, Thompson IM, Weidner W FCWWu Investigation. Treatment, and monitoring of late-onset hypogonadism in males: ISA, ISSAM, EAU, EAA, and ASA recommendations. J Androl. 2009;30:1–9. doi: 10.2164/jandrol.108.006486. [DOI] [PubMed] [Google Scholar]
- 29.Behre HM, Kliesch S, Leifke E, et al. Long-term effect of testosterone therapy on bone mineral density in hypogonadal men. J Clin Endocrinol Metab. 1997;82:2386–2390. doi: 10.1210/jcem.82.8.4163. [DOI] [PubMed] [Google Scholar]
- 30.Kapoor D, Goodwin E, Channer KS, et al. Testosterone replacement therapy improves insulin resistance, glycaemic control, visceral adiposity and hypercholesterolaemia in hypogonadal men with type 2 diabetes. Eur J Endocrinol. 2006;154:899–906. doi: 10.1530/eje.1.02166. [DOI] [PubMed] [Google Scholar]
- 31.Snyder PJ, Peachey H, Berlin JA, et al. Effects of testosterone replacement in hypogonadal men. Effects of testosterone replacement in hypogonadal men. 2000;85:2670–2677. doi: 10.1210/jcem.85.8.6731. [DOI] [PubMed] [Google Scholar]
- 32.McInnes KJ, Smith LB, Hunger NI, et al. Deletion of the androgen receptor in adipose tissue in male mice elevates retinol binding protein 4 and reveals independent effects on visceral fat mass and on glucose homeostasis. Diabetes. 2012;61:1072–1081. doi: 10.2337/db11-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schupp M, Cristancho AG, Lefterova MI, et al. Re-expression of GATA2 cooperates with peroxisome proliferator-activated receptor-gamma depletion to revert the adipocyte phenotype. J Biol Chem. 2009;284:9458–9464. doi: 10.1074/jbc.M809498200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rosen ED, Sarraf P, Troy AE, et al. PPAR gamma is required for the differentiation of adipose tissue in vivo and in vitro. Mol Cell. 1999;4:611–617. doi: 10.1016/s1097-2765(00)80211-7. [DOI] [PubMed] [Google Scholar]
- 35.Paic F, Igwe JC, Nori R, et al. Identification of differentially expressed genes between osteoblasts and osteocytes. Bone. 2009;45:682–692. doi: 10.1016/j.bone.2009.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim HJ, Minashima T, McCarthy EF, et al. Progressive ankylosis protein (ANK) in osteoblasts and osteoclasts controls bone formation and bone remodeling. J Bone Miner Res. 2010;25:1771–1783. doi: 10.1002/jbmr.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petryk A, Shimmi O, Jia X, et al. Twisted gastrulation and chordin inhibit differentiation and mineralization in MC3T3-E1 osteoblast-like cells. Bone. 2005;36:617–626. doi: 10.1016/j.bone.2005.01.018. [DOI] [PubMed] [Google Scholar]
- 38.Macsai CE, Georgiou KR, Foster BK, et al. Microarray expression analysis of genes and pathways involved in growth plate cartilage injury responses and bony repair. Bone. 2012;50:1081–1091. doi: 10.1016/j.bone.2012.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Parisi MS, Gazzerro E, Rydziel S, et al. Expression and regulation of CCN genes in murine osteoblasts. Bone. 2006;38:671–677. doi: 10.1016/j.bone.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 40.Wi S, Kang Q, Luu HH, et al. CCN1/Cyr61 is regulated by the canonical Wnt signal and plays an important role in Wnt3A-induced osteoblast differentiation of mesenchymal stem cells. Mol Cell Biol. 2006;26:2955–2964. doi: 10.1128/MCB.26.8.2955-2964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hessle L, Johnson KA, Anderson HC, et al. Tissue-nonspecific alkaline phosphatase and plasma cell membrane glycoprotein-1 are central antagonistic regulators of bone mineralization. Proc Natl Acad Sci USA. 2002;99:9445–9449. doi: 10.1073/pnas.142063399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nam HK, Liu J, Kragor A, et al. Ectonucleotide pyrophosphatase/phosphodiesterase-1 (ENPP1) protein regulates osteoblast differentiation. J Biol Chem. 2011;286:39059–39071. doi: 10.1074/jbc.M111.221689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tezuka K, Yasuda M, Watanabe N, et al. Simulation of osteoblastic cell differentiation by Notch. J Bone Miner Res. 2002;17:231–239. doi: 10.1359/jbmr.2002.17.2.231. [DOI] [PubMed] [Google Scholar]
- 44.Engin F, Yao Z, Yang T, et al. Dimorphic effects of Notch signaling in bone homeostasis. Nat Med. 2008;14:299–305. doi: 10.1038/nm1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reinholt FP, Hultenby K, Oldberg A, et al. Osteopontin-a possible anchor of osteoclasts to bone. Proc Natl Acad Sci USA. 1990;87:4473–4475. doi: 10.1073/pnas.87.12.4473. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.