Abstract
Purpose
To identify genetic predictors of diabetes-associated ED using genome wide and candidate gene approaches in a cohort of men with type I diabetes.
Methods
We examined 528 white men with T1D (125 with ED) from the DCCT and its observational follow up EDIC Study. ED was defined from a single item of the IIEF. An Illumina Human1M BeadChip was used for genotyping. 867,125 single nucleotide polymorphisms (SNPs) were subjected to analysis. Whole genome and candidate gene approaches tested the hypothesis that genetic polymorphisms may predispose men with T1D to ED. Univariate and multivariate models were used controlling for age, HbA1c, diabetes duration, and prior randomization to intensive or conventional insulin therapy during DCCT. A stratified false discovery rate was used to perform the candidate gene approach.
Results
Two SNPs located on chromosome 3 in one genomic loci were associated with ED with p < 1×10−6. rs9810233 had a p-value of 7 × 10−7 and rs1920201 had a p-value of 9×10−7 The nearest gene to these two SNPs is ALCAM. The genetic association results at these loci were similar in univariate and multivariate analysis. No candidate genes met criteria for statistical significance.
Conclusions
Two SNPs, rs9810233 and rs1920101, which are 25 kb apart, are both associated with ED, albeit not meeting the standard GWAS significance criteria of p < 5 × 10−8. Other studies with larger sample sizes will be required to determine whether ALCAM represents a novel gene in the pathogenesis of diabetes associated ED.
MESH Key Words: Erectile Dysfunction, Diabetes, Genetics
Introduction
More than 40% of men over age 40 report erectile dysfunction (ED).1 Currently, if all men in the U.S. with ED received treatment the cost would exceed $10 billion.2 While early detection and prevention therapies have the potential to significantly decrease the costs associated with ED, little is known about the genetic modulators of ED. A twin study in middle aged male veterans that included men with diabetes found that about one third of the variance in the ED phenotype could be explained by additive genetic factors.3 The G894T polymorphism (rs1799983) in endothelial nitric oxide synthase (eNOS or NOS3)) and G-protein GNB3 C825T (rs5443) have been jointly associated with ED, hypertension and coronary artery disease but results are inconsistent.4–6 The Angiotensin Converting Enzyme (ACE) DD genotype is significantly more frequent in men with ED than those without, and Angiotensin II when injected into the corpora cavernosa in mice can induce detumescence. 4, 7
Prior studies of the genetic contributors to ED have been limited by reliance on model in-vitro systems to generate a priori hypotheses that made identification of novel pathways unlikely and the failure to control adequately for ethnicity. On the other hand, the success of Genome Wide Association Study (GWAS) studies in identifying novel biologic mechanisms for diseases such as diabetes and prostate cancer suggests a role for a whole-genome approach for deciphering the genetic underpinnings of diseases such as ED.8, 9 Consequently, the aim of this study was to identify specific genetic markers for ED in a high-risk cohort of patients with type 1 diabetes using both a whole genome approach and a candidate gene analysis.10
Materials & Methods
Overview
Our study population consisted of men enrolled in the Diabetes Control and Complications Trial (DCCT) and its observational follow up the Epidemiology of Diabetes Interventions and Complications (EDIC) study, which consists of over 1300 men and women with type 1 diabetes who have been followed since 1983.11 The DCCT (1983–93) was a randomized controlled clinical trial that examined the effects of near-normal glycemic control on the development and progression of diabetic complications. The presence of ED was self-reported at year 10 of EDIC (calendar year 2003) with a single item of the International Index of Erectile Function (IIEF)12, obtained as part of an ancillary study of urological complications of type 1 diabetes (UroEDIC).13 We performed a whole genome as well as a candidate gene approach to identify genetic predictors of ED in 528 white men with type 1 diabetes who met both phenotypic and genotypic quality control metrics.
Study Subjects
DCCT randomly assigned 761 male patients with type 1 diabetes to intensive or conventional therapy, treating them for a mean of 6.5 years between 1983 and 1993. (Figure 1) 14 The primary prevention cohort consisted of 378 male subjects with no detectable retinopathy by 7-field fundus photography, a urinary albumin excretion rate <40 mg per 24 hours, and diabetes duration 1 to 5 years at study entry. The secondary intervention cohort consisted of 383 male subjects who had mild to moderate non-proliferative retinopathy, urinary albumin excretion rate ≤200 mg per 24 hours, and diabetes duration of 1 to 15 years. Individuals were excluded from the DCCT if they had hypertension (systolic ≥ 140 or diastolic ≥ 90 mm Hg), or were taking antihypertensive medications symptomatic ischemic heart disease, or symptomatic peripheral neuropathy requiring treatment.
Figure 1.
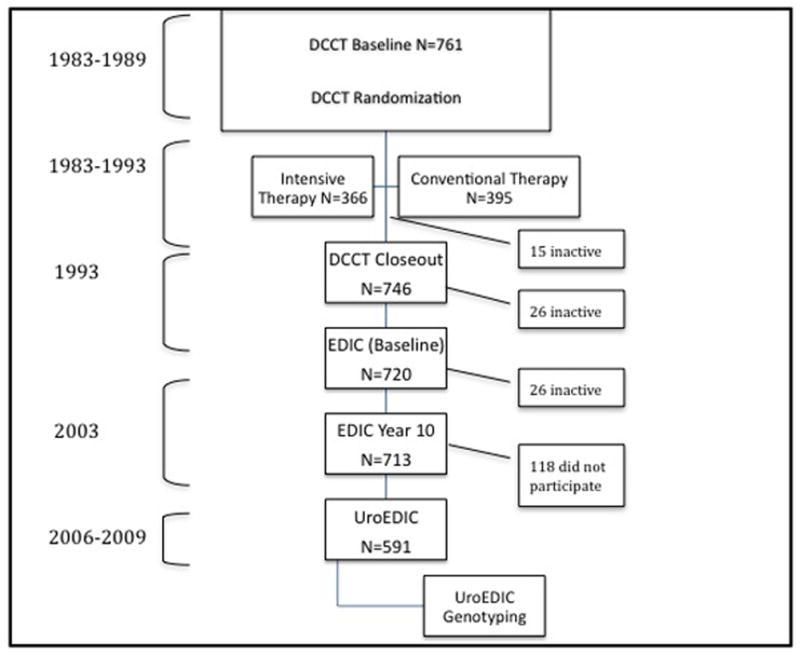
Of the 761 men enrolled, 746 completed DCCT, of whom 720 (97%) elected to participate in the first of annual examinations of the Epidemiology of Diabetes Interventions and Complications (EDIC) follow-up study in 1994. In the 10th year of EDIC, of the 713 men still active in the trial, 591 (83%) men agreed to participate in UroEDIC, an ancillary study of urological symptoms and sexual function. Of these, 571 (80%) men provided data on erectile function and comprise the study cohort for the current analyses. Figure 1 shows the flow of participants through the study. The institutional review board of each participating center approved the study, and a Certificate of Confidentiality was issued by the Federal Government.
DCCT interventions and ED measurements
Intensive glycemic therapy during DCCT consisted of insulin administered three or more times per day by injection or by continuous subcutaneous infusion with an external pump, with dose adjustments based on at least four self-monitored glucose measurements per day. Daily glucose goals were 70 to 120 mg/dl (3.9 to 6.7 mmol/l) before meals and peak levels of less than 180 mg/dl (10.0 mmol/l) after meals. The goal for HbA1c was <6.05%. Intensively treated subjects also received frequent dietary counseling. Conventional therapy consisted of one or two daily injections of insulin and had no glucose goals beyond those needed to prevent symptoms of hyperglycemia and hypoglycemia.
Participants were asked the sixth question from the Erectile Function domain of the International Index of Erectile Function (IIEF)1, 15: “Over the past 4 weeks, how would you rate your confidence that you get and keep your erection?” If the participant answered ‘Very Low’ (1) or ‘Low’ (2) they were considered to have erectile dysfunction. If they answered ‘Moderate’ (3), ’High’ (4), or ‘Very High’ (5), they were considered to have no erectile dysfunction.
Genotyping and quality control
Genotyping was performed for 571 white male DCCT/EDIC subjects with the Illumina Human1M Beadchip (Illumina Inc., San Diego, CA, USA), of which 528 met quality control measures and had ED data available. Quality control was performed according to previously established metrics from other GWAS papers from EDIC.16 Genotypes were called using BeadStudio using all individuals at once. Data from three probands was excluded because of discrepancies between reported sex and genotype data and no individuals were removed because of low genotype call rate (minimum call rate threshold was 0.988).
Genotypes from 24 duplicate samples had an agreement rate of 99.9995%. Sample contamination was assessed by calculation of the mean heterozygosity across the genome for each individual, and using a range of 0.25–0.32, none were removed. To detect cryptically related individuals and/or sample mix-ups, identical-by-state estimates between all pairs of individuals were performed, and two probands were removed. A total of 841,342 SNPs with a minor allele frequency >1% were subsequently analyzed statistically.
Autosomal SNPs showing significant association with sex (P<10−8) or deviating from the Hardy-Weinberg equilibrium (P<10−8) were excluded from the analysis. To reduce the possibility of population stratification, we limited the analysis to individuals who self-identified as white and excluded individuals who were determined to be admixed between Caucasian and other ethnic groups through population genetic approaches, seeding with genotype data from the three major populations genotyped in HapMap Phase II.17 This resulted in the exclusion of 3 patients from the study as their racial admixture was determined to be non-caucasian.
Statistical analysis
Initial descriptive analysis examined the distribution of basic demographic variables such as age, marital status, smoking status and markers of diabetes control and treatment in those with and without ED. Analyses employed hemoglobin A1c at DCCT eligibility, mean HbA1c from DCCT/EDIC) as well as the age of the participants at the time of the IIEF evaluation. The impact of medications on ED was not investigated. Population structure was assessed to ensure that all samples were of European ancestry. In our GWAS analysis we used logistic regression to examine the association of the 840,354 SNPs with ED using PLINK (v1.07) with genotypic and additive models. Two models were run for each SNP: one not adjusting for any covariates, and a multivariate model adjusting for explanatory covariates.
Based on our previous work1, which showed that the only a priori predictors of developing ED for DCCT/EDIC patients were age, HbA1c at eligibility, and and randomization to intensive glycemic therapy (in the secondary prevention cohort only), we initially began with a simple model (M1) that consisted of all the significant exposure variables from the previous study. Our next model, Model 2 (M2) included the terms of our previous model as well as other biological predictors of ED in type 1 diabetes: duration of diabetes, baseline HbA1c and mean HbA1c from DCCT/EDIC.
Assessment of the distribution of GWAS SNP p values was assessed with a Quantile-Quantile (QQ) plot derived from the unadjusted genotypic and trend models as well as the logistic regression models (M1 and M2). A Manhattan plot was also generated for the unadjusted results.
A candidate gene list was generated a priori from previously published work (PubMatrix)18, gene ontology classification and the results of microarray expression genechips that assayed gene expression in streptozotocin-induced diabetic rat cavernosa and human cavernosal endothelial cells. (Supplemental Table 1, url: https://edic.bsc.gwu.edu/).19, 20 These gene locations were used to define regions of 100 kilobases up and downstream of the location of these genes. A stratified False Discovery Rate (FDR) approach was used as previously described by Sun et al. and q-values were calculated.10
Results
UroEDIC
The UroEDIC ancillary study of DCCT/EDIC assessed male participants for ED in 2003 and found that 403 participants did not have ED and 125 were identified as having ED (Table 1). Those having ED were older (48.2 yrs vs. 43.5 yrs, p<0.001), and more likely to smoke (24.8% vs. 16.6%, p=0.040). Those with ED had higher hemoglobin A1c at DCCT baseline (9.0 vs. 8.6, p=0.016) and higher DCCT/EDIC mean hemoglobin A1c (8.6 vs. 8.0, p<0.001).
Table 1.
Demographics of erectile dysfunction in the UroEDIC genotyped cohort
| CHARACTERISTIC | No Erectile Dysfunction (n=403) | Erectile Dysfunction (n=125) | p-value |
|---|---|---|---|
| Sociodemographic Attained Age(yr)* | 43.5 (6.5) | 48.2 (5.9) | 0.00098 |
| Diabetes Treatment and Control Intervention No. (%) | 0.11 | ||
| Intensive therapy | 207 (51.3) | 54 (43.2) | |
| Conventional therapy | 196 (48.7) | 71 (56.8) | |
| Cohort No.(%) | 0.30 | ||
| Primary | 205 (50.9) | 57 (45.6) | |
| Secondary | 198 (49.1) | 68 (54.4) | |
| HemoglobinA1c at DCCT baseline (%) | 8.6 (1.5) | 9.0 (1.5) | 0.016 |
| DCCT/EDIC mean Hemoglobin A1c | 8.0 (1.3) | 8.6 (1.4) | 0.00076 |
| Insulin dose (units/kg/day)* | 0.66 (0.26) | 0.63 (0.23) | 0.40 |
Assessed at EDIC year 10. All data are expressed as mean (SD) unless noted otherwise. P-values calculated based on chi-square for differences in proportions and linear regression for continuous measures.
Multiple models were generated based on previous work and tested for trend and genotypic models, M1 and M2 (Table 2,3). The univariate trend model had the most statistically significant values.
Table 2.
Adjusted logistic regression modeling of risk factors related to erectile dysfunction in the UroEDIC cohort
| Factors | OR (95% CI) | SE | p-value† |
|---|---|---|---|
| Primary Prevention Cohort* (intensive vs. conventional) | 1.20 (0.64, 2.24) | .38 | 0.57 |
| Secondary Intervention Cohort* (intensive vs. conventional) | 0.33 (0.17, 0.61) | .10 | <0.001 |
| Hemoglobin A1c at Eligibility (per HbA1c%) | 1.30 (1.12, 1.50) | .14 | <0.001 |
| Diabetes Duration (per month) | 1.00 (0.99, 1.01) | .0031 | 0.65 |
| Age (per year) | 1.13 (1.09, 1.17) | .021 | <0.001 |
| Primary v. Secondary Cohort | 1.10 (0.60, 2.02) | .34 | 0.75 |
Logistic regression adjusted for DCCT intervention, HbA1c at eligibility, diabetes duration, age, primary vs. secondary cohort, interaction term for prevention cohort status and intensive vs. conventional glycemic control.
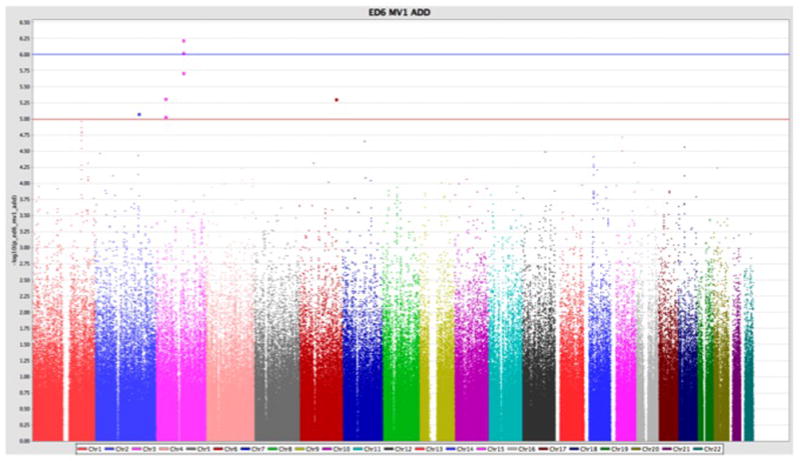
Both the unadjusted trend model and the M1 model implicated a novel SNP associated with ED pathogenesis in men with type 1 diabetes, in or near Activated Leukocyte Cell Adhesion Molecule (ALCAM) on chromosome 3 (Table 3). The G allele of rs1920201 with minor allele frequency (MAF)=0.245 was associated with increased risk of ED in the univariate trend model (p=8.6e-07). Similar borderline significant results were obtained for rs9810233, ~25 kb away, (p=6.98e-07). Further, the G risk allele was also implicated in the multivariate model for SNPs rs1920201 (OR=2.26, 95% CI 1.63–3.14, p=9.4 × 10−7 and SNP rs9810233 (OR=2.32, 95% CI 1.67–3.22, p=5.9 × 10−7). Although no SNPs achieved genome-wide significance (p <1×10−8), the Manhattan Plot (Figure 2) revealed several signals of interest, such as ALCAM.
Table 3.
Unadjusted and multivariate adjusted logistic regression of SNPs with erectile dysfunction for P<10−6
| A. Unadjusted Results
| ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Chromosome | SNP | Position | A1 | A2 | MAF** | Missing count | Hardy-Weinberg equilibrium P | Location of SNP to the nearest gene | Nearest Gene | p-value* |
| 3 | rs9810233 | 106493580 | G | A | 0.270 | 1 | 0.725 | flanking_5UTR | ALCAM | 6.98E-07 |
| 3 | rs1920201 | 106518787 | G | T | 0.245 | 5 | 0.653 | flanking_5UTR | ALCAM | 8.60E-07 |
| 2 | rs836589 | 173201324 | A | G | 0.186 | 8 | 0.521 | flanking_3UTR | PDK1 | 1.63E-06 |
| 3 | rs1438552 | 106545285 | G | A | 0.247 | 2 | 0.603 | flanking_5UTR | ALCAM | 1.72E-06 |
| 6 | rs6931865 | 144121547 | A | G | 0.336 | 1 | 0.901 | intron | PHACTR2 | 3.57E-06 |
| 3 | rs950146 | 36655103 | C | A | 0.491 | 5 | 0.698 | flanking_3UTR | STAC | 4.32E-06 |
| 3 | rs9850224 | 142057977 | A | G | 0.010 | 1 | 1 | flanking_5UTR | SLC25A36 | 4.80E-06 |
| 1 | rs2990510 | 195287281 | G | T | 0.305 | 0 | 0.556 | intron | F13B | 5.89E-06 |
| 1 | rs10754210 | 195278734 | A | G | 0.304 | 1 | 0.513 | intron | F13B | 5.89E-06 |
| 2 | rs7566884 | 173189446 | G | A | 0.177 | 1 | 0.393 | flanking_3UTR | PDK1 | 6.39E-06 |
| 1 | rs4520444 | 195408611 | T | G | 0.296 | 5 | 0.842 | intron | ZBTB41 | 7.77E-06 |
| 1 | rs5998 | 195276421 | G | A | 0.454 | 0 | 0.696 | coding | F13B | 8.30E-06 |
| 1 | rs7518773 | 195278259 | G | A | 0.453 | 2 | 0.615 | intron | F13B | 8.30E-06 |
| 1 | rs1332668 | 195272359 | T | C | 0.455 | 1 | 0.655 | flanking_3UTR | F13B | 8.50E-06 |
| B. Multivariate
Results*
| ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chrom | SNP | Position | A1 | A2 | MAF# | missing | Hardy- Weinberg equilibrium P | Location of SNP to nearest gene | Nearest Gene | Model | OR | 95% CI | p-value | |
| 3 | rs9810233 | 106493580 | G | A | 0.270 | 1 | 0.725 | flanking_5UTR | ALCAM | M1 | 2.32 | 1.67 | 3.22 | 5.93E-07 |
| 3 | rs1920201 | 106518787 | G | T | 0.245 | 5 | 0.653 | flanking_5UTR | ALCAM | M1 | 2.26 | 1.63 | 3.14 | 9.39E-07 |
| 6 | rs6931865 | 144121547 | A | G | 0.336 | 1 | 0.901 | intron | PHACTR2 | M1 | 2.02 | 1.50 | 2.74 | 4.85E-06 |
| 3 | rs950146 | 36655103 | C | A | 0.491 | 5 | 0.698 | flanking_3UTR | STAC | M1 | 0.48 | 0.35 | 0.66 | 4.77E-06 |
| 2 | rs836589 | 173201324 | A | G | 0.186 | 8 | 0.521 | flanking_3UTR | PDK1 | M1 | 2.27 | 1.58 | 3.25 | 8.29E-06 |
| 3 | rs950146 | 36655103 | C | A | 0.491 | 5 | 0.698 | flanking_3UTR | STAC | M2 | 0.46 | 0.33 | 0.64 | 6.52E-06 |
| 6 | rs6931865 | 144121547 | A | G | 0.336 | 1 | 0.901 | intron | PHACTR2 | M2 | 2.48 | 1.73 | 3.54 | 6.89E-07 |
Fishers exact test.
Minor Allele Frequency A1 is the minor allele.
All models run with additive tests for significance in PLINK with covariates.
Adjusting for cohort, treatment, cohort*treatment. (M1)
Adjusting for cohort, treatment, cohort*treatment, diabetes duration, age, initial HgbA1c at eligibility. (M2)
Minor Allele Frequency
Figure 2.
We also performed a candidate gene approach according to the a priori derived candidate genes highlighted in supplemental Table 1 (url: https://edic.bsc.gwu.edu/). There were no significant results (Table 4).
Table 4.
Candidate Gene Results for Multivariate M2* model with Q Scores
| Candidate Gene | Chrom | SNP | Position | A1 | A2 | MAF** | Function | p-value | Q-Score |
|---|---|---|---|---|---|---|---|---|---|
| S100 Ca2-binding protein A4 | 1 | rs2274739 | 151878757 | C | T | 0.453 | intron(dbSNP) | 0.0065 | 0.6837 |
| S100 Ca2-binding protein A4 | 1 | rs12564925 | 151880169 | G | A | 0.454 | intron(dbSNP) | 0.0081 | 0.6837 |
| Ceruloplasmin | 3 | rs7634024 | 150475920 | G | A | 0.102 | intergenic(GVS) | 0.0131 | 0.6837 |
| Ceruloplasmin | 3 | rs13098532 | 150373682 | G | T | 0.012 | mrna-utr(dbSNP) | 0.0157 | 0.6837 |
| Lysyl Oxidase | 5 | rs890750 | 121463478 | C | A | 0.197 | coding- nonsynonymous (dbSNP) | 0.0435 | 0.9756 |
| Lysyl Oxidase | 5 | rs2731659 | 121498469 | A | G | 0.204 | intron(dbSNP) | 0.0524 | 1.0000 |
| eNOS/CD36 | 7 | rs1404313 | 79992399 | G | A | 0.429 | intergenic(GVS) | 0.0065 | 0.6837 |
| eNOS/CD36 | 7 | rs2366735 | 79994250 | G | A | 0.422 | intergenic(GVS) | 0.0066 | 0.6837 |
| Lipoprotein Lipase | 8 | rs2410622 | 19899053 | T | C | 0.155 | intergenic(GVS) | 0.0294 | 0.9027 |
| Lipoprotein Lipase | 8 | rs1561747 | 19777848 | G | T | 0.435 | intergenic(GVS) | 0.0401 | 0.9756 |
| Kruppel-like factor 5 | 13 | rs11544631 | 72437467 | G | A | 0.026 | coding-nonsynonymous (dbSNP) | 0.0237 | 0.8174 |
| Kruppel-like factor 5 | 13 | rs9530145 | 72543301 | T | C | 0.154 | intron(dbSNP) | 0.0334 | 0.9558 |
| Gremlin 1 | 15 | rs3743103 | 30812919 | G | A | 0.408 | mrna-utr(dbSNP) | 0.0398 | 0.9756 |
| Gremlin 1 | 15 | rs1258749 | 30873773 | C | T | 0.173 | intron(dbSNP) | 0.0421 | 0.9756 |
| ACE | 17 | rs17687734 | 58838777 | T | C | 0.124 | intron(dbSNP) | 0.0307 | 0.9027 |
| ACE | 17 | rs4968772 | 58835351 | G | A | 0.254 | intron(dbSNP) | 0.0778 | 1.0000 |
| O-linked GlcNAc transferase | 23 | rs5937104 | 70789692 | NA | NA | 0.139 | intron(GVS) | 0.0746 | 1.0000 |
| O-linked GlcNAc transferase | 23 | rs3736670 | 70693455 | NA | NA | 0.114 | intron(dbSNP) | 0.0932 | 1.0000 |
All models run with additive tests for significance in PLINK with covariates. Adjusting for cohort, treatment, cohort*treatment, diabetes duration, age, initial HgbA1c at eligibility. (M2)
Minor Allele Frequency
Discussion
The 528 men with type 1diabetes in UroEDIC included in this study represent a well-defined cohort that allowed us to rigorously adjust for non-genetic risk factors for ED such as age and diabetes control. Moreover, the exclusion of all non-whites allowed us to disregard the impact of ethnicity. 14, 21 Despite these advantages, the results of this small exploratory analysis with only 125 cases failed to reveal any robust new genetic associations with diabetic ED, although at least one SNP in linkage disequilibrium with ALCAM may be considered for further exploration in larger cohorts.
There were suggestive findings implicating the G allele of rs1920201 and rs9810233. Previous work in a cohort of 480 Danish families suggested that genetic variation in the exons of ALCAM may be involved in a mechanism whereby cytotoxic T lymphocyte antigen 4 modulates gene-gene interactions and confers genetic susceptibility of type 1 diabetes.22 The magnitude of the association for SNPs near ALCAM in our study was much larger than that typically found in GWAS studies for complex traits. This is suggestive of either involvement of these SNPs with diabetic ED or co-segregation with other factors that modulate ED in this region.
While the sample size in the current study was larger than in many previous investigations, a major limitation is its small size. Another potential source of error in the study is the variability in the ED measurements. The IIEF, a validated clinical instrument, is nevertheless a subjective form of reporting. Unfortunately, there are no validated quantitative clinical tests for ED and some subjects might have been unwilling or embarrassed to report ED, even though questionnaires were identified only by a study ID. Finally, the relatively young age and well controlled HbA1cs of this population may have led to incomplete penetrance of ED at the time of the study.
We propose that the IIEF Erectile Function Domain or at the very least a single item question assessing ED be added into large cohort studies employing GWAS methodology such as the Wellcome Trust Sanger studies. A working group under the direction of the AUA or NIDDK could be formed to investigate genetics of benign urologic diseases such as ED through collaborative efforts to obtain the requisite sample sizes of 10–15,000 subjects needed to make definitive conclusions
Conclusion
We performed a novel GWAS and candidate gene study for genetic predictors of ED in men with type I diabetes and identified 2 SNPs near ALCAM of potential interest. These SNPs should be explored in other cohorts to increase the sample size to validate findings of this pilot study.
Supplementary Material
Acknowledgments
Funding:
NIH RO1 5R01DK083927-03
NIH T-32 DK007779-09
The DCCT/EDIC project is supported by contracts with the Division of Diabetes, Endocrinology, and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Diseases; the National Eye Institute; the National Institute of Neurological Disorders and Stroke; the General Clinical Research Centers Program and the Clinical and Translational Science Awards Program, National Center for Research Resources; and by Genentech through a Cooperative Research and Development Agreement with the National Institute of Diabetes and Digestive and Kidney Diseases.
References
- 1.Saigal CS, Wessells H, Pace J, et al. Predictors and prevalence of erectile dysfunction in a racially diverse population. Arch Intern Med. 2006;166:207. doi: 10.1001/archinte.166.2.207. [DOI] [PubMed] [Google Scholar]
- 2.Brotons FB, Campos JC, Gonzalez-Correales R, et al. Core document on erectile dysfunction: key aspects in the care of a patient with erectile dysfunction. Int J Impot Res. 2004;16 (Suppl 2):S26. doi: 10.1038/sj.ijir.3901240. [DOI] [PubMed] [Google Scholar]
- 3.Fischer ME, Vitek ME, Hedeker D, et al. A twin study of erectile dysfunction. Arch Intern Med. 2004;164:165. doi: 10.1001/archinte.164.2.165. [DOI] [PubMed] [Google Scholar]
- 4.Park JK, Kim W, Kim SW, et al. Gene-polymorphisms of angiotensin converting enzyme and endothelial nitric oxide synthase in patients with erectile dysfunction. Int J Impot Res. 1999;11:273. doi: 10.1038/sj.ijir.3900437. [DOI] [PubMed] [Google Scholar]
- 5.Brand E, Wang JG, Herrmann SM, et al. An epidemiological study of blood pressure and metabolic phenotypes in relation to the Gbeta3 C825T polymorphism. J Hypertens. 2003;21:729. doi: 10.1097/00004872-200304000-00016. [DOI] [PubMed] [Google Scholar]
- 6.Eisenhardt A, Sperling H, Hauck E, et al. ACE gene I/D and NOS3 G894T polymorphisms and response to sildenafil in men with erectile dysfunction. Urology. 2003;62:152. doi: 10.1016/s0090-4295(03)00137-7. [DOI] [PubMed] [Google Scholar]
- 7.Becker AJ, Uckert S, Stief CG, et al. Possible role of bradykinin and angiotensin II in the regulation of penile erection and detumescence. Urology. 2001;57:193. doi: 10.1016/s0090-4295(00)00881-5. [DOI] [PubMed] [Google Scholar]
- 8.Mohlke KL, Boehnke M, Abecasis GR. Metabolic and cardiovascular traits: an abundance of recently identified common genetic variants. Hum Mol Genet. 2008;17:R102. doi: 10.1093/hmg/ddn275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Easton DF, Eeles RA. Genome-wide association studies in cancer. Hum Mol Genet. 2008;17:R109. doi: 10.1093/hmg/ddn287. [DOI] [PubMed] [Google Scholar]
- 10.Sun L, Craiu RV, Paterson AD, et al. Stratified false discovery control for large-scale hypothesis testing with application to genome-wide association studies. Genet Epidemiol. 2006;30:519. doi: 10.1002/gepi.20164. [DOI] [PubMed] [Google Scholar]
- 11.The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. The Diabetes Control and Complications Trial Research Group. The New England journal of medicine. 1993;329:977. doi: 10.1056/NEJM199309303291401. [DOI] [PubMed] [Google Scholar]
- 12.Rosen RC, Riley A, Wagner G, et al. The international index of erectile function (IIEF): a multidimensional scale for assessment of erectile dysfunction. Urology. 1997;49:822. doi: 10.1016/s0090-4295(97)00238-0. [DOI] [PubMed] [Google Scholar]
- 13.Wessells H, Penson DF, Cleary P, et al. Effect of intensive glycemic therapy on erectile function in men with type 1 diabetes. The Journal of urology. 2011;185:1828. doi: 10.1016/j.juro.2010.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy. the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA. 2003;290:2159. doi: 10.1001/jama.290.16.2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wessells H, Penson DF, Cleary P, et al. Effect of Intensive Glycemic Therapy on Erectile Function in Men With Type 1 Diabetes. J Urol. 2011 doi: 10.1016/j.juro.2010.12.098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paterson AD, Waggott D, Boright AP, et al. A genome-wide association study identifies a novel major locus for glycemic control in type 1 diabetes, as measured by both A1C and glucose. Diabetes. 2010;59:539. doi: 10.2337/db09-0653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frazer KA, Ballinger DG, Cox DR, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Becker KG, Hosack DA, Dennis G, Jr, et al. BMC bioinformatics. 2003;4:61. doi: 10.1186/1471-2105-4-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan CJ, Teal TH, Luttrell IP, et al. Microarray analysis reveals novel gene expression changes associated with erectile dysfunction in diabetic rats. Physiol Genomics. 2005;23:192. doi: 10.1152/physiolgenomics.00112.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wessells H, Sullivan CJ, Tsubota Y, et al. Transcriptional profiling of human cavernosal endothelial cells reveals distinctive cell adhesion phenotype and role for claudin 11 in vascular barrier function. Physiol Genomics. 2009;39:100. doi: 10.1152/physiolgenomics.90354.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerns SL, Ostrer H, Stock R, et al. Genome-wide association study to identify single nucleotide polymorphisms (SNPs) associated with the development of erectile dysfunction in African-American men after radiotherapy for prostate cancer. International journal of radiation oncology, biology, physics. 2010;78:1292. doi: 10.1016/j.ijrobp.2010.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yokoi N, Komeda K, Wang HY, et al. Cblb is a major susceptibility gene for rat type 1 diabetes mellitus. Nat Genet. 2002;31:391. doi: 10.1038/ng927. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


