Abstract
Sirtuins play an essential role in the cellular response to environmental stress, promoting DNA repair, telomere stability, cell cycle arrest, cellular senescence, and apoptosis. Much attention has been given to the role of sirtuins in aging and cancer development; however, less is known about their role in stem cell regulation. This review focuses in this topic and discusses the possible implications in adult stem cell aging.
Keywords: sirtuins, stem cells, development, aging
Introduction
Epigenetic mechanisms are essential elements for the regulation of cellular differentiation and the maintenance of cell type–specific gene expression patterns. They manipulate gene expression directly through modification of DNA (DNA methylation) or indirectly via modification of chromatin. Chromatin functionality and structure are tightly linked to covalent modification in histones. These modifications are generally classified as repressing or activating, correlating with gene silencing and gene induction, respectively. Among them, histone acetylation is associated with gene expression and is regulated by the action of histone acetyltransferases (HATs) and histone deacetylases (HDACs). Thus, the balanced recruitment of HATs and HDACs to targeted loci is an essential determinant of chromatin functionality.
HDACs are grouped into classes I, II, and III based on homology with their yeast orthologs Rdp3, HdaI, and Sir2, respectively, and class IV, which has only 1 member (HDAC11).1,2 The silent information regulator (SIR) family of proteins was first described in Saccharomyces cerevisiae.3 In yeast, these proteins exhibit deacetylase activity and are involved in cell cycle regulation, DNA repair, and chromatin silencing.4 The mammalian homologs of SIR genes are the sirtuins (class III HDACs), a family of proteins composed of 7 members, SIRT1 to SIRT7, with a NAD-dependent protein deacetylase activity coupled with the ability to form O-acetyl-ADP-ribose.5 These enzymes are involved in a broad range of biological functions that includes the regulation of chromatin structure and gene expression, metabolic homeostasis, apoptosis, senescence, DNA repair, and cell differentiation.6-8 In addition, sirtuins are sensitive to environmental stimuli and thus act as stress sensors that help to organize the stress response in the cell.9 In order to achieve this, sirtuins have evolved to have a wide range of protein targets and cellular locations, including the nucleus, cytoplasm, and mitochondria, and to translocate from one compartment to another in response to certain stress conditions.
Because sirtuins act as sensors of environmental stimuli and coordinate the stress response of cells, it is not surprising that deregulation of these proteins is associated with cancer.9,10 Also, some sirtuins seem to be involved in aging and cell senescence,7,11-15 although some of the experimental models reported are controversial. Here, we will review the evidence linking sirtuins to development and cell differentiation and will also discuss how these processes could be related to “stemness” and aging.
Sirtuins and Their Targets
As mentioned above, sirtuins are involved in many cellular functions and can target a wide array of proteins, both histone and nonhistone. The closest homolog to the yeast Sir2 gene, and the most widely studied, is SIRT1. Many biological functions have been associated with it, although its main function seems to be linked to gene silencing through heterochromatin formation. SIRT1 preferentially deacetylates lysine 6 of histone H4 (H4K6) and lysines 9 and 56 of histone H3 (H3K9 and H3K56), promoting the formation of facultative heterochromatin.16,17 Additionally, SIRT1 can acetylate and recruit histone H1 to the chromatin, increasing local compaction.18 In addition, it can deacetylate other nonhistone targets that also contribute to heterochromatin formation, such as the histone methyltransferase Suv39h1, thereby promoting dimethylation and trimethylation of lysine 9 in histone H319 and contributing to heterochromatin formation as the transition from acetylation to methylation in H3K9 spreads over gene-coding regions. Besides its role in gene expression, SIRT1 contributes to cellular homeostasis in response to stress. For instance, in response to genotoxic or oxidative stress, SIRT1 can acetylate proteins such as FOXO, p53, and the transcription factor NF-κB, all involved in cell cycle progression, DNA repair, and apoptosis.20-24
SIRT2 is a cytoplasmic tubulin deacetylase that can localize in the nucleus during G2/M transition. It is a protein that has been well conserved throughout evolution, and its main function is related to cell cycle regulation. In vivo, it can deacetylate microtubules and chromatin.25-28 Acetylation in α-tubulin was reported to stabilize microtubules, although SIRT2−/− knockout mice do not show clear defects in microtubule organization, and consequently, the role of SIRT2 in microtubule function and organization remains unclear.29 In chromatin, SIRT2 deacetylates H4K16 at the global level to increase chromatin compaction during G2/M transition30; the molecular mechanism that drives this process is, however, unknown.
SIRT3 is involved in metabolism and mitochondrial function.31-33 It is primarily located in the mitochondria but can be translocated to the nucleus in response to genotoxic stress and calorie restriction.34 In the nucleus, it can deacetylate H4K16 and H3K9, although specific loci have not been described so far. In the mitochondria, SIRT3 deacetylates and activates acetyl-CoA synthetase 2 (AceCS2), increasing the metabolic rate in the organelle.35,36 It can also increase the expression of mitochondrial factors such as ATP synthetase, cytochrome C oxidase subunits, and the transcription factor PGC1α.37 Additionally, SIRT3 can promote cell survival thorough deacetylation of Ku70, involved in DNA repair, in response to genotoxic agents.38,39
SIRT6 function has been related to genomic stability, DNA repair, and gene silencing.15,40 It can deacetylate histones in H3K9Ac, a residue located in the histone tail, and H3K56, which is in the core of histone H3, in gene promoters and telomere chromatin, contributing to heterochromatin formation and telomere stability.40-42
Not much is known about the remaining sirtuins: SIRT4 is a mitochondrial sirtuin without deacetylase activity and is apparently involved in the insulin metabolism of pancreatic β cells.43 SIRT5 is another mitochondrial sirtuin that targets cytochrome C and carbamoyl phosphate synthetase 1.44,45 Interestingly, it has been observed, both in vitro and in vivo, that SIRT5 has a very strong demalonylase and desuccinylase activity, which suggests additional functions beyond lysine deacetylation.46 Finally, SIRT7 is a protein localized in the nucleolus that interacts with and activates RNA polymerase I but does not deacetylate Pol I, and its molecular mechanism is currently unknown.47
Developmental Role of Sirtuins
Sirtuins have been linked to development and cell differentiation through 2 different mechanisms: 1) gene expression modulation through histone deacetylation in targeted loci and 2) deacetylation of nonhistone proteins involved in differentiation.
The role of sirtuins in development is not surprising given the important role of histone acetylation in the regulation of gene expression. Indeed, histone acetylation is an important determinant of cell fate, and it undergoes dynamic changes starting at the early preimplantation development stage. Just after fertilization, protamines in the male pronucleus are exchanged by highly acetylated histones.48 Immediately, some of the acetylated residues are substituted by monomethyl groups such as H3K4me1, H3K9me1, and H3K27me1, a process that is in part mediated by HDACs.49 After syngamy and during the cleavage stages, global histone acetylation dynamics have been observed, although it is not clear how this is correlated with chromatin function.50 During this period, until the blastocyst stage, the principal deacetylase has been found to be HDAC1. Nonetheless, HDAC1 knockdown in mouse embryos does not affect the global transcription rate, indicating that the role of HDACs in early preimplantation embryos is probably restricted to specific loci.51 Moreover, no developmental effect associated with sirtuins has been described during preimplantation, and all sirtuin knockout mice reported on to date pass through early embryonic stages.15,33,43,46,52-56
In embryonic stem (ES) cells, derived from the inner mass of the blastocyst, the levels of histone acetylation appear to be higher than in lineage-restricted stem cells or differentiated cells, in accordance with the higher levels of transcription observed in these cells.57 Thus, histone deacetylation plays an important role in tissue-specific gene silencing during cell differentiation. Of all sirtuins, SIRT1 seems to be the most clearly involved in this process since SIRT1 knockout mice die during fetal development or soon after birth and show severe developmental defects that include growth impairment, defects in the retina and heart, and abnormal cephalic development.52-54 In ES cells, SIRT1 is highly expressed but is quickly downregulated upon differentiation by a mechanism mediated, at least partially, by miRNA.58-60 Interestingly, SIRT1 disruption does not affect cell proliferation and does not induce spontaneous differentiation under normal conditions, suggesting that this protein is not an essential player in stemness maintenance.58 However, under mild oxidative conditions, SIRT1 mediates dependent apoptosis by blocking the nuclear translocation of p53 and promoting mitochondrial translocation instead.61 Since nuclear translocation of p53 suppresses Nanog expression, which in turn is required to maintain stemness in ES cells, SIRT1 may play an indirect role in preventing differentiation under certain stress conditions. SIRT1 is also involved in the establishment of development programs upon differentiation of ES cells. Chip-on-chip analysis has revealed that, in undifferentiated ES cells, SIRT1 binds to the promoter region of many genes linked to the differentiation and developmental processes, such as PAX6, WNT6, BMP1, and HOXA5.58 Additionally, SIRT1 is a component of Polycomb repressive complex 4 (PRC4), which is specific to ES cells.62 PRC4 adds methyl groups to lysine 27 of histone H3 (H3K27), a histone mark associated with the repression of developmental genes, and to lysine 26 in histone H1 (H1K26). Interestingly, H1K26 is a known target of SIRT1,18 suggesting that histone deacetylation in H1 is required before methylation by PRC4 can proceed. Taken together, these data suggest that SIRT1 may contribute to gene silencing of developmental genes in undifferentiated cells and that SIRT1 downregulation during differentiation may contribute to the epigenetic reactivation of these genes.
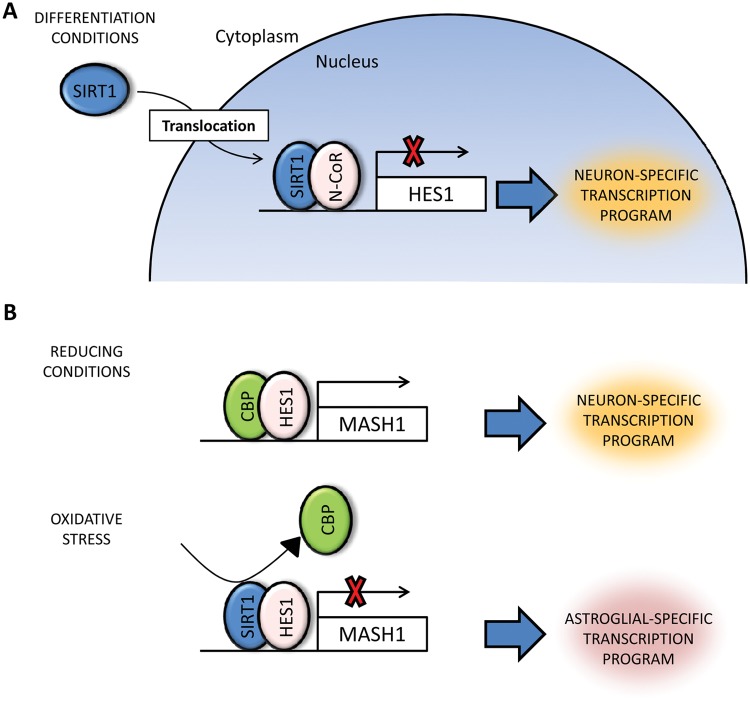
Beyond its role in gene silencing and pluripotency maintenance in ES cells, SIRT1 has been also associated with cell differentiation. The first evidence came from the finding that SIRT1 knockout mice show severe encephalic and retinal defects, indicating SIRT1’s putative role in ectodermal patterning.52-54 During the initial steps in neural specification in the embryo, SIRT1 downregulation may be required to allow the expression of key transcription factors, such as PAX6,58 although a direct role in this process has not been reported so far. Furthermore, there is strong evidence that links SIRT1 with the differentiation process of neural stem cells into astrocytes and neurons. It has been observed that SIRT1 is principally located in the cytoplasm in neural progenitors.63 When these cells are cultivated under differentiation conditions, SIRT1 quickly migrates to the nucleus, where it interacts with the nuclear receptor co-repressor (N-CoR), promoting neuronal differentiation63 (Fig. 1). However, under oxidative stress conditions, SIRT1 interacts with Hairy and enhancer of split (Hes1), inducing astroglial differentiation and inhibiting neuronal differentiation64 (Fig. 1). Thus, in such situations, SIRT1 may promote astroglial differentiation in order to facilitate astrogliosis and healing, which results in scarring at the damaged site, which is commonly observed in brain and spinal cord injuries. Interestingly, other differentiation pathways are also affected by SIRT1 in stress conditions. Under fasting conditions, SIRT1 inhibits myogenin and myosin heavy chain genes in myocytes through deacetylation of PCAF and MyoD, inhibiting muscle differentiation.65 Under the same conditions, SIRT1 also interacts with N-CoR, suppressing adipogenesis via downregulation of PPAR-γ.66 Finally, SIRT1 has been linked to normal hematopoiesis, although the current data are somewhat controversial. It has been observed that differentiation of human ES cells towards hematoendothelial phenotypes is severely affected by SIRT1 disruption.67 Additionally, it was recently reported that fetal liver hematopoietic progenitors isolated from SIRT1−/− mice have a significantly lower hematopoietic potential than normal progenitors, suggesting that SIRT1 may be necessary to maintain stem cell pools.68 In a recent report, however, no adverse effect on the adult hematopoietic stem cell pool was observed when bone marrow–derived cells from SIRT1−/− mice were transplanted into irradiated animals.69 Thus, the role of SIRT1 during fetal and adult hematopoiesis remains unclear, although it is unlikely that SIRT1 is essential in this process since knockout mice display an almost normal hemogram result and are not immunocompromised, at least under normal conditions.69
Figure 1.
SIRT1 during neurogenesis. SIRT1 disruption alters the differentiation potential of neural progenitor cells (NPCs). Two different models have been proposed. (A) In NPCs under differentiation conditions, SIRT1 transiently translocates to the nucleus, where it interacts with N-CoR. The N-CoR/SIRT1 complex binds to the promoter region of the Hes1 gene, a downstream target of the Notch signaling pathway, promoting neuronal differentiation. (B) Differentiation potential of NPCs is affected by the redox status via SIRT1 activation. In a reducing environment, Hes1 recruits transcription activators such as CREB binding protein (CBP) to the Mash1 promoter, a key transcription factor during neuronal differentiation. Under oxidizing conditions, Hes1 recruits SIRT1 to the Mash1 promoter, inducing histone deacetylation. This represses the transcription of Mash1 and promotes astroglial differentiation.
SIRT2 is also clearly associated with neural and fat differentiation and has been shown to function as an α-tubulin deacetylase and key regulator of cell division.25-28 It is expressed in all tissues but is particularly abundant in the brain, especially in myelin sheaths and in mature and premyelinating oligodendrocytes.70 During the differentiation of glial progenitors into premyelinating oligodendrocytes, both levels of SIRT2 and microtubule acetylation increase, which appears to be contradictory since SIRT2 is the major microtubule deacetylase in oligodendrocytes.71 Nonetheless, SIRT2 does seem to be involved in the differentiation process since siRNA silencing reduces tubulin acetylation and enhances the morphological differentiation of oligodendrocyte precursors, while overexpression suppresses oligodendroglial differentiation.71 Thus, the role of SIRT2 in this process is not clear, although it may be required to counterbalance microtubule acetylation during differentiation in order to keep morphological changes under control. In addition to its role in oligodendroglial differentiation, SIRT2, like SIRT1, is a suppressor of adipogenesis under fasting conditions, although SIRT2-dependent suppression is mediated by FOXO1 deacetylation, which in turn represses PPAR-γ expression.72,73 Finally, it has been reported that SIRT2 is a regulator of NF-κB, through deacetylation of p65.74 Since NF-κB is a major regulator of hematopoiesis,75 it is likely that SIRT2 is also involved in this process; this possible role has nevertheless not been reported to date.
Less is known about the other sirtuins during development and cell differentiation. Knockout mice have been reported for SIRT2, SIRT3, SIRT4, SIRT5, SIRT6, and SIRT7, but none of them is born with any clear developmental defect.15,33,43,46,55,56 However, some of these animals do develop significant pathologies before adulthood. These are especially severe in the case of SIRT6−/− mice, which are smaller at birth and quickly develop lymphopenia, loss of subcutaneous fat, lordokyphosis, and metabolic defects.15 This phenotype is similar to some progeria syndromes, and it has been linked to defects in DNA repair and gene instability rather than with cell differentiation.40-42 In in vitro models, only SIRT3 has been observed to be associated with differentiation. Although SIRT3-deficient mice show normal adaptive thermogenesis and are metabolically unremarkable, SIRT3 is apparently necessary for in vitro differentiation of brown adipocytes.76 Thus, SIRT3 might be a positive regulator of adipogenesis, in contrast to SIRT1 and SIRT2, which are negative regulators. Finally, in the particular case of SIRT7−/− mice, severe heart hypertrophy has been observed, but this defect was associated with inflammation and apoptosis.56 Nevertheless, SIRT7 can deacetylate p53 in a similar fashion to SIRT1,56 which suggests that the developmental role of p53 could also be affected by this protein, although further studies are necessary to fully explore the biological function of SIRT7.
Sirtuins, Life Span, and Aging: Are Adult Stem Cells Involved?
The aging process involves a complex combination of genetic, environmental, and stochastic factors, whose relative contributions are still not clearly defined. The stem cell theory of aging suggests that an important mechanism of aging involves a progressive decline in the self-renewal of adult stem cells and their potential to differentiate into specific cell types in order to replenish the tissues of an organism77,78 (Fig. 2). Although the age-dependent loss of function of different types of adult stem cells, such as hematopoietic, mesenchymal, neural, muscular, or melanocytic, has been reported, the molecular mechanisms involved in this process are not yet fully understood. It has been suggested that stem cell attrition produced by DNA damage and telomere shortening could play a significant role.78 Moreover, not only genetic but also epigenetic mechanisms could be implicated in stem cell dysfunction over time,79-81 such that the accumulation of epigenetic alterations might favor their aging.82
Figure 2.
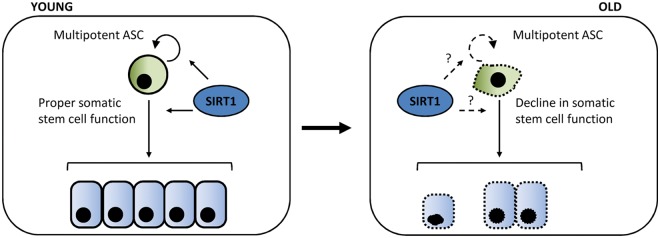
The stem cell theory of aging. With age, adult stem cells accumulate DNA, and chromosomal damage results in the failure of proper stem cell functions and, consequently, tissue regeneration. It is suggested that SIRT1 participates in both self-renewal and differentiation processes in adult stem cells and therefore could be an important component of the mechanisms that cause adult stem cells to grow old.
Sirtuins were first proposed to play a role in aging in studies of Saccharomyces cerevisiae, where Sir2, Sir3, and Sir4 showed a negative regulation function.11 Subsequently, several works reporting on different species, such as Caenorhabditis elegans83 and Drosophila melanogaster,84 showed associations at different levels between sirtuins and life span and aging. In mammals, sirtuins have also been associated with extending life spans. Evidence comes from several studies in mice, where lack of SIRT1 was related to a reduced life span,85,86 or conversely, SIRT1 overexpression led to an increased life span.87 It has been proposed that the effects of sirtuins on life span could be explained by calorie restriction, which increases life spans in lower organisms and mammals.88,89 Several studies have shown that sirtuins are activated in the calorie restriction response and play an important role in life-span regulation.84,90-92 This function of sirtuins is partially mediated by their capacity to detect changes in the cellular nutritional status based on the ratio of NAD+/NADH and, in effect, promote survival mechanisms.88,93 However, recent studies cast doubt on the associations of Sir2 with life span or at least suggest that the associations found so far had been overestimated.94,95 In mammals, the role of SIRT1 in regulating life span is also unclear,96 but new evidence has shown that SIRT6 might be a good candidate.97 A still poorly explored possibility is that the role of sirtuins in aging is also mediated by their role in adult stem cells. SIRT1 is the sirtuin member that has been most extensively studied and may control stem cell aging through the appropriate maintenance of telomeres and reactive oxygen species.98 Several studies in hematopoietic and neural stem cells suggest that SIRT1 regulates self-renewal and differentiation processes in response to environmental conditions and that this regulation becomes more important with age.81 SIRT1 activity is probably important to offset the aging process triggered by oxidative stress, but the implication of SIRT1 in cell differentiation also indicates that sirtuins are involved in stem cell function in adult tissues, especially under stress conditions (Fig. 2). Future studies, including more stem cell types and other sirtuin members, will help to elucidate the role of sirtuins in the maintenance of adult stem cell functions and, consequently, in the aging process.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors received the following financial support for the research, authorship, and/or publication of this article: Instituto Universitario de Oncología del Principado de Asturias (IUOPA), Obra social Cajastur (to R.M.R.), Fondo de Investigación Sanitaria/Fondo Europeo de Desarrollo Regional (FIS/FEDER) (PI11/01728) and Instituto de Salud Carlos III (ISCIII) (CP11/00131) (to A.F.F.), Spanish Ministry of Health (PI12/01080) and Fundación Ramon Areces (to M.F.F.). The Cancer Epigenetics Laboratory (IUOPA) also thanks Mutua Madrileña and Ramón Areces Foundations for financial support. The IUOPA is supported by the Obra Social Cajastur, Spain.
References
- 1. de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737-49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gregoretti IV, Lee YM, Goodson HV. Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338:17-31 [DOI] [PubMed] [Google Scholar]
- 3. Shore D, Squire M, Nasmyth KA. Characterization of two genes required for the position-effect control of yeast mating-type genes. EMBO J. 1984;3:2817-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Guarente L. Diverse and dynamic functions of the Sir silencing complex. Nat Genet. 1999;23:281-5 [DOI] [PubMed] [Google Scholar]
- 5. Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000;278:685-90 [DOI] [PubMed] [Google Scholar]
- 6. Saunders LR, Verdin E. Sirtuins: critical regulators at the crossroads between cancer and aging. Oncogene. 2007;26:5489-504 [DOI] [PubMed] [Google Scholar]
- 7. Rodriguez RM, Fraga MF. Aging and cancer: are sirtuins the link? Future Oncol. 2010;6:905-15 [DOI] [PubMed] [Google Scholar]
- 8. Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bosch-Presegue L, Vaquero A. The dual role of sirtuins in cancer. Genes Cancer. 2012;2:648-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carafa V, Nebbioso A, Altucci L. Sirtuins and disease: the road ahead. Front Pharmacol. 2012;3:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126-8 [DOI] [PubMed] [Google Scholar]
- 13. Wood JG, Rogina B, Lavu S, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686-9 [DOI] [PubMed] [Google Scholar]
- 14. Berdichevsky A, Viswanathan M, Horvitz HR, Guarente L. C. elegans SIR-2.1 interacts with 14-3-3 proteins to activate DAF-16 and extend life span. Cell. 2006;125:1165-77 [DOI] [PubMed] [Google Scholar]
- 15. Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315-29 [DOI] [PubMed] [Google Scholar]
- 16. Pruitt K, Zinn RL, Ohm JE, et al. Inhibition of SIRT1 reactivates silenced cancer genes without loss of promoter DNA hypermethylation. PLoS Genet. 2006;2:e40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yuan J, Pu M, Zhang Z, Lou Z. Histone H3-K56 acetylation is important for genomic stability in mammals. Cell Cycle. 2009;8:1747-53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93-105 [DOI] [PubMed] [Google Scholar]
- 19. Vaquero A, Scher M, Erdjument-Bromage H, Tempst P, Serrano L, Reinberg D. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440-4 [DOI] [PubMed] [Google Scholar]
- 20. Brunet A, Sweeney LB, Sturgill JF, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011-5 [DOI] [PubMed] [Google Scholar]
- 21. Giannakou ME, Partridge L. The interaction between FOXO and SIRT1: tipping the balance towards survival. Trends Cell Biol. 2004;14:408-12 [DOI] [PubMed] [Google Scholar]
- 22. Vaziri H, Dessain SK, Ng Eaton E, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149-59 [DOI] [PubMed] [Google Scholar]
- 23. Yeung F, Hoberg JE, Ramsey CS, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yuan Z, Zhang X, Sengupta N, Lane WS, Seto E. SIRT1 regulates the function of the Nijmegen breakage syndrome protein. Mol Cell. 2007;27:149-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173-85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nahhas F, Dryden SC, Abrams J, Tainsky MA. Mutations in SIRT2 deacetylase which regulate enzymatic activity but not its interaction with HDAC6 and tubulin. Mol Cell Biochem. 2007;303:221-30 [DOI] [PubMed] [Google Scholar]
- 27. North BJ, Verdin E. Mitotic regulation of SIRT2 by cyclin-dependent kinase 1-dependent phosphorylation. J Biol Chem. 2007;282:19546-55 [DOI] [PubMed] [Google Scholar]
- 28. Inoue T, Hiratsuka M, Osaki M, et al. SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene. 2007;26:945-57 [DOI] [PubMed] [Google Scholar]
- 29. Vaquero A, Reinberg D. Sirtuins in biology and disease. In: Esteller M, editor. Epigenetics in biology and medicine. Boca Raton, FL: CRC Press; 2009. p. 73-104 [Google Scholar]
- 30. Vaquero A, Scher MB, Lee DH, et al. SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev. 2006;20:1256-61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647-57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lombard DB, Alt FW, Cheng HL, et al. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560-7 [DOI] [PubMed] [Google Scholar]
- 38. Yang H, Yang T, Baur JA, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095-107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384-401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kawahara TL, Michishita E, Adler AS, et al. SIRT6 links histone H3 lysine 9 deacetylation to NF-kappaB-dependent gene expression and organismal life span. Cell. 2009;136:62-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Haigis MC, Mostoslavsky R, Haigis KM, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941-54 [DOI] [PubMed] [Google Scholar]
- 44. Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790-801 [DOI] [PubMed] [Google Scholar]
- 45. Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Du J, Zhou Y, Su X, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L. Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev. 2006;20:1075-80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Adenot PG, Mercier Y, Renard JP, Thompson EM. Differential H4 acetylation of paternal and maternal chromatin precedes DNA replication and differential transcriptional activity in pronuclei of 1-cell mouse embryos. Development. 1997;124:4615-25 [DOI] [PubMed] [Google Scholar]
- 49. Lepikhov K, Walter J. Differential dynamics of histone H3 methylation at positions K4 and K9 in the mouse zygote. BMC Dev Biol. 2004;4:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Worrad DM, Turner BM, Schultz RM. Temporally restricted spatial localization of acetylated isoforms of histone H4 and RNA polymerase II in the 2-cell mouse embryo. Development. 1995;121:2949-59 [DOI] [PubMed] [Google Scholar]
- 51. Ma P, Schultz RM. Histone deacetylase 1 (HDAC1) regulates histone acetylation, development, and gene expression in preimplantation mouse embryos. Dev Biol. 2008;319:110-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McBurney MW, Yang X, Jardine K, et al. The mammalian SIR2alpha protein has a role in embryogenesis and gametogenesis. Mol Cell Biol. 2003;23:38-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Cheng HL, Mostoslavsky R, Saito S, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang RH, Sengupta K, Li C, et al. Impaired DNA damage response, genome instability, and tumorigenesis in SIRT1 mutant mice. Cancer Cell. 2008;14:312-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim HS, Vassilopoulos A, Wang RH, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20:487-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Vakhrusheva O, Smolka C, Gajawada P, et al. Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res. 2008;102:703-10 [DOI] [PubMed] [Google Scholar]
- 57. Efroni S, Duttagupta R, Cheng J, et al. Global transcription in pluripotent embryonic stem cells. Cell Stem Cell. 2008;2:437-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Calvanese V, Lara E, Suarez-Alvarez B, et al. Sirtuin 1 regulation of developmental genes during differentiation of stem cells. Proc Natl Acad Sci U S A. 2010;107:13736-41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Saunders LR, Sharma AD, Tawney J, et al. miRNAs regulate SIRT1 expression during mouse embryonic stem cell differentiation and in adult mouse tissues. Aging (Albany NY). 2010;2:415-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Tarantino C, Paolella G, Cozzuto L, et al. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010;24:3255-63 [DOI] [PubMed] [Google Scholar]
- 61. Han MK, Song EK, Guo Y, Ou X, Mantel C, Broxmeyer HE. SIRT1 regulates apoptosis and Nanog expression in mouse embryonic stem cells by controlling p53 subcellular localization. Cell Stem Cell. 2008;2:241-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Kuzmichev A, Margueron R, Vaquero A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci U S A. 2005;102:1859-64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hisahara S, Chiba S, Matsumoto H, et al. Histone deacetylase SIRT1 modulates neuronal differentiation by its nuclear translocation. Proc Natl Acad Sci U S A. 2008;105:15599-604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Prozorovski T, Schulze-Topphoff U, Glumm R, et al. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat Cell Biol. 2008;10:385-94 [DOI] [PubMed] [Google Scholar]
- 65. Fulco M, Schiltz RL, Iezzi S, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12:51-62 [DOI] [PubMed] [Google Scholar]
- 66. Picard F, Kurtev M, Chung N, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ou X, Chae HD, Wang RH, et al. SIRT1 deficiency compromises mouse embryonic stem cell hematopoietic differentiation, and embryonic and adult hematopoiesis in the mouse. Blood. 2011;117:440-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Matsui K, Ezoe S, Oritani K, et al. NAD-dependent histone deacetylase, SIRT1, plays essential roles in the maintenance of hematopoietic stem cells. Biochem Biophys Res Commun. 2012;418:811-7 [DOI] [PubMed] [Google Scholar]
- 69. Leko V, Varnum-Finney B, Li H, et al. SIRT1 is dispensable for function of hematopoietic stem cells in adult mice. Blood. 2012;119:1856-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Southwood CM, Peppi M, Dryden S, Tainsky MA, Gow A. Microtubule deacetylases, SirT2 and HDAC6, in the nervous system. Neurochem Res. 2007;32:187-95 [DOI] [PubMed] [Google Scholar]
- 71. Li W, Zhang B, Tang J, et al. Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating alpha-tubulin. J Neurosci. 2007;27:2606-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jing E, Gesta S, Kahn CR. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007;6:105-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009;20:801-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Rothgiesser KM, Erener S, Waibel S, Luscher B, Hottiger MO. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123:4251-8 [DOI] [PubMed] [Google Scholar]
- 75. Hayden MS, Ghosh S. NF-kappaB in immunobiology. Cell Res. 2011;21:223-44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Giralt A, Hondares E, Villena JA, et al. Peroxisome proliferator-activated receptor-gamma coactivator-1alpha controls transcription of the Sirt3 gene, an essential component of the thermogenic brown adipocyte phenotype. J Biol Chem. 2011;286:16958-66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Krishnamurthy J, Sharpless NE. Stem cells and the rate of living. Cell Stem Cell. 2007;1:9-11 [DOI] [PubMed] [Google Scholar]
- 78. Sharpless NE, DePinho RA. How stem cells age and why this makes us grow old. Nat Rev Mol Cell Biol. 2007;8:703-13 [DOI] [PubMed] [Google Scholar]
- 79. Chambers SM, Shaw CA, Gatza C, Fisk CJ, Donehower LA, Goodell MA. Aging hematopoietic stem cells decline in function and exhibit epigenetic dysregulation. PLoS Biol. 2007;5:e201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Gonzalo S, Jaco I, Fraga MF, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416-24 [DOI] [PubMed] [Google Scholar]
- 81. Pollina EA, Brunet A. Epigenetic regulation of aging stem cells. Oncogene. 2011;30:3105-26 [DOI] [PubMed] [Google Scholar]
- 82. Fraga MF. Genetic and epigenetic regulation of aging. Curr Opin Immunol. 2009;21:446-53 [DOI] [PubMed] [Google Scholar]
- 83. Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227-30 [DOI] [PubMed] [Google Scholar]
- 84. Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci U S A. 2004;101:15998-6003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Boily G, Seifert EL, Bevilacqua L, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS One. 2008;3:e1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Li Y, Xu W, McBurney MW, Longo VD. SirT1 inhibition reduces IGF-I/IRS-2/Ras/ERK1/2 signaling and protects neurons. Cell Metab. 2008;8:38-48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bordone L, Cohen D, Robinson A, et al. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759-67 [DOI] [PubMed] [Google Scholar]
- 88. Cohen HY, Miller C, Bitterman KJ, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390-2 [DOI] [PubMed] [Google Scholar]
- 89. Mair W, Dillin A. Aging and survival: the genetics of life span extension by dietary restriction. Annu Rev Biochem. 2008;77:727-54 [DOI] [PubMed] [Google Scholar]
- 90. Frankel S, Rogina B. Sir2, caloric restriction and aging. Pathol Biol (Paris). 2006;54:55-7 [DOI] [PubMed] [Google Scholar]
- 91. Guarente L, Picard F. Calorie restriction: the SIR2 connection. Cell. 2005;120:473-82 [DOI] [PubMed] [Google Scholar]
- 92. Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech Ageing Dev. 2006;127:48-56 [DOI] [PubMed] [Google Scholar]
- 93. Chen D, Guarente L. SIR2: a potential target for calorie restriction mimetics. Trends Mol Med. 2007;13:64-71 [DOI] [PubMed] [Google Scholar]
- 94. Burnett C, Valentini S, Cabreiro F, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1-2 [DOI] [PubMed] [Google Scholar]
- 96. Baur JA, Chen D, Chini EN, et al. Dietary restriction: standing up for sirtuins. Science. 2010;329:1012-3, author reply 3-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Kanfi Y, Naiman S, Amir G, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218-21 [DOI] [PubMed] [Google Scholar]
- 98. Mantel C, Broxmeyer HE. Sirtuin 1, stem cells, aging, and stem cell aging. Curr Opin Hematol. 2008;15:326-31 [DOI] [PMC free article] [PubMed] [Google Scholar]