Abstract
Background
MicroRNAs (miRNAs) participate in various cellular processes such as cell growth, differentiation, cell death and play an important role in a variety of diseases, especially in cancer. Recently, a number of studies have investigated the association between single nucleotide polymorphisms (SNPs) on the hsa-miR-149 rs2292832 and susceptibility to cancer; however, the results remain inconclusive.
Methodology/Principal Findings
We carried out a meta-analysis of 12 studies including 5937 cases and 6081 controls from PubMed to assess the association between the hsa-miR-149 rs2292832 and cancer risk by pooled odds ratios (ORs) and 95% confidence intervals (CIs). However, our results showed that genotype distribution of the hsa-miR-149 rs2292832 was not associated with cancer risk in all genetic models. Subgroup analysis by cancer type, ethnicity or study design showed no significant association either.
Conclusion
Results of this meta-analysis suggest that the hsa-miR-149 rs2292832 polymorphism is not associated with cancer risk in spite of the potentially protective role of C allele in hepatocellular carcinoma and male gastric cancer.
Introduction
MicroRNAs (miRNAs) are an abundant class of small non-coding RNAs that negatively regulate gene expression by base pairing with the 3’-untranslated region of target mRNAs, resulting in either mRNA cleavage or translational repression [1,2]. Many studies have indicated that miRNAs are involved in regulating various biological processes, such as cellular differentiation, proliferation, angiogenesis, metabolism and cancer development [3–7]. As a crucial part of tumor formation, maintenance, and metastasis, dysregulated miRNAs, as either tumor suppressors or oncogenes, may play an important role in cancer [8].
Single nucleotide polymorphisms (SNPs) located at miRNA genes may be associated with changes in miRNA processing, thus leading to functional changes of miRNA by influencing interaction between miRNAs and their target mRNAs [9,10]. Many studies have explored association between SNPs of miRNA and susceptibility to various cancers. The hsa-miR-499 rs3746444 polymorphism might increase breast cancer risk [11]. Wang et al. showed that the miR-146a rs2910164 polymorphism was significantly associated with risk of papillary thyroid carcinoma, primary liver cancer and cervical cancer and the miR-196a2 rs11614913 polymorphism was associated with breast cancer, lung cancer, and colorectal cancer [12].
Recently, many studies have investigated the association between the hsa-miR-149 rs2292832 polymorphism and cancer risk. But the results were not conclusive and consistent. Considering the limits of the single study, we performed this meta-analysis of 12 published studies to derive a more powerful estimation of the association between the hsa-miR-149 rs2292832 polymorphism and cancer risk.
Methods
Publication search and inclusion criteria
Medical subheading (Mesh) terms: ‘microRNA’, ‘cancer’ and ‘polymorphism’ were used to search on PubMed for eligible studies (last search: April 28, 2013). The references of articles and reviews were also examined to explore potentially additional studies. Studies were eligible if they met the following criteria: (a) case-control studies; (b) investigating the association between the hsa-miR-149 rs2292832 polymorphism and cancer risk; (c) detailed genotype data for estimating of odds ratio (OR) and 95% confidence interval (CI); (d) full text articles in English. If multiple studies had overlapping or duplicate data, only those with complete data or recent studies were included.
Data extraction
Data were evaluated and extracted from the eligible studies by two investigators (Xu and Zhou) independently. The following items from each study were recorded: first author’s name, year of publication, country or area of origin, ethnicity, cancer type, source of controls, genotyping method, total number of cases and controls, genotype distributions of cases and controls, and Hardy-Winberg equilibrium (HWE), respectively. If discrepancies existed between two investigators, another investigator (Qiu) was invited to discuss and check the data until a consensus was reached.
Statistical analysis
HWE was evaluated for controls in each study by the chi-square test and a p<0.05 was considered as departure from HWE. Odds ratios (ORs) with 95% confidence intervals (CIs) were used to assess the strength of association between the hsa-miR-149 rs2292832 polymorphism and cancer risk. Pooled ORs were performed for allelic comparison (T vs. C), recessive model (TT vs. CT/CC), dominant model (TT/CT vs. CC), homozygote comparison (TT vs. CC), and heterozygote comparison (CT vs. CC), respectively. The statistic significance of pooled ORs was determined by Z-test and a p<0.05 was considered as statistically significant. A chi-square based Q-test was used to check the heterogeneity among the studies. A p<0.10 for Q-test suggested significant heterogeneity among studies, and the random-effects model (DerSimonian-Laird method) was conducted to calculate the pooled ORs [13]; Otherwise, the fixed-effects model (Mantel-Haenszel method) was used [14]. Subgroup analyses were also performed to test the effects of ethnicity, cancer type and source of controls. Sensitivity analysis was carried out to identify the effect of data from each study on pooled ORs. Begg’s funnel plot and the Egger’s linear regression test were performed to evaluate publication bias of literatures and a p<0.05 was considered significant [15]. Trim and fill method was used to assesses potential asymmetry in the funnel plot. All of the statistical tests were calculated with STATA software version 12.0 (STATA Corporation, College Station, TX, USA).
Results
Study characteristics
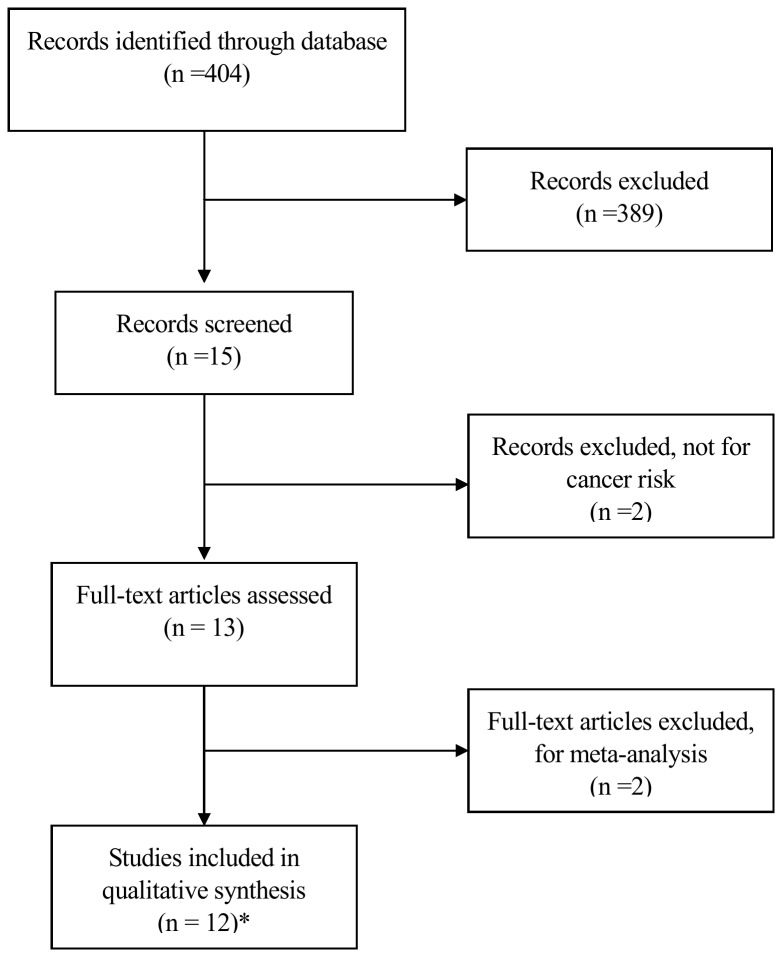
A total of 11 articles were retrieved from PubMed according to the inclusion criteria [16–26]. Figure 1 shows the detailed screening process. The study of Zhang et al. [21] presented separate OR by different cancer types (gastric cancer and colorectal cancer) and each of them was considered separately in this meta-analysis. Thus, a total of 12 studies involving 5937 cases and 6081 controls were analyzed in our meta-analysis. As shown in Table 1, 3 of 12 studies were Caucasians and the other 9 studies were Asians. Almost all cases were diagnosed histologically or pathologically. High Resolution Melting (HRM), TaqMan genotyping assay and polymerase chain reaction-restriction fragment length polymorphism (PCR–RFLP) were used as genotyping methods in 2, 1 and 9 studies respectively. All studies used blood sample for genotyping. Age and sex were matched for controls in almost all studies, of which 7 were hospital-based and 5 were population-based. The distribution of genotypes in the controls was in agreement with the HWE except the study of Chu et al. [17].
Figure 1. Flow chart of studies in the analysis.
*11 articles were retrieved and two separate studies were reported in one article, so 12 studies were eligible.
Table 1. Characteristics of eligible studies.
| Author | Year | Country | Ethnicity | Cancer type | Study design | Genotyping method | Phwe | Cases | Controls |
Cases
|
Controls
|
||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| TT | CT | CC | TT | CT | CC | ||||||||||
| Vinci[16] | 2013 | Italy | Caucasian | CRC | HB | HRM | 0.91 | 160 | 178 | 23 | 58 | 79 | 17 | 75 | 86 |
| Chu[17] | 2012 | Taiwan | Asian | OSCC | HB | TaqMan | <0.001 | 480 | 425 | 345 | 88 | 47 | 315 | 84 | 26 |
| Kim[18] | 2012 | Korea | Asian | HCC | HB | PCR–RFLP | 0.34 | 159 | 201 | 81 | 64 | 14 | 83 | 97 | 21 |
| Zhang[19] | 2012 | China | Asian | BC | PB | PCR–RFLP | 0.21 | 245 | 229 | 120 | 102 | 23 | 92 | 113 | 24 |
| Min[20] | 2012 | Korea | Asian | CRC | HB | PCR–RFLP | 0.95 | 446 | 502 | 221 | 177 | 48 | 232 | 219 | 51 |
| Zhang[21] | 2012 | China | Asian | CRC | PB | PCR–RFLP | 0.58 | 435 | 443 | 187 | 202 | 46 | 203 | 190 | 50 |
| Zhang[21] | 2012 | China | Asian | GC | PB | PCR–RFLP | 0.70 | 274 | 269 | 132 | 101 | 41 | 114 | 120 | 35 |
| Vinci[22] | 2011 | Italy | Caucasian | LC | HB | HRM | 0.97 | 101 | 129 | 16 | 41 | 44 | 11 | 53 | 65 |
| Liu[23] | 2010 | USA | Caucasian | HNSCC | HB | PCR–RFLP | 0.27 | 1109 | 1130 | 88 | 441 | 580 | 99 | 445 | 586 |
| Tian[24] | 2009 | China | Asian | LC | PB | PCR–RFLP | 0.86 | 1058 | 1035 | 463 | 472 | 123 | 470 | 453 | 112 |
| Hu[25] | 2009 | China | Asian | BC | PB | PCR–RFLP | 0.16 | 1009 | 1093 | 99 | 460 | 450 | 108 | 503 | 482 |
| Ahn[26] | 2012 | Korea | Asian | GC | HB | PCR–RFLP | 0.98 | 461 | 447 | 241 | 176 | 44 | 220 | 187 | 40 |
CRC: colorectal cancer; OSCC: oral squamous cell carcinoma; HCC: hepatocellular carcinoma; BC: breast cancer; GC: gastric cancer; LC: lung cancer; HNSCC: squamous cell carcinoma of the head and neck PB: population-based; HB: hospital-based. Phwe: Hardy-Winberg equilibrium
Meta-analysis results
The main results of this meta-analysis are listed in Table 2. We did not find any significant association between the hsa-miR-149 rs2292832 polymorphism and cancer risk in all genetic models (T versus C:OR = 1.01, 95% CI 0.95–1.06, Pheterogeneity = 0.286; TT vs. CC:OR= 0.98, 95% CI 0.86–1.11, Pheterogeneity = 0.451; CT vs. CC:OR= 0.96, 95% CI 0.86–1.05, Pheterogeneity = 0.851; TT/CT vs. CC:OR= 0.97, 95% CI 0.88–1.06, Pheterogeneity = 0.858; TT vs. CT/CC: OR= 1.05, 95% CI 0.96–1.14, Pheterogeneity = 0.118) (see Figure S1 to Figure S5 in File S1). Similarly, no significant association between the hsa-miR-149 rs2292832 polymorphism and cancer risk was found in our subgroup analysis by ethnicity (Asian and Caucasian), study design (hospital-based and population-based) or cancer type (squamous cancer, breast cancer, colorectal cancer, lung cancer and gastric cancer).
Table 2. Meta-analysis results.
| N | case/control |
T vs. C
|
TT vs. CC
|
CT vs. CC
|
TT/CT vs. CC
|
TT vs. CT/CC
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OR | Ph | OR | Ph | OR | Ph | OR | Ph | OR | Ph | |||
| Total | 12 | 5937/6081 | 1.01(0.95,1.06) | 0.286 | 0.98(0.86,1.11) | 0.451 | 0.95(0.86,1.05) | 0.851 | 0.97(0.88,1.06) | 0.858 | 1.05(0.96,1.14) | 0.118 |
| Cancer type | ||||||||||||
| SC | 2 | 1589/1555 | 0.93(0.83,1.05) | 0.218 | 0.8(0.62,1.04) | 0.191 | 0.82(0.49,1.37) | 0.07 | 0.81(0.51,1.3) | 0.066 | 0.89(0.73,1.1) | 0.978 |
| BC | 2 | 1254/1322 | 1.08(0.86,1.35) | 0.121 | 1.04(0.79,1.37) | 0.361 | 0.98(0.82,1.16) | 0.907 | 0.99(0.84,1.17) | 0.656 | 1.17(0.82,1.27) | 0.123 |
| CRC | 3 | 1041/1123 | 1.02(0.9,1.16) | 0.669 | 1.07(0.81,1.43) | 0.621 | 0.94(0.73,1.22) | 0.548 | 0.99(0.77,1.26) | 0.889 | 1.05(0.88,1.25) | 0.186 |
| LC | 2 | 1159/1164 | 1.09(0.76,1.56) | 0.073 | 1.26(0.55,2.9) | 0.058 | 0.99(0.76,1.27) | 0.562 | 0.99(0.78,1.27) | 0.238 | 1.23(0.6,2.54) | 0.071 |
| GC | 2 | 735/716 | 1.07(0.92,1.25) | 0.876 | 0.99(0.7,1.4) | 0.983 | 0.79(0.56,1.12) | 0.628 | 0.89(0.64,1.24) | 0.787 | 1.18(0.96,1.45) | 0.608 |
| Study design | ||||||||||||
| PB | 5 | 3021/3069 | 0.99(0.92,1.07) | 0.402 | 0.98(0.82,1.16) | 0.842 | 0.97(0.84,1.11) | 0.755 | 0.97(0.85,1.11) | 0.924 | 1.01(0.9,1.13) | 0.147 |
| HB | 7 | 2916/3012 | 1.02(0.94,1.11) | 0.182 | 0.98(0.82,1.18) | 0.147 | 0.94(0.82,1.07) | 0.631 | 0.96(0.85,1.09) | 0.505 | 1.1(0.97,1.24) | 0.172 |
| Ethnicity | ||||||||||||
| Asian | 9 | 4567/4644 | 1.01(0.94,1.07) | 0.26 | 0.96(0.83,1.1) | 0.648 | 0.93(0.82,1.05) | 0.742 | 0.95(0.84,1.06) | 0.786 | 1.05(0.96,1.14) | 0.166 |
| Caucasian | 3 | 1370/1437 | 1.01(0.9,1.14) | 0.22 | 1.05(0.81,1.38) | 0.102 | 0.99(0.85,1.16) | 0.688 | 1(0.86,1.16) | 0.567 | 1.29(0.76,2.17) | 0.082 |
N: number of studies included; OR: odds ratio; Ph: p value for heterogeneity; SC: squamous cell carcinoma CRC, colorectal cancer; BC: breast cancer; GC: gastric cancer; LC: lung cancer; PB: population-based; HB: hospital-based
Sensitivity analysis
One single study was deleted each time from pooled results to investigate the influence of individual study on the pooled ORs. Our results showed that pooled ORs were not altered which suggested that no individual study significantly affected the pooled results.
Publication bias
We used Begg’s funnel plot and the Egger’s linear regression test to assess publication bias. Publication bias was detected (T vs. C:P = 0.047 for Begg’s test and P=0.025 for Egger’s test; TT vs. CC:P = 0.011 for Begg’s test and P=0.028 for Egger’s test; TT vs. CT/CC: P=0.024 for Begg’s test and P=0.007 for Egger’s test). Thus, a trim and fill method was used and pooled ORs were recalculated with hypothetically non-published to evaluate the asymmetry in the funnel plot (Figure 2). The recalculated ORs did not change significantly (T vs. C:OR = 0.981, 95% CI= 0.914–1.053; TT vs. CC:OR = 0.912, 95% CI= 0.814–1.022; TT vs. CT/CC: OR= 1.031, 95% CI =0.917–1.16), indicating the stability of the results.
Figure 2. Funnel plot adjusted with trim and fill method (A:T vs. C; B: TT vs. CC; C: TT vs. CT/CC ).
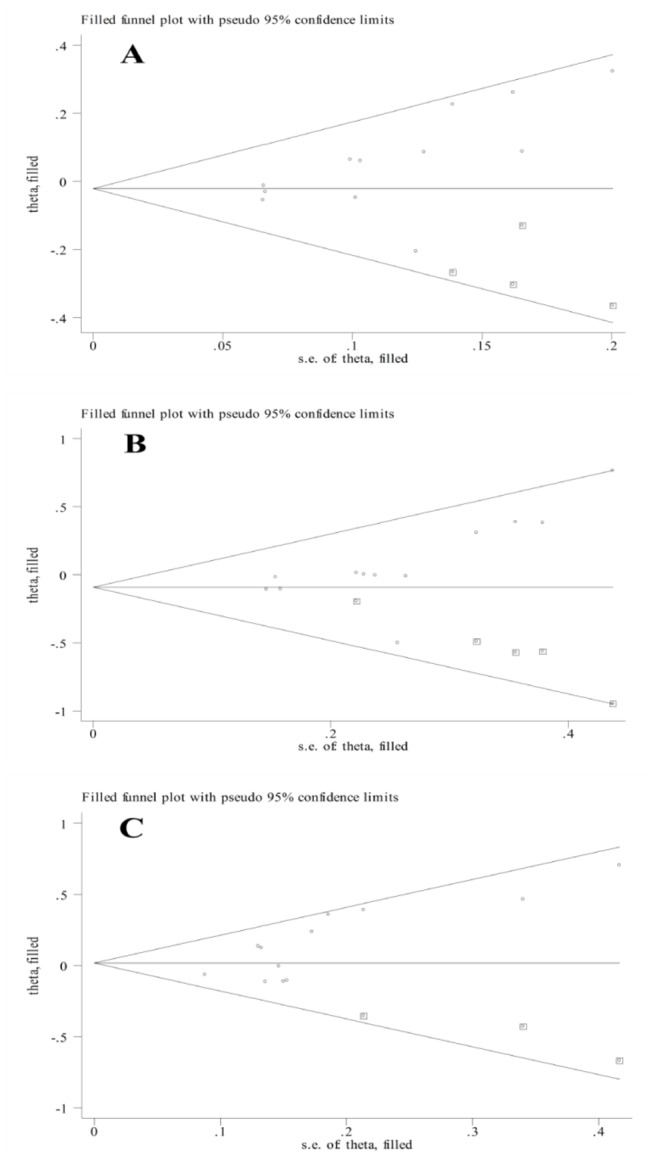
Circles: included studies. Diamonds: presumed missing studies.
Discussion
MiRNAs play important role in various cellular processes and are involved in many diseases including various cancers [2,27,28]. Genetic variations which arise in miRNA genes including their pri- and pre-miRNA regions, might affect processing and expression of miRNA such as occurs in the down regulation of mature let-7e by a G to A mutation at 19nt downstream of the pre-let-7e [29]. Recently, many studies have demonstrated the association between SNPs of miRNA gene and cancer risk. The miR-27a polymorphism (rs11671784) may decrease the expression of mature miR-27a and reduce susceptibility to gastric cancer [30]. A significantly increased risk of colorectal cancer was observed with the miR-196a2 (rs11614913) CT/CC genotype compared with the TT genotype [31].
As a tumor suppressor gene, miR-149 may inhibit proliferation and induce cell cycle arrest by targeting ZBTB2 in gastric cancer [32]. Pan et al. showed that miR-149 might be involved in the proliferation and invasion of glioma cells via blockade of AKT1 signaling [33]. Thus, alterations in miR-149 gene may contribute to cancer risk. Association between the hsa-miR-149 rs2292832 polymorphism and cancer risk has been explored in some studies. Kim et al. demonstrated that the risk of HCC was significantly lower for patients with the miR-149 CT or CT/CC genotypes [18]. A potentially protective role of C allele of hsa-miR-149 was observed in male gastric cancer [21,26]. But, no significant association between the hsa-miR-149 rs2292832 polymorphism and cancer risk was observed among other studies. As for the inconsistent results from individual studies for all types of cancers, this meta-analysis including 12 studies with 5937 cases and 6081 controls in total was built to assess the association. However, our results showed that no significant association between the hsa-miR-149 rs2292832 polymorphism and susceptibility to cancer. Similarly, subgroup analyses by cancer type, study design or ethnicity did not suggest a significantly different result. When we deleted Chu’s study [17] nonconformity to HWE in the control group in the sensitivity analysis, the pooled results did not change significantly. As publication bias was observed, we adopted trim and fill method to recalculate the adjusted ORs and did not find a different result, suggesting the stability of the statistic analysis.
Several limitations of the meta-analysis should be considered. Firstly, the studies searched on PubMed were full text in English. This may be partially responsible for the observed publication bias, though they do not change the results by using trim and fill method. Secondly, the number of studies for subgroup analysis was small that only one study investigated the association in oral squamous cell carcinoma [17], squamous cell carcinoma of the head and neck [23] and hepatocellular carcinoma [18] respectively, of which Kim’s study [18] revealed biologic significance. More studies with homogeneous cancer patients and controls were needed to confirm the results. Some covariates such as sex, age and residence area were also not available in all studies for adjusted ORs which need to be further considered, as we found the potentially protective role of C allele in male population of gastric cancer.
In conclusion, the pooled results of this meta-analysis suggested that the hsa-miR-149 rs2292832 polymorphism may not contribute to the susceptibility of all types of cancers in spite of the potentially protective role of C allele in hepatocellular carcinoma and male gastric cancer. However, our results should be considered with caution due to the observed publication bias and limitations listed above. To further confirm the results, large scale case-control studies with different ethnic groups and multiple cancer types are needed.
Supporting Information
Forest plots of the association between the hsa-miR-149 rs2292832 polymorphism and cancer risk.
(DOCX)
PRISMA checklist.
(DOC)
Funding Statement
This work was supported by Natural Science Foundation of Jiangsu Province (BK2010589, BK2011857), China. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manu script.
References
- 1. Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75: 843-854. doi:10.1016/0092-8674(93)90529-Y. PubMed: 8252621. [DOI] [PubMed] [Google Scholar]
- 2. Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297. doi:10.1016/S0092-8674(04)00045-5. PubMed: 14744438. [DOI] [PubMed] [Google Scholar]
- 3. Carrington JC, Ambros V (2003) Role of microRNAs in plant and animal development. Science 301: 336-338. doi:10.1126/science.1085242. PubMed: 12869753. [DOI] [PubMed] [Google Scholar]
- 4. Suárez Y, Sessa WC (2009) MicroRNAs as novel regulators of angiogenesis. Circ Res 104: 442-454. doi:10.1161/CIRCRESAHA.108.191270. PubMed: 19246688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hatley ME, Patrick DM, Garcia MR, Richardson JA, Bassel-Duby R et al. (2010) Modulation of K-Ras-dependent lung tumorigenesis by MicroRNA-21. Cancer Cell 18: 282-293. doi:10.1016/j.ccr.2010.08.013. PubMed: 20832755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xu P, Guo M, Hay BA (2004) MicroRNAs and the regulation of cell death. Trends Genet 20: 617-624. doi:10.1016/j.tig.2004.09.010. PubMed: 15522457. [DOI] [PubMed] [Google Scholar]
- 7. Bartel DP, Chen CZ (2004) Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet 5: 396-400. doi:10.1038/nrg1328. PubMed: 15143321. [DOI] [PubMed] [Google Scholar]
- 8. Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M (2012) microRNAs in cancer management. Lancet Oncol 13: e249-e258. doi:10.1016/S1470-2045(12)70073-6. PubMed: 22652233. [DOI] [PubMed] [Google Scholar]
- 9. Link A, Kupcinskas J, Wex T, Malfertheiner P (2012) Macro-role of microRNA in gastric cancer. Dig Dis 30: 255-267. doi:10.1159/000336919. PubMed: 22722550. [DOI] [PubMed] [Google Scholar]
- 10. Chen K, Song F, Calin GA, Wei Q, Hao X et al. (2008) Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis 29: 1306-1311. doi:10.1093/carcin/bgn116. PubMed: 18477647. [DOI] [PubMed] [Google Scholar]
- 11. Wang Y, Yang B, Ren X (2012) Hsa-miR-499 polymorphism (rs3746444) and cancer risk: a meta-analysis of 17 case-control studies. Gene 509: 267-272. doi:10.1016/j.gene.2012.08.008. PubMed: 22922391. [DOI] [PubMed] [Google Scholar]
- 12. Wang J, Wang Q, Liu H, Shao N, Tan B et al. (2012) The association of miR-146a rs2910164 and miR-196a2 rs11614913 polymorphisms with cancer risk: a meta-analysis of 32 studies. Mutagenesis 27: 779-788. doi:10.1093/mutage/ges052. PubMed: 22952151. [DOI] [PubMed] [Google Scholar]
- 13. DerSimonian R, Laird N (1986) Meta-analysis in clinical trials. Control Clin Trials 7: 177-188. doi:10.1016/0197-2456(86)90046-2. PubMed: 3802833. [DOI] [PubMed] [Google Scholar]
- 14. Mantel N, Haenszel W (1959) Statistical aspects of the analysis of data from retrospective studies of disease. J Natl Cancer Inst 22: 719-748. PubMed: 13655060. [PubMed] [Google Scholar]
- 15. Egger M, Davey Smith G, Schneider M, Minder C (1997) Bias in meta-analysis detected by a simple, graphical test. BMJ 315: 629-634. doi:10.1136/bmj.315.7109.629. PubMed: 9310563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Vinci S, Gelmini S, Mancini I, Malentacchi F, Pazzagli M et al. (2013) Genetic and epigenetic factors in regulation of microRNA in colorectal cancers. Methods 59: 138-146. doi:10.1016/j.ymeth.2012.09.002. PubMed: 22989523. [DOI] [PubMed] [Google Scholar]
- 17. Chu YH, Tzeng SL, Lin CW, Chien MH, Chen MK et al. (2012) Impacts of microRNA gene polymorphisms on the susceptibility of environmental factors leading to carcinogenesis in oral cancer. PLOS ONE 7: e39777. doi:10.1371/journal.pone.0039777. PubMed: 22761899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim WH, Min KT, Jeon YJ, Kwon CI, Ko KH et al. (2012) Association study of microRNA polymorphisms with hepatocellular carcinoma in Korean population. Gene 504: 92-97. doi:10.1016/j.gene.2012.05.014. PubMed: 22583825. [DOI] [PubMed] [Google Scholar]
- 19. Zhang M, Jin M, Yu Y, Zhang S, Wu Y et al. (2012) Associations of miRNA polymorphisms and female physiological characteristics with breast cancer risk in Chinese population. Eur J Cancer Care ( Engl) 21: 274-280 doi:10.1111/j.1365-2354.2011.01308.x. PubMed: 22074121. [DOI] [PubMed] [Google Scholar]
- 20. Min KT, Kim JW, Jeon YJ, Jang MJ, Chong SY et al. (2012) Association of the miR-146aC>G, 149C>T, 196a2C>T, and 499A>G polymorphisms with colorectal cancer in the Korean population. Mol Carcinog 51 Suppl 1: E65-E73. doi:10.1002/mc.21849. PubMed: 22161766. [DOI] [PubMed] [Google Scholar]
- 21. Zhang MW, Jin MJ, Yu YX, Zhang SC, Liu B et al. (2012) Associations of lifestyle-related factors, hsa-miR-149 and hsa-miR-605 gene polymorphisms with gastrointestinal cancer risk. Mol Carcinog 51 Suppl 1: E21-E31. doi:10.1002/mc.20863. PubMed: 21976437. [DOI] [PubMed] [Google Scholar]
- 22. Vinci S, Gelmini S, Pratesi N, Conti S, Malentacchi F et al. (2011) Genetic variants in miR-146a, miR-149, miR-196a2, miR-499 and their influence on relative expression in lung cancers. Clin Chem Lab Med 49: 2073-2080. PubMed: 21902575. [DOI] [PubMed] [Google Scholar]
- 23. Liu Z, Li G, Wei S, Niu J, El-Naggar AK et al. (2010) Genetic variants in selected pre-microRNA genes and the risk of squamous cell carcinoma of the head and neck. Cancer 116: 4753-4760. doi:10.1002/cncr.25323. PubMed: 20549817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian T, Shu Y, Chen J, Hu Z, Xu L et al. (2009) A functional genetic variant in microRNA-196a2 is associated with increased susceptibility of lung cancer in Chinese. Cancer Epidemiol Biomarkers Prev 18: 1183-1187. doi:10.1158/1055-9965.EPI-08-0814. PubMed: 19293314. [DOI] [PubMed] [Google Scholar]
- 25. Hu Z, Liang J, Wang Z, Tian T, Zhou X et al. (2009) Common genetic variants in pre-microRNAs were associated with increased risk of breast cancer in Chinese women. Hum Mutat 30: 79-84. doi:10.1002/humu.20837. PubMed: 18634034. [DOI] [PubMed] [Google Scholar]
- 26. Ahn DH, Rah H, Choi YK, Jeon YJ, Min KT et al. (2012) Association of the miR-146aC>G, miR-149T>C, miR-196a2T>C, and miR-499A>G polymorphisms with gastric cancer risk and survival in the Korean population. Mol Carcinog. PubMed: 23001871. [DOI] [PubMed] [Google Scholar]
- 27. Wang Z, Yao H, Lin S, Zhu X, Shen Z et al. (2013) Transcriptional and epigenetic regulation of human microRNAs. Cancer Lett 331: 1-10. doi:10.1016/j.canlet.2012.12.006. PubMed: 23246373. [DOI] [PubMed] [Google Scholar]
- 28. Chen PS, Su JL, Hung MC (2012) Dysregulation of microRNAs in cancer. J Biomed Sci 19: 90. doi:10.1186/1423-0127-19-90. PubMed: 23075324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wu M, Jolicoeur N, Li Z, Zhang L, Fortin Y et al. (2008) Genetic variations of microRNAs in human cancer and their effects on the expression of miRNAs. Carcinogenesis 29: 1710-1716. doi:10.1093/carcin/bgn073. PubMed: 18356149. [DOI] [PubMed] [Google Scholar]
- 30. Yang Q, Jie Z, Ye S, Li Z, Han Z et al. (2012) Genetic variations in miR-27a gene decrease mature miR-27a level and reduce gastric cancer susceptibility. Oncogene. PubMed: 23246964. [DOI] [PubMed] [Google Scholar]
- 31. Zhu L, Chu H, Gu D, Ma L, Shi D et al. (2012) A functional polymorphism in miRNA-196a2 is associated with colorectal cancer risk in a Chinese population. DNA Cell Biol 31: 350-354. doi:10.1089/dna.2011.1348. PubMed: 21815818. [DOI] [PubMed] [Google Scholar]
- 32. Wang Y, Zheng X, Zhang Z, Zhou J, Zhao G et al. (2012) MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLOS ONE 7: e41693. doi:10.1371/journal.pone.0041693. PubMed: 23144691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pan SJ, Zhan SK, Pei BG, Sun QF, Bian LG et al. (2012) MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. Int J Immunopathol Pharmacol 25: 871-881. PubMed: 23298478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Forest plots of the association between the hsa-miR-149 rs2292832 polymorphism and cancer risk.
(DOCX)
PRISMA checklist.
(DOC)