Abstract
The fluorescence spectroscopy of 7-azaindole (7aIn) incorporated in DNA oligonucleotides is investigated. Incorporation of 7aIn into DNA oligonucleotides is accomplished through standard solid-phase phosphoramidite chemistry. Fluorescence emission of the 7aIn chromophore shifts slightly to the red (from 386 nm to 388 nm) upon glycosylation at the N − 1 position, but its relative fluorescence quantum yield increases 23 times, from 0.023 to 0.53. Upon incorporation into DNA, the fluorescence emission of 7aIn is greatly quenched with fluorescence quantum yields of 0.020 and 0.016 in single and double strand DNA, respectively. The fluorescence emission for 7aIn in DNA oligonucleotides shifts to the blue with an emission maximum at 379 nm. Both the strong fluorescence quenching and the blue shift of the emission spectrum signify that 7aIn is stacked with neighboring DNA bases in both single and double strand DNA. As the duplex DNA melts due to temperature increase, the fluorescence of the 7aIn chromophore increases, indicating the transition from the less fluorescent duplex DNA to the more fluorescent single strand DNA. Since this fluorescent 7aIn is a structural analog of purine, its fluorescence property may be utilized as a probe for studying nucleic acid structure and dynamics.
Keywords: 7-Azaindole, DNA, Fluorescence, Synthesis
1. Introduction
The use of a non-natural ‘base’ to replace one of the natural nucleic acid bases in DNA has been an important tool for studying biological events such as DNA-protein interaction and DNA structure and dynamics [1-27]. Research findings in this area have provided some important information toward the understanding our living systems at the molecular level. There are mainly two groups of ‘bases’ synthesized and incorporated into DNA oligomers: those made by minor modification of the natural bases or the derivatives of cyclic aromatic compounds. While the former includes 2-aminopurine [1-8], inosine [9] and isoinosine [9-11], 3- and 7-deazaadenine [12-15], and 3- and 7-deazaguanine [14,16], the latter includes analogs of pteridine [17-19] and indole [20-24], benzene [23-26], naphthalene and pyrene derivatives [24], The first group of compounds, when replacing a natural base in duplex DNA, can still form hydrogen bonds with the base in the opposite strand, while the second group of compounds cannot form any hydrogen bonds.
Fluorescence has been one of the very useful techniques used for studying DNA structure and dynamics. The fluorescence properties of the above base analogs have been utilized for studying DNA structure and duplex stability [1,2,10,14,15,22,26], protein-DNA interactions [4-8,13,18,25], or simply as universal base analogs [20,27]. Some of the unusual ‘bases’ are used as anti-HIV or anti-cancer drugs [28-30].
One interesting fluorescent molecule used for studying protein structure and dynamics is 7-azaindole (7aIn) [31-38]. 7aIn is weakly fluorescent in water due to dimer formation [34-36]. The excitation energy of 7aIn is lost due to double proton transfer, and therefore quenches the fluorescence. After methylation of the N − 1 nitrogen to prevent dimer formation, 7aIn becomes strongly fluorescent. The fluorescence property of 7aIn, as well as its 3-alanyl derivative, 7-azatryptophan, is well understood. It has been incorporated into peptides to obtain peptide/protein structural information [31,33,39]. Due to the unique fluorescence properties of this molecule, we wish to report the incorporation of 7aIn into duplex DNA to examine its fluorescence properties in 2′-deoxyriboside, in single and double strand DNA, and the fluorescence change during DNA duplex thermomelting.
2. Experimental
2.1. Materials and instruments
Chemicals and solvents were purchased from either Aldrich Chemical Company or Fisher Scientific. All solid compounds were dried in vacuo over phosphorous pentoxide. Anhydrous solvents from Aldrich were used without further treatment, except for acetonitrile, which was dried with CaH2 and distilled under nitrogen atmosphere. 7aIn, obtained from ICN Biochemicals, was recrystallized from alcohol and dried in vacuo before use. 1-(α)-Chloro-3,5-di-O-(p-toluoyl)-2-deoxy-D-ribose was purchased from Berry and Associates, Inc. (Ann Arbor, MI) and used directly. 1H- and 31p-NMR spectra were recorded on a Bruker AM250 MHz NMR spectrometer at ambient temperature. The NMR spectra were in ppm with tetramethylsilane as internal standard. High-resolution mass spectrometric analyses (HRMS) were performed at Louisiana State University’s Mass Spectrometry Facility using the Fast Atomic Bombardment (FAB) technique. DNA oligonucleotides containing 7aIn in the place of either an adenine or a guanine were synthesized by Gemini Biotech (Alachua, FL) using the phosphoramidite 4. The 4-step synthesis of 4 is shown in Fig. 1.
Fig. 1.
Synthesis of 1′-(7-azaindolyl)-2′-deoxy-D-riboside and the incorporation into DNA via the phosphoramidite chemistry.
Fluorescence emission spectra were recorded using a Fluoromax-2 spectrofluorometer from Instruments SA Inc. (Edison, NJ). Sample solutions were prepared in 1 × TBE buffer in the presence of 0.2 M sodium chloride. The quantum yields were determined using indole as a standard. The excitation wavelength was set at 300 nm for 7aIn. The sample temperature was controlled by an external water bath and the actual solution temperature was determined using a thermometer with a temperature probe.
2.2. Synthesis of 7-azaindole phosphoramidite 4
2.2.1. Glycosylation of 7aIn with 1-(α)-chloro-3,5-di-O-(p-toluoyl)-2-deoxy-D-ribose (modified procedure from Refs. [11,20,40,41])
Sodium hydride (475 mg, 11.9 mmol, 60% in oil) was added to 86 ml of anhydrous acetonitrile containing 7aIn (1.30 g, 11.0 mmol). The mixture was stirred for 2 h at room temperature under nitrogen atmosphere. Then the mixture was cooled to − 10 °C using an ice salt bath before 1-(α)-chloro-3,5-di -O-(p-toluoyl)-2-deoxy-D-ribose (5.13 g, 13.2 mmol) was added portionwise while stirring. The solution was kept at − 10 °C under nitrogen atmosphere for 3 h before it was filtered to remove a small amount of insoluble material. The filtrate was evaporated to give an oily crude product, which was purified by silica gel chromatography (column: 33 × 345 mm2) eluting with ethyl acetate/hexanes (50:50). After evaporation of the solvent, 3.42 g white, foamy product, 1′-(7-azaindolyl)-3′,5′-di-O-(p-toluoyl)-2′-deoxy-D-riboside (1, 66%), was obtained. Rf = 0.75 (ethyl acetate/hexanes 50:50). 1H-NMR (DMSO-d6, ppm): 8.26 (1H, d, H-C6), 7.98 (1H, d, H-C4), 7.72 (1H, d, H-C2), 7.13 (1H, q, H-C5), 6.70 (1H, dd, H1′), 6.52 (1H, d, H-C3), 5.25 (0.5H, OH), 5.04 (0.5H, OH), 4.42 (1H, m, H3′), 3.85 (1H, m, H 4′), 3.55 (2H, m, H5′ and H5″), 2.61 (1H, m, H 2′) and 2.20 (1H, m, H2″). HRMS: MH+: 471.1935. Calc. 471.1935 (M + 1) for C28H26N2O5.
2.2.2. Synthesis of 2 by the hydrolysis of 1
Product 1 (3.42 g, 7.26 mmol) was added to 40 ml of methanol containing sodium methoxide prepared by dissolving 75 mg of sodium (3.3 mmol) in anhydrous methanol prior to the addition. The reaction mixture was stirred at room temperature for 3 h while large amounts of product precipitated. After completion of the reaction, solid NH4Cl was added to the solution to adjust its pH to basic (pH 8) before it was filtered to collect the majority of the product, 1′-(7-azaindolyl)-2′-deoxy-D-riboside (2), as a white powder. A further portion of the product was crystallized from the filtrate and the combined yield was 75% (1.28 g, 5.45 mmol). 1H-NMR (CDCl3, ppm): 8.30 (1H, d, H-C6), 7.89 (1H, d, H-C4), 7.38 (1H, d, H-C2), 7.32 (9H, m, Ph), 7.09 (1H, q, H-C5), 6.92 (1H, t, H1′), 6.82 (4H, m, Ph), 6.46 (1H, d, H-C3), 4.65 (1H, m, H3′), 4.08 (1H, m, H4′), 3.80 (6H, d, 2 × CH3O ), 3.38 (2H, m, H5′ and H5″), 2.68 (1H, m, H2′) and 2.48 (1H, m, H2″). HRMS: MH+: 235.1100; Calc. 235.1005 (M + 1) for C12H14N2O3.
2.2.3. Synthesis of 3 by the 4,4’-dimethoxytritylation of 2
4,4-Dimethoxytrityl chloride (745 mg, 2.20 mmol) was added to 14 ml of anhydrous pyridine containing 6 mg of 4-(N,N-dimethylamino)-pyridine (DMAP) and 469 mg of 2 (2.00 mmol) which was dried over P2O5 in vacuo overnight before use. The mixture was then stirred for 2.5 h at room temperature before 10 ml of methanol was added to quench the reaction. The resulting mixture was poured into 70 ml of 5% aqueous NaHCO3 solution and the mixture emulsified. The emulsion was extracted using CH2Cl2 (3 × 40 ml) and the combined organic layers were dried over anhydrous Na2SO4. After evaporation of the solvent, an oily product was obtained and purified by silica gel chromatography (column: 20 × 310 mm2) eluting with ethyl acetate/hexanes (70:30) to produce 1′-(7-azaindolyl)-5′-O-(4,4′-dimethoxytrityl)-2′-deoxy-D-riboside (3) as a yellowish foam in 75% yield (805 mg, 1.50 mmol). Rf= 0.59 (ethyl acetate/hexanes 70:30). lH-NMR (CDCl3, ppm): 8.30 (1H, d, H-C6), 7.89 (1H, d, H-C4), 7.38 (1H, d, H-C2), 7.32 (9H, m, Ph), 7.09 (1H, q, H-C5), 6.92 (1H, t, H1′), 6.82 (4H, m, Ph), 6.46 (1H, d, H-C3), 4.65 (1H, m, H3′), 4.08 (1H, m, H4′), 3.80 (6H, d, 2 × CH3O), 3.38 (2H, m, H5′ and H5″), 2.68 (1H, m, H2′) and 2.48 (1H, m, H2″). HRMS (FAB-MS): MH+: 537.2402; Calc. 537.2312 (M + 1) for C33H32N2O5.
2.2.4. Synthesis of phosphoramidite 4
2-Cyanoethyl diisopropylaminochlorophosphoramidite (620 μl, 2.76 mmol) was added to 10 ml of anhydrous CH2Cl2 containing 3 (370 mg, 0.69 mmol) and 1.2 ml of diisopropylethylamine (6.9 mmol) under nitrogen atmosphere. The reaction mixture was allowed to stir for 1 h at room temperature before it was poured into 30 ml of 5% NaHCO3 aqueous solution and extracted with CH2Cl2 (3 × 20 ml). The combined organic layers were dried over anhydrous Na2SO4 and concentrated to produce the crude product, which was purified by silica gel chromatography (column: 20 × 245 mm) eluting with ethyl acetate/hexanes (70:30). After evaporation of the solvent, the phosphoramidite 4, 1′-(7-azaindolyl)-5′-O-(4,4′-dimethoxytrityl)-2′-deoxy-D-riboside-3′-O-(2-cyanoethyl)-N,N-diisopropylphosphoramidite, was collected as a white foam in 77% yield (390 mg, 0.53 mmol). Rf= 0.76 (ethyl acetate/hexanes/triethylamine 7:3:1). It separated into two spots with nearly equal intensity on TLC with a less polar solvent: Rf = 0.38 and 0.44 (ethylacetate: hexanes: triethylamine 3:7:1). The broadband decoupled 31P-NMR (CDCl3) gives two signals at 149.67 and 149.49 ppm with about a 1:1 intensity ratio. The 1H-NMR spectrum is complex and shows a mixture of two co-existing diastereomers.
2.3. Preparation of DNA oligonucleotides containing 7aIn
Phosphoramidite 4 was used directly for automated DNA synthesis performed by scientists at Gemini Biotech (Alachua, FL). The synthesis was carried out on a 1.0 μmol scale. All of the syntheses were optimized for trityl release and all oligonucleotides were HPLC purified. The sequence of the synthetic DNA oligonucleotides are: Oligo-1: 5′-GCGCGC7aInCGCGCGC-3′ (self-complimentary); Oligo-2: 5′-CGC7aInGAATTC-3′; Oligo-3: 5′-CGG7aInAATTC-3′; (matching strand: 5′-GAATTCCCG-3′). The concentration of Oligos 1– 3 was determined by their absorption value at 260 nm using Beer’s Law: C = A260/εl, where A260 is the absorbance of the oligomer at 260 nm; ε is the molar absorption extinction coefficient at 260 nm and l is the cell path length (1 cm). The ε was calculated based upon the following equation: ε = 0.89 [(A × 15480) + (C × 7340) + (G × 11760) + (T × 8850)], where A, C, G, T = number of A, C, G, and T bases in the oligomer. The coefficient for 7aIn was determined to be 4400 at 260 nm. Double stranded Oligo-3 was prepared by matching equimolar amounts of the two complimentary strands followed by annealing. The buffer used for all DNA samples were the 1 × TBE buffer (89 mM Tris base, 89 mM boric acid, 0.1 mM EDTA, pH 8.2) with 200 mM NaCl.
3. Results and discussions
3.1. Synthesis of 1’-(7-azaindolyl)-2’-deoxy-D-riboside and its conversion into phosphoramidite
The 4-step synthesis and the respective reaction yield for each step are shown in Fig. 1. The glycosylation of 7aIn with l-(α)-chloro-3,5-di-O-(p-toluoyl)-2-deoxy-D-ribose was accomplished at − 10 °C with an admirable yield (66%). Prior to this procedure, several attempts at room temperature (20 °C) yielded multiple by-products, although most chemists were successful with the glycosylation of other bases at room temperature [11,20,40], The by-products were not characterized. Lowering the reaction temperature by 30 °C effectively eliminated the unwanted reactions to yield 1. 1H-NMR spectrum of purified 1 indicated that the β-isomer was the only product, there was no sign indicating the presence of the α-isomer. The glycosylation is at the N − 1, not the N − 7, as shown by Seela and Gumbiowski [41], Hydrolysis of 1 in methanol in the presence of a catalytic amount of sodium methoxide was completed in 3 h at room temperature. Compound 2 crystallized readily during the reaction and was easily isolated and purified. Tritylation of 2 with 1.5-equivalent of 4,4′-dimethoxytrityl chloride partially protected both the 5′- and 3′-hydroxyl groups of the sugar as revealed by 1H-NMR spectrum of the isolated main product (not shown), although such procedure performed on other modified bases protected only the 5′-hydroxyl group [10,15,16,20,22]. Therefore, only 1.1-equivalent of 4,4′-dimethoxytrityl chloride was used for the reaction and a 75% yield for 3 was achieved after chromatography purification. The reaction formula for the synthesis of phosphoramidite 4 from the literature also had to be modified. The optimized mole ratio was 1:4:10 for compound 3: chlorophosphoramidite: diisopropylethylamine under our conditions, compared to 1:2:4 by Seela and Chen [10] for isoinosine or 1:6:4 by Cosstick et al. [13] for 3-deazaadenosine. The large amount (10-equivalent) of diisopropylethylamine proved to be necessary in our case. 1H-NMR spectrum of 4 is very complex because compound 4 exists as a diastereomeric mixture since the phosphorous atom is chiral and it adopts both an R and S-configuration. Actually, the two diastereomers can be isolated on TLC (Rf = 0.38 and 0.44) when eluting with ethylacetate: hexanes: triethylamine = 3:7:1. The two spots were nearly equal in intensity and size (data not shown), indicating that both diastereomers exist in nearly equal amount. The broadband decoupled 31P-NMR spectrum (not shown) has two peaks at 149.67 and 149.49 ppm with a 1:1 intensity ratio for the single phosphorous atom in the molecule.
3.2. Incorporation of 7aIn into DNA oligonucleotide
In order to test whether the phosphoramidite 4 is suitable to be used for automated DNA synthesis, the following oligonucleotides containing 7aIn were prepared: Oligo-1: 5′ -GCGCGC7aInCGCGCGC-3′; Oligo-2: 5′-CGC7aInGAATTC-3′; Oligo-3: 5′-CGG 7aInAATTC-3′. Although 7aIn was put in different sequences, the coupling proceeded as planned. The coupling efficiency for 7aIn was about the same as that for the natural bases A, T, G, C as revealed by monitoring the trityl release at each coupling step (data not shown). After HPLC purification, the overall isolated yield for all oligonucleotides ranged from 6 to 22% including 10 other DNA oligonucleotides containing 7aIn prepared in a separate synthesis (not shown). This yield is comparable with the synthesis of DNA oligonucleotides comprised of only natural bases.
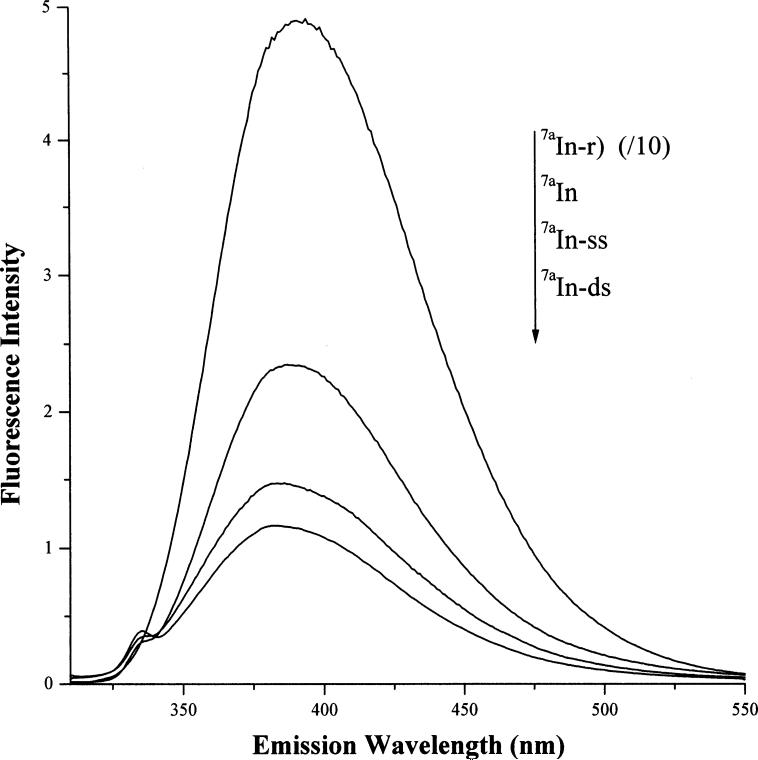
3.3. Fluorescence emission spectra for 7aIn, 7aIn 2-deoxyriboside (7aIn-r), 7aIn in single stranded DNA (7aIn-ss), and 7aIn in double stranded DNA (7aIn-ds)
The fluorescence emission spectra for 7aIn, 7aIn-r, 7aIn-ss, and 7aIn-ds were recorded and plotted together after normalization for the absorbance value at 300 nm (Fig. 2). In order to fit all the normalized spectra on the same graph, the fluorescence emission of 7aIn-r was lowered 10-fold. The glyco-linkage causes the fluorescence intensity of 7aIn to increase by about 23 times. This was expected because similar increase was observed if 7aIn was N − 1 methylated [31-38]. However, incorporation of 7aIn into DNA quenches its fluorescence significantly as the fluorescence intensity of 7aIn-ss and 7aIn-ds are significantly lower than that for 7aIn-r. The quenching is probably due to base stacking in DNA. This strong quenching effect was also seen for 2-aminopurine upon incorporation into DNA, in which the fluorescence quenching is attributed to both base stacking and hydrogen bonding [3]. Since 7aIn cannot form hydrogen bonds with the opposite base, the fluorescence quenching should be due to base stacking. In comparison, the fluorescence emission of both 7aIn-ss and 7aIn-ds are only slightly lower than the fluorescence for 7aIn itself (Fig. 2). Also, the fluorescence of 7aIn in double stranded DNA is weaker than in single stranded DNA as shown in Fig. 2. Using 0.023 as the fluorescence quantum yield for 7aIn determined using indole as a standard, the quantum yields for 7aIn-r, 7aIn-ss, and 7aIn-ds are 0.53, 0.020, 0.016, respectively. In summary, the fluorescence of 7aIn-r is 96% quenched upon incorporation into single stranded DNA, and further quenched when incorporated into double stranded DNA.
Fig. 2.
Fluorescence emission spectra (from top to bottom): (1) 7-azaindole 2′-deoxyriboside (7aIn-r) whose intensity is lowered 10-fold to fit to the scale, (2) 7-azaindole (7aIn), (3) 7-azaindole in single stranded DNA (7aIn-ss), and (4) in double stranded DNA (7aIn-ds). The excitation wavelength was 300 nm. The emission intensity for each derivative was normalized by dividing its absorbance value at this wavelength. The solvent was 1 × TBE buffer (89 mM TRIS, 89 mM boric acid, 0.1 mM EDTA) at pH 8.2.
The fluorescence emission maximum for 7aIn in water is at 286 nm. When it is glycosylated, its emission maximum shifts slightly to the red at 288 nm. Upon incorporation into DNA, it shifts to the blue at about 279 nm in both single and double strand DNA. This blue shift signifies that 7aIn is in a less polar environment in DNA due to base stacking.
3.4. Fluorescence change for the incorporated 7aIn during DNA thermomelting
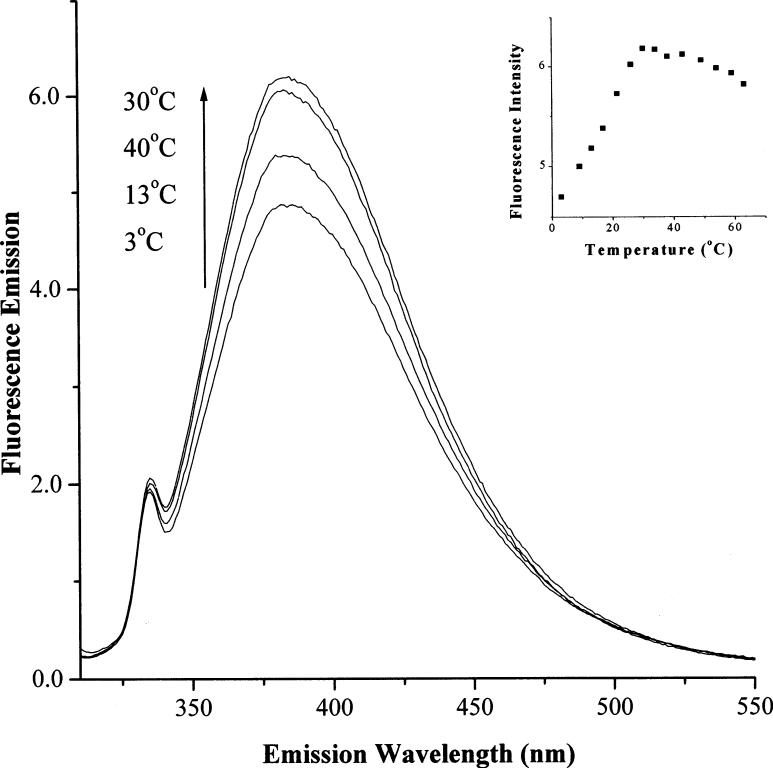
Since the fluorescence intensity for 7aIn is weaker in double stranded DNA than in single stranded DNA, it can be utilized to monitor DNA thermomelting. Fluorescence emission spectrum for Oligo-3 is recorded as a function of temperature and shown in Fig. 3. At 2 °C, the DNA is mostly double stranded where its fluorescence intensity is the lowest. As the temperature increases to 13 °C, the fluorescence increases 15%, and the increase continues until about 30 °C where the maximum fluorescence increase is 30% (Fig. 3 insert). If temperature continues to increase, a slight decrease of fluorescence is observed due to temperature related quenching processes. Since this fluorescent 7aIn is a structurally similar to a purine base, its fluorescence property may be utilized as a probe for studying nucleic acid structure and dynamics.
Fig. 3.
Change: of fluorescence emission spectra for 7-azaindole due to duplex melting as temperature increases. The DNA sequence was d(5′-CGG7aInAATTC-3′/d(3′-GCCCTTAAG-5′): The excitation wavelength was 300 nm. The concentration of the DNA was 10.7 μM in DNA strands. The spectra and their corresponding temperatures are shown in the graph. Insert: Fluorescence intensity change at 380 nm versus temperature plot for the duplex.
Acknowledgements
The authors wish to thank the National Institutes of Health for the NIH-MBRS grant # S06 GM08047 and NIH-RCMI 12RR13459.
References
- 1.McLaughlin LW, Leong T, Benseler F, Piel N. Nucleic Acids Res. 1988;16:5631. doi: 10.1093/nar/16.12.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eritja R, Kaplan BE, Mhaskar D, Sowers LC, Petruska J, Goodman MF. Nucleic Acids Res. 1986;14:5869. doi: 10.1093/nar/14.14.5869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hochstrasser RA, Carver TE, Sowers LC, Millar DP. Biochemistry. 1994;33:11971. doi: 10.1021/bi00205a036. [DOI] [PubMed] [Google Scholar]
- 4.Bloom LB, Otto MR, Beechem JM, Goodman MF. Biochemistry. 1993;32:11247. doi: 10.1021/bi00092a039. [DOI] [PubMed] [Google Scholar]
- 5.Bloom LB, Otto MR, Eritja R, Reha-Krantz LJ, Goodman MF, Beechem JM. Biochemistry. 1994;33:7576. doi: 10.1021/bi00190a010. [DOI] [PubMed] [Google Scholar]
- 6.Frey MW, Sowers LC, Millar DP, Bencovic SJ. Biochemistry. 1995;34:9185. doi: 10.1021/bi00028a031. [DOI] [PubMed] [Google Scholar]
- 7.Petrauskene OV, Schmidt S, Karyagina AS, Nikolskaya II, Gromova ES, Cech D. Nucleic Acids Res. 1995;23:2192. doi: 10.1093/nar/23.12.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raney KD, Sowers LC, Millar DP, Benkovic SJ. Proc. Natl. Aca. Sci. USA. 1994;91:6644. doi: 10.1073/pnas.91.14.6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes KT, Gaines PCW, Karlinsey JE, Vinayak R, Simon MI. EMBO J. 1992;11:2695. doi: 10.1002/j.1460-2075.1992.tb05335.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seela F, Chen Y. Nucleic Acids Res. 1995;23:2499. doi: 10.1093/nar/23.13.2499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seela F, Chen Y, Bindig U, Kazimierczuk Z. Helv. Chim. Acta. 1994;77:194. [Google Scholar]
- 12.Ono A, Ueda T. Nucleic Acids Res. 1987;15:3059. doi: 10.1093/nar/15.7.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cosstick R, Li X, Tuli DK, Williams DM, Connolly BA, Newman PC. Nucleic Acids Res. 1991;18:4771. [PMC free article] [PubMed] [Google Scholar]
- 14.Grein T, Lampe S, Mersmann K, Rosemeyer H, Thomas H, Seela F. Bioorg. Med. Chem. Lett. 1994;4:971. [Google Scholar]
- 15.Seela F, Grein T. Nucleic Acids Res. 1992;20:2297. doi: 10.1093/nar/20.9.2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Seela F, Lampe S. Helv. Chim. Acta. 1991;74:1790. [Google Scholar]
- 17.Driscoll SL, Hawkins ME, Balis FM, Pfeiderer W, Laws WR. Biophys. J. 1997;73:3277. doi: 10.1016/S0006-3495(97)78352-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hawkins ME, Pfeiderer W, Mazumder A, Pommier YG, Balis FM. Nucleic Acids Res. 1995;23:2872. doi: 10.1093/nar/23.15.2872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hawkins ME, Pfeiderer W, Balis FM, Porter D, Knutzen JR. Anal. Biochem. 1997;244:86. doi: 10.1006/abio.1996.9879. [DOI] [PubMed] [Google Scholar]
- 20.Loakes D, Brown DM. Nucleic Acids Res. 1994;22:4039. doi: 10.1093/nar/22.20.4039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Erzberger JP, Barsky D, Schrarer OD, Colvin ME, Wilson DMI. Nucleic Acids Res. 1998;26:2771. doi: 10.1093/nar/26.11.2771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schweitzer BA, Kool ET. J. Am. Chem. Soc. 1995;117:1863. doi: 10.1021/ja00112a001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweitzer BA, Kool ET. J. Org. Chem. 1994;59:7238. doi: 10.1021/jo00103a013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moran S, Ren RX-F, Sheils CJ, Rumney SI, Kool ET. Nucleic Acids Res. 1996;24:2044. doi: 10.1093/nar/24.11.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morales JC, Kool ET. Nature Struct. Biol. 1998;5:950. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- 26.Guckian KM, Krugh TR, Kool ET. Nature Struct. Biol. 1998;5:954. doi: 10.1038/2930. [DOI] [PubMed] [Google Scholar]
- 27.Smith CL, Simmonds AC, Felix IR, Hamilton AL, Kumar S, Nampolli S, Loakes P, Hill F, Brown DM. Nucleosides Nucleotides. 1998;17:541. doi: 10.1080/07328319808005198. [DOI] [PubMed] [Google Scholar]
- 28.Flexner CW, Hildreth JE, Kuncl RW, Drachman DB. The Lancet. 1992;339:438. doi: 10.1016/0140-6736(92)90133-n. [DOI] [PubMed] [Google Scholar]
- 29.Jurgensen CH, Huaber BE, Zimmerman TP, Wolberg G. J. Immunol. 1990;144:653. [PubMed] [Google Scholar]
- 30.Andreassen PR, Margolis RL. Proc. Natl. Acad. Sci. USA. 1992;89:2272. doi: 10.1073/pnas.89.6.2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smirnov AV, English DS, Rich RL, Lane J, Teyton L, Schwabacher AW, Luo S, Thornburg RW, Petrich JW. J. Phys. Chem. B. 1997;101:2758. [Google Scholar]
- 32.Rich RL, Chen Y, Neven D, Negrerie M, Gai F, Petrich JW. J. Phys. Chem. 1993;97:1781. [Google Scholar]
- 33.English DS, Rich RL, Petrich JW. Photochem. Photobiol. 1998;67:76. [PubMed] [Google Scholar]
- 34.Chen Y, Rich RL, Gai F, Petrich JW. J. Phys. Chem. 1993;97:1770. [Google Scholar]
- 35.Gordon MS. J. Phys. Chem. 1995;100:3974. [Google Scholar]
- 36.Chou P-T, Wei C-Y, Chang C-P, Kuo M-S. J. Phys. Chem. 1995;99:11994. [Google Scholar]
- 37.Negrerie M, Gai F, Bellefeuille, Petrich JW. J. Phys. Chem. 1991;95:8663. [Google Scholar]
- 38.Gai F, Rich RL, Petrich JW. J. Am. Chem. Soc. 1994;116:735. [Google Scholar]
- 39.Cioni P, Erijman L, Strambini GB. Biochem. Biophys. Res. Commun. 1998;248:347. doi: 10.1006/bbrc.1998.8939. [DOI] [PubMed] [Google Scholar]
- 40.Kazimierczuk Z, Cottam HB, Revankar GR, Robbins RK. J. Am. Chem. Soc. 1984;106:6379. [Google Scholar]
- 41.Seela F, Gumbiowski R. Heterocycles. 1989;29:795. [Google Scholar]